Abstract
Background
With the aggregation of clinical data and the evolution of computational resources, artificial intelligence-based methods have become possible to facilitate clinical diagnosis. For congenital heart disease (CHD) detection, recent deep learning-based methods tend to achieve classification with few views or even a single view. Due to the complexity of CHD, the input images for the deep learning model should cover as many anatomical structures of the heart as possible to enhance the accuracy and robustness of the algorithm. In this paper, we first propose a deep learning method based on seven views for CHD classification and then validate it with clinical data, the results of which show the competitiveness of our approach.
Methods
A total of 1411 children admitted to the Children’s Hospital of Zhejiang University School of Medicine were selected, and their echocardiographic videos were obtained. Then, seven standard views were selected from each video, which were used as the input to the deep learning model to obtain the final result after training, validation and testing.
Results
In the test set, when a reasonable type of image was input, the area under the curve (AUC) value could reach 0.91, and the accuracy could reach 92.3%. During the experiment, shear transformation was used as interference to test the infection resistance of our method. As long as appropriate data were input, the above experimental results would not fluctuate obviously even if artificial interference was applied.
Conclusions
These results indicate that the deep learning model based on the seven standard echocardiographic views can effectively detect CHD in children, and this approach has considerable value in practical application.
Keywords: cardiology, diagnostic Imaging, child health
WHAT IS ALREADY KNOWN ON THIS TOPIC
Currently, the detection accuracy of the mass testing methods commonly used worldwide for congenital heart disease (CHD) in children is low.
There have been attempts to use the standard five views of grayscale echocardiography as input to a deep learning model for the automatic detection of CHD in children.
To better visualize the anatomical details of the heart, we used the standard seven views of the echocardiography images as input to a deep learning model and added color modal images for the detection of CHD in children.
WHAT THIS STUDY ADDS
We found some variation in experimental results across image modalities, with our approach yielding excellent experimental results and resistance to artificial interference when appropriate input image modalities are selected.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study demonstrates the feasibility of the seven-view method and provides a new idea for the automatic detection of CHD in children by echocardiography.
Introduction
Congenital heart disease (CHD) is the most common congenital anomaly in children, and the reported incidence of CHD is approximately 0.69%–0.93%, accounting for one-third of all major congenital anomalies.1 With surgical intervention, the mortality for children with CHD can be reduced to as low as 3%.2 Therefore, early detection of CHD is very important.
At present, echocardiography reading mainly relies on manual labor, but the training cycle of echocardiographic doctors is long, and much experience is needed as the basis for accurate diagnosis. A European Union study recommended that beginners in echocardiography need to undergo >350 tests to achieve basic practical competence.3 This grim situation has led researchers to focus on the application of artificial intelligence.
Deep learning has revolutionized image classification and recognition because of its high accuracy; in some cases, it demonstrates performance comparable to or exceeding that of medical experts. There are many practical applications of deep learning in the medical field, especially in medical images, including image registration/localization, cell structure detection, disease diagnosis/prognosis, etc.4 The tissue boundary of ultrasonic images is fuzzy, the image has more noise interference, and the selection and interpretation of images is subjective. The processing and interpretation of ultrasonic images are always difficult in medical imaging. Therefore, the application of deep learning in echocardiography is relatively lagging compared with other medical imaging, and the application of this technology in CHD also started late.
Unlike computed tomography (CT), magnetic resonance imaging (MRI), and other medical images, the selection of echocardiographic views is quite subjective. Therefore, the selection and standardization of echocardiographic views are crucial for the application of deep learning. In 2013, a work US government-led work called fetal intelligent navigation echocardiography (FINE) succeeded in automating the selection of standard fetal echocardiographic views, which can automatically select nine stamdard echocardiographic views including four-chamber, five-chamber, left ventricular outflow tract, etc. Subsequently, 54 fetuses were tested between 18.6 and 37.2 weeks of gestation were examined, demonstrating that the FINE system can automatically select nine standard views for both normal fetuses and fetuses with CHD and can better visualize the abnormal features of complex CHD.5 Studies conducted by several centers in the past 10 years have shown that the detection rate and specificity of fetal CHD with echocardiography are <50% under the traditional screening method.6–8 A study at the University of California, San Francisco, used deep learning techniques to train, test, and verify 4108 fetuses (0.9% CHD) with more than one million echocardiographic images, and the results of the deep learning model were exceptional, with 95% sensitivity (95% confidence interval (CI)=84% to 99%) and 96% specificity (95% CI=95% to 97%) in distinguishing normal from abnormal hearts.9 Other studies have shown that the accuracy of CHD diagnosis can be significantly improved by convolutional neural network (CNN) preprocessing and segmentation of fetal echocardiographic images and then using a deep learning model for image diagnosis.10
There are relatively few studies on the application of deep learning in pediatric echocardiography, and the research progress in the whole field is relatively slow compared with that in fetal echocardiography. Diller et al proved that deep learning technology can effectively remove artifacts and noise in both normal and CHD images.11 After processing specific pediatric echocardiographic images by deep learning technology, the average individual leaflet segmentation accuracy in children with hypoplastic left heart syndrome is close to the limit of human eye resolution.12 A research center has developed an unsupervised deep learning model named DGACNN in view of the characteristics of echocardiographic images, and its ability to automatically label and screen fetal echocardiographic images (four-chamber view images only) has surpassed that of middle-level professional doctors in related fields.13 Capital Medical University and Carnegie Mellon University designed a five-channel CNN with a single-branch that can diagnose negative samples and ventricular septal defect (VSD) or atrial septal defect (ASD) classifications with an accuracy over 90%.14 Most of the deep learning studies in pediatric echocardiography are basic technical studies with fewer clinical applications. Major clinical studies tend to achieve classification with five (four) views or even a single view.
This paper aims to use a new method of view selection named the seven views15 approach, and apply the deep learning model to analyze pediatric echocardiographic images, to achieve mass detection of pediatric CHD.
Method
Standard view selection
Compared with CT, X-ray, and other examinations, ultrasound examination is highly subjective. Therefore, the existence of the standard view is of great significance to echocardiography examinations. At present, there are five standard views commonly used in pediatric echocardiography: (1) parasternal long-axis view; (2) parasternal short-axis view; (3) apical four (five)-chamber view; (4) subxiphoid four-chamber view; (5) suprasternal long-axis view. Each view corresponds to specific anatomical structures of the heart, and abnormal cardiac anatomy is the essence of CHD. Previous deep learning studies have been based on one or more of these views, for example, a study on transposition of great arteries (TGA) conducted by Imperial College London used apical four-chamber and parasternal short-axis view as input data in its deep learning model.16
There are various types of CHD. To improve the accuracy, the input images of the deep learning model need to cover as many anatomical structures of the heart as possible, and show the relationship between each anatomical structure in more detail. Therefore, we proposed using seven views instead of the traditional five views as the standard view of pediatric echocardiography, and verified its feasibility with the deep learning model. The seven views15 are as follows:
Parasternal long-axis view;
Parasternal short-axis view;
Parasternal four-chamber view;
Parasternal five-chamber view;
Subxiphoid four-chamber view;
Subxiphoid biatrial view;
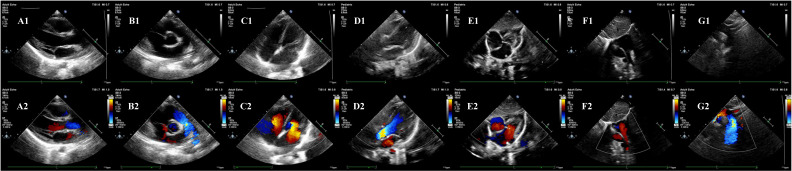
Suprasternal long-axis view (figure 1).
Figure 1.
Seven standard views of echocardiography. (A1) Parasternal long-axis view (grayscale modality); (A2) parasternal long-axis view (color modality); (B1) parasternal short-axis view (grayscale modality); (B2) parasternal short-axis view (color modality); (C1) parasternal four-chamber view (grayscale modality); (C2) parasternal four-chamber view (color modality); (D1) parasternal five-chamber view (grayscale modality); (D2) parasternal five-chamber view (color modality); (E1) subxiphoid four-chamber view (grayscale modality); (E2) subxiphoid four-chamber view (color modality); (F1) subxiphoid biatrial view (grayscale modality); (F2) subxiphoid biatrial view (color modality); (G1) suprasternal long-axis view (grayscale modality); (G2) suprasternal long-axis view (color modality).
Compared with the traditional five views, the main transformations are as follows: (1) parasternal four (five)-chamber view is adopted instead of apical four (five)-chamber view, and both four-chamber view and five-chamber view are selected instead of one of them; (2) add subxiphoid biatrial view. The former shows better performance in the diagnosis of ASD and can always reveal part of the morphology of the aorta, while the latter shows good results in the performance of the atrial septum and the superior and inferior vena cava, and reveals part of the pulmonary veins and the connection between the superior and inferior vena cava and the right atrium.
This improved view selection approach can more stably reflect the anatomic abnormalities of aortic-related CHDs such as coarctation of the aorta, and reveal the anatomic details around the atrial septum carefully, which is of great benefit for the detection of ASDs.
Data acquisition and dealing
The study participants were 1411 children admitted to our hospital between 2018 and 2020, including 336 normal children and 1075 children with CHD. A small number of children were excluded due to lost to follow-up or poor echocardiographic video quality, and the final number of children included was 1376. The distribution is shown in table 1. All echocardiographic videos were obtained by Philips iE 33, Philips EPIQ5, Philips EPIQ 7C color Doppler ultrasonography with probe model S8-3 (probe frequency range 3–8 MHz). S5-1 (probe frequency range 1–5 MHz). Seven standard view images (including color images, grayscale images, and bimodal images) were selected and marked from the echocardiographic videos by professional echocardiographic doctors. The number of images obtained for the first time was 16 088, and the final number of images was 14 838 after screening and cleaning. In this study, the echocardiography data set is mainly divided into three parts: training data (for model training), validation data (for model parameter adjustment), and test data (for the final evaluation of the model), and the data volume ratio of each part is 8:1:1. Vertical flip transformation is used to increase the size of the training data set. To improve the generalization ability of the model and its robustness to adversarial examples, Mix Up17 is used for data enhancement. Finally, the amplified images are resized to 224×224×3.
Table 1.
The distribution of CHD
| Category | Initial children count | Final children count |
| Normal | 336 | 330 |
| PFO | 63 | 62 |
| PDA | 113 | 112 |
| VSD | 428 | 411 |
| ASD | 433 | 423 |
| Complex | 38 | 38 |
| Total | 1411 | 1376 |
Complex, which means having more than one CHD, or complex CHD (which is quite rare) such as pulmonary stenosis.
ASD, atrial septal defect; CHD, congenital heart disease; PDA, patent ductus arteriosus; PFO, patent foramen ovale; VSD, ventricular septal defect.
Since there are two image modalities of bimodal images at the same time, when they are used as input data, it is necessary to segment the bimodal images into grayscale images and color images in advance, and then the two types of images are input. We found that the grayscale images obtained by this segmentation as the input result will be different from the result obtained after running of simple grayscale images, and the same is true for color images. Thus, for these three different types of echocardiographic images, the following different image input combinations can be constructed: (A) single grayscale echocardiographic images; (B) single color echocardiographic images; (C) single bimodal echocardiographic images; (D) bimodal echocardiographic images combined with grayscale echocardiographic images; (E) bimodal echocardiographic images combined with color echocardiographic images. It should be noted here that the bimodal images and the single modal images come from different groups of participants, which can reduce the duplicate information in groups D and E and enhance the value of input data.
Model development and training
The classical CNN model mainly relies on the superposition of the convolution layer and the pooling layer. However, with the increase in convolution layers and pooling layers, the learning effect of the model does not improve as expected, and the vanishing/exploding gradients and degradation problems appear instead. To solve these problems, Kaiming et al proposed the Residual Network (ResNet) model,18 which alleviates the problem of network degradation and is beneficial to the extraction of deep image features. In this paper, ResNet50 is used as the feature extraction network.
In the training process, the AdamW optimizer was employed with a batch size of 12, and the initial learning rate was set to 5e-4. The network was trained on an NVIDIA GeForce 3060 GPU. The experimental model was built in Python 3.7 and Pytorch 1.7.1. Each sample is labeled as negative and positive inputs to the neural network. Negative represents normal, and positive represents patients with CHD. All echocardiographic seven-view images are evaluated using a single model.
To judge the antinoise ability of the deep learning model when echocardiographic seven-view images are used as input, in addition to the previously mentioned data enhancement methods, each group forms a self-contrast based on whether shear transformation (regardless of whether the shear transformation is performed, all of the previously described data enhancement methods have been applied), shear transformation is performed on all images in the group (instead of regrouping within a group). Taking group A as an example, all the images in group A are first input into the deep learning model for operation to obtain a result. Then, all the images in group A undergo shear transformation, and are input into the deep learning model again to obtain another result. Thus, one group will end up with two results.
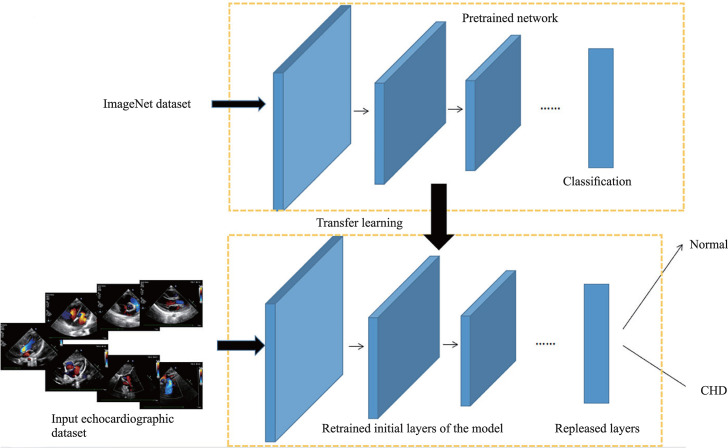
Transfer learning
Transfer learning is an effective mechanism for image classification under limited data scenarios. Some natural pictures are first pretrained as objects, and then the echocardiographic data set is input into the pretrained network for retraining, and further validation and testing. Finally, a deep learning model with detection ability is obtained (figure 2).19
Figure 2.
Visualization for transfer learning using pretrained models.19 CHD, congenital heart disease.
Result
Since the deep learning model can only process the image of one modality at a time, the bimodal images need to be divided into grayscale images and color images before input. Group A and group B are single modal images, which only need to be input into the deep learning model for running. The images of group C are bimodal images that need to be divided into grayscale and color modal images and input into the deep learning model separately for running. Since the grayscale part and the color part of the bimodal images are from the same child, the above two calculation results are comparable and can be used to evaluate the examination effect of deep learning technology on echocardiographic images of different modalities. Group D consists of bimodal images and grayscale images. Before input, the bimodal images are similarly segmented, but the color modal images are discarded, and all grayscale images are taken as input. Group E is to discard the grayscale images, and the rest of the process is the same as group D. Combined with whether shear transformation is carried out, there are altogether 12 groups of results.
Shear transformation or not
The difference between the experimental results is not obvious whether shear transformation is performed or not. The maximum difference in accuracy is 5.6%, the minimum difference is 0.7%, the maximum difference in the area under the curve (AUC) value is 0.02, and the minimum difference <0.01 (in expressing the results of the AUC values, we have taken the approach of retaining two digits after the decimal point, in this way the seemingly identical AUC values are actually different after three decimal places, and therefore this difference should be expressed as <0.01).
Results of grayscale modality
According to the composition of group D, group D can be compared with group A, and the grayscale modality in group C can also be compared. However, because the data for groups A and C are from different participants, they cannot be directly compared. After shear transformation, the accuracy and the AUC value of group D are 84.1% and 0.84, respectively, which are 15% and 0.08 higher than the grayscale modality of group C. Compared with group D, the accuracy of group A is improved by 8.2%, but the AUC value is decreased by 0.02. The results before the shear transformation are approximate. The accuracy and the AUC value of group D are 84.8% and 0.85, respectively, which are 10.1% and 0.08 higher than the grayscale modality of group C. The accuracy of group A is 4.7% higher, but the AUC value is 0.03 lower than that of group D.
Results of color modality
Similarly, group E can be compared with groups B and C. After shear transformation, the accuracy of group E is 84.8%, which is 1.9% lower than the color modality of group C, and the AUC values of both groups are 0.91. Compared with group E, the accuracy of group B is improved by 0.9%, but the AUC value is decreased by 0.03. Before shear transformation, the accuracy of group E is 87.2% and the AUC value is 0.91, which are 1.8% and 0.02 higher than the color modality of group C, and the difference values between groups E and B are 4.9% and 0.05, respectively.
Comparison between grayscale modality and color modality
As mentioned previously, the two modal parts of a bimodal image came from the same child. Therefore, the results of the grayscale modality and the color modality of group C are comparable. After shear transformation, the accuracy and the AUC value of group C in the color modality are better, the accuracy is 86.7% and the AUC value is 0.91, which are 17.6% and 0.15 higher than those in the grayscale modality, respectively. The results before shear transformation are similar, the accuracy is 85.4%, and the AUC value is 0.89 in the color modality, which are 10.7% and 0.12 higher than those in the grayscale modality, respectively. All the accuracy and the AUC values are shown in table 2.
Table 2.
Results of different groups
| Group to be input | Shear transformation | Accuracy (%) | AUC value |
| Group A | No | 89.5 | 0.82 |
| Yes | 92.3 | 0.82 | |
| Group B | No | 82.3 | 0.86 |
| Yes | 85.7 | 0.88 | |
| Group C (grayscale modality) | No | 74.7 | 0.77 |
| Yes | 69.1 | 0.76 | |
| Group C (color modality) | No | 85.4 | 0.89 |
| Yes | 86.7 | 0.91 | |
| Group D | No | 84.8 | 0.85 |
| Yes | 84.1 | 0.84 | |
| Group E | No | 87.2 | 0.91 |
| Yes | 84.8 | 0.91 |
AUC, area under the curve.
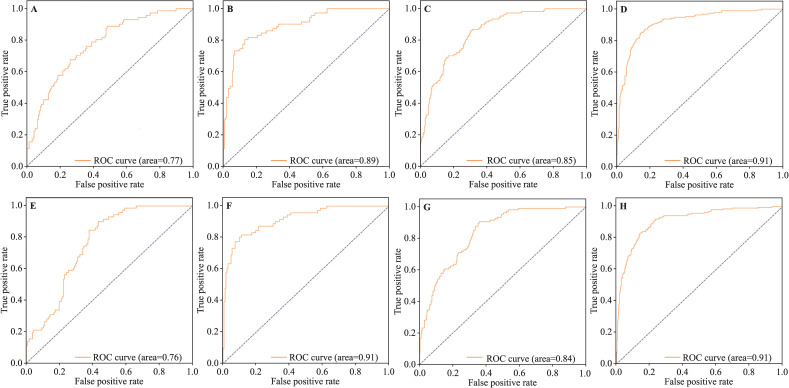
In addition, group D can form a contrast with group C, and the same for group E. In summary, the receiver operating characteristic curves and the AUC values of operation results of groups C, D, and E (with or without shear transformation) were plotted for visual comparison (figure 3).
Figure 3.
The ROC curves and the AUC values of groups C, D, and E. (A) Group C in grayscale modality (without shear transformation); (B) group C in color modality (without shear transformation); (C) group D (without shear transformation); (D) group E (without shear transformation); (E), group C in grayscale modality (with shear transformation); (F) group C in color modality (with shear transformation); (G) group D (with shear transformation); (H) group E (with shear transformation). AUC, area under the curve; ROC, receiver operating characteristic.
Discussion
CHD is still an important cause of death in children, which has created a worldwide socioeconomic burden that cannot be ignored20; therefore, early diagnosis of CHD is of great significance to prognosis. Quite a number of children with CHD have difficulty with sellf-healing during growth, and surgery is almost the only treatment for them. The mortality rate of children with CHD who are suitable for surgery can be reduced to a very low level.6 20 21 However, late detection of CHD is more likely to progress to pulmonary hypertension, increasing the risk of surgery and mortality. Some patients develop Eisenmanger’s syndrome and miss out on surgery.22 23
Status of congenital heart disease detection
The false negative rate of CHD diagnosis in children is high worldwide. For critical CHD with more obvious clinical manifestations, there is still a false negative rate of >10% in high-income countries, and even >40% in some hospitals.24 With such a high false negative rate, the accuracy is definitely difficult to guarantee. It is difficult to be optimistic in the low-income and middle-income countries, where healthcare is even poorer.
Some studies suggest that pulse oximetry can be used to detect CHD. However, for major CHDs such as ASD and VSD, when pulse oximetry is used as the detecting method, even in high-income countries with high medical levels, the sensitivity is <50%.25 A study has shown that the sensitivity of pulse oximetry as a screening method for children’s pneumonia can exceed 90%.26 Obviously, similar respiratory diseases may also reduce blood oxygen saturation, which also explains pulse oximetry’s poor screening performance for major congenital heart defects. With the addition of traditional methods such as auscultation of cardiac murmur, the sensitivity of screening children with CHD can be improved to 98%, but the specificity is only 39%,27 and such a high false positive rate will undoubtedly increase the social and economic burden.
Echocardiography is a non-invasive, safe imaging modality that is frequently used to evaluate the heart function and structure. A large retrospective study including 1258 patients with prenatally diagnosed CHD found that the postnatal diagnoses were discrepant in 29.3% of cases.28 In another study by Aguilera and Dummer, 106 patients were included, with a discrepancy rate of 30.2%.29 Thus, fetal CHD cannot be fully detected with echocardiography, so echocardiography in children is crucial. Because of the long training period of echocardiographic doctors, the application of deep learning technology has gradually become a trend.
Analysis of result
This is the first time that the seven-view approach has been applied to the detection of CHD using deep learning technology. From the results, many details are presented, which are worth further in-depth analysis.
Obviously, the experimental results of color modality are better, which may be related to the relatively large amount of information contained in color echocardiographic images. Blood flow is definitely significant for the interpretation of echocardiographic images. Color echocardiographic images can better reveal the abnormal shunt and abnormal manifestations of normal blood flow, which can reflect the abnormal anatomical structure of the heart and thus detect CHD. This may be due to the absence of this important diagnostic point that the performance of grayscale modality detection of echocardiographic images is worse.
With proper image input and a correct modal model, the accuracy rate can reach 92.3%, which is better than general medical institutions,24 and it performs better than the routine screening that is now widely available.25 27
Limitation
There are several limitations in this study. First, inherent bias due to the retrospective nature of this study always exists. Second, to ensure the comparability and consistency of the data, all echocardiographic images are obtained from the same brand of ultrasound machines, which may not be conducive to the adaptability of the deep learning model, because ultrasound machines in different medical institutions are different in reality. Third, despite the excellent performance of the accuracy and the AUC values in experimental results, it is still difficult to know the specific diagnostic details of the deep learning model in detecting CHD. Fourth, a study has shown that there are gender differences in cardiac anatomy of children with CHD.30 However, the present study does not further explore the possible influence of gender factor because our main objective is to find a universal method for CHD detection. Finally, the number of cases of complex CHD in this study was low because the seven-view approach of echocardiography is a new form of view selection and has been in use for a relatively short time, and the incidence of complex CHD is very low.31 We have partially addressed this problem through data enhancement, and further dissemination of the seven-view approach of echocardiography is needed to fundamentally solve this problem.
Dialectics of potential methods
The training model adopted in this paper inputs seven standard echocardiographic views from all participants into a single deep learning model for training, and the same applies to validation and testing. The advantages of this approach are as follows: (1) it can effectively expand the number of training images, which is directly related to the training effect; (2) it can enable the deep learning model to uniformly extract all view features. In this way, in the process of extracting the seven standard views described in this article during the diagnosis and treatment of echocardiographic doctors, even if the view extraction is incomplete due to the poor cooperation of children, the remaining views can also be used for CHD diagnosis.
In the preliminary experiment, we tried to train a separate model for each standard view and finally summarize the diagnosis, but the experimental results were not satisfactory. The reason may be that this treatment method will significantly reduce the number of training samples for each model, thus making the experimental results worse than expected. In addition, this mode will certainly increase the complexity of the algorithm structure, potentially lengthening the training time and the time that it takes to diagnose individual children. Moreover, when applied to actual diagnosis, it is difficult to determine the final conclusion if different echocardiographic views of the same patient obtain opposite feedback, and structural abnormalities in CHD are often not reflected in all echocardiographic views. Assuming that all positive results are judged as CHD, in this mode, although the sensitivity can be improved, the false positive rate will also increase, which also significantly increases the social and medical burden. Setting diagnostic weights separately for each view may solve part of this problem, but new problems are created, such as how to design the specific value of weight. In addition, the same echocardiographic view has different diagnostic value for different types of CHD. Therefore, the weight value of the same echocardiographic view will vary for different CHDs, which makes it difficult to choose the weight allocation method when diagnosing an undiagnosed echocardiographic image.
The result of this paper is bivariate: normal and CHD. Because the purpose of this study is to detect children with CHD, this bivariate outcome is appropriate. If further exploration is carried out in the future, it may be possible to take multiclassification research as a target, that is, output results for specific diseases, such as ASD, VSD, PDA, etc. The deep learning model can be used as a detection tool for pediatric CHD, and as a reference for clinicians to diagnose. However, with the increase in classifications, new problems will arise. To ensure that the sensitivity, specificity, accuracy, and other indicators are similar to those of the bivariate experiment, the sample size of the multiclassification model needs to be significantly increased. It is possible to collect a large number of echocardiographic images for relatively common CHD types such as ASD and VSD, but for rare CHDs such as TGA and total anomalous pulmonary venous connection, collecting a large amount of echocardiographic images is quite difficult. The deficiency of images may lead to awful experimental results, and the false positive rate and false negative rate may rise. However, the prognosis of these rare CHDs is often poor, and they should be more accurately screened and diagnosed in the early stage. If these serious CHDs fail to obtain a high diagnostic rate, the significance of such a deep learning model will be greatly reduced. To solve this problem, it is probably necessary to seek more extensive regional medical cooperation, or more in-depth research on deep learning model technology to seek breakthroughs.
In summary, the deep learning model based on the input of the seven standard echocardiographic views can effectively detect the CHD in children. Further exploration showed that the use of color modal echocardiographic images is relatively better. Moreover, the deep learning model with the input of seven standard echocardiographic views has certain anti-interference ability, which means that the detection method adopted in this paper also has considerable value in practical applications.
Footnotes
XJ and JY contributed equally.
Contributors: XJ: conceptualization, data curation, methodology, software, investigation, visualization, formal analysis, writing—original draft, project administration; JYu (should be considered as co-first author): conceptualization, data curation, methodology, investigation, visualization, formal analysis, writing—original draft; JYe: validation, investigation, data curation; WJ: software, formal analysis; WX (should be considered as co-corresponding author): conceptualization, funding acquisition, resources, supervision, writing—review and editing. QS (corresponding author): conceptualization, funding acquisition, resources, supervision, validation, writing—review and editing. QS is the guarantor.
Funding: This study was supported by the Public Welfare Technology Application Research Project of Zhejiang Science and Technology Department (LGF22H180002); Key Research and Development Plan of Zhejiang Province (No. 2022C03087).
Competing interests: Author QS is the Editor-in-Chief for World Journal of Pediatric Surgery. The paper was handled by the other Editor and has undergone rigrous peer review process. Author QS was not involved in the journal's review of, or decisions related to, this manuscript.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Ethics Committee of Children’s Hospital, Zhejiang University School of Medicine (2021-IRB-286). Participants gave informed consent to participate in the study before taking part.
References
- 1.van der Linde D, Konings EEM, Slager MA, et al. Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J Am Coll Cardiol 2011;58:2241–7. 10.1016/j.jacc.2011.08.025 [DOI] [PubMed] [Google Scholar]
- 2.Holcomb RM, Ündar A. Are outcomes in congenital cardiac surgery better than ever? J Card Surg 2022;37:656–63. 10.1111/jocs.16225 [DOI] [PubMed] [Google Scholar]
- 3.BA P, MJ A, LP B, et al. European Association of echocardiography recommendations for training, competence, and quality improvement in echocardiography. Eur J 2009;10:893–905. 10.1093/ejechocard/jep151 [DOI] [PubMed] [Google Scholar]
- 4.Shen D, Wu G, Suk H-I. Deep learning in medical image analysis. Annu Rev Biomed Eng 2017;19:221–48. 10.1146/annurev-bioeng-071516-044442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo L, Romero R. Fetal intelligent navigation echocardiography (FINE): A novel method for rapid, simple, and automatic examination of the fetal heart. Ultrasound Obstet Gynecol 2013;42:268–84. 10.1002/uog.12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donofrio MT, Moon-Grady AJ, Hornberger LK. Diagnosis and treatment of fetal cardiac disease: a scientific statement from the American heart Association. Circulation 2014;129:2183–242. 10.1161/01.cir.0000437597.44550.5d [DOI] [PubMed] [Google Scholar]
- 7.The American Institute of Ultrasound . Practice Bulletin No. 175: ultrasound in pregnancy. Obstet Gynecol 2016;128:e241–56. 10.1097/AOG.0000000000001815 [DOI] [PubMed] [Google Scholar]
- 8.Sekar P, Heydarian HC, Cnota JF, et al. Diagnosis of congenital heart disease in an era of universal prenatal ultrasound screening in Southwest Ohio. Cardiol Young 2015;25:35–41. 10.1017/S1047951113001467 [DOI] [PubMed] [Google Scholar]
- 9.Arnaout R, Curran L, Zhao Y, et al. An ensemble of neural networks provides expert-level Prenatal detection of complex congenital heart disease. Nat Med 2021;27:882–91. 10.1038/s41591-021-01342-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han G, Jin T, Zhang L, et al. Adoption of compound echocardiography under artificial intelligence algorithm in fetal Congenial heart disease screening during gestation. Appl Bionics Biomech 2022;2022:1–8. 10.1155/2022/6410103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Diller G-P, Lammers AE, Babu-Narayan S, et al. Denoising and Artefact removal for transthoracic echocardiographic imaging in congenital heart disease: Utility of diagnosis specific deep learning Algorithms. Int J Cardiovasc Imaging 2019;35:2189–96. 10.1007/s10554-019-01671-0 [DOI] [PubMed] [Google Scholar]
- 12.Herz C, Pace DF, Nam HH, et al. Segmentation of tricuspid valve leaflets from transthoracic 3D Echocardiograms of children with hypoplastic left heart syndrome using deep learning. Front Cardiovasc Med 2021;8:735587. 10.3389/fcvm.2021.735587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gong Y, Zhang Y, Zhu H, et al. Fetal congenital heart disease Echocardiogram screening based on DGACNN: Adversarial one-class classification combined with Video transfer learning. IEEE Trans Med Imaging 2020;39:1206–22. 10.1109/TMI.2019.2946059 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Liu X, Wang F, et al. Automated interpretation of congenital heart disease from multi-view echocardiograms. Med Image Anal 2021;69:1–12. 10.1016/j.media.2020.101942 [DOI] [PubMed] [Google Scholar]
- 15.Ye J, Xu W, Yu J. Seven-step Echocardiographic screening for pediatric congenital heart disease (Xiao ER Xian Tian Xing Xin Zang Bing Chao Sheng Qi BU Shai CHA FA, Chinese). In: Chinese medical multimedia press. Beijing, 2022: 1–42. [Google Scholar]
- 16.Diller G-P, Babu-Narayan S, Li W, et al. Utility of machine learning Algorithms in assessing patients with a systemic right ventricle. Eur Heart J Cardiovasc Imaging 2019;20:925–31. 10.1093/ehjci/jey211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Cisse M, Dauphin YN. Mixup: beyond empirical risk Minimization[J]. ArXiv:171009412 [Preprint] October 2017. 10.48550/arXiv.1710.09412 [DOI] [Google Scholar]
- 18.He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016:770–8. 10.1109/CVPR.2016.90 [DOI] [Google Scholar]
- 19.Kaur T, Gandhi TK. Classifier fusion for detection of COVID-19 from CT scans. Circuits Syst Signal Process 2022;41:3397–414. 10.1007/s00034-021-01939-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Bom T, Zomer AC, Zwinderman AH, et al. The changing epidemiology of congenital heart disease. Nat Rev Cardiol 2011;8:50–60. 10.1038/nrcardio.2010.166 [DOI] [PubMed] [Google Scholar]
- 21.Romley JA, Chen AY, Goldman DP, et al. Hospital costs and inpatient mortality among children undergoing surgery for congenital heart disease. Health Serv Res 2014;49:588–608. 10.1111/1475-6773.12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das BB. Perioperative care of children with Eisenmenger syndrome undergoing non-cardiac surgery. Pediatr Cardiol 2015;36:1120–8. 10.1007/s00246-015-1184-7 [DOI] [PubMed] [Google Scholar]
- 23.Saxena A. Congenital cardiac surgery in the less privileged regions of the world. Expert Rev Cardiovasc Ther 2009;7:1621–9. 10.1586/erc.09.141 [DOI] [PubMed] [Google Scholar]
- 24.Hoffman JIE. It is time for routine neonatal screening by pulse oximetry. Neonatology 2011;99:1–9. 10.1159/000311216 [DOI] [PubMed] [Google Scholar]
- 25.Ewer AK, Middleton LJ, Furmston AT, et al. Pulse Oximetry screening for congenital heart defects in newborn infants (Pulseox): A test accuracy study. Lancet 2011;378:785–94. 10.1016/S0140-6736(11)60753-8 [DOI] [PubMed] [Google Scholar]
- 26.McCollum ED, King C, Deula R, et al. Pulse Oximetry for children with pneumonia treated as outpatients in rural Malawi. Bull World Health Organ 2016;94:893–902. 10.2471/BLT.16.173401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodríguez-González M, Alonso-Ojembarrena A, Castellano-Martínez A, et al. Heart murmur in children less than 2 years-old: looking for a safe and effective referral strategy. An Pediatr (Engl Ed) 2018;89:286–93. 10.1016/j.anpedi.2018.01.007 [DOI] [PubMed] [Google Scholar]
- 28.Bensemlali M, Stirnemann J, Le Bidois J, et al. Discordances between pre-natal and post-natal diagnoses of congenital heart diseases and impact on care strategies. J Am Coll Cardiol 2016;68:921–30. 10.1016/j.jacc.2016.05.087 [DOI] [PubMed] [Google Scholar]
- 29.Aguilera M, Dummer K. Concordance of fetal echocardiography in the diagnosis of congenital cardiac disease utilizing updated guidelines. J Matern Fetal Neonatal Med 2017:1–6. 10.1080/14767058.2017.1297791 [DOI] [PubMed] [Google Scholar]
- 30.Sarikouch S, Boethig D, Beerbaum P. Gender-specific Algorithms recommended for patients with congenital heart defects: Review of the literature. Thorac Cardiovasc Surg 2013;61:79–84. 10.1055/s-0032-1326774 [DOI] [PubMed] [Google Scholar]
- 31.Chang R-K, Gurvitz M, Rodriguez S. Missed diagnosis of critical congenital heart disease. Arch Pediatr Adolesc Med 2008;162:969–74. 10.1001/archpedi.162.10.969 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on reasonable request.