Abstract
To understand how sleep-wakefulness cycles are regulated, it is essential to disentangle structural and functional relationships between the preoptic area (POA) and lateral hypothalamic area (LHA), since these regions play important yet opposing roles in the sleep-wakefulness regulation. GABA- and galanin (GAL)-producing neurons in the ventrolateral preoptic nucleus (VLPO) of the POA (VLPOGABA and VLPOGAL neurons) are responsible for the maintenance of sleep, while the LHA contains orexin-producing neurons (orexin neurons) that are crucial for maintenance of wakefulness. Through the use of rabies virus-mediated neural tracing combined with in situ hybridization (ISH) in male and female orexin-iCre mice, we revealed that the vesicular GABA transporter (Vgat, Slc32a1)- and galanin (Gal)-expressing neurons in the VLPO directly synapse with orexin neurons in the LHA. A majority (56.3 ± 8.1%) of all VLPO input neurons connecting to orexin neurons were double-positive for Vgat and Gal. Using projection-specific rabies virus-mediated tracing in male and female Vgat-ires-Cre and Gal-Cre mice, we discovered that VLPOGABA and VLPOGAL neurons that send projections to the LHA received innervations from similarly distributed input neurons in many brain regions, with the POA and LHA being among the main upstream areas. Additionally, we found that acute optogenetic excitation of axons of VLPOGABA neurons, but not VLPOGAL neurons, in the LHA of male Vgat-ires-Cre mice induced wakefulness. This study deciphers the connectivity between the VLPO and LHA, provides a large-scale map of upstream neuronal populations of VLPO→LHA neurons, and reveals a previously uncovered function of the VLPOGABA→LHA pathway in the regulation of sleep and wakefulness.
SIGNIFICANCE STATEMENT We identified neurons in the ventrolateral preoptic nucleus (VLPO) that are positive for vesicular GABA transporter (Vgat) and/or galanin (Gal) and serve as presynaptic partners of orexin-producing neurons in the lateral hypothalamic area (LHA). We depicted monosynaptic input neurons of GABA- and galanin-producing neurons in the VLPO that send projections to the LHA throughout the entire brain. Their input neurons largely overlap, suggesting that they comprise a common neuronal population. However, acute excitatory optogenetic manipulation of the VLPOGABA→LHA pathway, but not the VLPOGAL→LHA pathway, evoked wakefulness. This study shows the connectivity of major components of the sleep/wake circuitry in the hypothalamus and unveils a previously unrecognized function of the VLPOGABA→LHA pathway in sleep-wakefulness regulation. Furthermore, we suggest the existence of subpopulations of VLPOGABA neurons that innervate LHA.
Keywords: circuit, hypothalamus, preoptic area, sleep, wakefulness
Introduction
The hypothalamus has been thought to play a significant role in sleep-wakefulness regulation since von Economo's seminal study (von Economo, 1930). Particularly, the preoptic area (POA) plays a crucial role in the initiation and maintenance of sleep (Saper et al., 2010; Weber and Dan, 2016; Scammell et al., 2017; Sulaman et al., 2023). For instance, studies in cats and rats demonstrated that cell destruction in the POA led to a decrease in non-rapid eye movement (NREM) and rapid eye movement (REM) sleep amounts (Szymusiak and McGinty, 1986; Sallanon et al., 1989; Lu et al., 2000). Likewise, excitatory manipulations of POA neurons increased sleep (Saito et al., 2013; Chung et al., 2017; Kroeger et al., 2018; Ma et al., 2019). This area contains neurons that fire at high frequency during sleep and cease firing during wakefulness, as shown by immunostaining (Sherin et al., 1996; Gaus et al., 2002; Gong et al., 2004) or electrophysiological recordings (Kaitin, 1984; Szymusiak et al., 1998; K. Takahashi et al., 2009).
Neurons in and around the ventrolateral preoptic nucleus (VLPO) in the POA are known to play a significant role in sleep regulation (Tsuneoka and Funato, 2021). These neurons send GABAergic projections to wakefulness-promoting neuronal populations (Weber and Dan, 2016; Scammell et al., 2017). Neurons with other chemical identities in the VLPO, including galanin (GAL)-expressing neurons (VLPOGAL neurons), have also been implicated in sleep regulation (Kroeger et al., 2018; Ma et al., 2019).
Recent studies, however, suggest that the role of the VLPO in sleep-wakefulness regulation is more complex than previously thought. It has been reported that not all GABAergic VLPO neurons (VLPOGABA neurons) participate in sleep promotion (Chung et al., 2017; Vanini et al., 2020; Yamagata et al., 2021; de Luca et al., 2022), and it has been shown that glutamatergic VLPO neurons rather induce wakefulness (Chung et al., 2017; Vanini et al., 2020).
The lateral hypothalamic area (LHA) also plays a prominent role in the regulation of sleep-wakefulness states. Especially, orexin (also known as hypocretin)-producing neurons in the LHA (orexin neurons) play a crucial role in maintenance of wakefulness (Sakurai, 2007; Carter et al., 2009), highlighted by the finding that loss of orexin neurons resulted in narcolepsy in several mammalian species (Lin et al., 1999; Peyron et al., 2000; Thannickal et al., 2000; Hara et al., 2001; Tabuchi et al., 2014; Branch et al., 2016). Other types of neurons in the LHA are also involved in control of sleep-wakefulness states. For instance, excitatory manipulations of GABAergic and neurotensin-producing neurons in the LHA induced arousal (Venner et al., 2016; Naganuma et al., 2019), while melanin-concentrating hormone-producing neurons in the LHA have been implicated in the regulation of REM sleep (Jego et al., 2013; Tsunematsu et al., 2014; Vetrivelan et al., 2016).
The intrahypothalamic connectivity between the VLPO and LHA is suggested to be important for sleep-wakefulness regulation (Scammell et al., 2017). Notably, previous studies showed that the POA provides direct inhibitory synaptic input to orexin neurons in the LHA (Sakurai et al., 2005; Yoshida et al., 2006; Saito et al., 2013, 2018). Additionally, a recent study showed that orexin induced arousal partially through acting on the VLPO (de Luca et al., 2022).
Despite the prominent roles of the POA and LHA in the regulation of sleep-wakefulness states, the functional and structural connectivity between them has only been partially examined. In addition, how VLPO neurons might be regulated, and which types of neuronal populations make synaptic contact with these neurons, has not been shown so far.
Here, we confirmed and chemically defined neuronal populations in the VLPO that made monosynaptic input to orexin neurons in the LHA, and precisely localized these input neurons in subregions of the VLPO. We also identified upstream neurons that made monosynaptic contact with VLPOGABA and VLPOGAL neurons that send projections to the LHA. Moreover, we showed a functional relevance of the GABAergic pathway from the VLPO to the LHA using optogenetic manipulation.
Materials and Methods
Animals
All experiments involving animals were approved by the Animal Experiment and Use Committee of the University of Tsukuba and were therefore in accordance with National Institutes of Health guidelines. Animals were maintained under a 12:12 h light/dark cycle (9:00 lights on, 21:00 lights off for tracing experiments and 06:00 lights on, 18:00 lights off for sleep recordings). Food and water were available ad libitum. Experiments using genetically modified mice were conducted on heterozygotes and their WT littermates with C57BL/6J genetic background. Eight- to 24-week-old male and female mice were used for tracing experiments, while only male mice of the same age were used for optogenetic experiments.
The following mice were used in the study: Rosa26dreaddm4 (T. M. Takahashi et al., 2020); orexin-Cre Tg (Matsuki et al., 2009), Vgat-ires-Cre (Vong et al., 2011), Gal-Cre Tg (strain name: STOCK Tg(Gal-cre)KI87Gsat/Mmucd, RRID:MMRRC_031060-UCD) and orexin-iCre.
To generate orexin-iCre knock-in mice, we used CRISPR-Cas9 technology. We selected a sequence (5′-GAA GGA AAG TTC ATG GTG CC-3′) that contains the start codon of the Hcrt gene as the sgRNA target. We inserted this sequence into the pX330-mC plasmid, which carried both guide RNA and Cas9-mC expression units (Mizuno-Iijima et al., 2021). A chimeric intron, iCre, and rabbit globin polyadenylation sequences (Yamamoto et al., 2015) were inserted between the 5′ and 3′ homology arms of the donor DNA yielding piCre-Hcrt. chr11:100762842-100764478 and chr11:100761367-100762838 genomic regions (GRCm38/mm10) were used as the 5′ and 3′ homology arms in the donor DNA, respectively. Female C57BL/6J mice were intraperitoneally injected with pregnant mare serum gonadotropin (5 units) and human chorionic gonadotropin (5 units) at a 48 h interval, and mated with male C57BL/6J mice. We collected zygotes from the oviducts of the mated females, and a mixture of pX330-mC (circular, 5 ng/μl) and piCre-Hcrt (circular, 10 ng/μl) was microinjected into zygotes. Subsequently, the injected zygotes were transferred into the oviducts of pseudo-pregnant ICR females, and newborns were obtained.
Viruses
Adeno-associated virus (AAV) vectors were produced using a triple-transfection, helper-free method in HEK293T cells (Mieda et al., 2015). SADΔG-GFP(EnvA) and SADΔG-GFP-ERT2CreERT2 were made by transfecting pcDNA-SADB19L, pcDNA-SADB19G, pcDNA-SADB19N, pcDNA-SADB19P, and pSADΔG-GFP-F2 or pSADΔG-GFP-ERT2CreERT2 (Addgene #32 649) into B7GG cells, followed by pseudotyping in BHK-RGCD-EnvA cells and ultracentrifugation according to the method previously described (Osakada and Callaway, 2013; Saito et al., 2018).
The titers of recombinant AAV vectors (genome copies/ml) were determined by real-time PCR: AAV2-EF1a-FLEX(loxP)-TVA-mCherry, 3.74 × 1012; AAV2-CAG-FLEX(loxP)-RG, 1.91 × 1011; AAV10-EF1a-FLEX(loxP)-TVA-mCherry, 4.4 × 1014, 7.98 × 1013; AAV10-CAG-FLEX(loxP)-RG, 1.7 × 1014, 1.72 × 1013; AAV2-EF1a-DIO-ChR2-EYFP, 8.0 × 1012; AAV2-EF1a-DIO-GFP, 8.0 × 1012; AAV2-EF1a-DIO-SSFO-EYFP, 6.0 × 1012. Titers of SADΔG-GFP(EnvA) vectors were determined by infecting HEK293-TVA cell line and were found to be 1.2 × 109, 9.8 × 108, 6.8 × 108 infectious units/ml (three different batches were used throughout the study). Titer of SADΔG-GFP-ERT2CreERT2 vector was 3.0 × 108 infectious units/ml.
Surgery
Mice were anesthetized with isoflurane and positioned in a stereotaxic frame (David Kopf Instruments). Viral vectors were delivered using a glass microcapillary pipette operated through an air pressure injection system (Picospritzer III, Parker). After each injection, the pipette was kept in the injection site for 5-10 min to prevent retrograde flow of virus.
For validation of orexin-iCre mice, AAV2-EF1a-DIO-GFP was delivered at the coordinates (1) −1.4 mm AP, ±0.9 mm ML, 5.5 mm DV; and (2) −1.8 mm AP, ±0.9 mm ML, 5.7 mm DV from the skull surface (bregma) bilaterally, 500 nl/injection.
For retrograde rabies virus-mediated tracing from orexin neurons in the LHA, AAV10-EF1a-FLEX(loxP)-TVA-mCherry and AAV10-CAG-FLEX(loxP)-RG were delivered at the coordinates: (1) −1.4 mm AP, +0.9 mm ML, 5.5 mm DV; and (2) −1.8 mm AP, +0.9 mm ML, 5.7 mm DV from the bregma unilaterally (right hemisphere). Volume of AAV mix was 500 nl/injection with TVA:RG = 1:1 proportion by volume.
For projection-specific retrograde rabies virus-mediated tracing from GABA- or galanin-producing neurons in the VLPO projecting to the LHA, AAV2-EF1a-FLEX(loxP)-TVA-mCherry and AAV2-CAG-FLEX(loxP)-RG were delivered at the coordinates: +0.15 mm AP, +0.54 mm ML for Vgat-ires-Cre mice and +0.6 mm ML for Gal-Cre mice, 5.7 mm DV from the bregma unilaterally (right hemisphere). Volumes of AAV mix were 66-86 nl/injection with TVA:RG = 1:3 proportion by volume. After a 14 d recovery period in all tracing experiments, SADΔG-GFP(EnvA) was injected at the coordinates: (1) −1.4 mm AP, +0.9 mm ML, 5.5 mm DV; and (2) −1.8 mm AP, +0.9 mm ML, 5.7 mm DV from the bregma unilaterally (right hemisphere), at 500 nl/injection.
For dual-color retrograde tracing from the LHA and tuberomammillary nucleus (TMN), 1 mg/ml AlexaFluor-488-cholera toxin subunit B (Fisher Scientific #34775) was delivered into the LHA at the coordinates: −1.58 mm AP, +0.9 mm ML, 5.5 mm DV from the bregma unilaterally, at 500 nl/injection; and 2× diluted Red RetroBeads (RetroBeads IX Red 530; Nacalai Tesque #1RX) were delivered into TMN at the coordinates: −2.45 mm AP, +1.0 mm ML, 5.3 mm DV from the bregma unilaterally (right hemisphere), at 100 nl/injection.
For optogenetic manipulation in Vgat-ires-Cre mice, AAV2-EF1a-DIO-ChR2-EYFP, AAV2-EF1a-DIO-SSFO-EYFP, or AAV2-EF1a-DIO-GFP were injected at the coordinates: +0.15 mm AP, ±0.54 mm ML, 5.7 mm DV from the bregma bilaterally, at 44 nl/injection. Fiber optic cannulas (Kyocera) were then implanted at the coordinates: −1.58 mm AP, ±1.6 mm ML, 4.34 mm DV from the bregma bilaterally at an angle of ±10° to a perpendicular line. EEG/EMG electrodes were implanted as described below. Two stainless-steel screws for EEG recording connected to the electrode were placed each at +1.0 mm AP, +1.5 mm ML from the bregma and at +1.0 mm AP, +1.5 mm ML from lambda. Another screw serving as a ground and connected to the electrode was placed behind lambda over the opposite hemisphere. Two insulated silver wires for EMG recording (Cooner Wire #AS633) connected to the electrode were inserted into the trapezius muscles bilaterally. An anchor screw was then positioned into the skull. The whole construction was fixed to the skull with UV light-curable dental cement (3M #56818) and then covered with black nail polish.
For optogenetic manipulation in Gal-Cre mice, AAV2-EF1a-DIO-ChR2-EYFP or AAV2-EF1a-DIO-GFP were injected at the coordinates: +0.15 mm AP, ±0.6 mm ML, 5.7 mm DV from the bregma bilaterally, at 132 nl/injection. The rest of the procedure was performed as described for Vgat-ires-Cre mice.
Rabies virus-mediated tracing from orexin neurons in LHA
Rabies virus-mediated tracing was conducted at two different time points (two batches of experiments). The first batch of experiments was conducted, starting from injection of AAV10-EF1a-FLEX(loxP)-TVA-mCherry (4.4 × 1014 genome copies/ml) and AAV10-CAG-FLEX(loxP)-RG (1.7 × 1014 genome copies/ml) vectors and followed by injection of SADΔG-GFP(EnvA), for examination of colocalization of Vgat and Gal mRNA in input neurons in the VLPO. The second batch of experiments was conducted, starting from injection of AAV10-EF1a-FLEX(loxP)-TVA-mCherry (7.98 × 1013 genome copies/ml) and AAV10-CAG-FLEX(loxP)-RG (1.72 × 1013 genome copies/ml) vectors and followed by injection of SADΔG-GFP-ERT2CreERT2, for examination of colocalization of Gal and Vglut2 or Vglut2 and Vgat mRNA in input neurons in the VLPO.
EEG/EMG recording
Sleep recording was performed in specially designed recording chambers between 09:00 and 15:00 (ZT3-ZT9; 06:00 lights on, 18:00 lights off) during 2 consecutive days for each animal for channelrhodopsin-2 (ChR2)-mediated optogenetic manipulations or between 07:00 and 10:00 (ZT1-ZT4) during 1 d for stabilized step-function opsin (SSFO)-mediated optogenetic manipulations. Implanted EEG/EMG electrodes were connected to slip rings via flexible customized wires at least 2 d before the photoactivation experiments. EEG/EMG signals were amplified and filtered (EEG, 0.5-64 Hz; EMG, 16-64 Hz) using an amplifier AB-611J (Nihon Kohden), digitized at a sampling rate of 128 Hz, and recorded using VitalRecorder software (Kissei Comtec). EEG/EMG signals were analyzed by characterizing 4 or 10 s epochs into wakefulness, NREM sleep, and REM sleep according to standard criteria (Oishi et al., 2016) using SleepSign software (Kissei Comtec). Defined vigilance states were then examined visually and corrected as necessary. Latency to wakefulness was analyzed manually with 1 s time resolution.
Optogenetic manipulation
Vgat-ires-Cre and Gal-Cre mice were placed into recording chambers 2-3 weeks after surgery and connected to EEG/EMG wires and optical patch cords at least 2 d before the start of the optogenetic stimulation experiments for habituation and to facilitate a physiological activity rhythm. For optogenetic stimulation, blue light (462 nm) was generated by a laser (Shanghai Laser) and applied to mice through optical fibers (Thorlabs). Blue light power intensity at the tip of the plastic fiber was ∼10 mW mm−2. All optogenetic manipulations were conducted bilaterally.
For photostimulation using ChR2, 10 Hz (for Vgat-ires-Cre mice) (Chung et al., 2017; Yamagata et al., 2021) or 1 Hz (for Gal-Cre mice) (Kroeger et al., 2018) light pulses with 10 ms width were applied for 20 s through an optic cannula with recording of EEG/EMG and video within ZT3-ZT9 time frame (09:00-15:00) for 2 consecutive days. The interstimulation period was at least 40 min (6-8 photostimulations/animal/day). Each photostimulation was applied after at least 40 s of NREM sleep. On the second day of the experiment, the last photostimulation was applied with parameters 10 Hz, 10 ms, 20 s light on/40 s light off, 10 min for Vgat-ires-Cre mice and with parameters 1 Hz, 10 ms, 20 s light on/40 s light off, 10 min for Gal-Cre mice (1 photostimulation/animal). To perform photostimulation with SSFO, we applied two 1 s light pulses starting from ZT2 (08:00) 30 min apart (2 photostimulations per animal) (Hasegawa et al., 2022).
Brain tissue processing
Mouse brains were sampled 7 d after rabies virus injection and dual-color retrograde tracing, 14 d after orexin-iCre validation experiment, or 90 min after ChR2-mediated 10 min pulse stimulation and the second SSFO-mediated 1 s stimulation. Mice under anesthesia with 2% 2,2,2-tribromoethanol (Sigma-Aldrich #T48402-25G), 1.2% 2-methyl-2-butanol (Fujifilm Wako #014-03706), 8% ethanol (99.5; Fujifilm Wako #057-00456) in 1× phosphate-buffered saline (PBS) (avertin composition) were intracardially perfused with prechilled 1× PBS (unless noted otherwise) followed by freshly prepared 4% PFA (Nacalai Tesque #09154-85) in 1× PBS. Brains were postfixed in 4% PFA solution overnight at 4°C, then cryoprotected using 20% sucrose solution in 1× PBS (unless noted otherwise) overnight. All brains were frozen using liquid nitrogen. The brains were sectioned (four series/brain, thickness 40 μm, unless noted otherwise) using cryostat (Leica Biosystems) throughout the entire brain length (from ∼+3.0 mm AP to ∼−6.0 mm AP) for retrograde rabies virus-mediated tracing or through ROIs for other experiments.
Immunohistochemistry
Sections were incubated with 3% BSA (Nacalai Tesque #01863-48), 0.2v/v% Triton X-100 (MP Biomedicals #807426) solution in 1× PBS (blocking solution) at room temperature for 30 min. Sections were then incubated with primary antibodies in blocking solution at 4°C overnight. Next, sections were washed 3 times with 1× PBS at room temperature followed by application of secondary antibodies and DAPI (stock 1 mg/1 ml, 1:1000, Dojindo #D523) in blocking solution overnight at 4°C or for 3 h at room temperature. Sections were then washed the same way and mounted on slides.
The primary antibodies used in the study were as follows: rat anti-GFP (1:1000; Nacalai Tesque #04404-84), goat anti-mCherry (1:1000; SICGEN #AB0040-500, RRID:AB_2333092), rabbit anti-orexin A (1:2000; Phoenix Pharmaceuticals #H-003-30, RRID:AB_2315019), rabbit anti-cFos (1:1000; EnCor Biotechnology #RPCA-c-Fos-AP, RRID:AB_2572236), and guinea pig anti-cFos (1:500; Synaptic Systems #226308, RRID:AB_2905595). The secondary antibodies used in the study were as follows: AlexaFluor-488 donkey anti-rat (1:1000; Fisher Scientific #A-21208, RRID:AB_2535794), AlexaFluor-594 donkey anti-goat (1:1000; Fisher Scientific #A-11058, RRID:AB_2534105), AlexaFluor-647 donkey anti-goat (1:1000; Fisher Scientific #A-21447, RRID:AB_2535864), AlexaFluor-488 donkey anti-rabbit (1:1000; Fisher Scientific #A-21206, RRID:AB_2535792), AlexaFluor-594 donkey anti-rabbit (1:1000; Fisher Scientific #A-21207, RRID:AB_141637), AlexaFluor-647 donkey anti-rabbit (1:1000; Fisher Scientific #A-31573, RRID:AB_2536183), and AlexaFluor-647 donkey anti-guinea pig (1:500; Jackson ImmunoResearch Laboratories #706-605-148, RRID:AB_2340476). Representative images were obtained using microscope Axio Zoom.V16 (Carl Zeiss) or TCS SP8 STED 3X (Leica Biosystems).
FISH
FISH was conducted using RNAscope Fluorescent Multiplex Reagent Kit (Advanced Cell Diagnostics #320850) in accordance with the manufacturer's protocol. Mice were intracardially perfused with Dulbecco's PBS (–) without Ca and Mg (1×) (Nacalai Tesque #14249-24) followed by freshly prepared 4% PFA in 1× PBS. Brains and sections were maintained in RNase-free conditions. After fixation in 4% PFA solution and cryoprotection in 20% sucrose solution in Dulbecco's PBS, brains were frozen and then sectioned (four series/brain, thickness 20 μm) using cryostat (Leica Biosystems). Sections were mounted on slides and used immediately or stored at −80°C. Probes used in the study were Mm-Gal (#400961), Mm-Slc32a1 (#319191), Mm-Slc17a6 (#319171), Mm-Slc32a1-C2 (#319191-C2), Mm-Slc17a6-C2 (#319171-C2), and Mm-Adcyap1-C2 (#405911-C2).
For combination with immunohistochemistry, sections mounted on slides were washed 3 times with 1× PBS at room temperature immediately after hybridization with amplifiers. Sections were then incubated with 1v/v% Triton X-100 solution in 1× PBS followed by 10% Blocking One (Nacalai Tesque #03953-66), 0.3 v/v% Triton X-100 solution in 1× PBS at room temperature for 30 min each. Further steps included application of primary antibodies followed by washing in 1× PBS, application of secondary antibodies and a third wash, as described for immunohistochemistry. Sections were then coverslipped using ProLong Diamond Antifade Mountant with DAPI (Fisher Scientific #P36962). Fluorescence images were obtained using confocal microscope TCS SP8 STED 3X (Leica Biosystems).
Data acquisition and analysis for rabies virus-mediated retrograde tracing
For rabies virus-mediated retrograde tracing, we counted GFP-positive input neurons as well as GFP- and mCherry-double-positive starter neurons in an every fourth 40 (or 20) μm section of the whole brain based on the mouse brain map by Franklin and Paxinos (2007) using the anatomic classification by ARA (Lein et al., 2007). As the absolute numbers of input neurons and of starter neurons in the whole brain varied among individual animals, we calculated the percentages of input neuron number and of starter neuron number for each brain region to ensure unbiased representation of their distribution.
Experimental design and statistical analysis
Pearson correlation and linear regression were conducted to analyze the relationship between the numbers of input neurons and starter neurons in rabies virus-mediated tracing experiments (see Figs. 2B, 3B, 4B).
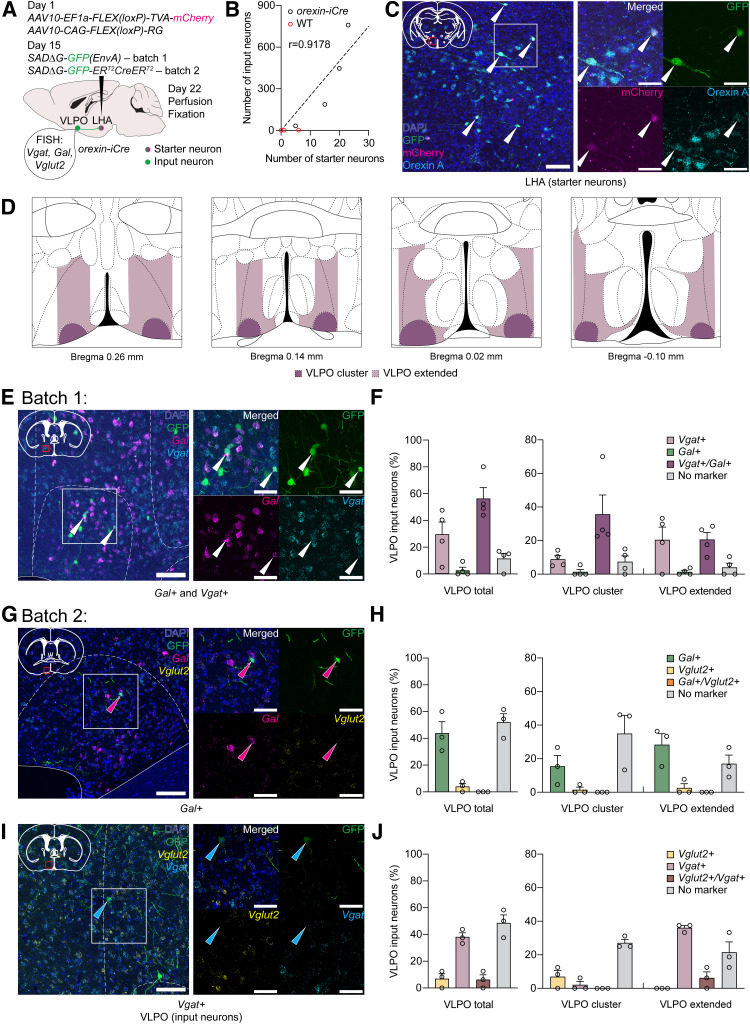
Figure 2.
Monosynaptic retrograde rabies virus-mediated tracing from orexin neurons. A, Scheme of trans-synaptic retrograde tracing experiment. Two weeks after injection of AAV10-EF1a-FLEX(loxP)-TVA-mCherry and AAV10-CAG-FLEX(loxP)-RG into LHA, SADΔG-GFP(EnvA) or SADΔG-GFP-ERT2CreERT2 were injected into the same sites in orexiniCre/+ mice. Mouse brains were examined by immunohistochemical study and FISH. Starter neurons were identified by expression of both mCherry and GFP, while input neurons expressed only GFP. B, Number of starter neurons positively correlates with number of produced input neurons in orexiniCre/+ mice (Pearson correlation, r = 0.9178, p = 0.0822; n = 4), while almost no cells were positive for mCherry and GFP in WT mice (n = 3). Linear regression: F(1,3) = 27.43, p = 0.0135, R2 = 0.8424. Several starter neurons were observed at the injection sites in WT mice despite extremely low number of input neurons. C, Representative images of starter neurons in LHA in orexiniCre/+ mice. Starter neurons were immunostained with anti-mCherry and anti-orexin A antibodies. Right, Magnified image of the boxed area in the left image. Arrowheads indicate starter neurons. Scale bars: left, 100 μm; right, 50 μm. D, Scheme depicting established borders of VLPO region. Brain outlines were taken from the Franklin and Paxinos (2007) mouse brain atlas. E, Representative images of Vgat- and Gal-double-positive input neurons in VLPO in orexiniCre/+ mice visualized by FISH. Right, Magnified image of the boxed area in the left image. Arrowheads indicate Vgat/Gal-double-positive input neurons. Scale bars: left, 100 μm; right, 50 μm. F, Percentages of input neurons in VLPO of orexin neurons categorized as Vgat+/Gal+, Vgat+, Gal+ or cells without expression of these markers (38±10 input neurons, n = 4). G, Representative images of input neurons in VLPO in orexiniCre/+ mice visualized by FISH using Gal and Vglut2 probes. Right, Magnified image of the boxed area in the left image. Magenta arrowhead indicates a Gal-positive, but Vglut2-negative, input neuron. Scale bars: left, 100 μm; right, 50 μm. H, Percentages of input neurons in VLPO of orexin neurons categorized as Gal+/Vglut2+, Gal+, Vglut2+ or cells without expression of these markers (17 ± 3 input neurons, n = 3). I, Representative images of input neurons in VLPO in orexiniCre/+ mice visualized by FISH using Vgat and Vglut2 probes. Right, Magnified image of the boxed area in the left image. Cyan arrowhead indicates a Vgat-positive, but Vglut2-negative, input neuron. Scale bars: left, 100 μm; right, 50 μm. J, Percentages of input neurons in VLPO of orexin neurons categorized as Vglut2+/Vgat+, Vglut2+, Vgat+ or cells without expression of these markers (12 ± 2 input neurons, n = 3). Circles represent data from individual animals. Data are mean ± SEM.
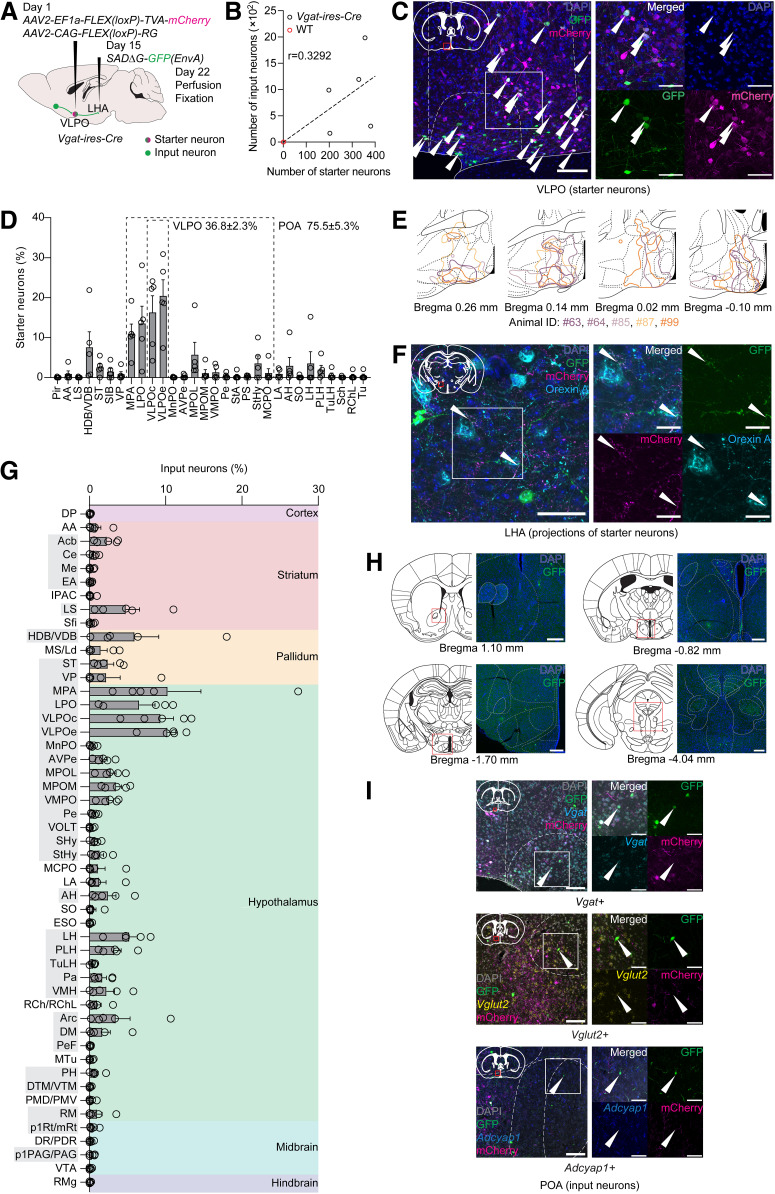
Figure 3.
Monosynaptic retrograde projection-specific rabies virus-mediated tracing from VLPOGABA neurons sending projections to LHA. A, Scheme of projection-specific tracing experiment. Two weeks after injection of AAV2-EF1a-FLEX(loxP)-TVA-mCherry and AAV2-CAG-FLEX(loxP)-RG into VLPO of Vgat-ires-Cre mice, SADΔG-GFP(EnvA) was injected into LHA. Mouse brains were examined by immunohistochemical study. B, Number of starter neurons has positive correlation with number of input neurons in Vgat-ires-Cre mice (Pearson correlation, r = 0.3292, p = 0.5885; n = 5). Linear regression: F(1,4) = 9.413, p = 0.0374, R2 = 0.1084. In WT mice, several starter and input neurons were observed close to VLPO only in 1 animal (n = 3). C, Representative images of starter neurons in VLPO of Vgat-ires-Cre mice. Starter neurons were immunostained with anti-GFP and anti-mCherry antibodies. Right, Magnified image of the boxed area in the left image. Arrowheads indicate starter neurons. Scale bars: left, 100 μm; right, 50 μm. D, Distribution of starter VLPOGABA→LHA neurons in POA and adjacent regions. Highlighted areas are POA and VLPO. Areas with starter neurons in all examined animals without cut-off are shown. E, Scheme depicting outlines of distribution of starter neurons in cross-sections containing VLPO. Brain outlines were taken from the Franklin and Paxinos (2007) mouse brain atlas. Colors represent individual animals. If there were no matching brain sections to the presented schemes in some animals, outlines were not shown. F, Representative images of axonal terminals of VLPOGABA→LHA neurons making close appositions to bodies of orexin A-expressing neurons in LHA. Immunostaining with anti-GFP, anti-mCherry, and anti-orexin A antibodies. Right, Magnified image of the boxed area in the left image. Arrowheads indicate GFP- and mCherry-double-positive neural fibers. Scale bars: left, 50 μm; right, 20 μm. G, Whole-brain distribution of monosynaptic input neurons of VLPOGABA→LHA neurons in Vgat-ires-Cre mice. Only brain areas with input neurons detected in at least 3 of 5 animals are shown. Colored areas represent major brain divisions. Abbreviations of input brain areas that are shared with VLPOGAL→LHA neurons (Fig. 4G) are highlighted by gray background. Circles represent data from individual animals. Nomenclature of brain regions was taken from the Franklin and Paxinos (2007) mouse brain atlas. Data are mean ± SEM; n = 5. H, Representative images of input neurons of VLPOGABA→LHA neurons. Scale bars, 200 μm. I, Representative images of monosynaptic input neurons in POA of VLPOGABA→LHA neurons analyzed by FISH using Vgat, Vglut2, and Adcyap1 probes. For each molecular marker: right, magnified image of the boxed area in the left image. Arrowheads indicate GFP-positive neuronal bodies coexpressing examined molecular signatures. Scale bars: left, 100 μm; right, 50 μm.
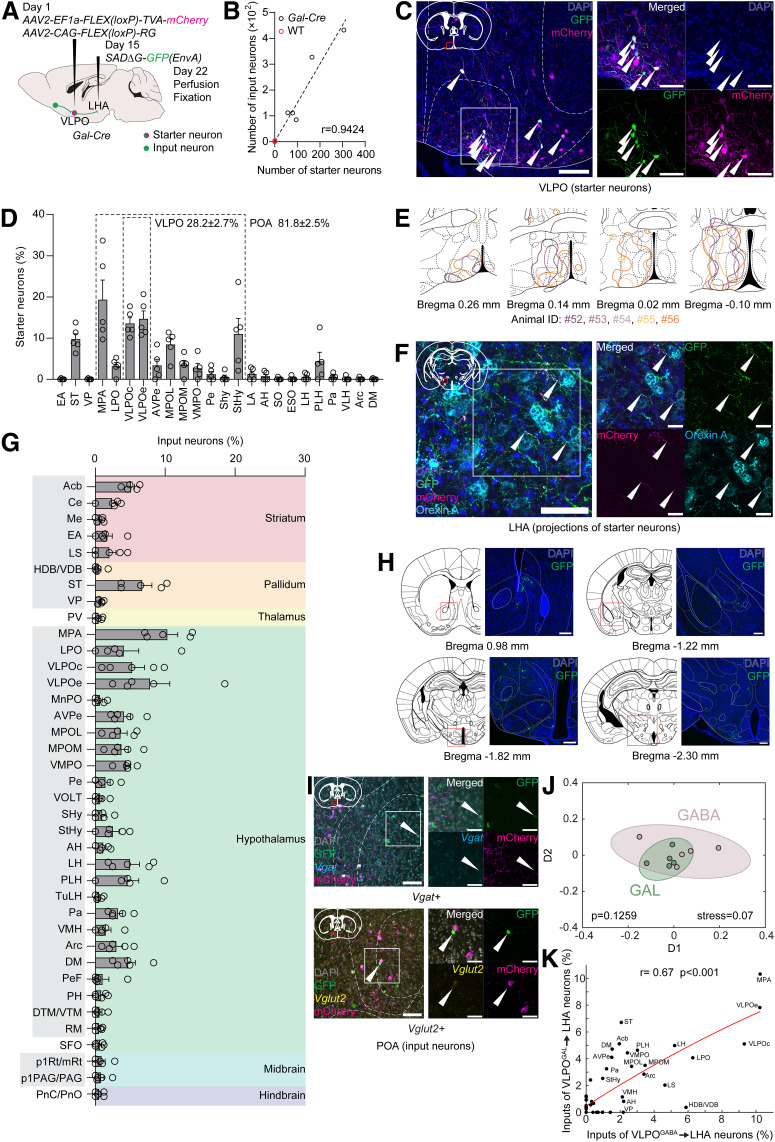
Figure 4.
Monosynaptic retrograde projection-specific rabies virus-mediated tracing from VLPOGAL→LHA neurons and examination of traced connections of two neuronal populations. A, Scheme of projection-specific tracing experiment. B, Number of starter neurons has strong positive correlation with number of input neurons in Gal-Cre mice (Pearson correlation, r = 0.9424, p = 0.0164; n = 5). Linear regression: F(1,4) = 114.0, p = 0.0004, R2 = 0.8882. In WT mice, several input neurons were observed only in 1 animal (n = 3). C, Representative images of VLPO starter neurons in Gal-Cre mice. Starter neurons were immunostained with anti-GFP and anti-mCherry antibodies. Right, Magnified image of the boxed area in the left image. Arrowheads indicate starter neurons. Scale bars: left, 100 μm; right, 50 μm. D, Distribution of starter VLPOGAL→LHA neurons in POA and neighboring regions. Highlighted areas represent POA and VLPO. Areas with starter neurons detected in all examined animals without cut-off are shown. E, Scheme depicting outlines of distribution of starter neurons in cross-sections containing VLPO. Colors represent individual animals. If there were no matching brain sections to the presented schemes in some animals, outlines were not shown. F, Representative images of axonal terminals of VLPOGAL→LHA neurons making close appositions to bodies of orexin A-expressing neurons in LHA. Immunostaining was performed using anti-GFP, anti-mCherry, and anti-orexin A antibodies. Right, Magnified image of the boxed area in the left image. Arrowhead indicates GFP- and mCherry-double positive neural fibers. Scale bars: left, 50 μm; right, 20 μm. G, Whole-brain distribution of monosynaptic inputs of VLPOGAL→LHA neurons in Gal-Cre mice. Only brain areas with input neurons in at least 3 of 5 animals are shown. Colored areas represent major brain divisions. Abbreviations of input brain areas that are shared with VLPOGABA→LHA neurons (Fig. 3G) are highlighted by gray background. Circles represent data from individual animals. Data are mean ± SEM (n = 5). H, Representative images of presynaptic partners of VLPOGAL→LHA neurons. Scale bars, 200 μm. I, Representative images of presynaptic partners of VLPOGAL→LHA neurons in POA. FISH with Vgat, Vglut2, and Adcyap1 probes. For each molecular marker: right, magnified image of the boxed area in the left image. Arrowheads indicate GFP-positive neuronal bodies expressing examined molecular signatures. Scale bars: left, 100 μm; right, 50 μm. J, Multidimensional scaling plot of monosynaptic input neurons of VLPOGABA→LHA (shown as GABA) and of VLPOGAL→LHA (shown as GAL) neurons (n = 5 for each group). Colored circles represent individual animals. Semitransparent circles represent closely located clusters formed of individual animals in each neuronal population, suggesting high similarity of presynaptic partners between them (F score = 1.5117, p = 0.1259, stress = 0.07). K, Spearman correlation plot comparing distribution of input neurons of VLPOGABA→LHA and VLPOGAL→LHA neurons. Distribution of presynaptic partners significantly correlates between neuronal groups (r = 0.67, p < 0.001).
To conduct multidimensional scaling analysis (see Fig. 4J), the numbers of input neurons of VLPOGABA→LHA and VLPOGAL→LHA neurons were first normalized to percentages to ensure a nonbiased representation of each mouse. Observations that were too low to be reliably detected across all mice were considered to be 0. This was done by defining a threshold equal to 100/minN, where minN is the smallest number of counted cells across mice. This threshold represents the smallest percentage of neurons that can be reliably detected across all mice. Connectivity landscapes of VLPOGABA→LHA and VLPOGAL→LHA neurons were compared using two-way analysis of variance with permutation (PERMANOVA). This differs from regular two-way ANOVA in that the significance of F scores is estimated by permutation of mice within groups (1000 permutations) rather than by a normal approximation, therefore disregarding normality assumptions. Multidimensional scaling analysis and PERMANOVA were performed in MATLAB using custom-made codes.
Spearman correlation was conducted to analyze the relationship between the distributions of input neurons of GABA- and galanin-producing neurons in the VLPO projecting to the LHA (see Fig. 4K).
Statistical significances between control and experimental mouse groups were calculated using the following tests. Unpaired two-tailed t test was used for latency to wakefulness in Vgat-ires-Cre and Gal-Cre mice (see Fig. 5E and Fig. 5L, respectively); transition probability to wakefulness within 20 s photostimulation in Gal-Cre mice (see Fig. 5N); number of wakefulness episodes during 10 min pulse stimulation in Vgat-ires-Cre and Gal-Cre mice; percentage of cFos- and orexin A-double-positive neurons in the LHA of Vgat-ires-Cre mice; and number of cFos-positive cells in the LHA of Vgat-ires-Cre mice. Mann–Whitney test was used for total amount of NREM sleep during 2 min photostimulation in Vgat-ires-Cre mice; transition probability to wakefulness within 20 s photostimulation in Vgat-ires-Cre mice (see Fig. 5G); number of ≥240 s wakefulness bouts observed during 1 h starting from the first 1 s light pulse in Vgat-ires-Cre mice (see Fig. 6D); and length of wakefulness episodes during 10 min pulse stimulation in Vgat-ires-Cre and Gal-Cre mice. Two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test was used for total amount of wakefulness before, during, and after 10 min pulse photostimulation in Vgat-ires-Cre and Gal-Cre mice (see Fig. 5H and Fig. 5O, respectively); time course of 5 min-bin wakefulness dynamics during 1 h after the first 1 s light pulse in Vgat-ires-Cre mice (see Fig. 6C); and total amount of wakefulness in percentages during 1 h before the first 1 s light pulse (ZT1), 1 h after the first light pulse (ZT2), and 1 h following them (ZT3) in Vgat-ires-Cre mice (see Fig. 6E). Ordinary two-way ANOVA followed by Sidak multiple comparisons test was used for EEG power density of NREM sleep during the last quarter of 1 h starting from the first 1 s light pulse in Vgat-ires-Cre mice (see Fig. 6F).
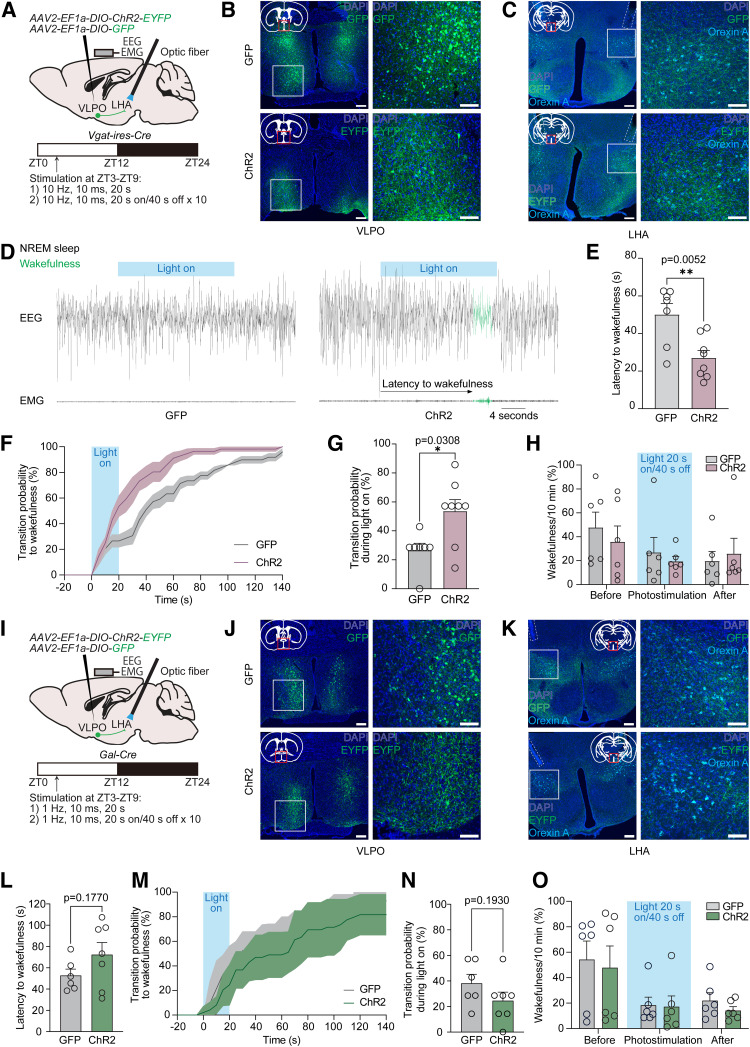
Figure 5.
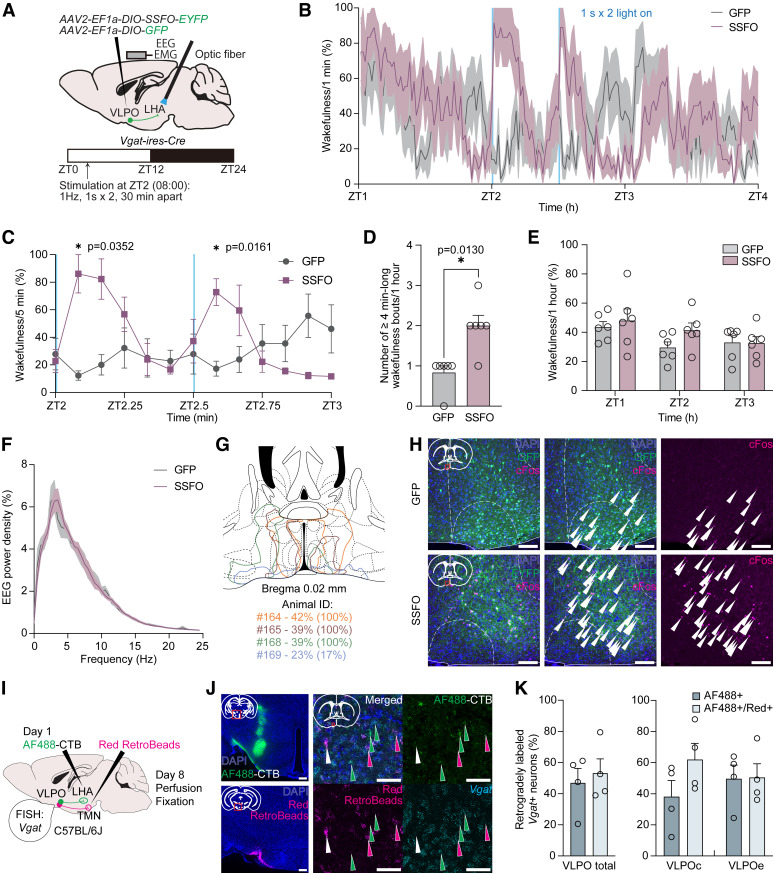
Acute optogenetic stimulation of VLPOGABA→LHA and VLPOGAL→LHA pathways. A, Scheme of pathway-specific optogenetic experiment in Vgat-ires-Cre mice. AAV2-EF1a-DIO-ChR2-EYFP or AAV2-EF1a-DIO-GFP was injected bilaterally into VLPO, and optical fibers were implanted bilaterally into LHA of Vgat-ires-Cre mice for manipulation of axons of GABA-producing VLPO neurons. At the same time, an EEG- and EMG-recording electrode was secured onto the skull. Mice were maintained under a 12:12 h light/dark cycle (06:00 lights on, 18:00 lights off), and photoactivation was conducted for 2 d within the ZT3-ZT9 time frame with at least 40 min interstimulation periods. B, J, Representative images of VLPO neurons expressing GFP (top) or ChR2 (bottom), visualized by immunostaining with anti-GFP antibody, in Vgat-ires-Cre (B) and Gal-Cre (J) mice. Right images, Magnified images of the boxed areas in the left images. Scale bars: left images, 200 μm; right images, 100 μm. C, K, Representative images of GFP- (top) or ChR2-expressing (bottom) axons of VLPO neurons in LHA located close to orexin A-positive neurons in Vgat-ires-Cre (C) and Gal-Cre (K) mice. Immunostaining was performed using anti-GFP and anti-orexin A antibodies. Right images, Magnified images of the boxed areas in the left images. Scale bars: left images, 200 μm; right images, 100 μm. D, Representative EEG and EMG wave traces measuring 40 s (10 s before, 20 s during, and 10 s after optogenetic stimulation) from 2 animals injected with AAV2-EF1a-DIO-GFP (GFP, left) or AAV2-EF1a-DIO-ChR2-EYFP (ChR2, right). Blue line indicates laser stimulation (10 Hz, 10 ms, 20 s). Black represents NREM sleep. Green represents wakefulness. Latency to wakefulness was calculated as time in seconds from the start of each photostimulation to the onset of wakefulness. E, Latency to wakefulness after photostimulation. Significant difference was observed between GFP (n = 7) and ChR2 (n = 8) animals (first 7 stimulations per animal; p = 0.0052, unpaired two-tailed t test). F, Probability of transition to wakefulness in GFP and ChR2 mice within 140 s after start of photoactivation. Blue field represents optogenetic stimulation. G, Probability of transition to wakefulness during 20 s of photostimulation. Significant difference was observed between GFP and ChR2 animals (p = 0.0308, Mann–Whitney test). H, Amount of wakefulness in 10 min time bins in percentages before, during, and after 10 min optogenetic stimulation. No significant difference between GFP (n = 6) and ChR2 (n = 6) mice was observed (two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). I, Scheme of pathway-specific optogenetic experiment in Gal-Cre mice. L, Latency to wakefulness after photostimulation. No significant difference was observed between GFP- (n = 6) and ChR2-expressing (n = 7) animals (first 7 stimulations per animal; p = 0.1770, unpaired two-tailed t test). M, Probability of transition to wakefulness in GFP and ChR2 mice within 140 s after start of photoactivation. Blue field represents optogenetic stimulation. N, Probability of transition to wakefulness during 20 s of photostimulation. No significant difference was observed between GFP and ChR2 animals (p = 0.1930, unpaired two-tailed t test). O, Amount of wakefulness in 10 min time bins in percentages before, during, and after 10 min optogenetic stimulation. No significant difference between GFP (n = 6) and ChR2 (n = 6) mice was observed (two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). Circles represent data from individual animals. Data are mean ± SEM. *p < 0.05. **p < 0.01.
Figure 6.
Long-term SSFO-mediated optogenetic stimulation of VLPOGABA→LHA pathway. A, Scheme of the pathway-specific optogenetic experiment. AAV2-DIO-SSFO-EYFP or AAV2-EF1a-DIO-GFP were injected bilaterally into VLPO, and optical fibers were implanted bilaterally into LHA of Vgat-ires-Cre mice. An EEG- and EMG-recording electrode was secured onto the skull. Mice were maintained under a 12:12 h light/dark cycle (06:00 lights on, 18:00 lights off), and photoactivation was applied 2 h after the onset of light phase (at ZT2 or 08:00; 1 s light pulse × 2, with 30 min interstimulation period). B, Time course of the amount of wakefulness during 1 h before the first 1 s light pulse (ZT1-ZT2; 07:00-08:00), 1 h after the first light pulse (ZT2-ZT3; 08:00-09:00), and 1 h following them (ZT3-ZT4; 09:00-10:00). Shown is the amount of wakefulness in 1 min time bins in percentages. Two blue lines indicate photostimulation. C, Time course of the amounts of wakefulness in 5 min bin during 1 h after the first light pulse. Significant differences were observed in the amount of wakefulness during the first 5 min after the light pulses between GFP- (n = 6) and SSFO-expressing (n = 6) mice (p = 0.0352 after the first light pulse and p = 0.0161 after the second light pulse, two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). D, Number of ≥4-min-long (240 s-long) wakefulness bouts, observed during 1 h from the first light pulse. Significant difference was observed between GFP and SSFO mice (p = 0.0130, Mann–Whitney test). E, Total amount of wakefulness in percentages during 1 h before the first 1 s light pulse (ZT1; 07:00), 1 h after the first light pulse (ZT2; 08:00), and 1 h following them (ZT3; 09:00). No significant difference was observed (two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). F, Relative amplitude of average EEG power density of NREM sleep during the last quarter (ZT2.75-ZT3; 08:45-09:00) of 1 h starting from the first light pulse (ZT2; 08:00). No significant difference was observed between GFP control and SSFO mice (ordinary two-way ANOVA followed by Sidak multiple comparisons test). G, Scheme depicting outlines of distribution of SSFO-expressing neurons in a cross-section containing VLPO. Brain outlines were taken from the Franklin and Paxinos (2007) mouse brain atlas. Colors represent individual animals. Four animals with the most distinct vector expression patterns are shown. Percentage following animal ID shows the amount of wakefulness during an hour since the first light pulse. Percentage in parentheses shows the amount of wakefulness during the first 5 min after the first light pulse. H, Representative images of cFos-positive VLPO neurons expressing GFP (top) or SSFO (bottom), visualized by immunostaining with anti-GFP and anti-cFos antibodies. Right images, Single-channel images showing cFos signal in cell nuclei. Scale bars, 100 μm. I, Scheme of dual-color retrograde tracing experiment. Recombinant cholera toxin subunit B conjugated with AlexaFluor-488 (AF488-CTB) was delivered into LHA, and Red RetroBeads was delivered into TMN of WT C57BL/6J mice. Retrogradely labeled neurons in VLPO were visualized for Vgat using FISH, and Vgat-expressing neurons solely labeled with AF488-CTB or colabeled with Red RetroBeads were counted. J, Representative images of injection sites of retrograde traces (left images) and neurons in VLPO containing the tracers (right). Green arrowheads indicate AF488-CTB single-positive neurons in the VLPO. Magenta arrowheads indicate Red RetroBeads single-positive neurons. White arrowhead indicates an AF488-CTB and Red RetroBeads double-positive neuron. Scale bars: left images, 200 μm; right, 50 μm. K, Percentages of Vgat-expressing VLPO neurons categorized as AF488-CTB+ (AF488+) and AF488-CTB/Red RetroBeads-double positive (AF488+/Red+) (n = 4). Circles represent data from individual animals. Data are mean ± SEM. *p < 0.05.
All statistical analyses were performed using GraphPad Prism 9.4.0 (unless noted otherwise). Data are presented as mean ± SEM.
Results
Vgat-, Gal-, and Vglut2-expressing neurons in VLPO make direct synaptic contact with orexin neurons
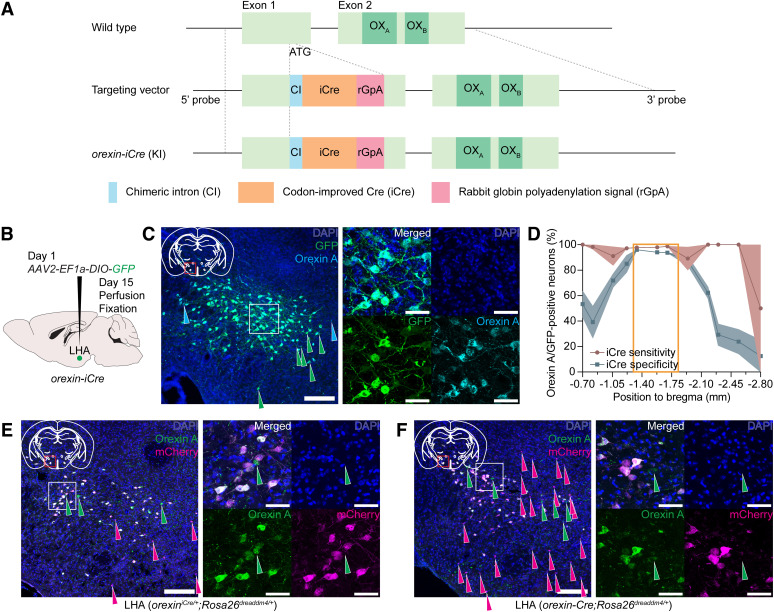
To precisely target orexin-producing neurons, we newly generated orexin-iCre knock-in mice (orexiniCre/+), in which codon-improved Cre recombinase (iCre) expression is driven by the endogenous orexin promoter (Fig. 1A). To validate this iCre driver line, we delivered AAV2-EF1a-DIO-GFP into the LHA of orexiniCre/+ mice and examined sensitivity and specificity of iCre recombinase activity (Fig. 1B). The sensitivity was defined as a percentage of orexin A/GFP-double positive neurons out of all orexin A-positive neurons, which were identified by immunohistochemistry using anti-orexin A antibody, and was found to be 97.4 ± 0.7% (n = 3). The specificity was defined as a percentage of orexin A/GFP-positive neurons out of all GFP-positive neurons and was determined to be 89.4 ± 0.7% (n = 3; Fig. 1C). Both sensitivity and specificity were >90% at the craniocaudal position where the highest density of orexin A-positive neurons was observed (Fig. 1D). Further, to compare the sensitivity between the orexiniCre/+ mice and previously generated orexin-Cre transgenic (Tg) mice (Matsuki et al., 2009), we crossed orexiniCre/+ mice with Rosa26dreaddm4 mice as a reporter, in which the CAG promoter and a loxP-flanked STOP sequence followed by the hM4Di-mCherry coding sequence are knocked into the Gt(ROSA)26Sor locus (T. M. Takahashi et al., 2020). Similarly to the results of our AAV experiment (Fig. 1B–D), we found that 95.8 ± 1.0% of orexin A-positive neurons expressed mCherry in orexiniCre/+;Rosa26dreaddm4/+ mice (Fig. 1E; n = 3). On the other hand, 79.4 ± 6.8% of orexin A-positive neurons expressed mCherry in the orexin-Cre Tg mice when crossed with Rosa26dreaddm4/+ mice (orexin-Cre Tg;Rosa26dreaddm4/+; Fig. 1F; n = 3). These results demonstrated higher sensitivity of iCre activity in orexiniCre/+ mice compared with orexin-Cre Tg mice (Matsuki et al., 2009). Although we found a small number of ectopic mCherry-expressing neurons in the tuber cinereum and amygdala regions, we did not think it caused a major problem as long as this line was used for delivery of Cre-activatable AAV vectors in the LHA locally.
Figure 1.
Generation of orexin-iCre knock-in mice. A, Structures of the targeting vector and targeting allele of orexin-iCre (orexiniCre/+) mice. B, Scheme of the validation experiment of orexin-iCre knock-in mice. AAV2-EF1a-DIO-GFP was injected bilaterally into the LHA. Two weeks later, mice were killed, and mouse brains were examined by immunohistochemical study. C, Representative images demonstrating colocalization of GFP and orexin A in the LHA that were immunostained with anti-GFP and anti-orexin A antibodies, respectively, in orexiniCre/+ mice. Right, Magnified image of the boxed area in the left image. Green and cyan arrowheads indicate GFP- and orexin A-single-positive neurons, respectively. Scale bars: left, 200 μm; right, 50 μm. D, Graph represents sensitivity and specificity of iCre activity in the craniocaudal axis in orexiniCre/+ mice. Orange rectangle represents an area with densely located orexin A-positive neurons in a mouse brain. Data are mean ± SEM; n = 3. E, F, Representative images demonstrating colocalization of hM4Di (dreaddm4)-mCherry and orexin A in LHA immunostained with anti-mCherry and anti-orexin A antibodies in orexiniCre/+;Rosa26dreaddm4/+ mice (E) and orexin-Cre;Rosa26dreaddm4/+ mice (F). Right images, Magnified images of the boxed areas in the left images. Green and magenta arrowheads indicate orexin A- and mCherry-single positive neurons, respectively. Scale bars: left images, 200 μm; right images, 50 μm.
We first examined the synaptic connectivity between neurons in the VLPO and orexin neurons. We conducted two batches of monosynaptic retrograde tracing experiments using rabies virus vectors SADΔG-GFP(EnvA) in the first batch and SADΔG-GFP-ERT2CreERT2 in the second batch at different time points (Fig. 2A). We used GFP-ERT2CreERT2 as a tracer in the second batch because it can be distributed in the nucleus with tamoxifen administration, in case it was difficult to confirm coexistence with Gal, Vglut2, or Vgat mRNA. However, this turned out to be unnecessary.
To minimize ectopic expression of AAV10-EF1a-FLEX(loxP)-TVA-mCherry (TVA-mCherry) and to restrict numbers of starter neurons, we conducted a series of preliminary experiments with several dilutions of TVA-mCherry vectors, as suggested by previous studies (Lavin et al., 2019, 2020), for each batch of experiments. Using the most successful dilutions, we found moderate numbers of starter neurons in the LHA (GFP- and mCherry-double-positive; 16 ± 4 cells/brain in the first batch, n = 4; 10 ± 2 cells/brain in the second batch, n = 3) and of input neurons (GFP-single-positive; 357 ± 159 cells/brain in the first batch, n = 4) in orexiniCre/+ mice, with a clear positive correlation between the numbers of these two subsets of neurons (Fig. 2B, black circles; Pearson correlation, r = 0.9178, p = 0.0822, n = 4; Fig. 2C, data from the first batch). Only a few GFP- and/or mCherry-positive neurons were observed at the injection sites in WT control mice (Fig. 2B, red circles, n = 3). The relatively small number of starter neurons obtained by dilution of TVA-mCherry allowed us to examine connectivity of orexin neurons precisely.
In both experiments, the majority of starter neurons in the LHA were orexin A-positive, and their distribution was consistent throughout all animals (87.7 ± 8.2% with 12.3 ± 8.2% of orexin A-negative neurons, n = 7). Their distribution was similar to that described in rodent brains (Peyron et al., 1998; Nambu et al., 1999). The specificity of starter neurons was lower than that of iCre activity, calculated in Figure 1C, D, presumably because of the toxicity of the rabies virus and its impact on gene expression in the infected neurons, as previously reported (Sun et al., 2019).
Input neurons of orexin neurons were distributed across the entire brain in a consistent pattern with earlier studies (González et al., 2016; Giardino et al., 2018; Saito et al., 2018). We here focused on the VLPO and precisely characterized the input neurons in this region (Fig. 2D–J). We adopted classification of this region into the VLPO cluster (VLPOc) and the dorsomedial extended VLPO (VLPOe), both of which have been shown to participate in NREM sleep regulation (Kroeger et al., 2018). The VLPOe was also suggested to contribute to REM sleep regulation (Lu et al., 2000, 2002; Scammell et al., 2017). We demarcated the VLPO subdivisions according to criteria described in the previous reports and to the location of sleep-active neurons that were obtained from the past electrophysiological and immunohistochemical studies (Fig. 2D, colored areas) (Sherin et al., 1996; Szymusiak et al., 1998; Lu et al., 2000, 2002; Wagner et al., 2000; Gaus et al., 2002; K. Takahashi et al., 2009; Alam et al., 2014; Kroeger et al., 2018).
Further, we determined whether the input neurons in the VLPO contained GABA, galanin, and glutamate, since these are the molecular signatures of sleep- and wakefulness-implicated neurons in this area (Chung et al., 2017; Kroeger et al., 2018; Vanini et al., 2020; Yamagata et al., 2021). We conducted a FISH study to visualize the expression of vesicular GABA transporter (Vgat, Slc32a1) and vesicular glutamate transporter 2 (Vglut2, Slc17a6), which are the molecular markers of GABA- and glutamate-producing neurons, respectively, as well as expression of galanin (Gal) in the VLPO input neurons (Fig. 2E,G,I). We examined colocalization of Vgat and Gal in the first batch of the tracing experiments (Fig. 2E), and colocalization of Gal and Vglut2 or Vglut2 and Vgat in the second batch (Fig. 2G,I). As a result, we found that more than half of the input neurons (56.3 ± 8.1%) were double positive for Vgat- and Gal (VLPOVgat/Gal neurons; Fig. 2F, left; 38 ± 10 VLPO input neurons in total, n = 4). VLPOVgat/Gal neurons were more concentrated in the VLPOc compared with the VLPOe (Fig. 2F, middle, right). A small population of the input neurons was solely positive for Gal (2.8 ± 2.3%), while Vgat-positive neurons (85.7 ± 5.4%) constituted the great majority of the total number of input neurons in the VLPO (Fig. 2F). We also investigated colocalization of Gal and Vglut2 in the VLPO input neurons; however, we did not find any Gal- and Vglut2-double-positive inputs in the examined samples (Fig. 2G,H; 17 ± 3 VLPO input neurons in total, n = 3). Notably, we found a small population of Vglut2- and Vgat-copositive input neurons (6.3 ± 3.6%, 12 ± 2 VLPO input neurons in total, n = 3), suggesting high molecular heterogeneity of presynaptic partners in the VLPO that send innervations to orexin neurons (Fig. 2I,J). Together, we precisely delineated connectivity of orexin neurons with major sleep- and wakefulness-associated subpopulations of VLPO neurons, and determined the chemical identities of VLPO neurons that make direct synaptic contact with orexin neurons in the LHA.
VLPOGABA→LHA neurons receive input from multiple brain areas
We next conducted a projection-specific rabies virus-mediated tracing experiment using Vgat-ires-Cre mice (Fig. 3A) (Piñol et al., 2018). We first injected a Cre-dependent AAV mixture for expressing TVA and rabies glycoprotein (RG) into the VLPO. Two weeks later, we delivered SADΔG-GFP(EnvA) into the LHA to infect TVA-expressing VLPOGABA neurons through their axonal terminals, allowing this vector to exclusively infect VLPOGABA neurons that send innervations to the LHA (VLPOGABA→LHA neurons) and to trans-synaptically spread in a retrograde direction.
We found a considerable number of TVA-mCherry- and GFP-double-positive GABA-producing starter neurons in the POA (294 ± 39 neurons/brain, n = 5), which were positively correlated with the number of their input neurons (GFP-single positive) in Vgat-ires-Cre mice (Fig. 3B, black circles; Pearson correlation, r = 0.3292, p = 0.5885; n = 5). Only a few starter and input neurons were found around the injection site in the VLPO of WT control mice (Fig. 3B, red circles, n = 3). Starter neurons were distributed across the POA (75.5 ± 5.3% of all starter neurons) with some involvement of adjacent regions (Fig. 3C–E). The VLPOc and VLPOe contained 36.8 ± 2.3% of all starter neurons, constituting nearly half of them in the POA (Fig. 3D; 49.2 ± 3.2% of POA starter neurons). We also observed mCherry- and GFP-double-positive axons making appositions to cell bodies of orexin A-positive neurons in the LHA, suggesting that some starter cells sent projections to orexin neurons (Fig. 3F).
A large number (928 ± 328 neurons/brain, n = 5) of input neurons to VLPOGABA→LHA neurons were widely distributed among various brain areas (Fig. 3G,H). The biggest number (≥10%) of input neurons was found in the medial preoptic area (MPA) and the VLPO, while regions with less concentrated input neurons (≥5%) were the diagonal band nucleus, lateral preoptic area (LPO), medial preoptic nucleus, and LHA. These results suggest that the MPA and VLPO regions constitute major inputs of the VLPOGABA→LHA neurons. LHA also contains a considerable number of input neurons, suggesting the presence of reciprocal connectivity between the VLPO and LHA. This result is also consistent with previous studies that reported on the arousal-promoting LHA→POA circuits (Venner et al., 2019; de Luca et al., 2022).
A moderate number of input neurons (≥2.5%) were located in the lateral septal nucleus, ventromedial preoptic nucleus, and arcuate hypothalamic nucleus. Small numbers of input neurons (<2.5%) were also found in multiple regions, such as the nucleus accumbens (Acb), amygdala, bed nucleus of stria terminalis (ST), ventral pallidum, and paraventricular, ventromedial, and dorsomedial hypothalamic nuclei. Despite apparent variability in the number and distribution of input and starter neurons between individual animals, presumably because of variability in expression of viral vectors, input neurons in all exhibited areas were consistently represented in at least three independent experiments (Fig. 3G). Such stability of the landscape of presynaptic partners of the VLPOGABA→LHA neurons might reflect the functional significance of these monosynaptic connections for the regulation of physiological processes.
We also characterized the local preoptic input neurons of the VLPOGABA→ LHA neurons using FISH to disentangle connectivity within the POA. We visualized the expression of Vgat and Vglut2 in input neurons because neurons expressing these molecular markers are proposed to be implicated in sleep-wakefulness regulation (Arrigoni and Fuller, 2022). This study found substantial numbers of Vgat- or Vglut2-positive input neurons in the POA (Fig. 3I, top, middle, n = 2). We also examined whether the POA input neurons of the VLPOGABA→ LHA neurons express Pacap (Adcyap1), which was shown to be expressed in neurons involved in thermoregulation (Tan et al., 2016). We found Pacap-expressing input neurons in the MPA (Fig. 3I, bottom, n = 2). These results suggest that the VLPOGABA→LHA neurons are interconnected with cells related to different functional units within the POA.
Input neurons of VLPOGABA→LHA and VLPOGAL→LHA neurons show similar distribution
Next, we identified monosynaptic inputs of galanin neurons in the VLPO projecting to the LHA (VLPOGAL→LHA neurons; Fig. 4A) using Gal-Cre mice (Wu et al., 2014). Implementing projection-specific retrograde rabies virus-mediated tracing, we labeled an appropriate number of starter neurons (141 ± 45 neurons/brain, n = 5) within the POA and neighboring areas (Fig. 4B, black circles; Fig. 4C–E). While 28.2 ± 2.7% of all starter neurons were located in both VLPOc and VLPOe, 81.8 ± 2.5% of neurons resided in the POA (Fig. 4D). We also found that the absolute numbers of input neurons and VLPOGAL→LHA starter neurons were positively correlated, while only several input neurons were detected in WT mice (Fig. 4B, black and red circles; Pearson correlation, r = 0.9424, p = 0.0164; n = 5; WT mice n = 3). As for the VLPOGABA→LHA neurons, we found that some axons of the starter VLPOGAL→LHA neurons made close appositions to the cell bodies of orexin A-positive neurons (Fig. 4F).
Many monosynaptic input neurons (213 ± 70 neurons/brain, n = 5) were found in various brain areas, with a similar distribution pattern to input neurons of the VLPOGABA→LHA neurons (Fig. 4G,H; Fig. 3G, shared input brain areas are highlighted by gray background). The highest number of input neurons (≥10%) was found in the MPA and the VLPO. Brain areas with less abundant input neurons (≥5%) were the Acb, ST, medial preoptic nucleus, and LHA. An intermediate number of input neurons (≥2.5%) were located in the LPO, anteroventral periventricular and ventromedial preoptic nuclei, and striohypothalamic nucleus, as well as in the paraventricular, arcuate, and dorsomedial hypothalamic nuclei. Input cells were also found in the central nucleus of the amygdala, lateral septal complex, and anterior hypothalamus, with <2.5% of the total number of input neurons in each region.
We also examined whether Vgat, Vglut2, and Pacap (Adcyap1) are contained in the preoptic monosynaptic input neurons of the VLPOGAL→LHA neurons, and found that Vgat- and Vglut2-positive POA neurons made direct synaptic inputs to these cells (Fig. 4I, n = 2). However, we did not observe any Pacap-expressing presynaptic partners in the POA at least in the two samples that we examined here.
Further, we compared input neuron landscapes of the VLPOGABA→LHA and VLPOGAL→LHA neurons. Using multidimensional scaling analysis, we identified that both neuronal populations were clustered together according to their inputs (Fig. 4J; p = 0.1259, stress = 0.07, n = 5 for each mouse group). In addition, we found that the distributions of the presynaptic partners were strongly correlated (Fig. 4K; Spearman correlation, r = 0.67, p < 0.001). These results were consistent with our observation that the majority of VLPO input neurons to orexin neurons were Vgat- and Gal-double-positive (Fig. 2F, left) and with findings in a recent study (de Luca et al., 2022), therefore suggesting that the VLPOGABA→LHA and VLPOGAL→LHA neurons constitute a common neuronal population. However, there were some differences in the input brain areas between VLPOGABA→LHA and VLPOGAL→LHA neurons. For instance, regions in cortex, extended amygdala, septum, anterior and posterior hypothalamus, as well as dorsal raphe nucleus and VTA in midbrain, were exclusive to VLPOGABA→LHA neurons (Fig. 3G). On the other hand, paraventricular thalamic nucleus, subfornical organ in hypothalamus, and reticular pontine nucleus were consistently found among upstream brain areas only for VLPOGAL→LHA neurons (Fig. 4G).
Acute optogenetic activation of VLPOGABA→LHA and VLPOGAL→LHA pathways produces distinct phenotypes with regard to wakefulness induction
We next examined the function of the VLPO→LHA pathway in the regulation of sleep-wakefulness behavior by optogenetic excitation of axon fibers of VLPOGABA neurons in the LHA. We delivered Cre-dependent AAVs expressing ChR2 fused with EYFP or expressing GFP (AAV2-EF1a-DIO-ChR2-EYFP or AAV2-EF1a-DIO-GFP, respectively) into the VLPO and implanted optical fibers into the LHA of Vgat-ires-Cre mice.
We first conducted 2 min photostimulation (10 Hz, 10 ms) 2 h after the onset of the dark phase (ZT14) during the wakefulness state, which lasted for 1 min. We conducted the experiment in the dark period because we initially hypothesized that the VLPOGABA→LHA pathway would have a role in inducing sleep. However, we observed that only one ChR2-delivered mouse was able to enter NREM sleep during photostimulation (total amount of NREM sleep during 2 min photostimulation 0.7 ± 0.7% in ChR2 mice, n = 8 vs 4.0 ± 2.8% in GFP mice, n = 7; p = 0.3231, Mann–Whitney test), suggesting that the VLPOGABA→LHA pathway does not play a major role in inducing sleep. Previous studies proposed that VLPOGABA neurons are composed of distinct subpopulations, presumably defined by the projection site, receptor expression, and expression of neuronal modulators, and that these subpopulations contribute to sleep or wakefulness, driving in an opposite manner (Liu et al., 2010; Chung et al., 2017; Liang et al., 2021; Yamagata et al., 2021; de Luca et al., 2022). We therefore examined whether the VLPOGABA→LHA and VLPOGAL→LHA pathways are rather involved in inducing wakefulness, using Vgat-ires-Cre and Gal-Cre mice, respectively (Fig. 5A, I).
We conducted the same surgical procedure and confirmed that orexin A-positive neurons were surrounded by the projections of ChR2- or GFP-expressing VLPOGABA neurons (Fig. 5B,C) in Vgat-ires-Cre mice. We applied photostimulation (10 Hz, 10 ms, 20 s) of ChR2- or GFP-expressing axonal fibers in the LHA during NREM sleep within the ZT3-ZT9 time window (12:12 h light/dark cycle; 06:00 lights on, 18:00 lights off), using similar optogenetic parameters (Chung et al., 2017; Yamagata et al., 2021) and experiment protocol to previous studies (Kodani et al., 2017; Venner et al., 2019) (Fig. 5A).
We observed that 20 s stimulation of the axons of VLPOGABA neurons in the LHA evoked short-lasting wakefulness episodes in ChR2-expressing mice (Fig. 5D; Movies 1 and 2). Latency to the wakefulness episodes after optogenetic stimulation in ChR2 mice was significantly shorter than that in GFP control mice (Fig. 5E; 27.0 ± 3.9 s in ChR2 mice, n = 8 vs 50.1 ± 5.9 s in control mice, n = 7; p = 0.0052, unpaired two-tailed t test). We also found that probability of transition from NREM sleep to wakefulness was significantly higher in ChR2-expressing mice on photostimulation (Fig. 5F,G; 53.6 ± 8.0% in ChR2 mice vs 26.5 ± 4.9% in GFP mice by the end of 20 s photostimulation; p = 0.0308, Mann–Whitney test). We also conducted 10 min pulse photostimulation (10 Hz, 10 ms, 20 s light on/40 s light off × 10), as described previously (Kodani et al., 2017). We did not observe a significant difference in the length of wakefulness episodes between mouse groups (21.6 ± 12.3 s in ChR2 mice, n = 6 vs 147.4 ± 138.5 s in GFP mice, n = 6; p = 0.9091, Mann–Whitney test), while the number of wakefulness episodes was slightly greater in ChR2 mice (8.2 ± 1.0 vs 5.5 ± 1.7 in GFP group; p = 0.2161, unpaired two-tailed t test). The total amount of wakefulness before, during, and after the optogenetic stimulation was also indistinguishable between the mouse groups (Fig. 5H; two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). These results indicate that activation of the VLPOGABA→LHA pathway using ChR2 induces transition from NREM to wakefulness, although it was very short-lasting.
Video clip of GFP-expressing Vgat-ires-Cre mice with EEG/EMG tracing during 10 s pre-photostimulation, 20 s photostimulation, and 10 s post-photostimulation periods. N, NREM sleep.
ChR2-expressing Vgat-ires-Cre mice with EEG/EMG tracing during 10 s pre-photostimulation, 20 s photostimulation, and 10 s post-photostimulation periods. A short episode of wakefulness was observed. N, NREM sleep; W, wakefulness.
On the other hand, in Gal-Cre mice, we did not observe such a wakefulness-promoting effect. We performed surgery as described above and conducted histologic verification of viral vectors expression and optical fiber implantation, similarly to Vgat-ires-Cre mice (Fig. 5I–K). In this experiment, we have modified frequency of the stimulation to 1 Hz (1 Hz, 10 ms, 20 s) because it had been previously determined as optimal for galanin-producing neurons in the VLPO, while 10 Hz frequency resulted in their hyperpolarization (Kroeger et al., 2018). We applied photostimulation to axonal fibers in the LHA of the ChR2- or GFP-expressing VLPOGAL neurons during NREM sleep. However, there were no significant differences in the latency to wakefulness (72.3 ± 11.4 s in ChR2 mice, n = 7 vs 52.6 ± 6.1 s in GFP mice, n = 6; p = 0.1770, unpaired two-tailed t test) and probability to transition to wakefulness (24.5 ± 6.8% in ChR2 mice vs 38.1 ± 7.1% in control mice by the end of the photostimulation; p = 0.1930, unpaired two-tailed t test) between ChR2 and GFP mice (Fig. 5L–N). Similarly, 10 min pulse stimulation did not produce significant differences in the amount of wakefulness before, during, and after the photostimulation between two mouse groups (Fig. 5O; 17.1 ± 8.5% of wakefulness in ChR2 mice, n = 6 vs 18.3 ± 6.4% in GFP mice, n = 6; p = 0.9993, two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test), as well as in the length (65.6 ± 55.7 s in ChR2 mice, n = 6 vs 49.9 ± 39.2 s in GFP mice, n = 6; p = 0.9740, Mann–Whitney test) and the number (4.2 ± 1.1 in ChR2 mice, n = 6 vs 6.2 ± 1.4 in GFP mice, n = 6; p = 0.2841, unpaired two-tailed t test) of wakefulness episodes. Therefore, we concluded that the VLPOGAL→LHA pathway does not participate in promotion of wakefulness.
Additionally, we identified how many galanin-expressing neurons overlapped with ChR2-expressing GABA-producing neurons in Vgat-ires-Cre mice that underwent the 20 s and 10 min photostimulations (Fig. 5A) by FISH using Gal probe. We found that only 29.6 ± 3.2% of Gal-positive neurons colocalized with ChR2-expressing neurons (n = 4), suggesting that the phenotypic differences between Vgat-ires-Cre and Gal-Cre mice were primarily because of the function of GABA neurons that do not produce galanin.
Together, we found that acute optogenetic activation of the VLPOGABA→LHA and VLPOGAL→LHA pathways results in different phenotypes, where the VLPOGABA→LHA pathway induces a short-lasting wakefulness, while the VLPOGAL→LHA pathway does not significantly contribute to regulation of wakefulness and sleep.
Long-term optogenetic activation of VLPOGABA→LHA pathway increases wakefulness
Since we found that activation of the VLPOGABA→LHA pathway provoked short bouts of wakefulness, we next examined an effect of a more long-term stimulation using SSFO (Yizhar et al., 2011) that induces membrane depolarization for ∼30 min in targeted neurons on photoactivation. We conducted the same surgical procedure, as above, using AAV2-EF1a-DIO-SSFO-EYFP and AAV2-EF1a-DIO-GFP vectors, and applied photostimulation (1 Hz, 1 s light pulse, 2 times, 30 min interval) of SSFO- or GFP-expressing axonal fibers in the LHA 2 h after the onset of the light phase (ZT2; Fig. 6A). We found that 1 s light pulses resulted in immediate transient increases in wakefulness in SSFO-expressing mice (Fig. 6B,C). Wakefulness was significantly increased in the first 5 min after the first and second light pulse applications in SSFO mice compared with control GFP mice (Fig. 6C; 86.1 ± 13.9% in SSFO mice, n = 6 vs 12.2 ± 3.4% in GFP mice, n = 6 after the first light pulse, p = 0.0352, and 72.8 ± 9.8% in SSFO mice vs 17.2 ± 5.4% in GFP mice after the second light pulse, p = 0.0161, two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). Additionally, the number of long wakefulness bouts (≥240 s) was significantly increased in SSFO mice (Fig. 6D; 2.0 ± 0.3 in SSFO mice vs 0.8 ± 0.2 in GFP mice; p = 0.0130, Mann–Whitney test). Interestingly, the increase in wakefulness that was observed after the second light pulse was weaker than that after the first light pulse (Fig. 6B,C), suggesting desensitization of neurons or homeostatic regulation of sleep. We also observed that photostimulation-induced increases in wakefulness were followed by consisted decreases in this state (Fig. 6B,C). As a result, the total amount of wakefulness during 1 h since the first light pulse (ZT2) was not significantly increased in SSFO mice compared with GFP mice (Fig. 6E; 41.4 ± 5.0% in SSFO mice vs 29.5 ± 3.9% in GFP mice; p = 0.2500, two-way repeated-measures ANOVA with Geisser–Greenhouse correction followed by Sidak multiple comparisons test). This result was similar to that observed on the 10 min pulse photostimulation in ChR2-expressing mice (Fig. 5H). As this phenotype might have resulted from homeostatic regulation of sleep, which was shown to be conducted by LPOGAD2 neurons (glutamate decarboxylase 2-expressing neurons in the LPO) (Yamagata et al., 2021), we also examined EEG power density of NREM sleep during the last quarter (ZT2.75-ZT3) of 1 h starting from the first light pulse (ZT2). However, we did not observe any significant differences between SSFO and control mice (Fig. 6F; ordinary two-way ANOVA followed by Sidak multiple comparisons test).
We next confirmed the expression of SSFO in the VLPO (VLPOc and VLPOe) and found that animals with abundant SSFO expression in the VLPOe produced more pronounced increases in wakefulness than the animals with expression mostly limited to the VLPOc (Fig. 6G). We also observed that optogenetic activation induced a robust increase in cFos in SSFO-expressing neurons compared with GFP-expressing neurons (Fig. 6H, white arrowheads). To examine the effect of activation of the VLPOGABA→LHA pathway on orexin neurons in the LHA, we compared percentages of cFos- and orexin A-double-positive neurons between SSFO and GFP mice. However, we did not find a significant difference between them (28.6 ± 4.7% in SSFO mice, n = 4 vs 25.3 ± 5.2% in GFP mice, n = 4; p = 0.6525, unpaired two-tailed t test). Similarly, there was no significant difference in the number of cFos-positive cells in 1 mm2 areas just ventrally to the tips of optical fibers in the LHA between SSFO and control mice (216.5 ± 29.3 cells in SSFO mice vs 172.3 ± 34.6 cells in GFP mice; p = 0.3674, unpaired two-tailed t test), as this area was previously reported to be affected during photostimulations using optic cannulas (Kroeger et al., 2018).
Together, these results indicate that photoactivation of the VLPOGABA→LHA pathway results in an increase in wakefulness without involving activation of orexin neurons.
Vgat-expressing VLPO neurons projecting to LHA and TMN partially overlap
A previous study identified that GAD2-expressing preoptic neurons projecting to the tuberomammillary nucleus (TMN; POAGAD2→TMN neurons) contribute to the induction of NREM and REM sleep (Chung et al., 2017). GAD2 is another molecular marker of GABA-producing neurons; thus, VLPO possibly contains a GABA-producing neuronal population projecting to the TMN that has a sleep-promoting effect. We therefore examined whether the VLPOGABA neurons projecting to the LHA belong to the same neuronal population originating the POAGAD2→TMN pathway using dual-color retrograde tracing (Fig. 6I). As a first step, we delivered recombinant cholera toxin subunit B conjugated with AlexaFluor-488 into the LHA and Red RetroBeads into the TMN of the same mice. Next, we conducted FISH using Vgat probe on the neurons in the VLPO to examine whether the VLPOGABA→LHA neurons overlap with the VLPOGABA→TMN neurons.
We found that Vgat-positive neurons in the VLPO constituted approximately half of all retrogradely labeled neurons projecting to the LHA (50.6 ± 2.6%, n = 4), which is similar to the results of our rabies virus-mediated tracing experiment of orexin neurons (Fig. 2F,J). Slightly more than half of the Vgat-positive VLPO neurons sending their axons to the LHA overlapped with Vgat-positive VLPO neurons projecting to the TMN (Fig. 6J, white arrowhead; Fig. 6K, left; 53.1 ± 9.2%, n = 4). Neurons in the VLPOc had a greater percentage of overlapping than the neurons in the VLPOe (Fig. 6K, right). Notably, we also observed a substantial number of Vgat-positive VLPO neurons containing only Red RetroBeads (i.e., projecting to the TMN; Fig. 6J, magenta arrowheads). Together, these results indicate that the VLPOGABA→LHA and VLPOGABA→TMN pathways originate from only partially shared, but largely distinct neuronal populations in the VLPO.
Discussion
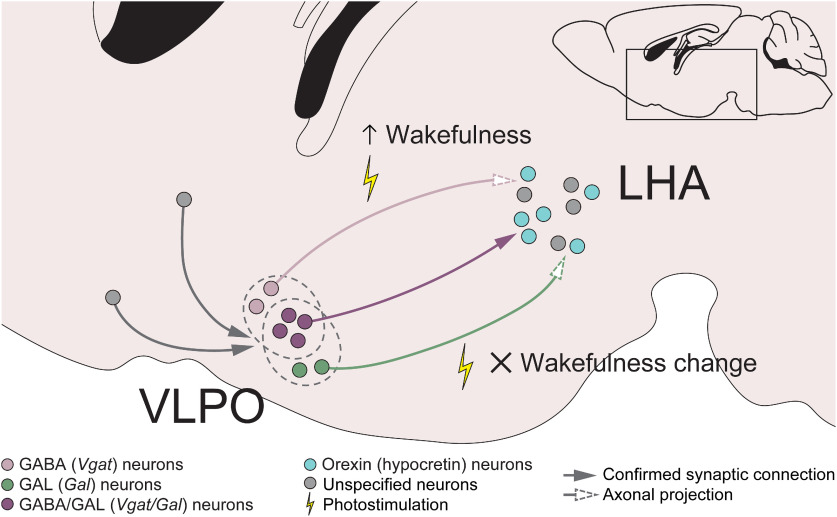
In this study, we analyzed structural and functional connectivity between the VLPO and LHA. We carefully examined the molecular identities of neurons comprising the VLPO→LHA pathway. We first determined that over half (56.3 ± 8.1%) of VLPO monosynaptic input neurons to orexin neurons expressed both Vgat and Gal, with smaller populations expressing Vglut2 and Vgat/Vglut2 (Fig. 2E–J). Further, we identified monosynaptic input neurons of the VLPOGABA→LHA and VLPOGAL→LHA neurons in the entire brain (Figs. 3G, 4G). We compared distributions of these input neurons and found that VLPOGABA→LHA and VLPOGAL→LHA neurons are largely composed of a common neuronal population (Fig. 4J–K). However, we found that acute photoactivation of the VLPOGABA→LHA pathway evoked wakefulness, while photoactivation of the VLPOGAL→LHA pathway did not (Fig. 5E–G,L–N), suggesting their different physiological functions or existence of subpopulations in these pathways (Fig. 7). In addition, long-term photoactivation of the VLPOGABA→LHA pathway increases wakefulness (Fig. 6B–D).
Figure 7.
Regulation of wakefulness by VLPOGABA→LHA and VLPOGAL→LHA pathways. GABA (Vgat)- and GAL (Gal)-expressing neurons in the VLPO make monosynaptic inputs to orexin neurons in the LHA, with approximately half of the VLPO inputs being double positive for these markers. VLPOGABA→LHA and VLPOGAL→LHA neurons receive innervations from similarly distributed input neurons in many brain regions, suggesting that at least some of the GABA- and GAL-producing VLPO neurons projecting to the LHA compose a common neuronal population. Optogenetic excitation of the VLPOGABA→LHA pathway results in induction of short-lasting wakefulness episodes and in increase in wakefulness, while photostimulation of the VLPOGAL→LHA pathway does not significantly affect wakefulness, suggesting distinct physiological roles of the VLPOGABA→LHA and VLPOGAL→LHA pathways.
Input neurons of VLPOGABA and VLPOGAL neurons that project to LHA
The distributions of monosynaptic inputs of the VLPOGABA→LHA and VLPOGAL→LHA neurons demonstrated marked similarity (Fig. 4J,K). Most input areas of the VLPOGABA→LHA and VLPOGAL→LHA neurons were overlapping (Figs. 3G, 4G; brain areas on gray background). Both neuronal populations also demonstrated high level of reciprocal connectivity with neurons in the LHA. Interestingly, a recent study proposed that a polysynaptic connection between orexin neurons and VLPOGABA/GAL neurons plays a role in inducing arousal (de Luca et al., 2022). Therefore, deciphering of the molecular identities of input LHA neurons will shed light on the interaction between LHA and VLPO neurons, and on its contribution to the regulation of sleep-wakefulness states.
We identified input neurons of VLPOGABA→LHA and VLPOGAL→LHA neurons in a variety of brain regions, including regions that participate in the regulation of sleep-wakefulness states (Oishi et al., 2017; Chen et al., 2018; Kodani et al., 2019). We also confirmed that at least some of the starter VLPOGABA→LHA and VLPOGAL→LHA neurons projected directly to orexin A-positive neurons in the LHA (Figs. 3F, 4F). The presence of reciprocal connectivity between the POA and LHA, as well as intra-POA connectivity, suggests the importance of these connections and underscores the complexity of the POA and LHA interaction. Further studies are needed to examine identified neuronal circuits and the GABA- and GAL-coproducing neuronal population in the VLPO.
Effect of optogenetic excitation of VLPOGABA→LHA pathway on sleep-wakefulness regulation
Reciprocal interaction between GABA-producing preoptic and wakefulness-promoting neurons has been described in the flip-flop model of sleep regulation (Saper et al., 2001, 2010). Because it has been generally believed that VLPOGABA neurons play a role in inducing sleep, we first tested whether the pathway connecting VLPOGABA and LHA neurons participates in the regulation of NREM sleep. However, we rather observed an opposite effect when we applied optogenetic excitation during the dark period. We therefore examined whether this pathway contributes to modulation of wakefulness, by applying optogenetic excitation during NREM sleep. Our results demonstrated that acute ChR2-mediated stimulation of the VLPOGABA→LHA pathway evoked short episodes of wakefulness (Fig. 5D–G) and long-term SSFO-mediated stimulation increased wakefulness (Fig. 6B–D). Consistently, more recent studies suggested that VLPO neurons comprise at least two subpopulations oppositely regulating sleep-wakefulness behavior (Liu et al., 2010; Liang et al., 2021). Another report also showed that optogenetic stimulation of GAD2-expressing preoptic neurons increased wakefulness, while stimulation of the POAGAD2→TMN pathway increased NREM and REM sleep (Chung et al., 2017), suggesting that bulk soma stimulation of VLPO neurons resulted in a dominant wakefulness-promoting effect. Additionally, our dual-color retrograde tracing identified that VLPOGABA→LHA and VLPOGABA→TMN pathways only partially shared Vgat-positive VLPO neurons (Fig. 6J,K). These results suggest that the GABA-producing preoptic neurons, which promote sleep and wakefulness, may be differentiated based on their projection site. GABA-producing VLPO neurons that do not express galanin were also proposed to have a wakefulness-inducing effect (de Luca et al., 2022). However, while chemogenetic stimulation of GAD1-positive POA neurons resulted in a small increase in NREM sleep (Saito et al., 2013), chemogenetic activation of VLPOGABA neurons did not affect sleep-wakefulness states (Vanini et al., 2020). Together, these studies underline the intricate neuronal composition and functional heterogeneity of the POA in terms of sleep-wakefulness governance.
SSFO-mediated optogenetic excitation of the VLPOGABA→LHA pathway evoked long wakefulness episodes (Fig. 6B–D). However, although SSFO was reported to produce a long-term depolarization, which generally lasts ∼30 min (Yizhar et al., 2011), the wakefulness-promoting effect was only observed for ∼15 min in our experiment (Fig. 6B,C). Interestingly, wakefulness episodes that were evoked after the second light pulse were shorter and less prominent than those after the first light pulse (Fig. 6B,C), while both of them were followed by consistent decreases in wakefulness. Consequently, the total amount of wakefulness during 1 h since the first light pulse did not differ between SSFO and control mice (Fig. 6E). Similarly, ChR2-mediated 10 min pulse stimulation was unable to produce differences in the amount of wakefulness (Fig. 5H), indicating that photoactivation of the pathway induced wakefulness episodes that did not last for a long time. A counteraction between wakefulness-promoting VLPOGABA→LHA neurons and presumably sleep-promoting VLPOGABA/GAL→LHA neurons on photostimulation might have resulted in this phenotype (de Luca et al., 2022). Alternatively, activity of VLPOGABA→LHA neurons might have been under strong homeostatic control (Yamagata et al., 2021), although we did not observe any significant differences in EEG power density of NREM sleep in the last quarter of the hour starting from the first light pulse (Fig. 6F). We should also consider inactivation or exhaustion of the VLPOGABA→LHA neurons on their photostimulation.
Another important question to be elucidated is how the excitation of the VLPOGABA→LHA pathway induces wakefulness. Further deciphering of the subpopulations of VLPOGABA neurons and of the identities of target LHA neurons comprising the VLPO→LHA pathway is needed for deeper understanding of its functional significance.
Distinct roles of VLPOGABA→LHA and VLPOGAL→LHA pathways in sleep-wakefulness regulation
VLPOGAL and VLPOGABA/GAL neurons have also been suggested to play a role in induction of NREM sleep (Kroeger et al., 2018; Ma et al., 2019; Arrigoni and Fuller, 2022; de Luca et al., 2022). We observed a wakefulness-promoting effect on the acute optogenetic stimulation of the VLPOGABA→LHA pathway and found that >50% of input neurons in the VLPO to orexin neurons coexpressed Vgat and Gal (Fig. 2F). Therefore, we examined a function of the VLPOGAL→LHA pathway using optogenetics. However, we did not observe any impact of the acute photostimulation of this pathway on wakefulness (Fig. 5L–O). We speculate that a subpopulation of Vgat-single-positive neurons (i.e., only GABA-producing) might be responsible for a wakefulness-inducing effect. Additionally, because we did not target a specific neuronal population in the LHA during pathway-specific optogenetic manipulation, the proportion between GABA-producing, GAL-producing, and GABA- and GAL-coproducing neuronal populations might have been shifted more toward GABA-producing neurons that might have resulted in a distinct phenotype observed on excitation of the VLPOGABA→LHA pathway. Further investigation is required to elucidate the role of the VLPOGAL→LHA pathway in the regulation of wakefulness and sleep.
Our present study has unveiled connectivity of the sleep- and wakefulness-regulating hypothalamic circuitry at a whole-brain scale. Functional examination of the identified VLPOGABA→LHA, VLPOGAL→LHA, and VLPOGABA/GAL→LHA pathways will shed further light on the complexity of sleep-wakefulness regulation. Exploration of the physiological roles of the neuronal circuits comprising GABA and/or galanin VLPO neurons and their input neurons in different brain areas would clarify how sleep and wakefulness are regulated at the level of interaction between different functional units within the brain in physiological and pathologic conditions. Indeed, we found that the limbic regions and septal regions contain considerable numbers of input neurons of the VLPOGABA→LHA neurons. It might be possible that limbic input to the wakefulness-promoting VLPOGABA→LHA pathway plays a role in evoking arousal in response to emotionally salient information coming from these regions. This study therefore provides a comprehensive and exhaustive background for precise elucidation of multiple components of the intricate sleep/wake circuitry and demonstrates a different insight into the physiological role of the VLPO→LHA neural pathway in regulation of sleep and wakefulness states.
Footnotes
This work was supported by Japan Society for the Promotion of Science KAKENHI Grant-in-Aid for Scientific Research (B) (JP 18H02595) to T.S.; AMED Grant JP21zf0127005 to T.S.; Japan Science and Technology Agency CREST Grant JPMJCR1655 Japan to T.S.; and TMFC Japan Foundation for applied Enzymology to A.H. We thank Dr. P. Vergara for conducting multidimensional scaling and Spearman correlation analyses; Dr. Y. Cherasse for preparing virus vectors; and M. Fukatsu and Y. Katsuyama for technical assistance.
The authors declare no competing financial interests.
References
- Alam MA, Kumar S, McGinty D, Alam MN, Szymusiak R (2014) Neuronal activity in the preoptic hypothalamus during sleep deprivation and recovery sleep. J Neurophysiol 111:287–299. 10.1152/jn.00504.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni E, Fuller PM (2022) The sleep-promoting ventrolateral preoptic nucleus: what have we learned over the past 25 years? Int J Mol Sci 23:2905. 10.3390/ijms23062905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AF, Navidi W, Tabuchi S, Terao A, Yamanaka A, Scammell TE, Behn CD (2016) Progressive loss of the orexin neurons reveals dual effects on wakefulness. Sleep 39:369–377. 10.5665/sleep.5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Adamantidis A, Ohtsu H, Deisseroth K, de Lecea L (2009) Sleep homeostasis modulates hypocretin-mediated sleep-to-wake transitions. J Neurosci 29:10939–10949. 10.1523/JNEUROSCI.1205-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KS, Xu M, Zhang Z, Chang WC, Gaj T, Schaffer DV, Dan Y (2018) A hypothalamic switch for REM and non-REM sleep. Neuron 97:1168–1176.e4. 10.1016/j.neuron.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Chung S, Weber F, Zhong P, Tan CL, Nguyen TN, Beier KT, Hörmann N, Chang WC, Zhang Z, Do JP, Yao S, Krashes MJ, Tasic B, Cetin A, Zeng H, Knight ZA, Luo L, Dan Y (2017) Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 545:477–481. 10.1038/nature22350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luca R, Nardone S, Grace KP, Venner A, Cristofolini M, Bandaru SS, Sohn LT, Kong D, Mochizuki T, Viberti B, Zhu L, Zito A, Scammell TE, Saper CB, Lowell BB, Fuller PM, Arrigoni E (2022) Orexin neurons inhibit sleep to promote arousal. Nat Commun 13:15. 10.1038/s41467-022-31591-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economo CV (1930) Sleep as a problem of localization. J Nerv Ment Dis 71:249–259. 10.1097/00005053-193003000-00001 [DOI] [Google Scholar]
- Franklin KB, Paxinos G (2007) The mouse brain in stereotaxic coordinates (Ed 3). Amsterdam: Elsevier. [Google Scholar]
- Gaus SE, Strecker RE, Tate BA, Parker RA, Saper CB (2002) Ventrolateral preoptic nucleus contains sleep-active, galaninergic neurons in multiple mammalian species. Neuroscience 115:285–294. 10.1016/s0306-4522(02)00308-1 [DOI] [PubMed] [Google Scholar]
- Giardino WJ, Eban-Rothschild A, Christoffel DJ, Li SB, Malenka RC, de Lecea L (2018) Parallel circuits from the bed nuclei of stria terminalis to the lateral hypothalamus drive opposing emotional states. Nat Neurosci 21:1084–1095. 10.1038/s41593-018-0198-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, McGinty D, Guzman-Marin R, Chew KT, Stewart D, Szymusiak R (2004) Activation of c-fos in GABAergic neurones in the preoptic area during sleep and in response to sleep deprivation. J Physiol 556:935–946. 10.1113/jphysiol.2003.056622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JA, Iordanidou P, Strom M, Adamantidis A, Burdakov D (2016) Awake dynamics and brain-wide direct inputs of hypothalamic MCH and orexin networks. Nat Commun 7:11395. 10.1038/ncomms11395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara J, Beuckmann CT, Nambu T, Willie JT, Chemelli RM, Sinton CM, Sugiyama F, Yagami KI, Goto K, Yanagisawa M, Sakurai T (2001) Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron 30:345–354. 10.1016/s0896-6273(01)00293-8 [DOI] [PubMed] [Google Scholar]
- Hasegawa E, Miyasaka A, Sakurai K, Cherasse Y, Li Y, Sakurai T (2022) Rapid eye movement sleep is initiated by basolateral amygdala dopamine signaling in mice. Science 375:994–1000. 10.1126/science.abl6618 [DOI] [PubMed] [Google Scholar]
- Jego S, Glasgow SD, Herrera CG, Ekstrand M, Reed SJ, Boyce R, Friedman J, Burdakov D, Adamantidis AR (2013) Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat Neurosci 16:1637–1643. 10.1038/nn.3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitin KI (1984) Preoptic area unit activity during sleep and wakefulness in the cat. Exp Neurol 83:347–357. 10.1016/S0014-4886(84)90103-1 [DOI] [PubMed] [Google Scholar]
- Kodani S, Soya S, Sakurai T (2017) Excitation of GABAergic neurons in the bed nucleus of the stria terminalis triggers immediate transition from non-rapid eye movement sleep to wakefulness in mice. J Neurosci 37:7164–7176. 10.1523/JNEUROSCI.0245-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani S, Soya S, Sakurai T (2019) Optogenetic manipulation of neural circuits during monitoring sleep/wakefulness states in mice. J Vis Exp 148:e58613. [DOI] [PubMed] [Google Scholar]
- Kroeger D, Absi G, Gagliardi C, Bandaru SS, Madara JC, Ferrari LL, Arrigoni E, Münzberg H, Scammell TE, Saper CB, Vetrivelan R (2018) Galanin neurons in the ventrolateral preoptic area promote sleep and heat loss in mice. Nat Commun 9:4129. 10.1038/s41467-018-06590-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin TK, Jin L, Wickersham IR (2019) Monosynaptic tracing: a step-by-step protocol. J Chem Neuroanat 102:101661. 10.1016/j.jchemneu.2019.101661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin TK, Jin L, Lea NE, Wickersham IR (2020) Monosynaptic tracing success depends critically on helper virus concentrations. Front Synaptic Neurosci 12:6. 10.3389/fnsyn.2020.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Liang Y, Shi W, Xiang A, Hu D, Wang L, Zhang L (2021) The NAergic locus coeruleus-ventrolateral preoptic area neural circuit mediates rapid arousal from sleep. Curr Biol 31:3729–3742.e5. 10.1016/j.cub.2021.06.031 [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E (1999) The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98:365–376. 10.1016/s0092-8674(00)81965-0 [DOI] [PubMed] [Google Scholar]
- Liu YW, Li J, Ye JH (2010) Histamine regulates activities of neurons in the ventrolateral preoptic nucleus. J Physiol 588:4103–4116. 10.1113/jphysiol.2010.193904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Greco MA, Shiromani P, Saper CB (2000) Effect of lesions of the ventrolateral preoptic nucleus on NREM and REM sleep. J Neurosci 20:3830–3842. 10.1523/JNEUROSCI.20-10-03830.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Bjorkum AA, Xu M, Gaus SE, Shiromani PJ, Saper CB (2002) Selective activation of the extended ventrolateral preoptic nucleus during rapid eye movement sleep. J Neurosci 22:4568–4576. 10.1523/JNEUROSCI.22-11-04568.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Nomiyama M, Takahira H, Hirashima N, Kunita S, Takahashi S, Yagami KI, Kilduff TS, Bettler B, Yanagisawa M, Sakurai T (2009) Selective loss of GABAB receptors in orexin-producing neurons results in disrupted sleep/wakefulness architecture. Proc Natl Acad Sci USA 106:4459–4464. 10.1073/pnas.0811126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Miracca G, Yu X, Harding EC, Miao A, Yustos R, Vyssotski AL, Franks NP, Wisden W (2019) Galanin neurons unite sleep homeostasis and α2-adrenergic sedation. Curr Biol 29:3315–3322.e3. 10.1016/j.cub.2019.07.087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T (2015) Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85:1103–1116. 10.1016/j.neuron.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Mizuno-Iijima S, et al. (2021) Efficient production of large deletion and gene fragment knock-in mice mediated by genome editing with Cas9-mouse Cdt1 in mouse zygotes. Methods 191:23–31. 10.1016/j.ymeth.2020.04.007 [DOI] [PubMed] [Google Scholar]
- Naganuma F, Kroeger D, Bandaru SS, Absi G, Madara JC, Vetrivelan R (2019) Lateral hypothalamic neurotensin neurons promote arousal and hyperthermia. PLoS Biol 17:e3000172. 10.1371/journal.pbio.3000172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu T, Sakurai T, Mizukami K, Hosoya Y, Yanagisawa M, Goto K (1999) Distribution of orexin neurons in the adult rat brain. Brain Res 827:243–260. 10.1016/s0006-8993(99)01336-0 [DOI] [PubMed] [Google Scholar]
- Oishi Y, Takata Y, Taguchi Y, Kohtoh S, Urade Y, Lazarus M (2016) Polygraphic recording procedure for measuring sleep in mice. J Vis Exp 107:e53678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Xu Q, Wang L, Zhang BJ, Takahashi K, Takata Y, Luo YJ, Cherasse Y, Schiffmann SN, de Kerchove d'Exaerde A, Urade Y, Qu WM, Huang ZL, Lazarus M (2017) Slow-wave sleep is controlled by a subset of nucleus accumbens core neurons in mice. Nat Commun 8:734. 10.1038/s41467-017-00781-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Callaway EM (2013) Design and generation of recombinant rabies virus vectors. Nat Protoc 8:1583–1601. 10.1038/nprot.2013.094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron C, Tighe DK, van den Pol AN, de Lecea L, Heller HC, Sutcliffe JG, Kilduff TS (1998) Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci 18:9996–10015. 10.1523/JNEUROSCI.18-23-09996.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron C, et al. (2000) A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med 6:991–997. [DOI] [PubMed] [Google Scholar]
- Piñol RA, Zahler SH, Li C, Saha A, Tan BK, Škop V, Gavrilova O, Xiao C, Krashes MJ, Reitman ML (2018) Brs3 neurons in the mouse dorsomedial hypothalamus regulate body temperature, energy expenditure, and heart rate, but not food intake. Nat Neurosci 21:1530–1540. 10.1038/s41593-018-0249-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito YC, Tsujino N, Hasegawa E, Akashi K, Abe M, Mieda M, Sakimura K, Sakurai T (2013) GABAergic neurons in the preoptic area send direct inhibitory projections to orexin neurons. Front Neural Circuits 7:192. 10.3389/fncir.2013.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito YC, Maejima T, Nishitani M, Hasegawa E, Yanagawa Y, Mieda M, Sakurai T (2018) Monoamines inhibit GABAergic neurons in ventrolateral preoptic area that make direct synaptic connections to hypothalamic arousal neurons. J Neurosci 38:6366–6378. 10.1523/JNEUROSCI.2835-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T (2007) The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci 8:171–181. 10.1038/nrn2092 [DOI] [PubMed] [Google Scholar]
- Sakurai T, Nagata R, Yamanaka A, Kawamura H, Tsujino N, Muraki Y, Kageyama H, Kunita S, Takahashi S, Goto K, Koyama Y, Shioda S, Yanagisawa M (2005) Input of orexin/hypocretin neurons revealed by a genetically encoded tracer in mice. Neuron 46:297–308. 10.1016/j.neuron.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Sallanon M, Denoyer M, Kitahama K, Aubert C, Gay N, Jouvet M (1989) Long-lasting insomnia induced by preoptic neuron lesions and its transient reversal by muscimol injection into the posterior hypothalamus in the cat. Neuroscience 32:669–683. 10.1016/0306-4522(89)90289-3 [DOI] [PubMed] [Google Scholar]
- Saper CB, Chou TC, Scammell TE (2001) The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci 24:726–731. 10.1016/s0166-2236(00)02002-6 [DOI] [PubMed] [Google Scholar]
- Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE (2010) Sleep state switching. Neuron 68:1023–1042. 10.1016/j.neuron.2010.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scammell TE, Arrigoni E, Lipton JO (2017) Neural circuitry of wakefulness and sleep. Neuron 93:747–765. 10.1016/j.neuron.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherin JE, Shiromani PJ, McCarley RW, Saper CB (1996) Activation of ventrolateral preoptic neurons during sleep. Science 271:216–219. 10.1126/science.271.5246.216 [DOI] [PubMed] [Google Scholar]
- Sulaman BA, Wang S, Tyan J, Eban-Rothschild A (2023) Neuro-orchestration of sleep and wakefulness. Nat Neurosci 26:196–212. 10.1038/s41593-022-01236-w [DOI] [PubMed] [Google Scholar]
- Sun L, Tang Y, Yan K, Yu J, Zou Y, Xu W, Xiao K, Zhang Z, Li W, Wu B, Hu Z, Chen K, Fu ZF, Dai J, Cao G (2019) Differences in neurotropism and neurotoxicity among retrograde viral tracers. Mol Neurodegener 14:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymusiak R, Alam N, Steininger TL, McGinty D (1998) Sleep-waking discharge patterns of ventrolateral preoptic/anterior hypothalamic neurons in rats. Brain Res 803:178–188. 10.1016/s0006-8993(98)00631-3 [DOI] [PubMed] [Google Scholar]
- Szymusiak R, McGinty D (1986) Sleep suppression following kainic acid-induced lesions of the basal forebrain. Exp Neurol 94:598–614. 10.1016/0014-4886(86)90240-2 [DOI] [PubMed] [Google Scholar]
- Tabuchi S, Tsunematsu T, Black SW, Tominaga M, Maruyama M, Takagi K, Minokoshi Y, Sakurai T, Kilduff TS, Yamanaka A (2014) Conditional ablation of orexin/hypocretin neurons: a new mouse model for the study of narcolepsy and orexin system function. J Neurosci 34:6495–6509. 10.1523/JNEUROSCI.0073-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Lin JS, Sakai K (2009) Characterization and mapping of sleep–waking specific neurons in the basal forebrain and preoptic hypothalamus in mice. Neuroscience 161:269–292. 10.1016/j.neuroscience.2009.02.075 [DOI] [PubMed] [Google Scholar]
- Takahashi TM, Sunagawa GA, Soya S, Abe M, Sakurai K, Ishikawa K, Yanagisawa M, Hama H, Hasegawa E, Miyawaki A, Sakimura K, Takahashi M, Sakurai T (2020) A discrete neuronal circuit induces a hibernation-like state in rodents. Nature 583:109–114. 10.1038/s41586-020-2163-6 [DOI] [PubMed] [Google Scholar]
- Tan CL, Cooke EK, Leib DE, Lin YC, Daly GE, Zimmerman CA, Knight ZA (2016) Warm-sensitive neurons that control body temperature. Cell 167:47–59.e15. 10.1016/j.cell.2016.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thannickal TC, Moore RY, Nienhuis R, Ramanathan L, Gulyani S, Aldrich M, Cornford M, Siegel JM (2000) Reduced number of hypocretin neurons in human narcolepsy. Neuron 27:469–474. 10.1016/s0896-6273(00)00058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T, Ueno T, Tabuchi S, Inutsuka A, Tanaka KF, Hasuwa H, Kilduff TS, Terao A, Yamanaka A (2014) Optogenetic manipulation of activity and temporally controlled cell-specific ablation reveal a role for MCH neurons in sleep/wake regulation. J Neurosci 34:6896–6909. 10.1523/JNEUROSCI.5344-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneoka Y, Funato H (2021) Cellular composition of the preoptic area regulating sleep, parental, and sexual behavior. Front Neurosci 15:649159. 10.3389/fnins.2021.649159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanini G, Bassana M, Mast M, Mondino A, Cerda I, Phyle M, Chen V, Colmenero AV, Hambrecht-Wiedbusch VS, Mashour GA (2020) Activation of preoptic GABAergic or glutamatergic neurons modulates sleep-wake architecture, but not anesthetic state transitions. Curr Biol 30:779–787.e4. 10.1016/j.cub.2019.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner A, Anaclet C, Broadhurst RY, Saper CB, Fuller PM (2016) A novel population of wake-promoting GABAergic neurons in the ventral lateral hypothalamus. Curr Biol 26:2137–2143. 10.1016/j.cub.2016.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venner A, de Luca R, Sohn LT, Bandaru SS, Verstegen AM, Arrigoni E, Fuller PM (2019) An inhibitory lateral hypothalamic-preoptic circuit mediates rapid arousals from sleep. Curr Biol 29:4155–4168.e5. 10.1016/j.cub.2019.10.026 [DOI] [PubMed] [Google Scholar]
- Vetrivelan R, Kong D, Ferrari LL, Arrigoni E, Madara JC, Bandaru SS, Lowell BB, Lu J, Saper CB (2016) Melanin-concentrating hormone neurons specifically promote rapid eye movement sleep in mice. Neuroscience 336:102–113. 10.1016/j.neuroscience.2016.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S, Lowell BB (2011) Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71:142–154. 10.1016/j.neuron.2011.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Salin-Pascual R, Greco M, Shiromani P (2000) Distribution of hypocretin-containing neurons in the lateral hypothalamus and C-fos-immunoreactive neurons in the VLPO. Sleep Res Online 3:35–42. [PubMed] [Google Scholar]
- Weber F, Dan Y (2016) Circuit-based interrogation of sleep control. Nature 538:51–59. 10.1038/nature19773 [DOI] [PubMed] [Google Scholar]
- Wu Z, Autry AE, Bergan JF, Watabe-Uchida M, Dulac CG (2014) Galanin neurons in the medial preoptic area govern parental behaviour. Nature 509:325–330. 10.1038/nature13307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata T, Kahn MC, Prius-Mengual J, Meijer E, Sabanovi M, Guillaumin MC, van der Vinne V, Huang YG, McKillop LE, Jagannath A, Peirson SN, Mann EO, Foster RG, Vyazovskiy VV (2021) The hypothalamic link between arousal and sleep homeostasis in mice. Proc Natl Acad Sci USA 118:e2101580118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Ooshima Y, Nakata M, Yano T, Nishimura N, Nishigaki R, Satomi Y, Matsumoto H, Matsumoto Y, Takeyama M (2015) Efficient gene-targeting in rat embryonic stem cells by CRISPR/Cas and generation of human kynurenine aminotransferase II (KAT II) knock-in rat. Transgenic Res 24:991–1001. 10.1007/s11248-015-9909-1 [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, Ogshea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K (2011) Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477:171–178. 10.1038/nature10360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, McCormack S, España RA, Crocker A, Scammell TE (2006) Afferents to the orexin neurons of the rat brain. J Comp Neurol 494:845–861. 10.1002/cne.20859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video clip of GFP-expressing Vgat-ires-Cre mice with EEG/EMG tracing during 10 s pre-photostimulation, 20 s photostimulation, and 10 s post-photostimulation periods. N, NREM sleep.
ChR2-expressing Vgat-ires-Cre mice with EEG/EMG tracing during 10 s pre-photostimulation, 20 s photostimulation, and 10 s post-photostimulation periods. A short episode of wakefulness was observed. N, NREM sleep; W, wakefulness.