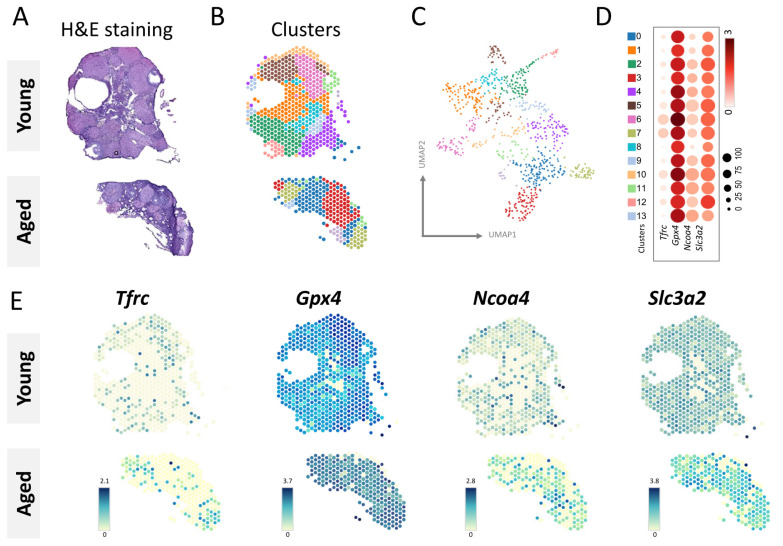
Figure 3.
Gene expression of spatial transcriptome-defined clusters in young and aging mouse ovaries. (A) By using spatial transcriptomics, tissue sections were analyzed to identify clusters, and the alignment with morphology was confirmed by hematoxylin and eosin staining and cluster mapping. (B) Thirteen unsupervised clusters of spatial transcription histology were overlaid on ovarian samples. (C) A UMAP projection of the spatial transcription histology data containing the 13 unsupervised clusters defined by Space Ranger was generated. (D) Gene expression levels in different clusters were presented using dot plots. (E) Visium spatial gene expression was used to show the spatial location and gene variation of Tfrc, Gpx4, Ncoa4, and Slc3a2 in different tissue sections.