Abstract
The 5′ UTR of c-myc mRNA contains an internal ribosome entry segment (IRES) and consequently, c-myc mRNAs can be translated by the alternative mechanism of internal ribosome entry. However, there is also some evidence suggesting that c-myc mRNA translation can occur via the conventional cap-dependent scanning mechanism. Using both bicistronic and monocistronic mRNAs containing the c-myc 5′ UTR, we demonstrate that both mechanisms can contribute to c-myc protein synthesis. A wide range of cell types are capable of initiating translation of c-myc by internal ribosome entry, albeit with different efficiencies. Moreover, our data suggest that the spectrum of efficiencies observed in these cell types is likely to be due to variation in the cellular concentration of non-canonical translation factors. Interestingly, the c-myc IRES is 7-fold more active than the human rhinovirus 2 (HRV2) IRES and 5-fold more active than the encephalomyocarditis virus (EMCV) IRES. However, the protein requirements for the c-myc IRES must differ significantly from these viral IRESs, since an unidentified nuclear event appears to be a pre-requisite for efficient c-myc IRES-driven initiation.
INTRODUCTION
The proto-oncogene c-myc is required for both cell proliferation and programmed cell death (apoptosis), and de-regulated c-myc expression is associated with a wide range of cancers (1,2). It is therefore not surprising that c-myc gene expression is tightly controlled at multiple levels (3). The post-transcriptional regulation of c-myc involves alterations in the stability of both the mRNA and the protein (4–7), and the control of c-myc translation (8–12)
In common with many other genes involved in the regulation of cell growth, the c-myc mRNA has a long and potentially highly structured 5′ untranslated region (UTR, located in exon 1). Multiple transcription start sites exist within the gene, giving rise to four transcripts (P0, P1, P2 and P3, with sizes of ~3.1, 2.4, 2.25 and 2.0 kb respectively; 13–15), with the predominant mRNA (P2) having a 5′ UTR of ~400 nt. It has been suggested that mRNAs with structured 5′ UTRs, such as c-myc, are poorly translated due to their reduced ability to associate with the cap-binding complex, the eukaryotic initiation factor 4F (eIF4F). Indeed, over-expression of the cap-binding protein eIF4E, which is believed to be a limiting component of this complex, causes an increase in the translation of mRNAs with structured 5′ UTRs such as c-myc (16–18). Furthermore, in certain circumstances the translational regulation of c-myc is mediated by phosphorylation and inactivation of the eIF4E inhibitor protein 4EBP1 (19).
It has also been shown that the 5′ UTR of c-myc contains an internal ribosome entry segment (IRES) (11,12). IRESs were originally identified in the 5′ UTRs of picornaviral RNAs and these complex structural elements allow ribosomes to enter at a considerable distance (often >1000 nt) from the 5′ end of the mRNA (20–22). Several eukaryotic mRNAs have the potential to initiate translation by an internal ribosome entry mechanism and interestingly many of the mammalian IRESs identified to date have been found in genes whose protein products are associated with the control of cell growth, e.g. c-myc, fibroblast growth factor –2 (FGF-2), platelet derived growth factor (PDGF) and vascular endothelial growth factor (VEGF) (11,12,23–26).
The region of c-myc mRNA that contains the IRES is located downstream of the P2 promoter (12). Approximately 75–90% of c-myc transcripts are synthesised from this promoter (3). Therefore, the majority of c-myc mRNAs have the potential to initiate translation via internal ribosome entry. The c-myc IRES appears to function under conditions where cap-dependent translation is compromised. Indeed, we have recently shown that the c-myc IRES is utilised during apoptosis when cap-dependent translation is reduced due to cleavage of eIF4G (27). Furthermore, in poliovirus-infected HeLa cells, in which there is a substantial reduction in cap-dependent protein synthesis due to the proteolysis of eIF4G and sequestration of eIF4E, c-myc mRNAs remain associated with heavy polysomes (28). However, since there is some evidence that c-myc mRNA can also be translated by a cap-dependent mechanism, to date it has not been possible to assess the contribution that either mechanism makes to the synthesis of c-Myc polypeptides (12,19).
In this study we present further evidence for the existence of an IRES in the c-myc 5′ UTR. In addition our data confirm that c-myc mRNAs can also be translated by a cap-dependent mechanism. This has led us to propose that both mechanisms operate in vivo. We demonstrate that the c-myc IRES is active (with one exception) in all cell lines of human origin tested, although there is a wide variation in its efficiency, whereas the IRES is not active in cell lines of murine origin. When compared to IRESs of picornaviral origin, the c-myc IRES is 7- and 5-fold more active than the IRESs derived from HRV and EMCV, respectively. Finally we provide evidence that the c-myc IRES depends on a prior nuclear event for efficient initiation of translation.
MATERIALS AND METHODS
Cell culture
All cell lines were grown at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal calf serum, in a humidified atmosphere containing 5% CO2. The cell lines HeLa (Human cervical epitheloid carcinoma), HepG2 (Human hepatocyte carcinoma), HK293 (Human embryonic kidney cell line immortalised with adenovirus DNA), Balb/c-3T3 (Murine embryonic fibroblast cell line), MCF7 (Human breast carcinoma), Cos-7 [Monkey epithelial cell line (CV-1) immortalised with SV40 DNA] and MEL cells (murine erythroleukaemic cells) were purchased from the American type culture collection. The cell line MRC5 (human lung fibroblast) was a kind gift from Dr M. MacFarlane (MRC-Human Toxicology Unit, Leicester, UK). The human SV40 immortalised fibroblast cell line GM637 was obtained from NIGMS.
Plasmid constructs
The plasmids pGL3, pGML (formerly pGL3utr), pRF and pRMF (formerly pGL3R and pGL3Rutr) have been described previously (12). cDNA encoding the HRV2 IRES was obtained from the plasmid pXLJ(10–605) (a gift from Dr R. Jackson, University of Cambridge) and inserted into pRF between the PvuII and NcoI sites, thus creating pRhrvF. To obtain the sequence encoding the EMCV IRES, a polymerase chain reaction (PCR) was performed using the oligonucleotides 5′-GATGACTAGTCCGCCCCTCTCCCTCCCCCC-3′ and 5′-GATGCCATGGC-CATATTATCATCGTGTT-3′, with pCAGSIP (an expression vector that contains the EMCV-IRES; a gift from Dr S. Monkley, University of Leicester, UK) as a template. Subsequently, the PCR product was inserted into pRF between the SpeI and NcoI sites to generate pRemcvF.
A DNA fragment containing a 60 bp palindromic sequence was amplified from pGL3RutrH (12) in a PCR using the oligonucleotides 5′-ACCTCGAGAGATATCTGGTACCGAGCTC-3′ and 5′-ACAAGCTTAGATCTGGTACCGAGCTC-3′. This fragment was inserted into pGL3 and pGML at the SpeI site, thus creating pHpL and pHpML, respectively.
The c-myc P2 cDNA was obtained by reverse transcription and PCR amplification of HeLa cell total RNA, using Superscript reverse transcriptase and Taq DNA polymerase (Life Technologies Inc). The fragments encoding the P2 c-myc cDNA from –396 to +6 and +7 to +1320 were amplified using the primer sets 5′-TAATTCCAGCGAGAGGCAGA-3′ with 5′-GGGCATCGTCGCGGGAGGCTG-3′, and 5′-CTCAAC-GTTAGCTTCACCAAC-3′ with 5′-CGGAATTCTTACGCA-CAAGAGTTGCCGAT-3′, respectively. These sequences were inserted sequentially into pSK+-bluescript (Stratagene) using the SmaI and EcoRI sites thus recreating the entire P2 cDNA in the plasmid pSKMyc. The construct pSKMycΔ1 containing the P2 sequence from –56 to + 1320 bp, was obtained by inserting a 1381 bp PvuII–EcoRI fragment derived from pSKMyc into pSK+ bluescript between the SmaI and EcoRI sites. Both constructs were linearised with HindIII prior to performing in vitro transcription reactions.
To create the bicistronic plasmids, pCRF and pCRMF, DNA fragments containing the Firefly luciferase (luc) coding region or a 5′ UTR-luc fusion were excised from pSKL and pSKutrL, respectively. These sequences were inserted into pRL-CMV (Promega) downstream of the Renilla luciferase coding region at the XbaI site.
The constructs in the pSP64R(x)L Poly A series were generated in two stages. Initially, the Renilla luciferase coding region was obtained from pRL-CMV and inserted into pSP64 Poly A (Promega) at the XbaI site. Subsequently, DNA fragments containing the luciferase coding region, a c-myc 5′ UTR-luc fusion and a HRV2 IRES-luc fusion were excised from pGL3, pGML and pRhrvF, respectively, and blunt-end ligated into the SmaI site of pSP64RPoly A downstream of the Renilla luciferase sequence. Constructs in this series were digested with EcoRI prior to inclusion in an in vitro transcription reaction. The resulting transcripts have a 3′ terminal polyadenylate tail of 30 residues.
DNA transfections
Calcium phosphate-mediated DNA transfection of mammalian cells, with the exception of MRC5, MEL and GM637 cells, was performed essentially as described by Jordan et al. (29). The remaining cell lines were transfected with FuGene6 (Roche) according to the manufacturer’s protocols.
In vitro run-off transcription and in vitro translation reactions
Plasmid constructs were linearised and in vitro transcriptions were performed using either SP6 (pSP6R(x)L series) or T3 (pSKMyc and pSKMycΔ1) polymerase as previously described. Capped transcripts were synthesised in a reaction containing 2 mM m7(5′)ppp(5′)G, 0.5 mM GTP and 1 mM of the remaining nucleotides. All RNAs were purified using size exclusion chromatography and quantified using the absorbance at 260 nm. In addition, the integrity of each transcript was verified using agarose gel electrophoresis and ethidium bromide staining.
In vitro translation reactions were performed using rabbit reticulocyte lysate (Promega) according to the manufacturer’s recommendations. The translation products were fractionated by SDS–polyacrylamide gel electrophoresis and visualised using phosphorimager analysis (Molecular Dynamics).
Cationic liposome-mediated RNA transfection
Cationic liposome-mediated RNA transfection of mammalian cells was performed as described previously (30). Capped and polyadenylated transcripts were synthesised using in vitro run-off transcription on an EcoRI linearised pSP64R(x)L poly(A) template. Approximately 2 × 105 HeLa cells were transfected with 5 µg of RNA previously incubated with 12.5 µg of Lipofectin (Life Technologies Inc.). After 8 h of transfection, cells were harvested and processed for reporter gene analysis.
Reporter gene analysis
The activity of Firefly luciferase in lysates prepared from cells transfected with pGL3, pGML, pHpML and pHpL was measured using a luciferase reporter assay system (Promega). Light emission was measured either over 1 s using a 1253 luminometer (Bio-Orbit) or over 10 s using an Optocomp-1 Luminometer (MGM instruments). The activity of both Firefly and Renilla luciferase in cell lysates with bicistronic luciferase plasmids was measured using the Dual-luciferase reporter assay system (Promega). Assays were performed according to the manufacturer’s recommendations. The activity of β-galactosidase in lysates prepared from cells transfected with pcDNA3.1/HisB/lacZ was measured using a Galactolight plus assay system (Tropix).
RESULTS
c-myc translation initiation can occur by internal ribosome entry and the conventional cap-dependent mechanism
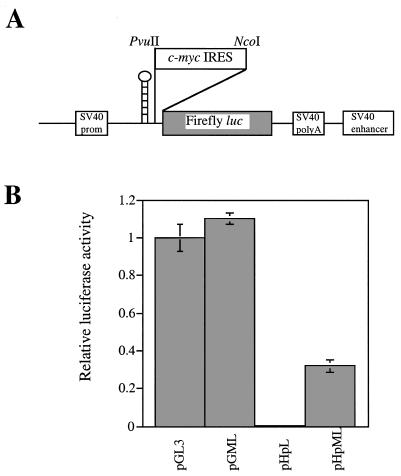
We and others have shown that c-Myc protein synthesis can occur in a cap-dependent manner and by internal ribosome entry (11,12,17,19). To assess the contribution that these two disparate mechanisms make to c-myc expression, a palindromic sequence capable of forming a stable RNA hairpin (–55 kcal/mol) was introduced into the control luciferase reporter construct, (pGL3) and the 5′ UTR containing construct (pGML, previously known as pGL3utr) at the SpeI site (Fig. 1A). As a consequence, ribosome scanning from the cap structure of the transcripts produced by the new constructs (pHpL and pHpML) should be severely impeded, whereas ribosomes entering at a site distal to the hairpin will be unaffected. HeLa cells were transfected with pGL3, pGML, pHpL or pHpML and in agreement with our previously published data, the c-myc IRES does not inhibit translation of the downstream Firefly luciferase reporter gene. Moreover, we consistently observe that there is a slight elevation in expression of this enzyme in the presence of the IRES (Fig. 1B). In cells transfected with the construct pHpL there is a 200-fold reduction in the amount of luciferase produced when compared to the control vector pGL3 (Fig. 1B). Hence, as expected the RNA hairpin structure inhibits cap-dependent translation initiation. However, in cells transfected with pHpML, in which the c-myc IRES lies downstream of the RNA hairpin, luciferase expression is stimulated by ~67-fold when compared to pHpL. These data demonstrate that the 5′ UTR can promote efficient translation initiation despite the presence of an RNA structure which blocks ribosome scanning from the 5′ end and thus provide further support for the presence of an IRES within this leader sequence. Nevertheless, it is notable that the RNA hairpin does reduce luciferase expression from a transcript containing the c-myc 5′ UTR by 3-fold. This observation would indicate that mRNAs originating from the P2 promoter must also support a cap-dependent scanning mechanism in addition to internal initiation.
Figure 1.
A comparison between the efficiency of IRES-mediated translation and scanning. (A) A diagrammatic representation of the monocistronic hairpin containing plasmids pHpL and pHpML. The hairpin was inserted into the SpeI site upstream of the PvuII site. (B) HeLa cells were transfected (in triplicate) with the plasmids shown and Firefly luciferase activity is expressed relative to the transfection control β-galactosidase. All experiments were performed on three independent occasions.
Comparison of c-myc IRES-mediated internal initiation in a range of cell types
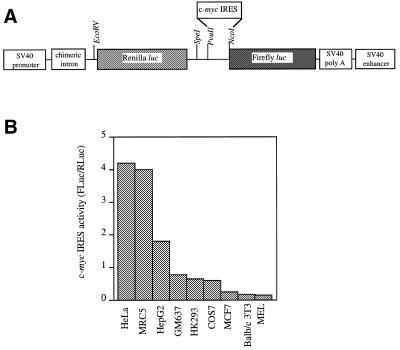
We have shown previously that the c-myc IRES is capable of promoting translation of the downstream cistron on a bicistronic mRNA in both HeLa and HepG2 cells. To investigate how widely the IRES is utilised, a range of cell types derived from different tissues, including Cos-7, MCF7, Balb/c-3T3, MEL, MRC5, HK293, GM637, HeLa and HepG2 were co-transfected with either pRF or pRMF and pcDNA3.1/HisB/lacZ (Fig. 2A). The expression from both Renilla and Firefly luciferase cistrons was assayed and normalised to the transfection control β-galactosidase. Between cell types, significant variation in the level of readthrough re-initiation was observed on the control bicistronic plasmid (data not shown). Accordingly, the efficiency of the IRES is represented as a ratio of FL to RL expressed from pRMF. In each cell line, the presence of the c-myc IRES in the mRNA did not significantly alter Renilla luciferase expression and indeed, the largest difference was observed in HeLa cells, in which the c-myc IRES reduced Renilla luciferase activity by ~11% (data not shown; 12). However, it is clear that the efficiency of c-myc IRES-driven translation varies widely between cell lines (Fig. 2B). Hence the IRES is most active in HeLa cells, followed by MRC5, HepG2, GM637, HK293 and Cos-7. Interestingly, the IRES is almost inactive in the MCF7 cells suggesting that these cells may lack a factor which is essential for IRES-mediated translation. Alternatively, these cells could express a higher level of a specific inhibitor of internal initiation. One possible explanation for the inactivity of the human c-myc IRES in cell lines of murine origin, Balb/c-3T3 and MEL cells, is that the function of the IRES displays species specificity. However, we have recently shown that this is not the case, since the c-myc IRES isolated from murine cells is active in HeLa cells and yet also relatively inactive in Balb/c-3T3 cells (data not shown).
Figure 2.
A comparison of the efficiency of c-myc IRES initiated translation in cell lines of different origin. (A) A schematic representation of the bicistronic reporter plasmids pRF and pRMF. (B) IRES activity is expressed using the ratio of downstream cistron expression to upstream cistron expression (Fluc/RLuc) with any differences in transfection efficiencies corrected for using the β-galactosidase transfection control. All experiments were performed in triplicate on three independent occasions.
c-myc P2 transcripts can be translated by a cap-dependent mechanism in Balb/c 3T3 cells, MCF-7 cells and in reticulocyte lysates
The relative inactivity of the c-myc IRES in Balb/c 3T3 and MCF7 cells enabled us to analyse the effect of the P2 5′ UTR on cap-dependent translation initiation. To this end, these cell lines were transfected with the monocistronic control construct, the 5′ UTR-containing constructs, pGL3 and pGML, and the c-myc 5′ UTR construct containing the hairpin pHpML respectively. The P2 5′ UTR does not inhibit cap-dependent translation initiation, at least in these cell lines (Fig. 3A). However, the additional presence of the hairpin structure was sufficient to prevent scanning demonstrating that the c-myc IRES is relatively inactive in these cell types and consequently c-myc is translated by a cap-dependent mechanism (Fig. 3A).
Figure 3.
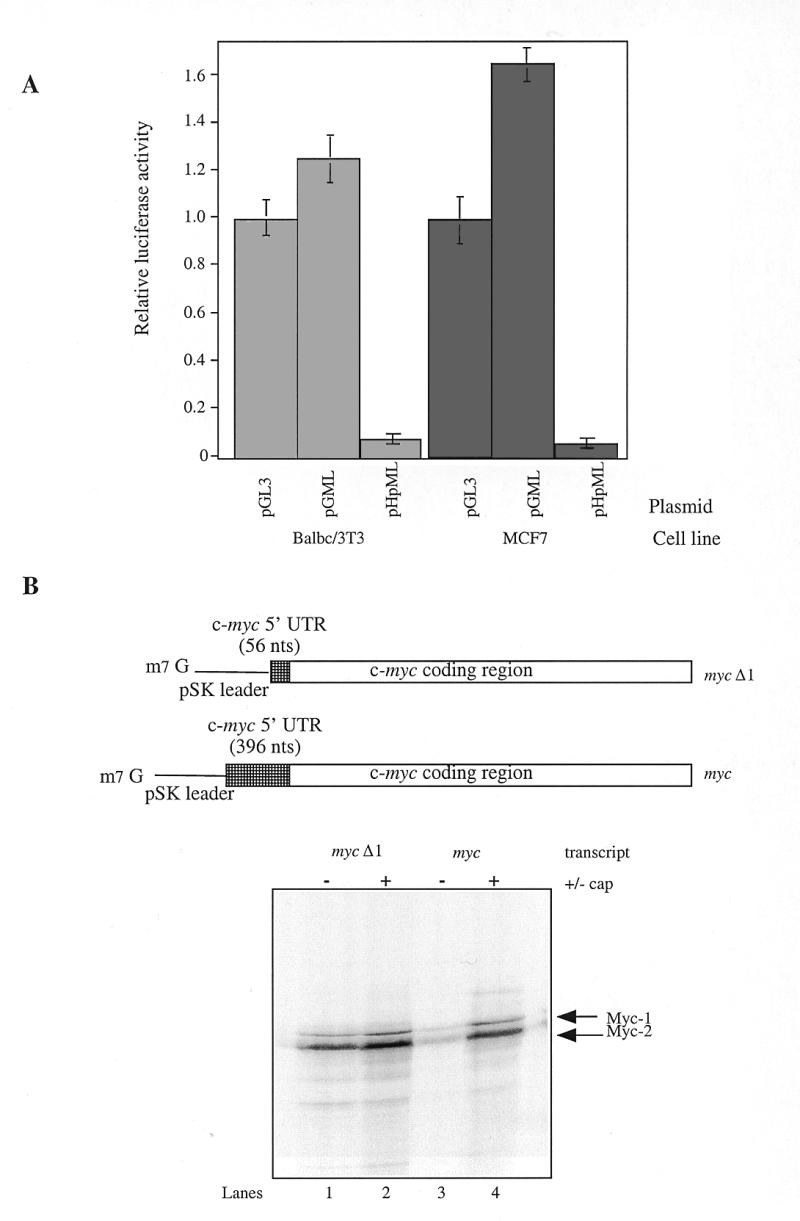
Cap-dependent translation of c-myc P2 transcripts in Balb/c 3T3, MCF7 cells and in rabbit reticulocyte lysates. (A) MCF7 and Balb/c cells were transfected with the plasmids pGL3, pGML or pHpML and the Firefly luciferase activity measured as described previously. (B) c-myc transcripts bearing 56 nt (myc Δ1) or 396 nt (myc) of the c-myc 5′ UTR were synthesised in vitro using linearised plasmids pSKMΔ1 or pSKM respectively. Rabbit reticulocyte lysate was programmed with 5 ng/µl of either capped (+) or uncapped (–) myc or myc Δ1 transcripts. Radiolabelled polypeptides synthesised in the reaction were then fractionated by SDS–PAGE and detected using phosphorimager analysis.
To further investigate the impact of the P2 5′ UTR on cap-dependent translation initiation we turned to reticulocyte lysate. This system cannot support internal ribosome entry on the c-myc leader sequence (our unpublished data; 31), therefore the contribution of the 5′ cap structure can be assessed directly. Thus, rabbit reticulocyte lysate was primed with capped or uncapped c-myc transcripts, either bearing the P2 5′ UTR sequence (myc) or lacking this element (mycΔ1) (Fig. 3B). Two species of c-myc protein can arise from the P2 transcripts by use of alternate translation initiation codon (CUG or AUG), which give rise to protein products with apparent molecular weights of 67 and 64 kDa respectively (32). As expected, capping the mycΔ1 RNA stimulated the synthesis of both Myc-1 and 2 polypeptides (Fig. 3B, lanes 1 and 2). This modest effect of 2–2.5-fold is consistent with the previously reported values for relatively unstructured RNAs using this system (33). In the absence of a cap structure, the c-myc 5′ UTR reduced the synthesis of both the AUG and CUG-initiated polypeptides by ~90% (Fig. 3B, lanes 1 and 3). It is likely that structural elements within the 5′ UTR are responsible for this effect since this element is GC-rich. However, the synthesis of both proteins was enhanced by 14–16-fold on capping of the myc transcript (Fig. 3B, lanes 3 and 4), with the result that the 5′ UTR inhibits translation initiation by only 50%. Hence, the P2 5′ UTR strongly attenuates the translational efficiency of uncapped c-myc transcripts. Nevertheless, much of this repression is relieved by the presence of a 5′ cap. Therefore, translation initiation on the P2 transcript is strongly cap-dependent in the reticulocyte lysate system.
Overexpression of bicistronic mRNAs inhibits the function of the c-myc IRES
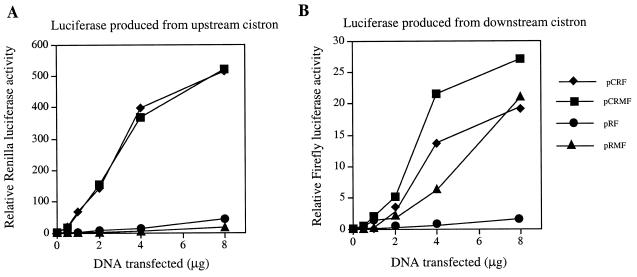
Thus far, we have demonstrated that in many cell lines c-myc translation can occur by the alternative mechanism of internal ribosome entry. However, c-myc can also be translated by the conventional cap-dependent mechanism in certain backgrounds. One model that would explain the cell-type specific variation in the efficiency of c-myc IRES-driven translation posits that non-canonical trans-acting factors are required for the recruitment of the 40S ribosome to this element. In this scenario, the activity of one or more of these factors is considerably reduced in the Balb/c-3T3 and MCF7 cell lines. Further evidence in support of this model was provided by experiments in which the bicistronic mRNAs were overexpressed using the powerful cytomegalovirus (CMV) promoter/enhancer region; this transcriptional element has been shown to result in significantly higher levels of expression than the SV40 promoter/enhancer (34). The Renilla luciferase activity measured in cells transfected with a CMV-based control bicistronic plasmid pCRF was significantly greater than that achieved with the analogous plasmid, pRF (~27-fold, Fig. 4A, compare pCRF Renilla luciferase to pRF Renilla luciferase). However, in cells transfected with the 5′ UTR-containing construct, pCRMF, there was not a corresponding increase in Firefly luciferase activity when compared to pRMF. Transfection with 4 or 8 µg of pCRMF produced only 4- or 1.25-fold more Firefly luciferase than pRMF, respectively (Fig. 4B). Consequently, using the CMV promoter/enhancer, the apparent activity of the c-myc IRES when calculated relative to readthrough is only 1.5–2 fold compared to 50-fold for the SV40 based constructs (Fig. 4B). These data suggest that a trans-acting factor, which is required for initiation of translation via the c-myc IRES, is present at a limiting concentration. A similar observation has been reported for the entero- and rhinovirus IRESs; the efficiency of translation mediated by these IRESs was considerably reduced when bicistronic mRNAs were expressed at high levels in vivo (35). This phenomenon correlates with a requirement for non-canonical factors, since it was not observed for either cap-dependent translation or translation driven by the cardio- and aphthovirus IRESs (35).
Figure 4.
The effect of the CMV promoter/enhancer on c-myc IRES directed internal initiation. HeLa cells were transfected with the CMV promoter/enhancer based plasmids pCRF or pCRMF or the SV40 promoter/enhanced based plasmids, pRF and pRMF. (A) Renilla and (B) Firefly luciferase activity was determined and normalised to that of the transfection control, β-galactosidase.
A comparison of the efficiency of the c-myc and viral IRESs
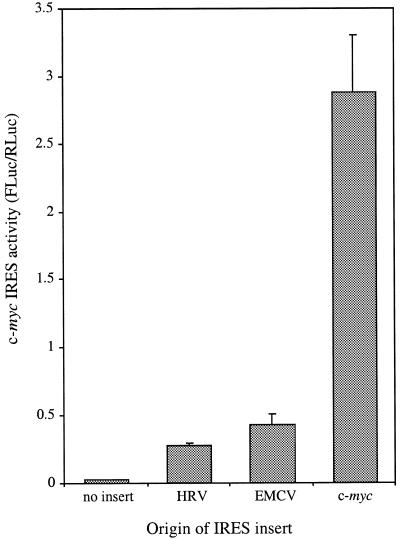
The previous data provided indirect evidence that the function of the c-myc IRES could depend on a non-canonical trans-acting factor. In this respect, it would be analogous to the IRESs of the entero- and rhinoviruses (36). To compare the efficiency of the c-myc, HRV and EMCV IRESs, HeLa cells were transfected with the plasmids pRF, pRMF, pRhrvF, pRemcvF. The activities of Renilla and Firefly luciferase were determined and normalised to that of the transfection control, β-galactosidase (Fig. 5). Expression of the upstream cistron, Renilla luciferase, was not greatly affected by the presence of the EMCV, HRV or the c-myc IRES in the intercistronic region (data not shown). A comparison of the downstream cistron activities revealed that all of these elements stimulated Firefly luciferase expression (Fig. 5). However, the extent to which expression from the downstream cistron was enhanced differed widely between these IRESs. In fact, the c-myc IRES elevated Firefly luciferase activity by 70.8-fold, whilst the HRV and EMCV IRESs caused a lesser stimulation of 9.6- and 14-fold, respectively. Thus, these data suggest that both of these IRESs are less efficient in this system at promoting internal ribosome entry than the c-myc IRES.
Figure 5.
A comparison of the efficiency of HRV, EMCV and c-myc IRES-initiated internal ribosome entry on bicistronic mRNAs transcribed in the nucleus. HeLa cells were transfected in triplicate with either the control plasmid pRF, the c-myc IRES containing plasmid pRMF, the HRV IRES containing plasmid pRhrvF or the EMCV IRES containing plasmid pRemcvF. Upstream cistron (Renilla luciferase) and downstream cistron (Firefly luciferase) activities were determined and normalised to that of the transfection control,
c-myc IRES-driven translation requires a nuclear event
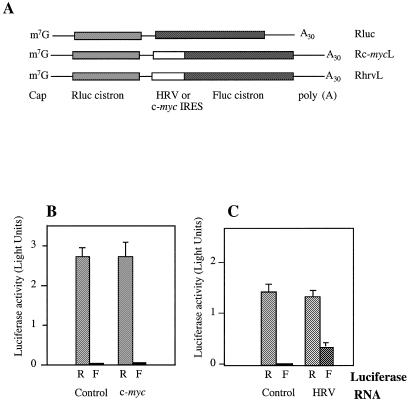
It has been suggested previously that efficient translation driven by the IRES located in the 5′ UTR of the immunoglobulin heavy chain binding protein (Bip) requires a nuclear event (37). Moreover, two specific nuclear protein factors have been identified which interact with the Bip IRES (38). To test whether the c-myc IRES also has such a requirement for nuclear factors, the plasmid constructs pSP64RL poly(A), pSP64R(c-myc)L poly(A) and pSP64R(hrv)L poly(A) were generated. Bicistronic transcripts containing an m7GpppG cap structure and a polyadenylated tail at the 5′ and 3′ termini, respectively, were synthesised from each of the plasmids in the pSP64RL(x)Lpoly(A) series by in vitro run-off transcription (Fig. 6A). Cationic liposomes were used to encapsulate equimolar quantities of each transcript and introduce them into the cytoplasm of HeLa cells. After a period of 8 h, the expression from the upstream and downstream cistrons was monitored (Fig. 6B and C). In cells transfected with the control bicistronic transcript, Rluc, the Renilla luciferase cistron was translated efficiently, whilst little expression of the downstream cistron was observed (Fig. 6B and C). Insertion of the HRV IRES between the two cistrons resulted in a 52-fold stimulation of Firefly luciferase activity when compared to the expression due to readthrough-re-initiation (Fig. 6C). In contrast, the expression of the downstream cistron was only enhanced by 1.4-fold on the Rc-mycL transcript (Fig. 6B). Thus, the c-myc IRES cannot stimulate the translation of the downstream cistron on a bicistronic mRNA introduced directly into the cytoplasm. To confirm these data, the plasmids pRF and pRMF were transfected into human TK143 cells previously infected with a recombinant vaccinia virus that expresses the T7 RNA polymerase (vTF7-3) (39). The presence of a T7 RNA polymerase promoter upstream of the Renilla luciferase cistron in pRF and pRMF results in the transcription of bicistronic mRNAs in the cytoplasmic compartment. However, the c-myc 5′ UTR did not promote internal initiation on mRNAs transcribed in the cytoplasm using the T7/vaccinia system (data not shown). In contrast, the IRESs of the entero- and rhinoviruses have been shown to function efficiently using bicistronic mRNAs expressed in this manner (35,40). These data appear to suggest a fundamental difference between the function of the entero- and rhinovirus IRESs and that of c-myc. The c-myc IRES is only able to promote internal initiation on transcripts expressed in the nucleus, however the HRV element is capable of performing this task on mRNAs that do not originate in this compartment. Therefore, we propose that a nuclear event is a pre-requisite for efficient c-myc internal initiation.
Figure 6.
A comparison of the efficiency of the c-myc IRES and HRV-IRES directed internal initiation on mRNAs introduced directly into the cytoplasm. (A) A diagrammatic representation of the control (Rluc), c-myc 5′ UTR containing (Rc-mycL) and HRV IRES containing (RhrvL) biscistronic RNAs. Transcripts were synthesised in vitro and possess both a 5′ cap structure and a 3′ terminal polyadenylate tail of 30 residues. (B) HeLa cells were transfected with Rluc (control) or Rc-mycL (c-myc) by lipofection. After 8 h Renilla (R) and Firefly (F) luciferase expression was determined. (C) Similarly, HeLa cells were transfected with RLuc (control) or RhrvL (HRV IRES) and Renilla and Firefly luciferase activities determined.
DISCUSSION
We and others have shown previously that the 5′ UTR of c-myc contains an IRES (11,12). We have investigated several features of the c-myc IRES and compared its activity in a range of cell lines and to IRESs of viral origin.
First, using a stable RNA structure to substantially impede ribosome scanning from the 5′ cap, we have demonstrated that efficient translation initiation can be restored by positioning the c-myc 5′ UTR downstream of this inhibitory element (Fig. 1). This observation provides further evidence that the P2 leader sequence can support internal entry of ribosomes via an IRES. In these experiments, internal initiation directed by the c-myc IRES is apparently 3-fold less efficient than cap-dependent translation initiation (but see later). However, reporter mRNAs are translated with comparable efficiency whether the 5′ UTR is present or not. Thus, we suggest that c-myc mRNAs originating from the P2 promoter are capable of being translated via a cap-dependent mechanism in addition to internal initiation. This hypothesis is strengthened by two observations. First, a reporter mRNA bearing the P2 leader sequence was translated efficiently in cell lines with a significantly reduced capacity to promote 5′ UTR-mediated internal initiation (Fig. 3A). Second, in reticulocyte lysate, a system in which the c-myc IRES is inactive (our unpublished data; 31), c-myc P2 transcripts are translated in a manner that is strongly dependent on the presence of a cap structure (Fig. 3B). In agreement with these data, Carter et al. (31) have recently shown that the considerable repression of translation initiation caused by the P1 5′ UTR in rabbit reticulocyte lysate can be relieved by the addition of eIF4F/E (31). Thus, we propose a dual mechanism for c-myc translation initiation. Under conditions where cap-dependent protein synthesis is compromised there is a shift from a cap-dependent to an IRES-directed mechanism of translation initiation. In accord with this hypothesis, we have recently shown that c-myc protein synthesis is maintained during apoptosis by virtue of the IRES, whereas overall cap-dependent translation is significantly inhibited (27).
We have also identified several factors that influence the efficacy of the c-myc IRES. Expression of bicistronic mRNAs containing the c-myc IRES in a panel of cell lines demonstrated that the activity of this element is critically dependent on cellular origin (Fig. 2). Although the IRES stimulated protein synthesis from the downstream cistron in all the cell lines tested, there was a 20-fold disparity between HeLa and MCF7 cells, the lines in which the IRES is most and least active, respectively. This cell-type specific variation in IRES activity implies that the function of this element could be modulated by non-canonical trans-acting factors. In this regard, we have recently demonstrated that ribonuclear protein complexes assembled on the c-myc 5′ UTR in vitro using cell extracts from different cell lines vary distinctly in composition (41). Furthermore, overexpression of bicistronic mRNAs using the powerful CMV promoter/enhancer drastically reduced the apparent efficiency of the c-myc IRES (Fig. 4). We speculate that the concentration of a trans-acting factor essential for c-myc IRES-driven translation initiation is limiting under these conditions. The low concentration of this factor could also explain why c-myc internal initiation appears to be 3-fold less efficient than cap-dependent translation (Fig. 1) since transcripts expressed from the monocistronic constructs (pGL3, pGML, pHpL and pHpML) accumulate to a level approximately an order of magnitude higher than those produced from the bicistronic constructs (pRF and pRMF) (our unpublished observations). Significantly, the characteristics described above are not unique to the c-myc IRES. Both cell-type specific variations in IRES activity and saturation of IRES function have also been described for the better defined IRESs of the entero- and rhinoviruses (35,40). The activity of these elements is known to be dependent on host-specific trans-acting factors suggesting that the c-myc IRES has similar requirements.
A comparison of the c-myc IRES to those of the human rhinovirus (HRV) and encephalomyocarditis virus (EMCV), using bicistronic mRNAs expressed in the nucleus, revealed that it is 7- and 5-fold more active, respectively (Fig. 5). However, the c-myc IRES differs markedly from those of viral origin, in that it is almost completely inactive when present in bicistronic mRNAs introduced directly into the cytoplasmic compartment (Fig. 6 and data not shown). Furthermore, it has also been observed that in contrast to the poliovirus IRES, the c-myc 5′ UTR could not promote internal initiation in HeLa cell extracts (42). Taken together, these data strongly suggest that a nuclear experience is an essential pre-requisite for internal initiation mediated by the c-myc IRES. The nature of this nuclear event is currently unknown. However, it is interesting to note that several nuclear factors have been shown to interact with the Bip IRES, the function of which is also dependent on a nuclear origin (37,38). Thus, factors recruited to these IRESs in the nucleus could subsequently promote internal initiation in the cytoplasm (37).
Carter et al. have recently suggested that the c-myc 5′ UTR does not contain an IRES (31). However, these experiments were performed in reticulocyte lysate, a specialised translation extract known to contain very limiting amounts of nuclear and cytoplasmic RNA binding proteins (33). We have also found that the c-myc IRES cannot function in reticulocyte lysate (data not shown). In this respect it is similar to the IRESs of the entero- and rhinoviruses, which function inefficiently or not at all in this system. Indeed, reticulocyte lysate must be supplemented with cytoplasmic extracts to support efficient entero/rhinovirus internal initiation (36). Most importantly, to our knowledge no eukaryotic cellular IRES has been shown to promote internal initiation in this system. Using bicistronic mRNAs expressed in the nucleus of cell lines, we and others identified an IRES in the c-myc 5′ UTR (11,12). This finding has been supported by the observation that c-myc mRNAs are efficiently translated in poliovirus-infected HeLa cells and in cells undergoing apoptosis (27,28). Here we present further evidence that c-myc mRNAs can be translated by internal initiation and we provide additional mechanistic insights. Our data support a model in which both non-canonical trans-acting factors and a nuclear experience participate in c-myc internal ribosome entry. In the light of these results, it is hardly surprising that the c-myc IRES does not function in the reticulocyte lysate system. Finally, we are currently attempting to identify the cytoplasmic and nuclear factors involved in the formation of ribonuclear protein complexes with the c-myc 5′ UTR. The effect of these factors on c-myc internal initiation can then be rigorously tested in cell-free extracts.
Acknowledgments
ACKNOWLEDGEMENTS
This work was funded by a grant from the Cancer Research Campaign (M.S. and T.S.). J.P.C.L.Q., M.J.C. and C.L.J. hold MRC studentships.
REFERENCES
- 1.Henriksson M. and Lüscher,B. (1996) Adv. Cancer Res., 68, 109–182. [DOI] [PubMed] [Google Scholar]
- 2.Prengergast G.C. (1999) Oncogene, 18, 2967–2987. [DOI] [PubMed] [Google Scholar]
- 3.Marcu K.B., Bossone,S.A. and Patel,A.J. (1992) Ann. Rev. Biochem., 61, 809–860. [DOI] [PubMed] [Google Scholar]
- 4.Ross J. (1995) Microbiol. Rev., 59, 16–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee C.H., Leeds,P. and Ross,J. (1998) J. Biol. Chem., 273, 25261–25271. [DOI] [PubMed] [Google Scholar]
- 6.Lüscher B. and Eisenmann,R.N. (1988) Mol. Cell. Biol., 8, 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shindo H., Tani,E., Matsumuto,T., Hashimoto,T. and Furuyama,J. (1993) Acta Neuropathol., 86, 345–352. [DOI] [PubMed] [Google Scholar]
- 8.Saito H., Hayday,A.C., Wiman,K., Hayward,W.S. and Tonegawa,S. (1983) Proc. Natl Acad. Sci. USA, 80, 7476–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butnick N.Z., Miyamoto,C., Chizzonite,R., Cullen,B.R., Ju,G. and Skalka,A.M. (1985) Mol. Cell. Biol., 5, 3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parkin N.T., Darveau,A., Nicholson,R. and Sonenberg,N. (1988) Mol. Cell. Biol., 8, 2875–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanbru C., Lafon,I., Audigier,S., Gensac,M.-G., Vagner,S., Huez,G. and Prats,A.-C. (1997) J. Biol. Chem., 272, 32061–32066. [DOI] [PubMed] [Google Scholar]
- 12.Stoneley M., Paulin,F.E.M., Le Quesne,J.P.C., Chappell,S.A. and Willis,A.E. (1998) Oncogene, 16, 423–428. [DOI] [PubMed] [Google Scholar]
- 13.Battey J., Moulding,C., Taub,R., Murphy,W., Stewart,T., Potter,H., Lenoir,G. and Leder,P. (1983) Cell, 34, 779–787. [DOI] [PubMed] [Google Scholar]
- 14.Bentley D.L. and Groudine,M. (1986) Mol. Cell. Biol., 6, 3481–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang J.-Q., Bauer,S.R., Mushinski,J.F. and Marcu,K.B. (1985) EMBO J., 4, 1441–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koromilas A.E., Lazaris-Karatzas,A. and Sonenberg,N. (1992) EMBO J., 11, 4153–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Benedetti A., Joshi,B., Graff,J.R. and Zimmer,S.G. (1994) Mol. Cell. Differ., 2, 347–371. [Google Scholar]
- 18.De Benedetti A. and Rhoads,R.E. (1990) Proc. Natl Acad. Sci. USA, 87, 8212–8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.West M.J., Stoneley,M. and Willis,A.E. (1998) Oncogene, 17, 769–780. [DOI] [PubMed] [Google Scholar]
- 20.Jackson R.J., Hunt,S.L., Reynolds,J.E. and Kaminski,A. (1995) Curr. Top. Microbiol. Immunol., 203, 1–29. [DOI] [PubMed] [Google Scholar]
- 21.Jackson R.J. and Kaminski,A. (1995) RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson R.J., Hunt,S.L., Gibbs,C.L. and Kaminski,A. (1994) Mol. Biol. Reports, 19, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagner S., Gensac,M.-C., Maret,A., Bayard,F., Amalric,F., Prats,H. and Prats,A.-C. (1995) Mol. Cell. Biol., 15, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernstein J., Sella,O., Le,S. and Elroy-Stein,O. (1997) J. Biol. Chem., 272, 9356–9362. [DOI] [PubMed] [Google Scholar]
- 25.Stein I., Itin,A., Einat,P., Skaliter,R., Grossman,Z. and Keshet,E. (1998) Mol. Cell. Biol., 18, 3112–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller D., Dibbens,J., Damert,A., Risau,W., Vadas,M. and Goodall,G. (1998) FEBS Lett., 434, 417–420. [DOI] [PubMed] [Google Scholar]
- 27.Stoneley M., Chappell,S.A., Jopling,C.L., Dickens,M., MacFarlane,M. and Willis,A.E. (2000) Mol. Cell. Biol., 20, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johannes G. and Sarnow,P. (1998) RNA, 4, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jordan M., Schallhorn,A. and Wurm,F.M. (1996) Nucleic Acids Res., 24, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwarki V.J., Malone,R.W. and Verma,I.M. (1993) Methods Enzymol., 217, 644–654. [DOI] [PubMed] [Google Scholar]
- 31.Carter P.S., Pardo-Jarqin,M. and DeBenedetti,A. (1999) Oncogene, 18, 4326–4335. [DOI] [PubMed] [Google Scholar]
- 32.Hann S.R., King,M.W., Bentley,D.L., Anderson,C.W. and Eisenman,R.N. (1988) Cell, 52, 185–195. [DOI] [PubMed] [Google Scholar]
- 33.Svitkin Y.V., Ovchinnikov,L.P., Dreyfuss,G. and Sonenberg,N. (1996) EMBO J., 15, 7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland L.C. and Williams,G.T. (1997) J. Immunol. Meth., 207, 179–183. [DOI] [PubMed] [Google Scholar]
- 35.Borman A.M., LeMercier,P., Girard,M. and Kean,K.M. (1997) Nucleic Acids Res., 25, 925–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belsham G.J. and Sonenberg,N. (1996) Microbiol. Rev., 60, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iizuka N., Chen,C., Yang,Q., Johannes,G. and Sarnow,P. (1995) Curr. Top. Microbiol. Immunol., 203, 156–177. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q. and Sarnow P. (1997) Nucleic Acids Res., 25, 2800–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuerst T.R., Niles,E.G., Studier,F.W. and Moss,B. (1986) Proc. Natl Acad. Sci. USA, 83, 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts L.O., Seamons,R.A. and Belsham,G.J. (1998) RNA, 4, 520–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulin F.E.M., Chappell,S.A. and Willis,A.E. (1998) Nucleic Acids Res., 26, 3097–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelletier J. and Sonenberg,N. (1988) Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]