Abstract
In recent years, ambient ionization mass spectrometry (AIMS) including laser ablation rapid evaporation IMS, has enabled direct biofluid metabolome analysis. AIMS procedures are, however, still hampered by both analytical, i.e., matrix effects, and practical, i.e., sample transport stability, drawbacks that impede metabolome coverage. In this study, we aimed at developing biofluid-specific metabolome sampling membranes (MetaSAMPs) that offer a directly applicable and stabilizing substrate for AIMS. Customized rectal, salivary, and urinary MetaSAMPs consisting of electrospun (nano)fibrous membranes of blended hydrophilic (polyvinylpyrrolidone and polyacrylonitrile) and lipophilic (polystyrene) polymers supported metabolite absorption, adsorption, and desorption. Moreover, MetaSAMP demonstrated superior metabolome coverage and transport stability compared to crude biofluid analysis and was successfully validated in two pediatric cohorts (MetaBEAse, n = 234 and OPERA, n = 101). By integrating anthropometric and (patho)physiological with MetaSAMP-AIMS metabolome data, we obtained substantial weight-driven predictions and clinical correlations. In conclusion, MetaSAMP holds great clinical application potential for on-the-spot metabolic health stratification.
Customized biofluid-specific MetaSAMPs enable superior metabolome coverage and stability for direct health stratification.
INTRODUCTION
To date, metabolomics of human biofluids is emerging as a promising biofocus as it provides advantages that classical diagnostics do not, following discovery of a suite of clinically relevant biomarkers that are simultaneously affected by disease while contributing to the elucidation of metabolic pathways underlying particular phenotypes of health and disease (i.e., metabotyping) (1–3). The most urgent application of metabolomics remains the identification of pathologies in children as early-infancy exposures, including nutrition and illness, severely affect adult health and functionality as well (4). In this respect, metabolite patterns offer a valid tool to profile individuals (5) at risk of developing e.g., the metabolic sequelae coinciding the global rise in overweight and obesity (6) by discerning between metabolically healthy and unhealthy phenotypes and predict future risks of developing comorbidities such as (pre)diabetes, metabolic syndrome, etc. (2, 7–10).
The urinary and fecal metabolomes have been popularized as they reflect both exogenous and endogenous metabolic products (11), as well as complex interactions between dietary intake, gut microbiome, and host (12). Salivary metabolites largely mirror those in blood and may thus reflect many pathophysiological and nutritional changes, as well as exposure to medication and environmental factors (13). Major bottlenecks in large metabolomics cohort studies remain the collection, transport, storage, and sample preparation of biofluids. Ongoing (bio)chemical reactions following sample collection, long-term preservation, and extraction may substantially introduce metabolome alteration (12, 14) and misleading interpretations, while the addition of chemical preservatives during shipping, storage, and extraction causes metabolome contamination by adduct formation and/or ion pairing (15).
The typical workflow used in conventional biofluid metabolomics comprises multiple time-consuming steps including sample collection and pretreatment such as lyophilization, extraction and chromatographic separation, etc., resulting in low sample throughput (ca. 60/day) and high costs (>200 euro per analysis) (16). During the past decades, instrumental advances have allowed the integration of near-real-time surface sampling or ambient ionization of matrices in their native analyte environment (17–19). Laser ablation coupled to rapid evaporative ionization mass spectrometry (REIMS), termed laser-assisted REIMS (LA-REIMS), is among the few ambient ionization-based techniques that have found their way into the clinic both in vivo and ex vivo (16, 20). LA-REIMS uses a sample-focused laser beam that excites the most intense vibrational band (oxygen-hydrogen stretching mode) of water molecules present in the sample, initiating ablation heat and causing evaporation. The resulting analyte-containing aerosol is aspirated toward the mass spectrometry (MS) under the instrument’s vacuum where it is mixed with a solvent and collides with a heated collision surface to form gas-phase ions for analysis (16, 20). To date, LA-REIMS is the only ambient ionization mass spectrometry (AIMS) platform that has been successfully used for the automated analysis of a range of crude biofluids, including feces, saliva, urine, etc. (1). LA-REIMS enables an analysis rate of only few min per sample and demonstrates substantial congruencies with mass spectra obtained by conventional metabolomics (16, 21, 22). Current challenges in direct LA-REIMS–based crude biofluid analysis are the existence of matrix effects and reduced ionization efficiency, resulting in a decreased selectivity and thus metabolome coverage (23). Moreover, biofluid collection may hamper a direct sampling-to-MS result workflow (1), because the need to wait for, e.g., defecation prevents immediate sample availability. A sampling device that allows the integration of sampling, sample preparation, and presenting the sample for direct metabolomics analysis would offer a substantial advance compared to crude biofluid analysis, because it would largely circumvent these challenges.
In this study, we aimed at developing, optimizing, and benchmarking biofluid-specific samplers (MetaSAMPs, WO/2021/191467) for direct rapid AIMS-based metabotyping (Fig. 1A). Our MetaSAMPs consist of (nano)fibrous membranes customized both in terms of polymer composition, ratio, and fiber diameter by electrospinning (ES) toward optimal biofluid-specific (feces, saliva, and urine) metabolome sampling, absorption and adsorption, and stabilization. ES is a simple and versatile technology that uses electrostatic forces to produce (nano)fibrous membranes with controllable compositions (24, 25). The typical porous bed structure of electrospun membranes enables analyte enrichment due to highly efficient mass transfer between biofluids and the polymerous (nano)fibers, which act as sorbents, and hence, diffusion of molecules into the porous membrane, thereby facilitating metabolite absorptive and adsorptive capabilities (26–28). The possibility of including specific chemical moieties offers a means to enhance the absorption and adsorption of analytes bearing a broad range of polarities. Moreover, biocompatible polymers such as polyacrylonitrile (PAN) are also suitable for ES, which allows for in vivo usage while supporting the exclusion of macromolecules (29). The MetaSAMPs may be configured as a sampling kit with an integrated electrospun membrane whether or not assisted by a medical swab that is directly amenable to our established rapid LA-REIMS platform (1) (Fig. 1A). We also confirmed the ability of MetaSAMPs to stabilize the biofluid-specific metabolome, increasing transport ease and duration. Last, the clinical applicability and superior performance of our MetaSAMPs relative to the analysis of crude biofluids were assessed ex vivo in two pediatric cohorts [Metabolomics research on Early Metabolic Disease (MetaBEAse) and Obesity Prevention through Emotion Regulation in Adolescents (OPERA)]. Hence, we have demonstrated that direct MetaSAMP-based biofluid metabotyping may have far-reaching potential as a future medical device.
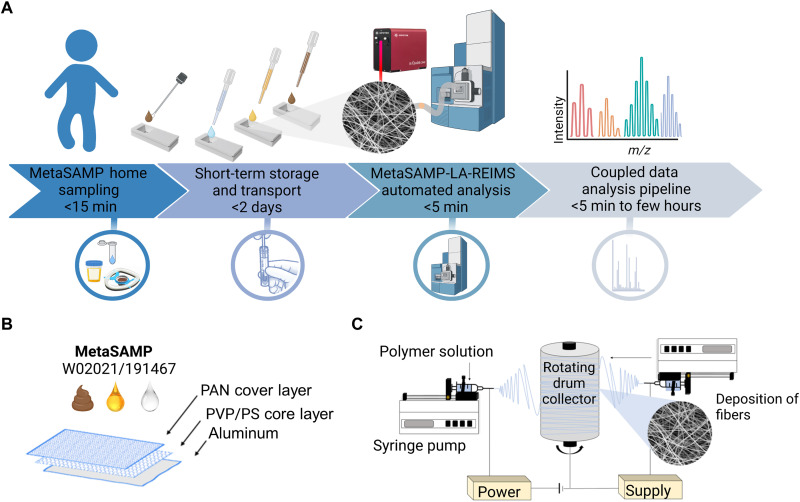
Fig. 1. MetaSAMP-LA-REIMS enables direct rectal, salivary, and urinary metabotyping.
(A) A straightforward workflow for MetaSAMP sampling hyphenated with LA-REIMS analysis. The analytes captured by MetaSAMP (configured as a sampling kit whether in combination with a medical swab) are desorbed, and the resulting analyte-rich aerosol is directly transferred through a vent line to the inlet capillary, where subsequent quadrupole time-of-flight analysis takes place. Data are visualized in real-time through MassLynx software or an in-house data analysis pipeline across the mass range installed, i.e., 50 to 1200 m/z range, after which metabolomic alterations can be quickly revealed by multivariate data analysis, leading to first-line segregation and risk classification based on distinctive fingerprints for decision-making in health care. (B) The three-layered MetaSAMP consists of a PAN cover layer, a PVP/PS core membrane, and an aluminum support layer as adhering electrically conductive collector. (C) The ES setup consists of a single-nozzle system that is duplicated to enable bidirectional ES of the loaded polymer blends onto a continuously rotating drum.
RESULTS
Optimized electrospun MetaSAMPs enable biofluid-specific metabolic fingerprinting
The development of our biofluid-specific metabolome sampling MetaSAMPs started by tuning the configuration and chemical composition of the core biofluid-specific membrane (feces, saliva, and urine; Fig. 1B) toward maximal metabolome coverage following LA-REIMS analysis (wide logP span and molecular feature count) and repeatability [as % of molecular features with a coefficient of variation (CV) ≤ 30%]. Hereto, the electrospun core membranes were fabricated using a rotating drum collector (Fig. 1C) on an aluminum support layer (Fig. 1B). Because the superiority of materials with a hydrophilic-lipophilic balance (HLB) as comprehensive absorptive and adsorptive sorbents has been reported (30–32), we selected two relevant HLB polymers [polyvinylpyrrolidone (PVP) and polystyrene (PS)] for our core membranes. As feces (and rectal content) are more heterogeneous, complex, and richer biofluids than saliva (13) or urine (11), the rectal MetaSAMP optimization was prioritized. Following impregnation with porcine rectal content, different core membrane polymer compositions were evaluated for their metabolome coverage using LA-REIMS analysis as detailed in note S1.1 and inspected through scanning electron microscopy (SEM) analysis (fig. S1). The use of porcine rectal content was rationalized on the basis of similarities in nutrition and metabolism between pigs and humans (33). It was observed that a balanced blend of PVP/PS (50/50, w/w) significantly outperformed [analysis of variance (ANOVA) false discovery rate (FDR)–adjusted P = 9.03 × 10−8, F = 41.44, df = 5] the other compositions tested [cross-linked PVP and PVP/PS (20/80)] in terms of metabolome coverage and signal intensity while providing acceptable repeatability (note S1.1 and fig. S2). Consequently, the HLB of the core membrane’s compositional fibers was demonstrated through modulated differential scanning calorimetry (mDSC). Separate glass transition temperatures (Tg) could be assigned to carbonyl- (PVP) and benzene-rich (PS) moieties (30) (note S2 and fig. S3). In addition, the LA-REIMS settings were further optimized for rectal MetaSAMP analysis using a design of experiments (DOE) approach (note S3.1). Significance (ANOVA FDR-adjusted P = 0.012 and P < 0.001, F = 14.62 and 25.35, df = 5) was detected for higher scan time and solvent flow rate. The optimal values were obtained by maximizing metabolome coverage, molecular feature signal intensity, and repeatability, resulting in a scan time of 0.7 scans s−1 and a solvent flow rate of 250 μl min−1 (table S1 and fig. S4).
Hereafter, electrospun PVP/PS membranes were tested for saliva and urine (note S1.2). When spiking the PVP/PS core membranes comprising 50 to 100% PS with saliva or urine, unacceptable long biofluid residence times were noted (>1 hour). This could be ascribed to the more aqueous nature of the biofluids saliva and urine as compared to the pig rectal content tested earlier. More homogeneous (nano)fiber size distributions were observed for the different core membranes’ composition with increasing hydrophilic PVP (10% or more) (fig. S5), which would benefit the reproducibility of LA-REIMS metabolomics analysis. Because these increased biofluid residence times were observed to bring about more variable impregnation times and thus unacceptable variation in obtained LA-REIMS metabolic fingerprints and because of the size distribution data (fig. S5), we decided to additionally evaluate the inclusion of the highly polar PAN polymer as a cover layer for our MetaSAMP and later in the core membrane blends for saliva and urine, as this polymer may increase the wettability (34) and thus speed of analysis upon inclusion (see below). Instrumental settings for LA-REIMS analysis of impregnated electrospun membranes with saliva and urine were the same as in our previously biofluid-specific optimized protocol (table S1) (1).
Next, we investigated the potential of a PAN cover layer considering the future rectal MetaSAMP‘s in vivo usage in a sampling kit or medical swab. On top of its high polarity and thus wetting capabilities for aqueous matrices (34), PAN is also a biocompatible polymer with documented in vivo applications (35, 36) that has previously been demonstrated to facilitate selective transport of small molecules to absorptive and adsorptive polymers by partially diminishing surface (bio)fouling mechanisms for molecules >1.5 kDa and completely preventing access to molecules >7 kDa (29, 37). As a result, a PAN cover layer was hypothesized to enhance core membrane stability as surface functionalities of the PVP/PS microporous core are expected to be less susceptible to oxygen or moisture in its presence. For this experiment, pooled quality control (QC) fecal samples were used, enabling the evaluation of our future target matrix and thus reproducibility of the entire analysis protocol best. Feces shows substantial similarity with rectal content with respect to bacterial community structure and functionality (38, 39). Hereto, PAN was tested at various concentrations (5 to 15%, w/w) and evaluated for its solution viscosity, quality of the PAN electrospun fiber network and fiber size distribution (table S2). Optimal small-molecule sampling and subsequent analysis were strived at by retaining the average fiber diameter and its distribution as low as possible in effectively excluding macromolecules like proteins and DNA. The combination of an acceptable average fiber diameter of 384 nm and a smooth fiber network without bead formation as observed by SEM (figs. S6 to S8 and table S2) rationalized our choice to use 10% (w/w) PAN in further experiments.
To confirm the optimal position of the electrospun PAN layer, we tested the effect of varying its position relative to the core membrane (PVP/PS). The effects of an electrospun PAN cover layer added to the PVP/PS electrospun core membrane both with and without including PAN as a bottom layer on the aluminum support were examined in terms of metabolic coverage (ranging from 50 to 1200 Da) and repeatability. To identify which metabolite classes were affected by the presence of PAN cover and bottom layers, we subdivided the measurable mass range into six subsets, including a subset covering 50 to 200 Da, encompassing low–molecular weight metabolites such as carbohydrates and short-chain fatty acids; two subsets covering 200 to 600 Da, encompassing medium-, long- and very long–chain fatty acids as well as amino acids, diacylglycerols, and sphingolipids; two subsets covering 600 to 1000 Da, encompassing phospholipids among others (16, 18, 40), and a subset covering 1000 to 1200 Da, mainly encompassing saccharolipids and glycosphingolipids (41). From a metabolic health perspective (2), metabolite classes covering all those mass ranges are relevant. The impregnated electrospun core rectal MetaSAMP layer covered by an additional electrospun PAN layer gave significantly (P < 0.001) higher metabolome coverage, ion intensities, and molecular feature count following LA-REIMS analysis compared to the analysis of the crude biofluid (fig. S9 and table S3). It was also observed that the electrospun PAN cover layer enhanced the spreading and therefore penetration of fecal water (fig. S10) as confirmed by smaller contact angle measurements (CAMs) (50° ± 2° versus 144° ± 4°) (fig. S11 and table S4). As a result, the increased hydrophilicity/wettability brought about by the electrospun PAN cover layer could promote beneficial chemical and physical interactions and hence efficient mass transfer between biofluid metabolites and the core membrane (fig. S12), as seen before in electrospun applications with high surface area and (micro)porosity (27, 42, 43).
Because the core membrane’s absorptive and adsorptive capabilities were expected to be affected by the PAN cover layer, a second optimization round was initiated for obtaining the final composition of each biofluid-specific MetaSAMP using a D-optimal experimental design (table S5). To this end, average fiber diameter and morphology, metabolome coverage, and repeatability were evaluated as end points. The core membrane composed of PVP/PS (60/40, w/w) with 8% (w/w) polymer wt % covered by PAN showed the highest metabolome coverage and repeatability upon LA-REIMS for fecal water (table S5 and fig. S13, A to C). For saliva, the PVP/PS (20/80, w/w) membranes showed the highest metabolome coverage and repeatability with the most stable fibers at 12% (w/w) polymer wt %, while for urine, the different membranes behaved similarly with a tendency toward better repeatability for the PVP/PS (20/80, w/w) as well (table S5). Because 20% PVP was the lowest tested, an additional experiment was performed for urine and saliva with 0 to 20% PVP while including 0 to 10% PAN in the blend (table S6). The core layers comprising PVP/PS (10/90) and PVP/PS/PAN (20/70/10) both with 12% (w/w) total polymer wt % covered with a PAN layer were lastly selected because higher salivary and similar urinary metabolome coverage and repeatability (95 and 97% of features with CV ≤ 30% for saliva and urine, respectively) (fig. S13, D to I) were achieved compared to crude analysis of saliva and urine, while no damage to the electrospun fibrous core blends was observed, suggesting morphological integrity even after prolonged biofluid exposure (>15 min) (fig. S14). Our three final optimized rectal, salivary and urinary MetaSAMPs consisted of, respectively, blends of electrospun PVP/PS 60/40, PVP/PS 10/90, and PVP/PS/PAN 20/70/10 covered with an electrospun PAN layer. In addition, the evaluation of different impregnation volumes (20 to 100 μl) and times pointed toward 30 μl or its equivalent in droplets (3) and 10 to 15 min as the optimal parameters (note S1.3 and fig. S15).
Biofluid-specific MetaSAMPs improve short-term metabolome stability and speed
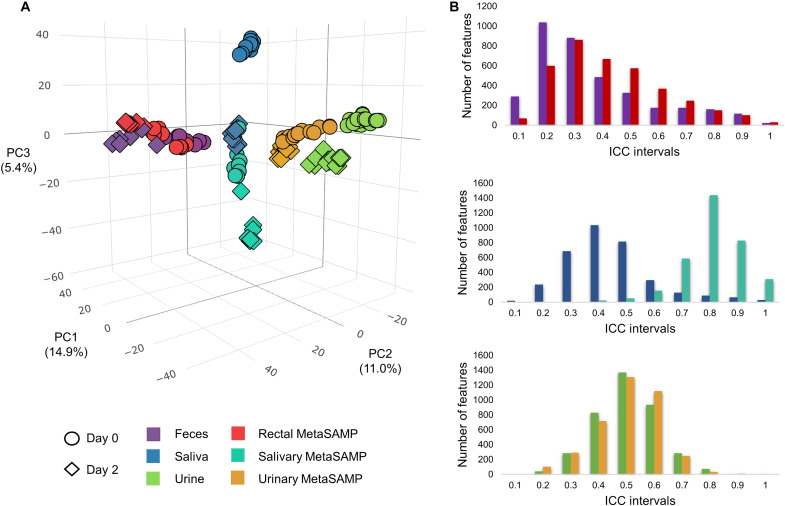
Biofluid collection and transport, especially in large cohorts, is not straightforward. In the case of fecal samples, the need to wait for defecation prevents the direct analysis, inferring the need of biobanking (and thus freezing). The addition of chemical preservatives to crude biofluids for shipping and storage stability negatively affects the metabolome’s accuracy (12, 14). Our optimized rectal, salivary and urinary MetaSAMPs were hypothesized to conserve a more accurate snapshot of the metabolome during storage and are directly amenable to sample preparation-free metabolomics analysis, as such lowering the individual time and cost per analysis. The storage period was set at 48 hours, which corresponds to the maximum transport duration from the patient’s home or physician’s office to the laboratory in Western countries (e.g., with courier services). To address this, metabolome coverage and repeatability based on crude biofluid fingerprinting (pooled QC samples, n = 6) and their corresponding impregnated MetaSAMPs were measured at room temperature (RT), i.e., 22° ± 2°C and 4°C and the total sampling and analysis times were recorded. Closer correlations in the LA-REIMS results were observed between 48 hours stored samples and those analyzed immediately after collection using the optimized MetaSAMPs stored at 4°C when compared to the corresponding correlations of the data obtained with their crude counterparts (Fig. 2A). At RT, however, both crude biofluids and MetaSAMPs brought about substantial metabolome changes as confirmed by low repeatability values (<50% of features with CV ≤30%). Subsequently, intraclass correlation coefficients (ICC) intervals were computed to investigate metabolome variation between time points, considering molecular features with ICC values ≤0.4 as more unstable over time (44). Upon storage at 4°C, 1371 (feces) versus 1850 (rectal MetaSAMP), 2460 (saliva) versus 3397 (salivary MetaSAMP), and 2677 (urine) versus 2721 (urinary MetaSAMP) molecular features remained stable (ICC >0.4) (Fig. 2B). Moreover, the total analysis time of the optimized biofluid-specific MetaSAMP-LA-REIMS methodology was recorded as <20 min per sample (<15 min for sampling incl. Impregnation and 5 min for the LA-REIMS analysis workflow) compared to <40 min per sample for the LA-REIMS analysis of crude biofluids, aside from the transport time for both methodologies (and lyophilization of feces for the LA-REIMS analysis of crude feces) (1, 12, 16).
Fig. 2. MetaSAMPs improve short-term (48 hours) biofluid-specific metabolome stability as compared to crude biofluid analysis.
(A) 3D-PCA (principal components analysis) score plots presenting metabolome fluctuations upon storage at 4°C (day 0 versus day 2). The molecular feature count per ICC interval of pooled QC (B) fecal, saliva, and urine samples, and the corresponding impregnated MetaSAMPs (n = 6 per biofluid) analyzed via LA-REIMS are depicted via histograms upon short-term storage at 4°C.
In addition, we successfully assessed whether the metabolome as covered by our optimized biofluid-specific MetaSAMPs was not compromised by any degradation products caused by the interaction between the laser ablation process and the membrane polymers, and hence its reliability in mirroring the metabolome of crude biofluid analysis (note S4 and figs. S16 and S17). In conclusion, our MetaSAMPs generally demonstrated a superior number of stable molecular features over time without any ion formation as caused by the interaction between the laser ablation process and the membrane polymers, providing an efficient means for rapid LA-REIMS–based metabotyping.
Physicochemically diverse clinically relevant metabolites can be reproducibly detected using rectal MetaSAMP-LA-REIMS
Last, in assessing the future clinical implementation of MetaSAMPs, a targeted approach was implemented using the rectal MetaSAMP on a selection of clinically relevant metabolites in (childhood) overweight and obesity (2) according to Food and Drug Administration (FDA) recommendations (45). The latter was performed with analytical standards that were selected on the basis of their plausible natural occurrence in feces (46) covering a broad polarity and mass range (50 to 1200 Da and logP of −4 to 13; table S7). The targeted detectability, technical precision, repeatability, and intermediate precision of these analytes were determined by applying three consecutive ablation events per membrane piece (n = 5) on two different days and calculating the number of molecular features upon LA-REIMS analysis (table S7). In addition, the standards were spiked directly onto the rectal MetaSAMP (table S8) and, hereafter, onto the impregnated rectal MetaSAMPs (table S9) using a pooled sample (from MetaBEAse, n = 3). Regarding the latter, intra- and interassay CVs were below the threshold (<30%) for respectively 9 and 7 (table S9) of the 11 metabolites that were initially detected when spiked into solvent. In addition, the recovery of the selected metabolites from MetaSAMP, and linearity of the method were evaluated by ultra-high performance liquid chromatography–high-resolution mass spectrometry (UHPLC-HRMS) and LA-REIMS analysis, respectively. We demonstrated that most (12 of 16) metabolites showed better detectability (quantitated as integrated peak area ratios relative to the analyte-specific internal standard) using our optimized rectal MetaSAMP as a sampling and stabilization tool as compared to crude feces extracted in a similar manner, as reflected by recovery values >100% (up to 150.70%) (note S5 and table S10). We also obtained an acceptable linear range of the MetaSAMP-LA-REIMS method upon untargeted (R2 ≥ 0.7) and targeted (R2 ≥ 0.9) analysis for >60% [740 of 1222 and 5 of 8 metabolites (0.75 to 0.99)], encouraging the future prospects of the MetaSAMP (note S5 and table S11). In conclusion, our results showcase that a diverse range of clinically relevant metabolites may be sufficiently reproducibly detected using rectal MetaSAMP-LA-REIMS.
Direct MetaSAMP-LA-REIMS offers a richer metabolic fingerprint than crude biofluid LA-REIMS in cohort samples
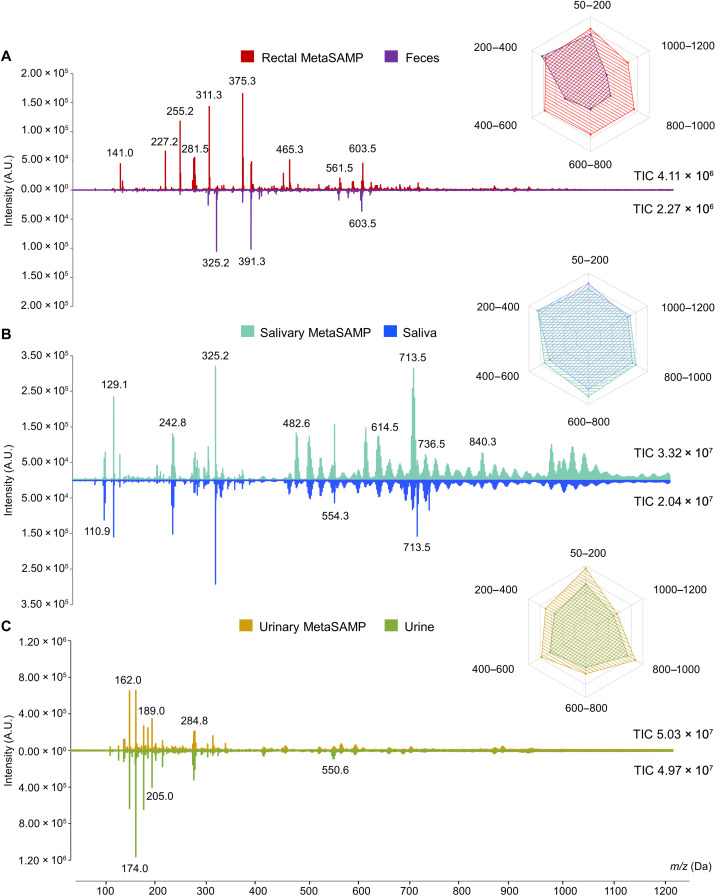
Following optimization, we aimed at demonstrating the advantages of direct MetaSAMP-LA-REIMS metabolic fingerprinting compared to crude biofluid LA-REIMS analysis in terms of metabolome coverage using impregnated rectal, salivary, and urinary MetaSAMPs from participant children covering all International Obesity Task Force (IOTF) body mass index cut-offs for overweight, obesity, and thinness scores [pooled QC samples, n = 3, MetaBEAse cohort for the rectal and urinary MetaSAMP and OPERA cohort (47) for the salivary MetaSAMP] (Table 1). Richer mass spectra were observed in impregnated MetaSAMPs compared to the crude biofluid LA-REIMS metabolic fingerprints (Fig. 3, A to C). The numbers of metabolic features counted (20 technical replicates), respectively, for the rectal, salivary, and urinary MetaSAMPs and feces, saliva, and urine were 2078 ± 11.05, 2150 ± 4.10, and 1697 ± 3.95 and 1896 ± 2.57, 1960 ± 3.12, and 1477 ± 8.31 (data S1 to S3, significantly different, independent sample t test P = 0.02, 5.60 × 10−6, and 2.08 × 10−4). Also, higher overall signal intensities (TIC) were measured for the respective MetaSAMPs (data S4 to S6). In addition, because metabolite classes covering the aforementioned mass ranges are clinically relevant with regard to the metabolic disturbances underlying overweight and obesity (2), metabolome coverage was also evaluated throughout the different mass ranges in further evincing higher overall signal intensities. Hence, palmitic acid [fatty acid, mass/charge ratio (m/z) value of 255.2 Da, logP 6.4], 1-heptadecanoyl-2-(9Z,12Z,15Z-octadecatrienoyl)-sn-glycerol (glycerolipid, m/z value of 603.5 Da, logP 13.4) (Fig. 3A), and a putatively identified lipid (ceramide sphingolipid or phosphoethanolamine glycerophospholipid, m/z value of 736.5 Da) (Fig. 3B) were detected by our MetaSAMP-LA-REIMS. As demonstrated via radar charts for each of the mass ranges, higher coverage was noted upon analysis with the biofluid-specific MetaSAMPs as compared to the crude biofluids (Fig. 3, A to C) for most mass ranges. In conclusion, our optimized biofluid-specific MetaSAMPs applied in cohort samples confirmed superior metabolome coverage across a broad mass range (50 to 1200 Da) and wide logP span compared to the analysis of crude biofluids.
Table 1. Demographic and anthropometric cohort findings.
Data from the MetaBEAse and OPERA (47) cohorts comprising overweight and normal-weight children and adolescents. LDL, low-density lipoprotein; HDL, high-density lipoprotein; HbA1c, hemoglobin A1c; TSH, thyroid-stimulating hormone; fT4, free thyroxine; (hs)-CRP, (highly sensitive) C-reactive protein; IL, interleukin; IFN, interferon; TNF, tumor necrosis factor; ALT, alanine transaminase; AST, aspartate transaminase; AF, alkaline phosphatase; IGF-1, insulin-like growth factor 1; SHBG, sex hormone-binding globulin; DHEAS, dehydroepiandrosterone sulfate; NA, not applicable.
| MetaBEAse | OPERA | |||
|---|---|---|---|---|
| MetaSAMP | #REIMS analyses | |||
| Rectal | 534 | NA | ||
| Salivary | NA | 372 | ||
| Urinary | 532 | NA | ||
| Demographics | Mean ± SD (or expressed in %) | |||
| No. of participants | No. of participants | |||
| Mean age (years) ± SD | 234 | 9.0 ± 2 | 101 | 14.0 ± 2 |
| Sex | 234 | 45% female | 101 | 55% female |
| Puberty | ||||
| Tanner stage ≤1 | 234 | 100% | 7 | 7% |
| Tanner stage >1 | 91 | 93% | ||
| Anthropometrics | ||||
| BMI-z scores | 234 | 101 | ||
| Range | −3–3 | −2–4 | ||
| ≤ −2.0 (underweight) | 5 (2.1%) | 1 (1.0%) | ||
| Between −2.0 and ≤ 1.0 (normal weight) | 136 (58.1%) | 69 (68.3%) | ||
| Between 1.0 and < 2.0 (overweight) | 36 (15.4%) | 19 (18.8%) | ||
| ≥ 2.0 (obese) | 57 (24.4%) | 12 (11.9%) | ||
| IOTF | 234 | 101 | ||
| Range | −3–3 | −1–2 | ||
| ≤ − 1 (underweight) | 19 (8.1%) | 4 (4.0%) | ||
| 0 (normal weight) | 125 (54.3%) | 66 (65.3%) | ||
| 1 (overweight) | 55 (23.5%) | 20 (19.8%) | ||
| ≥2 (obese) | 34 (14.1%) | 11 (10.9%) | ||
| Waist (cm) | 215 | 67.0 ± 12.6 | 101 | 74.6 ± 10.3 |
| WHR | 214 | 0.49 ± 0.07 | 101 | 0.45 ± 0.07 |
| Fat (%) | NA | 98 | 23.8 ± 8.7 | |
| Systolic BP (mmHg) | 215 | 105.0 ± 14.3 | NA | |
| Diastolic BP (mmHg) | 215 | 67.0 ± 9.6 | NA | |
| Heart rate (bpm) | 27 | 79.4 ± 12.2 | 86 | 77.3 ± 10.5 |
| Clinical laboratory tests | ||||
| LDL cholesterol (mg dl−1) | 55 | 97.0 ± 18.9 | 75 | 87.6 ± 21.0 |
| Triglycerides (mg dl−1) | 64 | 77.7 ± 35.5 | 75 | 81.9 ± 41.8 |
| HDL cholesterol (mg dl−1) | 63 | 50.3 ± 10.1 | 75 | 59.7 ± 13.7 |
| Fasting plasma glucose (mg dl−1) | 84 | 87.6 ± 6.7 | 75 | 88.1 ± 6.7 |
| Cholesterol (mg dl−1) | 64 | 154.9 ± 16.8 | 75 | 159.2 ± 28.1 |
| HbA1c (%) | 18 | 5.3 ± 0.2 | 76 | 5.3 ± 0.3 |
| Fasting plasma insulin (pmol liter−1) | 58 | 80.7 ± 35.7 | 75 | 13.3 ± 9.7 |
| TSH (mU liter−1) | 51 | 2.3 ± 0.8 | 75 | 2.4 ± 1.1 |
| fT4 (pmol liter−1) | 43 | 14.9 ± 2.4 | 75 | 15.4 ± 2.2 |
| (hs-)CRP (mg liter−1) | 55 | 2.1 ± 1.6 | 76 | 1.1 ± 1.5 |
| IL-10 (pg ml−1) | NA | 78 | 0.4 ± 0.7 | |
| IL-6 (pg ml−1) | NA | 78 | 0.5 ± 0.5 | |
| IL-8 (pg ml−1) | NA | 78 | 10.6 ± 5.2 | |
| IFN (pg ml−1) | NA | 78 | 13.8 ± 43.4 | |
| TNF (pg ml−1) | NA | 78 | 1.8 ± 0.6 | |
| Polymorphonuclear cells (%) | 52 | 49.0 ± 9.2 | NA | |
| Lymphocytes (%) | 62 | 36.3 ± 9.0 | NA | |
| ALT (U liter−1) | 69 | 19.2 ± 5.6 | NA | |
| AST (U liter−1) | 72 | 26.4 ± 5.2 | NA | |
| AF (U liter−1) | 29 | 272.0 ± 60.6 | NA | |
| Ureum (mg dl−1) | 65 | 28.1 ± 6.2 | NA | |
| Creatinine (mg dl−1) | 56 | 0.5 ± 0.1 | NA | |
| Cortisol (ng ml−1) | 47 | 79.0 ± 67.0 | NA | |
| 25-Hydroxyvitamin D (ng ml−1) | 47 | 33.5 ± 19.9 | NA | |
| Uric acid (mg dl−1) | 36 | 4.6 ± 0.9 | NA | |
| IGF-1 (μg liter−1) | 31 | 188.5 ± 55.8 | NA | |
| SHBG (nmol liter−1) | 42 | 49.1 ± 20.1 | NA | |
| DHEAS (μg dl−1) | 33 | 93.9 ± 61.0 | NA | |
Fig. 3. The use of MetaSAMPs increases the richness and intensity of fecal, salivary, and urinary metabolic fingerprints compared to those obtained with crude biofluid LA-REIMS–based metabotyping.
Head-to-tail comparison of LA-REIMS spectra obtained from impregnated (A) rectal, (B) salivary, and (C) urinary MetaSAMPs and feces, saliva, and urine, respectively, measured in negative polarity mode (m/z range of 50 to 1200 Da depicted). To accurately compare every crude biofluid and its respective MetaSAMP, binning of the inspected m/z range (0.1 Da) was performed. Average feature intensities [arbitrary units (A.U.)] are shown for the full spectra, and the highest intensity peaks were displayed with their specific m/z value. TIC values accompany each mass spectrum, representing the overall signal intensities. The selected mirrored mass spectra are representative of the biofluid metabolomes of children and adolescents covering all IOTF scores [MetaBEAse cohort for the rectal and urinary and OPERA cohort (47) for the salivary MetaSAMPs, pooled QC samples, n = 3]. Next to the mirrored mass spectra, radar charts visualize the averaged feature intensities of the crude biofluids (pooled QC samples originating from control children, n = 3) and their corresponding optimized biofluid-specific MetaSAMP. The metabolome coverage was subdivided into clinically relevant mass ranges with regard to the metabolic disturbances underlying overweight and obesity (2). The colored area represents the metabolome coverage, clearly showing an overlap for every crude biofluid and its specific MetaSAMP, yet indicating the richness of the latter.
Direct biofluid-specific MetaSAMP-LA-REIMS is a clinically valid metabotyping tool
Metabolomics analysis has substantial value in a plethora of metabolic and food-related diseases (2, 48). Metabolite patterns could therefore offer a valid tool to profile individuals at high risk of developing, e.g., obesity-related metabolic diseases, i.e., discerning between metabolically healthy and unhealthy phenotypes already at young age (2, 5). To provide proof of principle for the clinical potential of our MetaSAMP-LA-REIMS in childhood overweight and obesity, MetaSAMP-derived metabolic fingerprints of biofluids obtained in two pediatric cohorts: MetaBEAse (feces and urine of children aged 6 to 12 years) and OPERA (47) (saliva of children aged 6 to 16 years), were investigated in terms of discrimination and predictive potential. For this purpose, a large set of anthropometric and clinical measurements were performed (Table 1) to link potentially relevant metabolite discrepancies in overweight and obese children and pathophysiological processes.
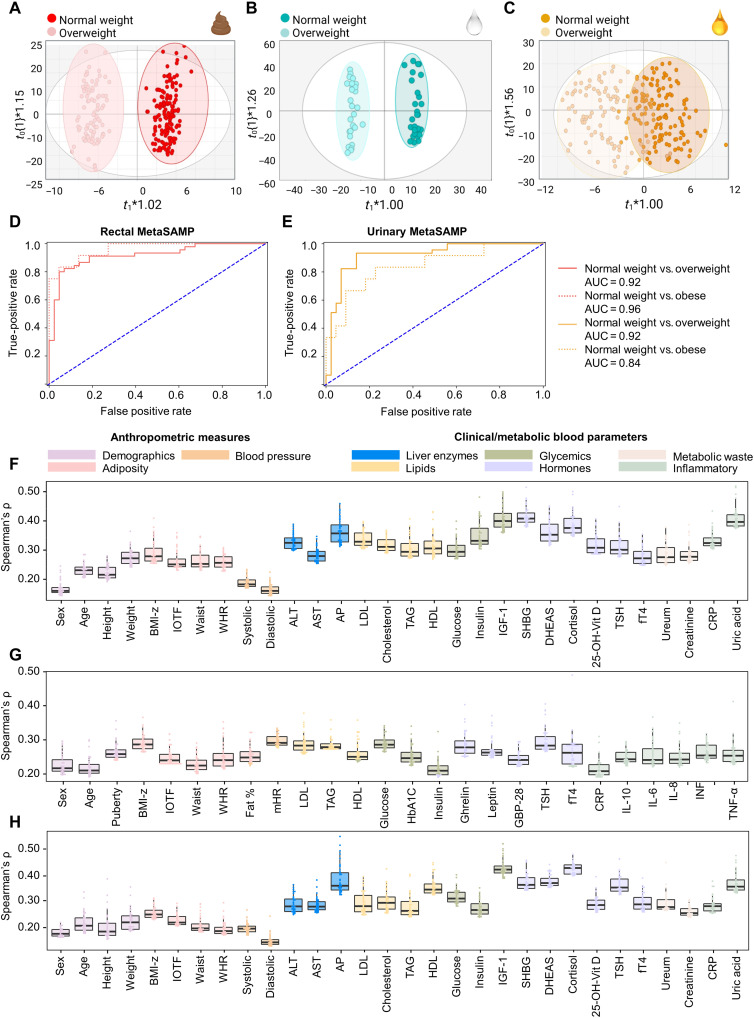
First, the discriminative and predictive performance of LA-REIMS analysis was compared between crude biofluids and their impregnated MetaSAMPs. By doing so, more subtle effects related to subclinical pathology, i.e., metabolic perturbations due to increased weight and/or adiposity, were visualized by valid orthogonal projection to latent structures discriminant analysis (OPLS-DA) models using the international obesity body mass index (BMI) cutoff for thinness, overweight, and obesity (IOTF) as a classifier (Fig. 4, A to C). A similar or even greater number of molecular features that were significantly differentiated on the basis of their value of variable importance in projection for the OPLS-DA models constructed (table S12) were retrieved for the analysis of the MetaSAMPs as compared to that of the crude biofluids. The same was noted for the validation parameters [CV-ANOVA P < 0.05, good permutation testing (n = 100), goodness-of-fit and predictive performance of the model reflected by R2(cum) > 0.8 and Q2(cum) > 0.4 for biological data], respectively (table S12). These findings provide evidence that discriminative categorization based on metabolic fingerprints was mostly superior using the biofluid-specific MetaSAMP-LA-REIMS methodology. Furthermore, its predictive potential for overweight classification using IOTF scores was assessed by logistic regression. This approach was only applied to the MetaBEAse (n = 234) cohort data because the sample size of the OPERA cohort (n = 101) did not sustain good model building (49). Metabolic fingerprints detected by rectal (Fig. 4D) and urinary (Fig. 4E) MetaSAMP-LA-REIMS analyses enabled predictive IOTF classification. We observed strong predictive potential [area under the curve (AUC) range 0.84 to 0.96] when all overweight (including obese) children as well as only obese children were included and compared to a balanced number of normal-weight children, suggesting the early onset, i.e., already in overweight (not yet obese) children, of metabolic divergences (2).
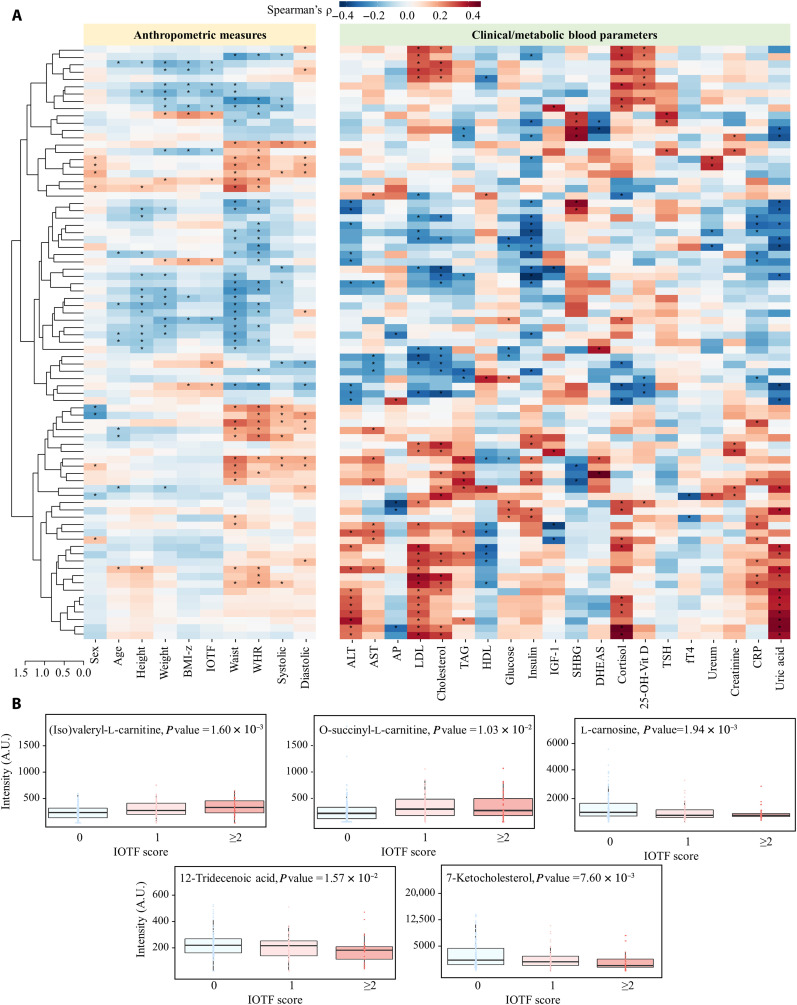
Fig. 4. Biofluid-specific MetaSAMP-LA-REIMS molecular features reflect anthropometric measures and clinical and metabolic blood markers.
(A to C) OPLS-DA score plots as obtained upon LA-REIMS fingerprinting of biofluid-specific MetaSAMPs with clustering according to weight classification in the patient cohorts, i.e., (A) feces [R2(cum) = 0.96, Q2(cum) = 0.63, CV-ANOVA P = 1.15 × 10−13], (B) saliva [R2(cum) = 0.98, Q2(cum) = 0.53, P = 4.05 × 10−5] and (C) urine [R2(cum) = 0.64, Q2(cum) = 0.58, P = 1.68 × 10−11]. Receiver operator characteristic curves are plotted using logistic regression analysis to visualize the predictive performance of LA-REIMS analysis in negative polarity mode with (D) rectal MetaSAMP and (E) urinary MetaSAMP based on IOTF classification (n = 125 with IOTF = 0, n = 55 with IOTF = 1 and n = 34 IOTF ≥ 2). Beeswarm boxplots showing the strongest observed links (anthropometrical measurements and clinical/metabolic blood metadata, horizontal) according to Spearman’s correlation (vertical), with molecular features measured using the (F) rectal, (G) salivary, and (H) urinary MetaSAMPs. Only the 50 strongest (highest modulus Spearman’s ρ value) and significant (P < 0.05) correlations of molecular features with anthropometric measures and/or clinical/metabolic blood parameters were included. The interior middle line in the beeswarm boxplots represents the median, lower, and upper bounds of the box represent the 25th and 75th percentile values, respectively. Whiskers are drawn from the corresponding box boundary to the most extreme data point located within the box bound ± 1.5 × interquartile range. Beeswarm dots represent Spearman’s correlations of individual molecular features with the anthropometric measure or clinical/metabolic blood parameter under investigation.
To further clinically validate our MetaSAMP-LA-REIMS metabotyping approach, Spearman’s correlations were calculated between normalized levels of molecular features captured with each biofluid-specific MetaSAMP and anthropometric and clinical measurements from both cohorts (MetaBEAse and OPERA). Among anthropometrics, adiposity measures [BMI-z, IOTF, waist circumference (Waist), and waist-to-height ratio (WHR)] that are related closely to overweight metabotypes (50) showed the highest Spearman's ρ values with fecal, salivary, and urinary metabolic profiles in both cohorts (Fig. 4, F to H). A number of clinically relevant end points, e.g., lipid, glycemic, inflammatory, and hormonal blood markers, showed moderate to good correlations (Spearman's ρ values 0.4 to 0.6) (51) with biofluid-specific MetaSAMP-derived metabolic fingerprints. For instance, glucose and insulin (−like growth factor 1) in blood, which have been associated with insulin resistance (52), and sex hormone binding globulin, which has been negatively associated with adiposity and low-grade inflammation in childhood obesity (53, 54), correlated well with rectal and urinary MetaSAMP-derived fingerprints (Spearman's ρ values up to 0.5; Fig. 4, F to H) (MetaBEAse cohort). Adipokine hormones with appetite suppressing and initiating effects, such as leptin and ghrelin, respectively (55), revealed moderate to good correlations (Spearman's ρ values up to 0.4; Fig. 4G) with metabolic fingerprints obtained by our salivary MetaSAMP (OPERA cohort), and hence were influenced by adiposity and insulin sensitivity (56). Collectively, these results suggest that our MetaSAMPs allow capturing multiple clinically relevant metabolites of energy, immune, and lipid metabolism.
As a final step in demonstrating the clinical potential of our MetaSAMP-LA-REIMS, the identification of molecular features that showed significant (P < 0.05, Wilcoxon rank-sum test with continuity correction) Spearman’s correlations with anthropometric and/or clinical parameters based on MetaSAMP-LA-REIMS analysis was pursued. We focused on the rectal MetaSAMP data (MetaBEAse cohort) because feces comprises the most complex matrix and is considered most relevant to capture gut-microbiome-diet interactions (12). That is, the role of the microbiome and its metabolites in obesity have been ubiquitously reported in the literature (2, 57–59). Hierarchical Ward-linkage clustering analysis was used to unveil unique molecular features clustering according to their shared correlation structure (Fig. 5A, Wilcoxon rank-sum test with continuity correction to assess which correlations were significant with P < 0.05). We found that 81 molecular features showed the highest correlations (moduli) and at least 4 significant (in-between comparisons adjusted P ≤ 0.01, using FDR correction) associations with anthropometric and/or clinical metadata were noted, as visualized in Fig. 5A. Of the 81 molecular features that were determined with rectal MetaSAMP-LA-REIMS (data S7) and which significantly correlated with and contributed to the discrimination of weight classification in children (Fig. 5A), 7 metabolites could be annotated as acylcarnitines, fatty acids, an amino acid, and a cholesterol derivate, i.e., (iso)valeryl-l-carnitine, O-succinyl-l-carnitine, l-carnosine, 12-tridecenoic acid, sphingosine, N-acetyl-l-glutamic acid, and 7-ketocholesterol. We observed that acylcarnitines were positively correlated with IOTF score, Waist, WHR, and systolic blood pressure (BP). Acylcarnitines have been previously linked to future metabolic risk (2). Furthermore, we observed similar positive correlations between the latter anthropometric measurements with 7-ketocholesterol as corroborated by literature, where cholesterol derivates were reported to be involved in macrophage foam cell formation associated with atherosclerosis (2). Subsequently, we assessed if those metabolites significantly (P < 0.05, Kruskal-Wallis test with Dunn’s post hoc test) changed between different weight groups. We found that five metabolites showed significant alterations between normal-weight children and children suffering from overweight (Fig. 5B). Furthermore, the 7 annotated metabolites could be detected by LA-REIMS analysis of both crude feces and rectal MetaSAMP, yet their normalized intensity values were generally higher when using the rectal MetaSAMP compared to crude fecal analysis (Welch two-sample t test or Wilcoxon rank-sum test, P < 0.05), and the metabolites were thus more reliably detected (fig. S18).
Fig. 5. Metabolites detected using rectal MetaSAMP-LA-REIMS enable individual stratification based on anthropometric and clinical blood markers.
(A) Heatmaps showing relative levels of LA-REIMS (negative ion mode)–derived molecular features that showed significant (Wilcoxon rank-sum tests with continuity correction, P < 0.05 marked with an asterisk) Spearman’s correlations with anthropometric measures and/or clinical/metabolic blood parameters from the MetaBEAse cohort. The molecular features (n = 81 unique correlation structures) with the highest correlations (moduli) and at least four significant associations (in-between comparisons adjusted P ≤ 0.01, using FDR correction). (B) Beeswarm boxplots of significant (P < 0.05, Kruskal-Wallis test with Dunn’s post hoc test) discriminatory features between normal-weight (IOTF < 1), overweight (IOTF = 1) and obese (IOTF > 1) children. The interior horizontal line represents the median value, lower and upper bounds of the box represent the 25th and 75th percentile values, respectively, and whiskers are drawn from the corresponding box boundary to the most extreme data point located within the box bound ± 1.5 × interquartile range. The y axis of the graphs shows the observed intensity for every adduct that was detected in replicated burns, while the x axis shows the groups compared. For the multiple comparisons, individual normalized metabolic feature intensities are shown for the different weight groups. Statistical analysis was performed using Kruskal-Wallis with Dunn’s post hoc test for multiple comparisons.
DISCUSSION
In this work, customized biofluid-specific metabolome samplers, called MetaSAMPs, were developed for rapid sampling, absorption and adsorption and stabilization of complex human biofluids including feces, saliva, and urine. The hyphenation of these optimized MetaSAMPs with our automated LA-REIMS platform (1) enables direct sampling and analysis of the respective metabolomes in less than 20 min per sample (Fig. 1A) and provides a richer and more stable reflection of the biofluid-specific metabolome as compared to crude biofluid analysis. The MetaSAMP-LA-REIMS metabotyping approach represents a superior alternative to crude biofluid LA-REIMS analysis regarding on-the-spot metabolic health stratification and holds great potential for usage in large cohort metabolomics studies. We have therefore successfully evaluated its clinical implementation in the context of the childhood obesity pandemic (6) using fecal, salivary, and urinary metabotyping in samples obtained from two pediatric cohorts (MetaBEAse and OPERA). Although our MetaSAMP-LA-REIMS methodology centers on untargeted metabolic fingerprinting using HRMS, we also demonstrated reproducible targeted analysis of a selection of physicochemically diverse metabolic disease-related metabolites.
The respective biofluid-specific rectal, salivary, and urinary MetaSAMPs comprised a core layer that was optimized toward maximal metabolome coverage as a blend of HLB electrospun (nano)fibrous polymers, i.e., PVP/PS 60/40, PVP/PS 10/90, and PVP/PS/PAN 20/70/10, covered with an electrospun PAN layer (Fig. 1B). The dual HLB properties of our MetaSAMP core membranes resulted in the enrichment of analytes with great physicochemical diversity in terms of size and polarity (m/z 50 to 1200 Da and logP of −4 to 13) (Fig. 3 and fig. S12). Several HLB-based absorptive and adsorptive sampling approaches have proven beneficial for subsequent MS-based analysis of a physiochemically wide range of metabolites (30, 32). As with SALDI, the underlying mechanism of signal amplification with our MetaSAMP-LA-REIMS methodology relies on increased laser energy transfer from the impregnated substrate to the analytes, leading to a rapid increase in surface temperature and contributing to enhanced desorption and ionization of the sample (60, 61), including in PVP- and PAN-based nanofibrous substrates (42, 43, 62). In particular, the smallest (fiber) size dimensions produced, namely, 1118 (±270) nm (8%, w/w), 3430 (±1100) nm (12%, w/w), 2730 (±700) nm (12%, w/w), and 300 to 600 nm (10%, w/w) for the rectal, salivary and urinary MetaSAMP and PAN cover layer, respectively, were demonstrated to generate mass spectra with increased metabolome coverage and signal intensities (figs. S2, S5 to S9 and tables S5 and S6). This can be attributed to the greater surface area and porosity in composites with smaller fiber sizes (28, 42, 62, 63), which allows for more efficient molecular interactions at the polymer level, as analyte molecules can partition into polymer (nano)fibers instead of adsorbing exclusively to the surface (27, 43, 62). Together, these structural characteristics inherent in electrospun membranes are particularly valuable when coupled with laser-based desorption and ionization (42). The observation of increased signal intensities in the various m/z mass ranges after MetaSAMP-LA-REIMS analysis compared to crude biofluid LA-REIMS analysis (Fig. 3 and figs. S2 and S9) confirms the rapid and efficient energy transfer and dissipation reported in the literature for structures with nano- to micrometer size range (42, 43, 60–62), resulting in improved laser-based desorption and ionization.
In contrast to the rectal and salivary core MetaSAMP membranes, we found that for the urinary core membrane, a blend of PVP, PS, and PAN was most favorable, given the very hydrophilic nature of the waste products that comprise the urinary metabolome (64). PAN is a polar polymer with wetting capabilities (34), thereby facilitating the impregnation of aqueous biofluids into the (relatively hydrophobic) core layer. Homogeneous sample distribution, as demonstrated through enhanced spreading of biofluids and smaller CAMs (figs. S10 and S11 and table S4), clearly served to enhance the interaction of the sample (biofluid) and the substrate (MetaSAMP). This is of particular interest in LA-REIMS–based analysis as the laser beam penetration depth and its energy dispersion is related to the optical absorption coefficient of water molecules (OH-vibrational bands) at the used laser wavelength (2.94 μm) (20). The markedly higher fraction of PS in the salivary and urinary MetaSAMPs compared to the rectal MetaSAMP‘s composition resulted in a more extensive metabolome coverage, especially for the higher m/z ranges in which more lipids reside that are relatively less concentrated and hence more difficult to selectively retain from such polar matrices (65). Furthermore, introducing the biocompatible polymer PAN (35, 36) as a cover layer (29) reduces biofouling due to restricting access to relatively large (macro)molecules such as DNA and proteins (29). PAN has proven advantageous in diminishing fragmentation and background noise for low molecular masses (42). In line herewith, we observed an increased metabolome coverage and precision upon introduction of an electrospun PAN cover layer. This may be ascribed to the advantageous complementary action of the large (micrometer scale), open pores of the core network of the (nano)fibrous PVP/PS(/PAN) membrane (43) that adsorbs and absorbs small molecules, and the nanofibrous PAN exclusion layer that prevents the interaction of macromolecules with this nanofibrous network (29), most likely resulting in reduced matrix interferences upon LA-REIMS analysis of the MetaSAMPs.
The fully optimized biofluid-specific MetaSAMPs increased the short-term biofluid metabolome stability (at 4°C) and the total sampling and analysis speed. These results point toward a decrease in storage-induced metabolome alterations when using our MetaSAMPs, as such decreasing the possible loss of clinically relevant metabolites in the duration between the home/practitioner’s office sampling and analysis in the laboratory. Significant salivary and urinary metabolome alterations when storing crude biofluids at 4°C have been reported to occur after 6 hours (66) and 5 days (14), respectively, confirming our observations. The stabilizing properties of our optimized biofluid-specific MetaSAMPs are hypothesized to result from the adsorption and absorption capabilities of the electrospun fibrous network and this both toward the different metabolites present in the biofluid and its aqueous content. In particular, this water content has been shown to provide a means for diverse degradation reactions (oxidation and hydrolysis) and microbial activity (enzymes) (12). In line herewith, the gastrointestinal matrices benefited most from MetaSAMP‘s stabilizing effects as an inhibition of the interaction between metabolites and microbes is expected. Moreover, the MetaSAMPs’ protective PAN cover layer also provides a certain degree of shielding of the biofluid metabolites (small molecules) captured via chemical and/or physical interactions (fig. S12) and may hence contribute to this reduced degradation and transformation. Alternative AIMS-based techniques, including other LA-REIMS–based approaches (1, 17), require relatively long collection procedures and/or sample preparation periods that take several hours up to days. The total analysis time of our optimized biofluid-specific MetaSAMP-LA-REIMS methodology was <20 min per sample.
Last, we demonstrated the targeted application of our rectal MetaSAMP-LA-REIMS approach of which the results indicated that potentially clinically relevant fecal metabolites may be detected with acceptable intra-assay and intermediate precision and linear range within a four-point dilution series (eight-point for untargeted metabolomics), and with improved recovery compared to LA-REIMS analysis of crude feces (45).
We successfully benchmarked the biofluid-specific MetaSAMPs in the context of a clinically relevant application using samples from two pediatric cohorts (MetaBEAse and OPERA). The ever-increasing rise in overweight and obesity urges the development of cost-effective ways to screen for elevated metabolic risk as early as possible, when metabolic impairments are still reversible (2). Our findings provided evidence that MetaSAMP-based metabotyping enables similarly to superior discriminative categorization of children based on weight classification compared to crude biofluid LA-REIMS analysis (1). This was confirmed by high predictive AUC values (>0.9), indicating good to excellent sensitivity and specificity of the metabolic fingerprints sampled and analyzed via MetaSAMP-LA-REIMS analysis regarding weight classification. Moreover, the obtained metabolic fingerprints exhibited meaningful pathophysiological correlations with a whole range of relevant anthropometric and clinical parameters for (childhood) overweight and obesity. The correlations found in this study showed similar (67, 68) or improved (58, 68) Spearman's ρ values (up to 0.5) compared to earlier work. For example, we observed a correlation between the fecal metabolome, BMI and blood lipid levels (Spearman's ρ values of up to 0.4), in line with previous results concerning the fecal metabolome of obese adults (68). Similarly, Spearman's ρ values of up to 0.3 for fecal microbiome diversity with BMI and glycemic and lipid measurements in adults were recently reported (58), while our MetaSAMPs reached ρ values of up to 0.6 regarding glycemic measurements such as glucose and insulin.
Despite the recent introduction of AIMS in biofluid metabolomics (1, 18), the direct sampling-to-MS result workflow used for these biological matrices still suffers from practical issues such as dependence on sample availability and laboratory pretreatment procedures such as (ultra)centrifugation for protein precipitation. In this aspect, the MetaSAMP cover layer introduced biocompatibility and reduced biofouling and hence matrix effects due to its selective filtering principle, i.e., hindering relatively large molecules, such as DNA and/or proteins, from their interaction with the open network of the MetaSAMPs (fig. S12). Consequently, superior absorption and adsorption for the direct sampling of complex matrices while eliminating interfering molecules and facilitating broader (Fig. 3) and more short-term stable (Fig. 2) metabolite recovery was evinced. With respect to metabolome coverage (the number of detected metabolic features), our biofluid-specific MetaSAMPs outperformed what has been previously reported in the literature for AIMS-based metabolomics analysis of human biofluids, including feces (1, 16, 18), saliva (1, 13, 69), and urine (1, 70, 71).
Our biofluid-specific MetaSAMP-LA-REIMS methodology offers a promising approach for first-line segregation of increased metabolic risk based on distinctive fingerprints that may overcome the limitations of conventional crude biofluid AIMS metabolomics analysis, such as costly and cumbersome analyses and matrix effects. Considering the role of membranes not only as biofluid samplers but also as substrates for stabilization and improved laser desorption and ionization in combination with the high throughput and versatility of the LA-REIMS platform for rapid analysis of biofluids (1), we strongly believe that the presented concept has a promising perspective to be installed as a prescreening approach in risk categorization and thus prevention and/or prognosis of metabolic disease from an early age as well as driving precision nutrition and/or lifestyle advice based on individual metabotype.
Despite the great clinical opportunities our MetaSAMPs could bring to the diagnostic, prognostic and/or preventive field of metabolic disease, we do acknowledge some flaws. We have assessed repeatability, reproducibility, linearity and recovery using a selection of clinically relevant targeted metabolites in the context of childhood obesity. The latter however requires a quantitative method and has been reported before in studies applying similar (semiquantitative and qualitative) principles (42, 72–75), implying the need for conventional hyphenated LC-MS methodologies for quantitative purposes. The main merits of AIMS are high throughput and direct clinical applicability compared to conventional metabolomics platforms. Such advantages have however mainly been restricted to untargeted metabolomics research (76) as there still is much to glean from the reproducibility, quantitation exquisiteness and ability to annotate potential differential marker molecules of more dedicated hyphenated analytical platforms (16). Further research is also warranted to follow large-scale cohorts of children with overweight and obesity longitudinally to consolidate the potential of MetaSAMP-based metabotyping as a tool for risk categorization in precision medicine. Such a longitudinal design will help reveal the predictive and/or prognostic value of differential marker molecules. Furthermore, future work should focus on in vivo sampling with the MetaSAMP to validate the ex vivo results presented in this paper.
MATERIALS AND METHODS
Chemicals and standards
Isopropyl alcohol (IPA), methanol (MeOH), and ethanol (EtOH) (all LC–MS-grade) were purchased from Fisher Scientific (UK). N,N-Dimethylformamide (DMF) and chloroform (CHCl3) were purchased from EMPLURA Merck Millipore (BE) and Sigma-Aldrich (US), respectively. Ultrapure water (UPW) was obtained through a Sartorius Arium 661 UV water purification system (Millipore, Belgium). Analytical standards [3-hydroxybutyric acid, linoleic acid, l-carnosine, 1,2-dioleoyl-sn-glycero−3-phospho-rac-(1-glycerol) (DOPG), 1-palmitoyl−2-oleoyl-sn-glycero−3-(phospho-rac-(1-glycerol)) (POPG), D-mannitol, myo-inositol, l-cysteine, l-arginine, l-kynurenine, and taurolitocholate-3-sulfate], and leucine enkephalin were purchased from Sigma-Aldrich (US), ICN Biomedicals Inc. (US), TLC Pharmchem (CA), or Waters Corporation (US) or as reported in (11, 77). Information on the purchased polymers and initial evaluation of different polymer solutions is given under note S1.
Electrospinning
PVP and PS were dissolved in DMF and CHCl3 (1:2, v/v) and stirred for a minimum of 4 hours at 60°C to obtain a clear solution. PAN (molecular weight = 150,000 g/mol, Sigma-Aldrich, US) was dissolved in DMF at 5, 8, 10, 12, and 15% (w/w), by stirring for a minimum of 2 hours at RT and subsequently 1 hour at 90°C until a clear, pale-yellow solution was obtained. The polymer blends (PVP/PS) were loaded in a 20-ml syringe and fed through a syringe KD Scientific KDS-100-CE Pump Series 100 (Sigma-Aldrich, US) using a capillary 18G needle of 0.80-mm inner diameter at a flow rate of 1 ml hour−1 and 1.5 ml min−1 for the core (PVP/PS for the salivary and rectal MetaSAMP and PVP/PS-PAN for the urinary MetaSAMP) and PAN cover layers, respectively. A Glassman High Voltage Series EH (High Bridge, US) was clamped to the tip of the needle and was used to apply a voltage between 15 and 17.5 kV. The tip-to-collector distance was averaged at 15 cm but slightly altered during the ES process toward attaining a stable, i.e., steady-state, Taylor cone (78). The (nano)fibrous network was collected on a grounded, thick aluminum foil–covered metal drum collector (Linari NanoTech, Italy) rotating at 125 RPM. All experiments were performed at ambient conditions, i.e., RT (22° ± 2°C) and relative humidity of 30 ± 10%, in a fumehood (78). MDSC measurements were performed (fig. S3) to determine the Tg of the polymers in the blend. Specifications of the viscosity measurements, SEM analyses, mDSC method, and CAMs are elaborated under note S2.
LA-REIMS instrumental analysis
All LA-REIMS analyses were performed using our previously biofluid-specific optimized protocols for urine and saliva (1), while for the rectal MetaSAMP-LA-REIMS analysis the protocol was optimized using a DOE design as described in note S3.1. The final optimized parameters are described in table S1. A mid-infrared laser system (Opolette 2940, Opotek, US) was used as a desorption and ionization source hyphenated by a polytetrafluoroethylene (PTFE) aerosol channeling tube to a Xevo G2-XS quadrupole time-of-flight mass spectrometer (Waters Corporation, US). A more comprehensive description of the instrumental analysis is described under note S3.2. LA-REIMS analysis of MetaSAMPs and crude biofluids was performed using our custom-built platform according to the manual and fully automated methodology, respectively, as described by Plekhova (1).
Clinical study samples
Fresh feces (Fecotainer, Excretas Medical BV, NL) and urine [6 to 12 years, MetaBEAse cohort, ClinicalTrials.gov (NCT04632511)] and saliva [6 to 16 years, OPERA cohort (47)] were collected. The Ghent University Hospital Ethics Committee approved both studies (BC-06939 and EC 2016/0673), and informed consent was obtained from every participant and their caretakers. Samples were stored at −80°C following collection and thawed at RT (22° ± 2°C) before analysis. Participant fecal samples underwent lyophilization (Christ 1-4 Alpha LSCplus), resulting in an average removal of 55.9% ± 6.4% water. Saliva samples were pretreated as described previously (13), and urine samples were used after thawing without additional processing.
The study participants were of both sexes (Table 1), were pseudonymized and did not receive antibiotic treatment for at least 3 months before sample donation or take any long-term medication (77). The classification was performed based on BMI z-scores (adjusted BMI for age and sex) (79) and IOTF score (80).
Sample preparation for optimization and short-term stability experiments
For initial optimization experiments with the rectal MetaSAMP, rectal sampling was mimicked by impregnating pieces (ca. 1 cm by 1 cm) of the electrospun PVP/PS membranes with the contents of freshly collected porcine colon and rectum (EC2018_70 and EC2018_91, Ghent University, Department of Translational Physiology, Infectiology and Public Health, Merelbeke, BE). For the primary experiments with the salivary and urinary MetaSAMP, sampling was performed using pooled QC samples (n = 6) (UGent-LCA biobank 210016). To evaluate the short-term stability of the rectal, salivary and urinary MetaSAMPs (nano)fiber composition, remaining aliquots from the samples of our MetaBEAse and OPERA pediatric cohorts (n = 6 for each biofluid) were combined to create pooled QC samples that aimed to reflect inherent biological variation. Pooled QC samples of the 3 biofluids were used to maximize metabolite coverage without substantially increasing the number of analyses (12, 81).
For the pooled QC samples used during optimization, short-term stability, and targeted experiments, fecal water was prepared by the addition of UPW to the fresh fecal sample at a 1:4 ratio (w/v %) followed by homogenization (Stomacher 400, Seward, Worthing, UK) for 10 min at the highest intensity (82) (see also note S5, fig. S19, and tables S10, S11, and S13) and subsequent centrifugation (Rotanta 460R, Hettich Zentrifugen, DE) at RT (500g, 2 min). The collected supernatant was divided into aliquots, which were stored at −80°C. Hence, freeze-thaw cycling, which may negatively affect metabolome stability (12), was limited since a new aliquot was used, after thorough vortexing (1 min at 400g, IKA vortex 3, IKA, and DE), for every optimization experiment performed. In line with the optimization (note S1.3), electrospun membrane pieces [ca. 1 cm by 1 cm in size, matching the dimensions of a 24-well plate (Greiner CELLSTAR, Greiner Bio One, DE)] were spiked with 30 μl of fecal water, saliva, or urine and impregnated for 10 to 15 min. For direct biofluid analysis (i.e., without MetaSAMP impregnation), 100 μl of each sample was added to a 96-well plate (Greiner 96-well microplates, Greiner Bio One, DE).
Compositional optimization, short-term stability, and laser ablation interaction evaluation of the biofluid-specific MetaSAMPs
The ratios at which PVP and PS were blended were controlled by varying the relative weight ratios of each polymer as well as the polymer weight relative to the total solution weight (w/w). Moreover, the addition of the PAN top layer (5 to 15%, w/w) was examined through the quality of the fiber network by SEM and evaluated in terms of metabolome coverage. To this end, both DOE matrices and response surface methodology models were constructed with the software program Modde (Sartorius, DE), whereby the compositions [polymer ratio (PVP%, w/w), polymer wt % (w/w)], duration of spinning the PAN cover layer (min) and duration of spinning the core membrane (h) for the rectal MetaSAMP (table S5 and fig. S13, A to C) and polymer ratio (PVP%, w/w), presence of PAN in the polymer blend (%), duration of spinning the PAN cover layer (min) and duration of spinning the core membrane (h) for the salivary and urinary MetaSAMPs were evaluated (table S6 and fig. S13, D to I). The goal was to maximize metabolome coverage, measurement repeatability (n = 5) (77), summarized normalized LA-REIMS intensity (83) and obtain good quality of the fiber network (by inspecting SEM images). Subsequently, the number of molecular features covered and their relative number (%) with CV ≤30% (77) were compared.
A short-term stability study of the impregnated optimized rectal, salivary and urinary MetaSAMPs was performed by using QC pool samples (see above) for up to 48 hours at RT (22° ± 2°C) and 4°C. As described above, biofluids were either applied on the respective MetaSAMP surface or analyzed as such directly upon collection (first time point) and after storage (second time point). For each condition eight sample aliquots were analyzed, of which three replicate measurements were performed. To assure reliable metabolome investigation following the application of LA-REIMS onto electrospun polymer networks, the interaction between substrate and analytical beam, i.e., the optimized MetaSAMPs and laser upon ablation, respectively, was assessed (note S4 and figs. S16 and S17).
Analytical and biological validation of the MetaSAMP-LA-REIMS methodology
The optimized MetaSAMP configurations were subjected to analytical (targeted and untargeted) and biological (untargeted) validation. To this end, feces and urine from children (MetaBEAse cohort, n = 88, IOTF ≥ 1 and n = 146, IOTF < 1) and saliva samples from children (OPERA cohort (47), n = 66, IOTF = 0, n = 29 IOTF ≥ 1) (13) were impregnated under the same experimental conditions as described above onto the optimized electrospun membranes.
The instrumental, intra-assay precision or repeatability, intermediate precision and linearity of the MetaSAMP-LA-REIMS methodology was assessed to confirm the analytical method as fit-for-purpose according to FDA recommendations (11, 45). This is, technical repeatability was assessed by LA-REIMS automated analysis (n = 3 burns of the same sample) (1) of the pooled QC samples and standard mixtures. For the intra-assay and intermediate precision tests, five pieces from an electrospun membrane were analyzed under repeatable experimental conditions by the same operator, and this setup was used again on a different day by another operator, respectively.
The target analytes (30 μl at 100 ng μl−1) were spiked using a micropipette onto the rectal MetaSAMP membranes in solvent (UPW and IPA, according to logP of the target analytes) as well as in fecal water into a 24-well plate (Greiner CELLSTAR, Greiner Bio One, DE) and subjected to our automatic platform for LA-REIMS analysis (1). The mass spectra generated in MassLynx and through our in-house data analysis pipeline as well as mass accuracy data for both platforms were investigated, and the repeatability of the targeted analysis was examined using the CV values. For this purpose, we evaluated the accurate m/z values of detectable adducts of the molecular ions to three decimal places after mass drift correction [LA-REIMS data and the monoisotopic mass from HMDB (46)].
To assess the applicability of the MetaSAMP-LA-REIMS platform, the established methodology was implemented in the clinical context of pediatric overweight and obesity (see above). The associations between anthropometric and clinical and metabolic blood data with molecular features were computed in those two independent cohorts [MetaBEAse and OPERA (47) cohorts] using Spearman’s rank correlation analysis (58) (absolute correlation values or moduli reported with ρ) and corresponding P values (Wilcoxon rank-sum tests with continuity correction) for each pair of metadata and molecular feature under investigation.
Ultra-high performance liquid chromatography coupled to HRMS analysis, a more comprehensive analysis technique that is conventionally used in metabolomics research [Tiers 1 to 3 according to metabolite identification standards (81)], was addressed to enable metabolite annotation of potentially clinically relevant molecular features. For this purpose, a selection of representative crude fecal patient samples [n = 4, extracted fecal samples of obese children (IOTF > 1), MetaBEAse cohort] and 525 fecal in-house analytical standards (data S7) were analyzed accordingly by combined complementary metabolomics (11) and lipidomics (77) analysis.
For every validation experiment, a pool of randomly selected clinical cohort samples (n ≥ 20) was prepared to include as QC sample during analysis to monitor instrument performance. For this purpose, QC samples were included at the beginning and end of the analytical batch (n = 6), as well as during analysis (n = 2) after every 10 samples, and samples were analyzed in a randomized order.
Data and statistical analysis
LA-REIMS molecular fingerprints were acquired by MassLynx (version 4.2, Waters, Corporation, US, stored as raw directories). To counteract plausible time-dependent instrumental variability, lock mass correction against the m/z value of leucine enkephalin (554.262 Da for negative ion mode data, 100-mDa mass tolerance window) was performed. Data were preprocessed to select representative burns and perform peak picking using Progenesis Bridge (version 1.0.29) and Progenesis QI (version 2.3., Waters Corporation, US), respectively, as described previously (1). Progenesis Bridge preprocesses the original raw data, acquired using the LA-REIMS setup, into bridged data by adaptive background subtraction according to the sample-specific total ion current (TIC) replicate threshold specified (applied thresholds of 2 × 106 to 5 × 107, depending on the experiments performed). Progenesis QI applies a peak-picking algorithm and performs TIC normalization, resulting in lists of m/z features. Further data analyses were conducted using R (version 3.4.3, AT) and Python (version 3.7.4, US) (table S14). Preprocessed data were subjected to pretreatment optimization, including a range of tested normalization strategies, which included QC-based robust locally estimated scatterplot smoothing signal correction (QC-RLSC algorithm) (13), internal QC correction, and TIC correction as well as their combinations. Molecular features, as addressed in terms of feature count and % of features below CV threshold, were assigned as relevant only when they occurred in at least 80% of QC replicates. For intensity analysis and the evaluation of replicate measures, mean, SD and CV values were calculated. All multivariate models were built using SIMCA (Sartorius, DE) after selection of the best raw data pretreatment strategy and the associated data. Unsupervised principal components analysis (PCA) was used for the identification of outliers, the evaluation of instrument stability based on QC clustering, and the assessment of the natural patterning of samples according to inherent metabolic fingerprints. Supervised multivariate statistical modeling using OPLS-DA was performed to assess the discriminative and predictive performance of the metabolic fingerprints. In addition, receiver operator characteristic analysis of the fecal and urinary metabolome coverage (MetaBEAse cohort) was executed on the basis of the logistic regression classification model (75-25 train-test split after balancing data, performed using fivefold cross-validation). Logistic regression is a popular method to analyze injury case-control studies, and has some advantages over linear regression analysis, using maximum likelihood estimation methods. To optimize the predictive power of our models, molecular features (1750 in total) were iteratively eliminated from the dataset while monitoring the accuracy of models trained on this reduced dataset. Quantification of AUC values and corresponding 95% CI was performed using bootstrap resampling (n = 200). To perform the Spearman’s correlation analysis, normalized, log-transformed and Pareto scaled datasets were used. In addition, univariate Wilcoxon rank-sum tests with continuity correction were performed with the normalized dataset (rectal MetaSAMP) to study which molecular features showed good and significant (P < 0.05) correlations with anthropometric and/or clinical parameters. Regarding the latter, the Spearman's ρ values were ranked on the basis of their moduli to select the highest correlations. However, for the clinical evaluation of the metabolite’s positive or negative correlation with a specific parameter, the signs of Spearman ρ values were considered. Targeted data processing of the significant molecular features was carried out with Xcalibur 3.0 software (Thermo Fisher Scientific, US), whereby compounds were identified based on their m/z value, C-isotope profile, and retention time relative to that of the internal standard, followed by putative identification (81) of significant and highly correlated LA-REIMS–MetaSAMP features (mass deviation below 50 ppm). Pairwise univariate and multiple comparisons were evaluated for the identified molecular features according to different IOTF groups based on Wilcoxon rank-sum test with continuity correction and Kruskal-Wallis test with Dunn’s post hoc test, respectively.
The R language was also used for preprocessing, data handling, and statistical analysis of data from the targeted analysis of spiked analytes representing clinically relevant metabolites in (childhood) overweight and obesity (2) with varying physicochemical properties (mass error tolerance set at a maximum value of 150 ppm), including a visualization tool generating mass spectra and graphics (data S8).
Acknowledgments
We thank S. Kasim and O. Verschatse, who helped during the experimental work using our LA-REIMS platform and executed SEM analyses, respectively. We thank the Laboratory of Virology (Faculty of Veterinary Medicine, Ghent University, H. Nauwynck) for providing colonic and rectal mucosa of slaughtered piglets. We thank UZ Antwerpen (K. Van De Maele and E. Vermeiren), UZ Brussel (J. De Schepper and I. Gies), UZ Leuven (K. Casteels, A. Jacobs, and A. Rochtus), AZ Jan-Palfijn (N. Baeck), AZ Alma (K. Vanneste), AZ Sint-Lucas (S. Deman), AZ Sint-Jan Brugge (S. Depoorter), and OLV van Lourdes Waregem (L. De Lille), for their active involvement in the recruitment of children with overweight and obesity. The Laboratory of Integrative Metabolomics (LIMET) research group is part of the Ghent University expertise center MSsmall.
Funding: This work was supported by the following grants: FWO 1S57922N (M.D.S.), FWO 1192622 N (K.W.), ERC FWO Runner-up G0G0119N (L.V. and M.D.G.), BOF GOA 2017/000102 (K.W., N.M., S.D.H., and L.V.), BOF 01 J07519 (L.V. and V.Pl.), FWO Hercules AUG/17/09 (L.V.) and IOF MetaSAMP F2020/IOF-StarTT/066 (LV, K.D.C., J.G., and E.S.).
Author contributions: Conceptualization: M.D.S., K.D.C,. and L.V. Methodology: M.D.S., V.Pl., J.G., E.S., B.P., V.S., K.D.W., and L.V. Investigation: M.D.S., K.W., and A.D.L. Visualization: M.D.S., V.Pa., M.D.G., and L.V. Supervision: I.G., N.M., S.D.H., K.D.C. and L.V. Writing-original draft: M.D.S. and L.V. Writing-review & editing: M.D.S., M.D.G. and L.V. Funding: L.V., K.D.C., S.D.H., N.M., M.D.S. and K.W.
Competing interests: A patent application has been submitted by Ghent University on 31 March 2020, authored by L.V., M.D.S., K.D.C., J.G,. and V.S., and has entered the national phase (WO/2021/191467, application number: PCT/EP2021/058162). The authors declare no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Notes S1 to S5
Figs. S1 to S19
Tables S1 to S14
Legends for data S1 to S8
Other Supplementary Material for this manuscript includes the following:
Data S1 to S8
REFERENCES AND NOTES
- 1.V. Plekhova, L. Van Meulebroek, M. De Graeve, A. Perdones-montero, M. De Spiegeleer, E. De Paepe, E. Van De Walle, Z. Takats, S. J. S. Cameron, L. Vanhaecke, Rapid ex vivo molecular fingerprinting of biofluids using laser-assisted rapid evaporative ionization mass spectrometry. Nat. Protoc. 16, 4327–4354 (2021). [DOI] [PubMed] [Google Scholar]
- 2.M. De Spiegeleer, E. De Paepe, L. Van Meulebroek, I. Gies, J. De Schepper, L. Vanhaecke, Paediatric obesity: A systematic review and pathway mapping of metabolic alterations underlying early disease processes. Mol. Med. 27, 145 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. F. Giskeodegard, S. K. Davies, V. L. Revell, H. Keun, D. J. Skene, Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci. Rep. 5, 14843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. McEniry, Early-life conditions and older adult health in low- and middle-income countries: A review. J. Dev. Orig. Health Dis. 4, 10–29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Liu, J. Xiao, C. Gijavanekar, K. L. Pappan, K. E. Glinton, B. J. Shayota, A. D. Kennedy, Q. Sun, V. R. Sutton, S. H. Elsea, Comparison of Untargeted Metabolomic Profiling vs Traditional Metabolic Screening to Identify Inborn Errors of Metabolism. JAMA Netw. Open 4, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization, “Who European Regional Obesity Report 2022” (2022).
- 7.R. L. Rosenfield, Identifying Children at Risk for Polycystic Ovary Syndrome. J. Clin. Endocrinol. Metab. 92, 787–796 (2007). [DOI] [PubMed] [Google Scholar]
- 8.A. Deeb, S. Attia, S. Mahmoud, G. Elhaj, A. Elfatih, Dyslipidemia and Fatty Liver Disease in Overweight and Obese Children. J. Obes. 2018, 1–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. C. Masquio, A. de Piano Ganen, R. M. Campos, P. de Lima Sanches, F. C. Corgosinho, D. Caranti, L. Tock, M. T. de Mello, S. Tufik, A. R. Dâmaso, Cut-off values of waist circumference to predict metabolic syndrome in obese adolescents. Nutr. Hosp. 31, 1540–1550 (2015). [DOI] [PubMed] [Google Scholar]
- 10.M. Propst, C. Colvin, R. L. Griffin, B. Sunil, C. M. Harmon, G. Yannam, J. E. Johnson, C. B. Smith, A. P. Lucas, B. T. Diaz, A. P. Ashraf, Diabetes and prediabetes are significantly higher in morbidly obese children compared with obese children. Endocr. Pract. 21, 1046–1053 (2015). [DOI] [PubMed] [Google Scholar]
- 11.E. De Paepe, L. Van Meulebroek, C. Rombouts, S. Huysman, K. Verplanken, B. Lapauw, J. Wauters, L. Y. Hemeryck, L. Vanhaecke, A validated multi-matrix platform for metabolomic fingerprinting of human urine, feces and plasma using ultra-high performance liquid-chromatography coupled to hybrid orbitrap high-resolution mass spectrometry. Anal. Chim. Acta 1033, 108–118 (2018). [DOI] [PubMed] [Google Scholar]
- 12.M. De Spiegeleer, M. De Graeve, S. Huysman, A. Vanderbeke, L. Van Meulebroek, L. Vanhaecke, Impact of storage conditions on the human stool metabolome and lipidome: Preserving the most accurate fingerprint. Anal. Chim. Acta 1108, 79–88 (2020). [DOI] [PubMed] [Google Scholar]
- 13.K. Wijnant, L. Van Meulebroek, B. Pomian, K. De Windt, S. De Henauw, N. Michels, L. Vanhaecke, Validated Ultra-High-Performance Liquid Chromatography Hybrid High-Resolution Mass Spectrometry and Laser-Assisted Rapid Evaporative Ionization Mass Spectrometry for Salivary Metabolomics. Anal. Chem. 92, 5116–5124 (2020). [DOI] [PubMed] [Google Scholar]
- 14.J. Laparre, Z. Kaabia, M. Mooney, T. Buckley, M. Sherry, B. Le Bizec, G. Dervilly-Pinel, Impact of storage conditions on the urinary metabolomics fingerprint. Anal. Chim. Acta 951, 99–107 (2017). [DOI] [PubMed] [Google Scholar]
- 15.W. Lu, X. Su, M. S. Klein, I. A. Lewis, O. Fiehn, J. D. Rabinowitz, Metabolite Measurement: Pitfalls to Avoid and Practices to Follow. Annu. Rev. Biochem. 86, 277–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.L. Van Meulebroek, S. Cameron, V. Plekhova, M. De Spiegeleer, K. Wijnant, N. Michels, S. De Henauw, B. Lapauw, L. Vanhaecke, Rapid LA-REIMS and comprehensive UHPLC-HRMS for metabolic phenotyping of feces. Talanta 217, 121043 (2020). [DOI] [PubMed] [Google Scholar]
- 17.P. Pruski, D. A. MacIntyre, H. V. Lewis, P. Inglese, G. D. S. Correia, T. T. Hansel, P. R. Bennett, E. Holmes, Z. Takats, Medical Swab Analysis Using Desorption Electrospray Ionization Mass Spectrometry: A Noninvasive Approach for Mucosal Diagnostics. Anal. Chem. 89, 1540–1550 (2017). [DOI] [PubMed] [Google Scholar]
- 18.S. J. S. Cameron, J. L. Alexander, F. Bolt, A. Burke, H. Ashrafian, J. Teare, J. R. Marchesi, J. Kinross, J. V. Li, Z. Takáts, Evaluation of Direct from Sample Metabolomics of Human Feces Using Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 91, 13448–13457 (2019). [DOI] [PubMed] [Google Scholar]
- 19.X. Li, R. Xu, X. Wei, H. Hu, S. Zhao, Y. M. Liu, Direct Analysis of Biofluids by Mass Spectrometry with Microfluidic Voltage-Assisted Liquid Desorption Electrospray Ionization. Anal. Chem. 89, 12014–12022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. Fatou, P. Saudemont, E. Leblanc, D. Vinatier, V. Mesdag, M. Wisztorski, C. Focsa, M. Salzet, M. Ziskind, I. Fournier, In vivo Real-Time Mass Spectrometry for Guided Surgery Application. Sci. Rep. 6, 25919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N. Abbassi-Ghadi, E. A. Jones, M. Gomez-Romero, O. Golf, S. Kumar, J. Huang, H. Kudo, R. D. Goldin, G. B. Hanna, Z. Takats, A Comparison of DESI-MS and LC-MS for the Lipidomic Profiling of Human Cancer Tissue. J. Am. Soc. Mass Spectrom. 27, 255–264 (2016). [DOI] [PubMed] [Google Scholar]
- 22.A. Alonso, S. Marsal, A. Julià, Analytical Methods in Untargeted Metabolomics: State of the Art in 2015. Front. Bioeng. Biotechnol. 3, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.T. H. Kuo, E. P. Dutkiewicz, J. Pei, C. C. Hsu, Ambient Ionization Mass Spectrometry Today and Tomorrow: Embracing Challenges and Opportunities. Anal. Chem. 92, 2353–2363 (2020). [DOI] [PubMed] [Google Scholar]
- 24.J. Geltmeyer, L. Van Der Schueren, F. Goethals, K. De Buysser, K. De Clerck, Optimum sol viscosity for stable electrospinning of silica nanofibres. J. Solgel Sci. Technol. 67, 188–195 (2013). [Google Scholar]
- 25.M. S. Islam, B. C. Ang, A. Andriyana, A. M. Afifi, A review on fabrication of nanofibers via electrospinning and their applications. SN Appl. Sci. 1, 1248 (2019). [Google Scholar]
- 26.N. Reyes-Garcés, E. Gionfriddo, G. A. Gómez-Ríos, M. N. Alam, E. Boyacl, B. Bojko, V. Singh, J. Grandy, J. Pawliszyn, Advances in Solid Phase Microextraction and Perspective on Future Directions. Anal. Chem. 90, 302–360 (2018). [DOI] [PubMed] [Google Scholar]
- 27.T. Lu, S. V. Olesik, Electrospun Nanofibers as Substrates for Surface-Assisted Laser Desorption/Ionization and Matrix-Enhanced Surface-Assisted Laser Desorption/Ionization Mass Spectrometry. Anal. Chem. 85, 4384–4391 (2013). [DOI] [PubMed] [Google Scholar]
- 28.N. Asano, S. Sugihara, S. I. Suye, S. Fujita, Electrospun Porous Nanofibers with Imprinted Patterns Induced by Phase Separation of Immiscible Polymer Blends. ACS Omega. 7, 19997–20005 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.B. Onat, H. Rosales-Solano, J. Pawliszyn, Development of a Biocompatible Solid Phase Microextraction Thin Film Coating for the Sampling and Enrichment of Peptides. Anal. Chem. 92, 9379–9388 (2020). [DOI] [PubMed] [Google Scholar]
- 30.S. Huysman, F. Vanryckeghem, E. De Paepe, F. Smedes, S. A. Haughey, C. T. Elliott, K. Demeestere, L. Vanhaecke, Hydrophilic Divinylbenzene for Equilibrium Sorption of Emerging Organic Contaminants in Aquatic Matrices. Environ. Sci. Technol. 53, 10803–10812 (2019). [DOI] [PubMed] [Google Scholar]
- 31.K. Dettmer, P. Aronov, B. D. Hammock, Mass spectrometry-based metabolomics. Mass Spectrom. Rev. 26, 51–78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Z. He, P. Wang, D. Liu, Z. Zhou, Hydrophilic-lipophilic balanced magnetic nanoparticles: Preparation and application in magnetic solid-phase extraction of organochlorine pesticides and triazine herbicides in environmental water samples. Talanta 127, 1–8 (2014). [DOI] [PubMed] [Google Scholar]
- 33.K. L. Nielsen, M. L. Hartvigsen, M. S. Hedemann, H. N. Lræke, K. Hermansen, K. E. Bach Knudsen, Similar metabolic responses in pigs and humans to breads with different contents and compositions of dietary fibers: A metabolomics study. Am. J. Clin. Nutr. 99, 941–949 (2014). [DOI] [PubMed] [Google Scholar]
- 34.P. Franco, I. De Marco, The Use of Poly(N-vinyl pyrrolidone) in the Delivery of Drugs: A Review. Polymers 12, 1114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.X. Liu, T. Lin, J. Fang, G. Yao, H. Zhao, M. Dodson, X. Wang, In vivo wound healing and antibacterial performances of electrospun nanofibre membranes. J. Biomed. Mater. Res. A 94, 499–508 (2010). [DOI] [PubMed] [Google Scholar]
- 36.J. Xue, T. Wu, Y. Dai, Y. Xia, Electrospinning and electrospun nanofibers: Methods, materials, and applications. Chem. Rev. 119, 5298–5415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. Niska-Blakie, L. Gopinathan, K. N. Low, Y. L. Kien, C. M. F. Goh, M. J. Caldez, E. Pfeiffenberger, O. S. Jones, C. B. Ong, I. V. Kurochkin, V. Coppola, L. Tessarollo, H. Choi, Y. Kanagasundaram, F. Eisenhaber, S. Maurer-Stroh, P. Kaldis, Knockout of the non-essential gene SUGCT creates diet-linked, age-related microbiome disbalance with a diabetes-like metabolic syndrome phenotype. Cell. Mol. Life Sci. 77, 3423–3439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Reyman, M. A. van Houten, K. Arp, E. A. M. Sanders, D. Bogaert, Rectal swabs are a reliable proxy for faecal samples in infant gut microbiota research based on 16S-rRNA sequencing. Sci. Rep. 9, 16072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R. B. Jones, X. Zhu, E. Moan, H. J. Murff, R. M. Ness, D. L. Seidner, S. Sun, C. Yu, Q. Dai, A. A. Fodor, M. A. Azcarate-Peril, M. J. Shrubsole, Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci. Rep. 8, 4139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.E. A. Jones, D. Simon, T. Karancsi, J. Balog, S. D. Pringle, Z. Takats, Matrix assisted rapid evaporative ionization mass spectrometry. Anal. Chem. 91, 9784–9791 (2019). [DOI] [PubMed] [Google Scholar]
- 41.P. T. Ivanova, S. B. Milne, H. A. Brown, Identification of atypical ether-linked glycerophospholipid species in macrophages by mass spectrometry. J. Lipid Res. 51, 1581–1590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Bian, S. V. Olesik, Surface-assisted laser desorption/ionization time-of-flight mass spectrometry of small drug molecules and high molecular weight synthetic/biological polymers using electrospun composite nanofibers. Analyst 142, 1125–1132 (2017). [DOI] [PubMed] [Google Scholar]
- 43.J. Bian, S. V. Olesik, Polyvinylpyrrolidone composite nanofibers as efficient substrates for surface-assisted laser desorption/ionization mass spectrometry. Int. J. Mass Spectrom. 448, 116253 (2020). [Google Scholar]
- 44.I. Agueusop, P. B. Musholt, B. Klaus, K. Hightower, A. Kannt, Short-term variability of the human serum metabolome depending on nutritional and metabolic health status. Sci. Rep. 10, 16310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Food and Drug Administration (FDA), Guidance for Industry: Bioanalytical Method Validation (2018); www.fda.gov/downloads/drugs/guidances/ucm070107.Pdf.
- 46.D. S. Wishart, A. Guo, E. Oler, F. Wang, A. Anjum, H. Peters, R. Dizon, Z. Sayeeda, S. Tian, B. L. Lee, M. Berjanskii, R. Mah, M. Yamamoto, J. Jovel, C. Torres-Calzada, M. Hiebert-Giesbrecht, V. W. Lui, D. Varshavi, D. Varshavi, D. Allen, D. Arndt, N. Khetarpal, A. Sivakumaran, K. Harford, S. Sanford, K. Yee, X. Cao, Z. Budinski, J. Liigand, L. Zhang, J. Zheng, R. Mandal, N. Karu, M. Dambrova, H. B. Schï Oth, R. Greiner, V. Gautam, HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 50, D622–D631 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. Wijnant, J. Klosowska, C. Braet, S. Verbeken, S. De Henauw, L. Vanhaecke, N. Michels, Stress Responsiveness and Emotional Eating Depend on Youngsters’ Chronic Stress Level and Overweight. Nutrients 13, 3654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.E. De Paepe, L. Van Gijseghem, M. De Spiegeleer, E. Cox, L. Vanhaecke, A Systematic Review of Metabolic Alterations Underlying IgE-Mediated Food Allergy in Children. Mol. Nutr. Food Res. 65, e2100536 (2021). [DOI] [PubMed] [Google Scholar]
- 49.J. D. Blume, Bounding Sample Size Projections for the Area Under a ROC Curve. J. Stat. Plan. Inference 139, 711–721 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Chavira-Suárez, C. Rosel-Pech, E. Polo-Oteyza, M. M. Ancira-Moreno, I. Ibarra-González, M. Vela-Amieva, N. Meraz-Cruz, C. Aguilar-Salinas, F. Vadillo-Ortega, Simultaneous evaluation of metabolomic and inflammatory biomarkers in children with different body mass index (BMI) and waist-to-height ratio (WHtR). PLOS ONE 15, e0237917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.P. Schober, C. Boer, L. A. Schwarte, Correlation coefficients: Appropriate use and interpretation. Anesth. Analg. 126, 1763–1768 (2018). [DOI] [PubMed] [Google Scholar]
- 52.American diabetes association , 2. Classification and diagnosis of Diabetes:Standards of medical care in Diabetes—2019. Diabetes Care 42, S13–S28 (2019). [DOI] [PubMed] [Google Scholar]
- 53.M. Ramon-Krauel, M. J. Leal-Witt, Ó. Osorio-Conles, M. Amat-Bou, C. Lerin, D. M. Selva, Relationship between adiponectin, TNFα, and SHBG in prepubertal children with obesity. Mol. Cell. Pediatr. 8, 3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.R. Santos-Silva, M. Fontoura, J. T. Guimarães, H. Barros, A. C. Santos, Association of dehydroepiandrosterone sulfate, birth size, adiposity and cardiometabolic risk factors in 7-year-old children. Pediatr. Res. 91, 1897–1905 (2022). [DOI] [PubMed] [Google Scholar]
- 55.M. D. Klok, S. Jakobsdottir, M. L. Drent, The role of leptin and ghrelin in the regulation of food intake and body weight in humans: A review. Obes. Rev. 8, 21–34 (2007). [DOI] [PubMed] [Google Scholar]
- 56.A.-V. Sitar-Taut, A. Cozma, A. Fodor, S.-C. Coste, O. H. Orasan, V. Negrean, D. Pop, D.-A. Sitar-Taut, New Insights on the Relationship between Leptin, Ghrelin, and Leptin/Ghrelin Ratio Enforced by Body Mass Index in Obesity and Diabetes. Biomedicine 9, 1657 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. Liu, J. Hong, X. Xu, Q. Feng, D. Zhang, Y. Gu, J. Shi, S. Zhao, W. Liu, X. Wang, H. Xia, Z. Liu, B. Cui, P. Liang, L. Xi, J. Jin, X. Ying, X. Wang, X. Zhao, W. Li, H. Jia, Z. Lan, F. Li, R. Wang, Y. Sun, M. Yang, Y. Shen, Z. Jie, J. Li, X. Chen, H. Zhong, H. Xie, Y. Zhang, W. Gu, X. Deng, B. Shen, X. Xu, H. Yang, G. Xu, Y. Bi, S. Lai, J. Wang, L. Qi, L. Madsen, J. Wang, G. Ning, K. Kristiansen, W. Wang, Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868 (2017). [DOI] [PubMed] [Google Scholar]
- 58.F. Asnicar, S. E. Berry, A. M. Valdes, L. H. Nguyen, G. Piccinno, D. A. Drew, E. Leeming, R. Gibson, C. L. Roy, H. A. Khatib, L. Francis, M. Mazidi, O. Mompeo, M. Valles-Colomer, A. Tett, F. Beghini, L. Dubois, D. Bazzani, A. M. Thomas, C. Mirzayi, A. Khleborodova, S. Oh, R. Hine, C. Bonnett, J. Capdevila, S. Danzanvilliers, F. Giordano, L. Geistlinger, L. Waldron, R. Davies, G. Hadjigeorgiou, J. Wolf, J. M. Ordovás, C. Gardner, P. W. Franks, A. T. Chan, C. Huttenhower, T. D. Spector, N. Segata, Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 27, 321–332 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Mar Rodríguez, D. Pérez, F. J. Chaves, E. Esteve, P. Marin-Garcia, G. Xifra, J. Vendrell, M. Jové, R. Pamplona, W. Ricart, M. Portero-Otin, M. R. Chacón, J. M. Fernández Real, Obesity changes the human gut mycobiome. Sci. Rep. 5, 14600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. K. M. Lai, H. W. Tang, K. C. Lau, K. M. Ng, Nanosecond UV Laser Ablation of Gold Nanoparticles: Enhancement of Ion Desorption by Thermal-Driven Desorption, Vaporization, or Phase Explosion. J. Phys. Chem. C 120, 20368–20377 (2016). [Google Scholar]
- 61.L. Chen, A. Ghiasvand, E. S. Rodriguez, P. C. Innis, B. Paull, Applications of nanomaterials in ambient ionization mass spectrometry. Trends Anal. Chem. 136, 116202 (2021). [Google Scholar]
- 62.J. Bian, S. V. Olesik, Ion desorption efficiency and internal energy transfer in polymeric electrospun nanofiber-based surface-assisted laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 412, 923–931 (2020). [DOI] [PubMed] [Google Scholar]
- 63.M. Zhang, L. Huang, J. Yang, W. Xu, H. Su, J. Cao, Q. Wang, J. Pu, K. Qian, Ultra-Fast Label-Free Serum Metabolic Diagnosis of Coronary Heart Disease via a Deep Stabilizer. Adv. Sci. 8, 2101333 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.S. Bouatra, F. Aziat, R. Mandal, A. Chi Guo, R. M. Wilson, C. Knox, T. Bjorndahl, R. Krishnamurthy, F. Saleem, P. Lu, Z. Dame, J. Poelzer, J. Huynh, F. Yallou, N. Psychogios, E. Dong, R. Bogumil, C. Roehring, D. Wishart, The Human Urine Metabolome. PLOS ONE 8, e73076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.T. Phornpimon, T. Visith, Urinary lipidomics, in Translational Bioinformatics 14: Lipidomics in Health & Disease, Ed. X. Wang. (Springer, 2018), vol. 14, pp. 97–112. [Google Scholar]
- 66.D. Duarte, B. Castro, J. Pereira, J. Marques, A. Costa, A. Gil, Evaluation of Saliva Stability for NMR Metabolomics: Collection and Handling Protocols. Metabolites 10, 515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.S. Liang, Z. Hou, X. Li, J. Wang, L. Cai, R. Zhang, J. Li, The fecal metabolome is associated with gestational diabetes mellitus. RSC Adv. 9, 29973–29979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Cui, A. Trimigno, J. L. Castro-Mejía, S. Reitelseder, J. Bülow, R. L. Bechshøft, D. S. Nielsen, L. Holm, S. B. Engelsen, B. Khakimov, Human fecal metabolome reflects differences in body mass index, physical fitness, and blood lipoproteins in healthy older adults. Metabolites 11, 717 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.X. Song, H. Chen, R. N. Zare, Conductive Polymer Spray Ionization Mass Spectrometry for Biofluid Analysis. Anal. Chem. 90, 12878–12885 (2018). [DOI] [PubMed] [Google Scholar]
- 70.F. G. Pinto, I. Mahmud, T. A. Harmon, V. Y. Rubio, T. J. Garrett, Rapid Prostate Cancer Noninvasive Biomarker Screening Using Segmented Flow Mass Spectrometry-Based Untargeted Metabolomics. J. Proteome Res. 19, 2080–2091 (2020). [DOI] [PubMed] [Google Scholar]
- 71.J. Zhang, T. Fuhrer, H. Ye, B. Kwan, D. Montemayor, J. Tumova, M. Darshi, F. Afshinnia, J. J. Scialla, A. Anderson, A. C. Porter, J. J. Taliercio, H. Rincon-Choles, P. Rao, D. Xie, H. Feldman, U. Sauer, K. Sharma, L. Natarajan, High-Throughput Metabolomics and Diabetic Kidney Disease Progression: Evidence from the Chronic Renal Insufficiency (CRIC) Study. Am. J. Nephrol. 53, 215–225 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.P. Pruski, G. D. S. Correia, H. V. Lewis, K. Capuccini, P. Inglese, D. Chan, R. G. Brown, L. Kindinger, Y. S. Lee, A. Smith, J. Marchesi, J. A. K. McDonald, S. Cameron, K. Alexander-Hardiman, A. L. David, S. J. Stock, J. E. Norman, V. Terzidou, T. G. Teoh, L. Sykes, P. R. Bennett, Z. Takats, D. A. MacIntyre, Direct on-swab metabolic profiling of vaginal microbiome host interactions during pregnancy and preterm birth. Nat. Commun. 12, 5967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. McCarthy, L. Saldana, D. N. Ackerman, Y. Su, J. V. John, S. Chen, S. Weihs, S. P. Reid, J. L. Santarpia, M. A. Carlson, J. Xie, Ultra-absorptive Nanofiber Swabs for Improved Collection and Test Sensitivity of SARS-CoV-2 and other Biological Specimens. Nano Lett. 21, 1508–1516 (2021). [DOI] [PubMed] [Google Scholar]
- 74.T. Gaissmaier, M. Siebenhaar, V. Todorova, V. Hüllen, C. Hopf, Therapeutic drug monitoring in dried blood spots using liquid microjunction surface sampling and high resolution mass spectrometry. Analyst 141, 892–901 (2016). [DOI] [PubMed] [Google Scholar]
- 75.C. Vega, C. Spence, C. Zhang, B. J. Bills, N. E. Manicke, Ionization Suppression and Recovery in Direct Biofluid Analysis Using Paper Spray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 27, 726–734 (2016). [DOI] [PubMed] [Google Scholar]
- 76.J. D. Odom, R. V. Sutton, Metabolomics in Clinical Practice: Improving Diagnosis and Informing Management. Clin. Chem. 12, 1606–1617 (2021). [DOI] [PubMed] [Google Scholar]
- 77.L. Van Meulebroek, E. De Paepe, V. Vercruysse, B. Pomian, S. Bos, B. Lapauw, L. Vanhaecke, Holistic Lipidomics of the Human Gut Phenotype Using Validated Ultra-High-Performance Liquid Chromatography Coupled to Hybrid Orbitrap Mass Spectrometry. Anal. Chem. 89, 12502–12510 (2017). [DOI] [PubMed] [Google Scholar]
- 78.G. Darko, A. Goethals, N. Torto, K. De Clerck, Steady state electrospinning of uniform polyethersulfone nanofibers using a non-heated solvent mixture. Appl. Nanosci. 6, 837–845 (2016). [Google Scholar]
- 79.M. Roelants, R. Hauspie, K. Hoppenbrouwers, References for growth and pubertal development from birth to 21 years in Flanders, Belgium. Ann. Hum. Biol. 36, 680–694 (2009). [DOI] [PubMed] [Google Scholar]
- 80.T. J. Cole, T. Lobstein, Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr. Obes. 7, 284–294 (2012). [DOI] [PubMed] [Google Scholar]
- 81.L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. M. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden, M. R. Viant, Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.D. Monleón, J. M. Morales, A. Barrasa, J. A. López, C. Vázquez, B. Celda, Metabolite profiling of fecal water extracts from human colorectal cancer. NMR Biomed. 22, 342–348 (2009). [DOI] [PubMed] [Google Scholar]
- 83.S. Huysman, L. Van Meulebroek, F. Vanryckeghem, H. Van Langenhove, K. Demeestere, L. Vanhaecke, Development and validation of an ultra-high performance liquid chromatographic high resolution Q-Orbitrap mass spectrometric method for the simultaneous determination of steroidal endocrine disrupting compounds in aquatic matrices. Anal. Chim. Acta 984, 140–150 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Notes S1 to S5
Figs. S1 to S19
Tables S1 to S14
Legends for data S1 to S8
Data S1 to S8