Abstract
Opioid use disorder (OUD) looms as one of the most severe medical crises facing society. More effective therapeutics will require a deeper understanding of molecular changes supporting drug-taking and relapse. Here, we develop a brain reward circuit-wide atlas of opioid-induced transcriptional regulation by combining RNA sequencing (RNA-seq) and heroin self-administration in male mice modeling multiple OUD-relevant conditions: acute heroin exposure, chronic heroin intake, context-induced drug-seeking following abstinence, and relapse. Bioinformatics analysis of this rich dataset identified numerous patterns of transcriptional regulation, with both region-specific and pan-circuit biological domains affected by heroin. Integration of RNA-seq data with OUD-relevant behavioral outcomes uncovered region-specific molecular changes and biological processes that predispose to OUD vulnerability. Comparisons with human OUD RNA-seq and genome-wide association study data revealed convergent molecular abnormalities and gene candidates with high therapeutic potential. These studies outline molecular reprogramming underlying OUD and provide a foundational resource for future investigations into mechanisms and treatment strategies.
Opioid intake drives persistent, brain-wide transcriptional reprogramming to promote drug-seeking behavior.
INTRODUCTION
Opioid use disorder (OUD) currently stands as one of the most severe public health crises facing North America. More than 70,000 people die each year to opioid-related overdose in the United States alone (1), and this mortality rate continues to rise, accelerated by the coronavirus 2019 pandemic (2). While tragic, overdose deaths represent the tip of the iceberg; for each person lost to opioid overdose, hundreds more are struggling with OUD, and thousands more are receiving opioids for pain management, which places them at risk for OUD. Unfortunately, abstinence alone is effectively impossible for most people, and available treatment strategies that improve daily functioning are limited in their ability to prevent relapse long-term (3, 4). To weather the growing opioid crisis, we desperately need new therapeutic approaches based on fundamental biological insights into how OUD develops and is maintained.
The primary barrier to treating OUD, as with other addictive disorders, is relapse (5). Patients struggling with OUD become trapped in a cycle of drug-taking and drug-seeking, which is fueled by craving during periods of abstinence. Craving not only can drive ongoing drug use but also can precipitate relapse even years after cessation. Drug use causes neuroplastic changes in the brain that promote craving by tuning motivation toward obtaining drugs of abuse and away from natural rewards and healthy goals (6, 7). These drug-induced maladaptive changes are thought to be supported in part by persistent molecular adaptations within cells that comprise the brain’s reward circuitry (8). Numerous molecular mechanisms that drive reward circuit dysfunction in addiction have been identified for psychostimulants, but comparably less is known for opioids (9). Much of our knowledge of molecular changes in animal models of OUD comes from noncontingent exposure conditions, the effects of which are known to differ markedly from self-administered opioids (10), and most studies have examined single brain regions [reviewed in (9)]. In addition, few studies have focused on identifying molecular changes that drive drug-seeking and relapse following protracted abstinence.
The goal of the present work was to identify transcriptional mechanisms supporting vulnerability to long-term opioid intake and relapse-like behavior using a mouse model of OUD. To achieve this, we performed genome-wide transcriptional profiling by RNA sequencing (RNA-seq) across six brain regions crucial for reward processing in multiple experimental conditions modeling distinct stages of OUD. Using bioinformatic approaches, we investigate transcriptional regulatory processes governing aspects of OUD and link behavioral profiles reflective of OUD with gene expression patterns. We then overlay these findings in mice to published RNA-seq and genome-wide association studies (GWAS) of human OUD and establish that the mouse models successfully recapitulate important swaths of the molecular pathology of the human syndrome and identify highly relevant interventional targets. Together, this study provides fundamental insights into the molecular underpinnings of OUD and opens several avenues for potential therapeutic development.
RESULTS
Heroin self-administration
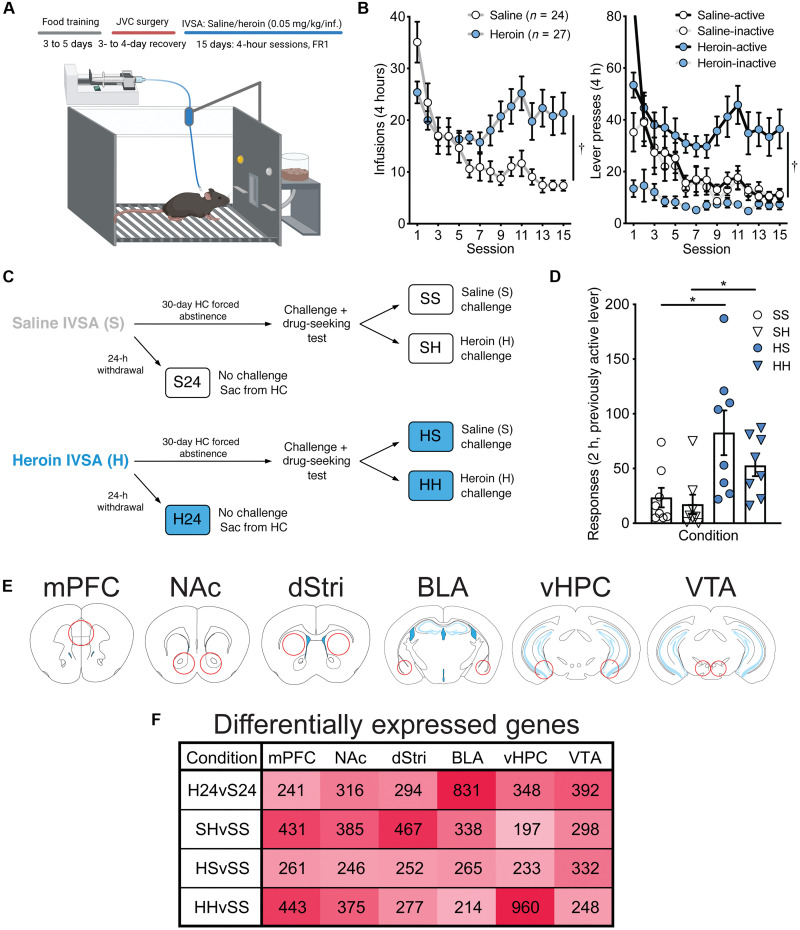
To model OUD, we used an intravenous self-administration (IVSA) paradigm in male mice (Fig. 1A). Mice were first briefly trained to perform a lever press for food reward and then received jugular vein catheterization. After recovery, mice were returned to operant boxes where they could now respond for IVSA of heroin (0.05 mg/kg per infusion; n = 27) or saline (n = 24) in 15 daily 4-hour sessions. Figure 1B demonstrates that heroin IVSA mice showed goal-directed lever pressing unlike saline mice, with more infusions earned (Group × Session: F14,686 = 5.84, P < 0.0001; Group: F1,49 = 7.85, P < 0.01) and more selective responding on the active lever compared to the inactive lever [two-way analysis of variance (ANOVA) Lever × Session; saline: no main effect of lever, F1,46 = 1.57, ns; heroin: Lever × Session, F14,728 = 52.62, P < 0.0001].
Fig. 1. Reward circuitry transcriptome profiling in a mouse model of volitional heroin intake and seeking.
(A) IVSA paradigm. Mice were first briefly trained to lever press for food and then received jugular vein catheterization (JVC) to enable intravenous infusions of saline or heroin contingent upon lever pressing on a fixed-ratio 1 (FR1) schedule of reinforcement. (B) Mice performing heroin IVSA took significantly more infusions (left; †P < 0.05, two-way ANOVA) and made significantly more active lever presses compared to inactive lever presses (right; †P < 0.05, two-way ANOVAs) compared to mice performing saline IVSA. (C) Schematic describing experimental conditions. After IVSA, mice were split into either 24-hour withdrawal (S24 and H24) or 30-day homecage (HC) forced abstinence conditions. Mice in the S24 and H24 groups were euthanized 24 hours after the final IVSA session. Mice in the 30-day group were further split to receive a challenge injection of either saline (SS and HS) or heroin (1 mg/kg) (SH and HH) and were placed back into operant boxes to measure drug-seeking under extinction conditions for 2 hours after which they were immediately euthanized. (D) Mice that previously self-administered heroin made significantly more responses on the previously active lever compared to saline IVSA mice (*P < 0.05, independent sample t test). (E) Six brain regions were extracted for RNA-seq: the medial prefrontal cortex (mPFC), nucleus accumbens (NAc), dorsal striatum (dStri), basolateral amygdala (BLA), ventral hippocampus (vHPC), and ventral tegmental area (VTA). (F) Summary of the number of DEGs induced across brain regions and experimental conditions (FC ± 30%, nominal P < 0.05).
Following the final IVSA session, mice from both saline and heroin groups were subdivided into short- and long-term homecage forced abstinence conditions of either 24 hours or 30 days, respectively (Fig. 1C). Mice in the 24-hour withdrawal condition (S24 and H24) were euthanized the following day, directly from the homecage. The 30-day condition was designed to model aspects of incubation of craving for opioids to promote relapse (11, 12). In this condition, mice were further subdivided into two groups in which they received a subcutaneous challenge injection of either saline [saline-saline (SS) and heroin-saline (HS)] or heroin (1 mg/kg) [saline-heroin (SH) and heroin-heroin (HH)] and were placed back into operant boxes for a 2-hour drug-seeking test. During this test, the levers were present in the chamber but were both inactive, enabling measurement of anticipatory responses for heroin. Animals with a history of heroin intake showed clear drug-seeking behavior compared to animals that self-administered saline (Fig. 1D; SS versus HS, t14 = 2.66, P < 0.05; SH versus HH, t14, 2.70, P < 0.05). This design enabled investigation of the transcriptomic landscape in multiple conditions that model distinct aspects of the OUD syndrome: first-ever heroin exposure (SH), ongoing/early withdrawal from heroin intake (H24), context-induced heroin-seeking following protracted abstinence (HS), and a combination of drug-induced and context-induced heroin-seeking representative of a relapse-like episode (HH).
Heroin exposure conditions engage separable gene expression programs in distinct cell types
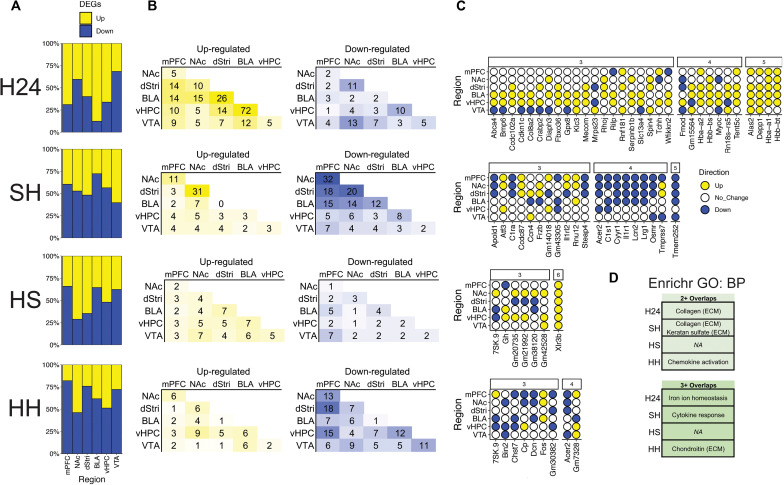
Immediately upon completion of the drug-seeking test, animals were euthanized and tissue was dissected for RNA-seq (Fig. 1E). Six brain regions that serve as critical hubs in reward-related behavior were extracted and processed for RNA-seq: medial prefrontal cortex (mPFC), nucleus accumbens (NAc), dorsal striatum (dStri), basolateral amygdala (BLA), ventral hippocampus (vHPC), and ventral tegmental area (VTA). Differential expression analysis (DESeq2) comparing each condition to its respective control (i.e., H24 versus S24 and SH/HS/HH versus SS) identified clear region- and condition-specific changes in transcriptional regulation [Fig. 1F; differentially expressed gene (DEG) lists available with associated raw data on the Gene Expression Omnibus (GSE228031)]. Across 24-hour and 30-day conditions, all brain regions exhibited hundreds of DEGs, demonstrating the ability of heroin intake to cause broad transcriptomic reprogramming throughout the brain’s reward circuitry that persist long after initial intake. We noted that different heroin exposure conditions exerted partly distinct region-specific effects on transcriptional regulation. For example, in the H24 condition, the number of DEGs in the BLA is much higher than in any other brain region studied. The first-ever heroin exposure (SH) elicited the weakest DEG response in the vHPC, but heroin re-exposure in animals with a distant (30 days) history of heroin intake (HH) caused robust transcriptional regulation in this brain region, indicating a priming effect of repeated drug exposure on the vHPC transcriptome. In contrast to SH and HH, fewer DEGs are observed across regions for the context-induced drug-seeking (HS) condition. The HS condition is unique compared to SH and HH, in that no drug is on board at the time of tissue harvesting. Thus, these results suggest that heroin exposure itself is a major disruptor to transcriptional homeostasis, while context-induced drug-seeking is associated with subtler transcriptional changes.
Gene biotype analysis (fig. S1A) found that the majority (34 to 88%) of DEGs are protein-coding, but heroin conditions induce particular shifts in biotype induction in a region- and condition-specific manner. The two conditions with the highest number of DEGs (H24, BLA; HH, and vHPC) involved a shift to predominantly protein-coding genes. In addition, heroin-primed drug-seeking (HH) caused a strong recruitment of noncoding genes in the dStri compared to other exposure conditions. Cell-type enrichment analysis (fig. S1B) revealed that in the BLA, the H24 condition uniquely engaged nonneuronal genes compared to other exposure conditions. In addition, in the mPFC, the SH condition elicited a strong induction of astrocytic genes, but a corresponding depletion of oligodendrocyte genes. In the VTA, we observed an expansion of microglial-enriched genes across exposure conditions peaking with re-exposure to heroin (HH). In the dStri, the first-ever exposure to heroin (SH) caused a strong induction of neuronal genes, which contrasts with heroin re-exposure conditions (HH) that exhibited higher astrocytic and oligodendrocytic gene enrichment. Notably, in the NAc, neuronal genes are enriched in the H24 condition, but this contracts for HS and HH conditions, suggesting a consolidation of neural transcriptional engagement through abstinence and re-exposure. Overall, these results demonstrate that heroin causes broad transcriptional responses throughout the reward circuitry affecting multiple biological domains, and these effects can shift with chronic intake and re-exposure.
Heroin intake induces coordinated transcriptional patterning across multiple brain regions
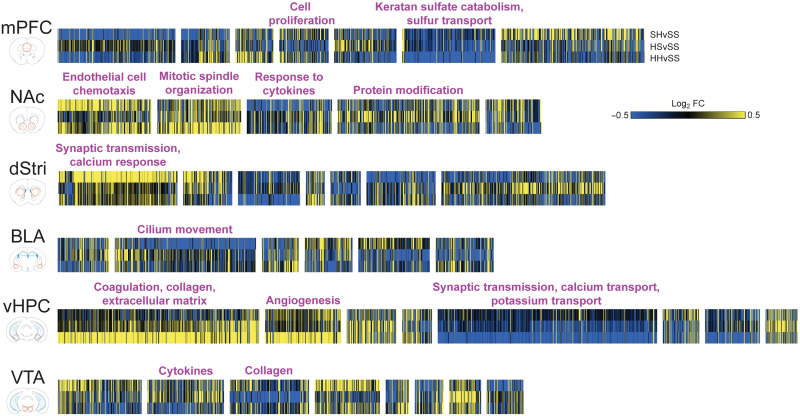
We next asked whether the broad DEG induction observed throughout the reward circuitry was mediated by the ability of heroin to cause coordinated regulation of gene expression across the brain regions examined. Examining the balance of up- and down-regulated genes in the six brain regions (Fig. 2A), we noted that the direction of gene expression changes varied for H24, SH, and HS conditions, but that HH generally suppressed gene expression across all regions. The H24 condition caused exceptionally strong up-regulation of genes in the BLA, which contrasts to the generalized down-regulation observed across all other conditions in this region. Furthermore, H24 exhibited high concordance of up-regulated genes in BLA and vHPC, with 72 genes commonly up-regulated in the two regions (Fig. 2B). In addition, many overlapping BLA and vHPC genes induced at H24 are up-regulated across other brain regions (Fig. 2C), including four genes (Hbb-bt, Hba-a1, Depp1, and Alas2) that are also up-regulated in five of six regions studied. The first-ever heroin exposure (SH) caused a coordinated transcriptional response across mPFC, NAc, dStri, and BLA, particularly for down-regulated genes (Fig. 2, B and C, and table S1). This coordinated response was less evident when animals had a history of heroin intake (HH), suggesting a potential weakening of coordinated transcriptional responses as a lasting consequence of repeated heroin intake. However, one gene, Acer2, stands out as being consistently suppressed in multiple regions in both SH and HH conditions. Tmem252 was also suppressed in SH across five brain regions and trended toward significant down-regulation in the vHPC (P = 0.077). This gene was also implicated as an addiction-relevant target spanning multiple brain regions in our previous paper examining cocaine self-administration (13). Despite context-induced drug-seeking (HS) exhibiting few overlapping DEGs, one gene, Xlr3b (X-linked lymphocyte-regulated protein 3B), was up-regulated across all six brain regions. Although not well characterized, Xlr3b is an X-linked gene that may be crucial for neurodevelopment and memory-related cognitive processes (14), making it a notable target to regulate context-driven drug-seeking behavior. Gene Ontology (GO) analysis of genes showing multiple overlaps across conditions identified enrichment of genes involved in multiple biological processes related to extracellular matrix (ECM) remodeling, as well as cellular trafficking (chemokines), immune responses (cytokines), and metal homeostasis (Fig. 2D). Together, these results demonstrate that heroin exposure, intake, seeking, and relapse each induce patterns of coordinated gene expression throughout the brain reward circuitry affecting overlapping biological domains.
Fig. 2. Heroin drives coordinated transcriptional patterning across brain reward regions that shifts with abstinence after a history of intake.
(A) Proportion of up-regulated (yellow) and down-regulated (blue) genes across brain regions for each experimental condition [heroin 24 hours (H24), saline-heroin (SH), heroin-saline (HS), and heroin-heroin (HH)]. (B) Number of shared DEGs identified across paired brain regions as being up-regulated (left) or down-regulated (right). (C) Genes identified as being regulated across three or more brain regions, with many showing coordinated expression changes across all regions identified. (D) GO analysis reveals that genes expressed across multiple brain regions enrich for biological processes related to various ECM processes including collagen, keratan sulfate, and chondroitin, in addition to iron homeostasis and cytokine and chemokine activity.
Early withdrawal from heroin intake is associated with alterations in ECM biology
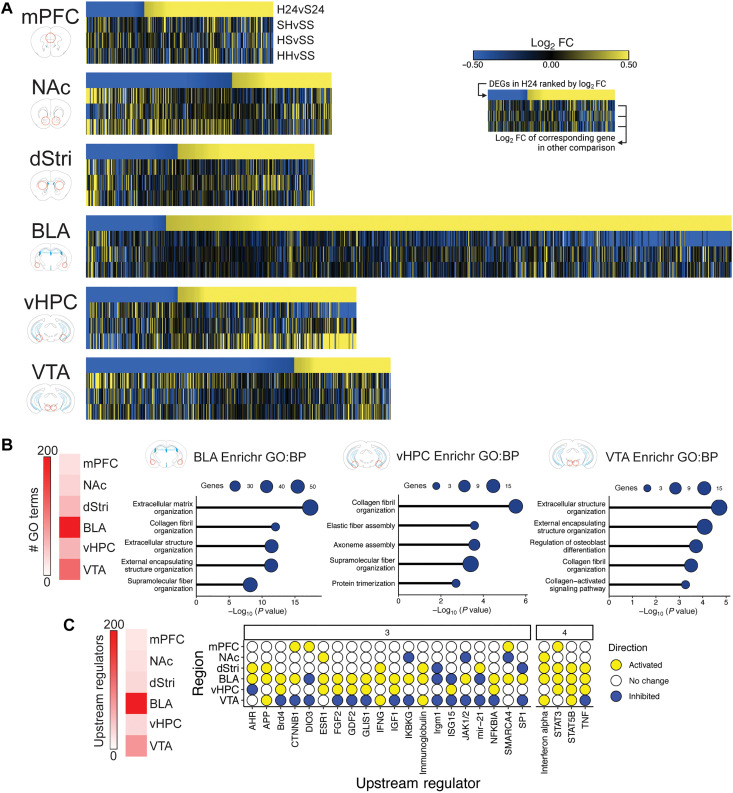
We next explored how transcriptional signatures varied across heroin intake and exposure conditions. The H24 condition, reflecting ongoing heroin intake with short-term withdrawal, was considered first based on the distinct control group used. Generally, H24 elicited a transcriptional response that showed minimal overlapping DEGs with the three forced abstinence conditions (SH, HS, and HH; Fig. 3A). This is likely due to the contrasting experience of the groups (and their respective controls) immediately before euthanasia: animals in the 24-hour condition were euthanized directly from their homecage, while animals in the forced abstinence groups experienced a subcutaneous injection and 2 hours of behavioral testing. These two experiences may initiate distinct activity/arousal-based transcriptional patterns unrelated to heroin exposure per se. Nonetheless, we noted that certain patterns of gene expression in the H24 condition were either recapitulated or inversed in the forced abstinence conditions. Particularly evident was transcriptional patterning in the BLA and vHPC, where we noted that the same genes up-regulated in short-term withdrawal (H24) are suppressed by the first-ever exposure to heroin (SH) in both of these brain regions. In the vHPC, these genes reverted to being strongly up-regulated upon re-exposure to heroin in the HH condition.
Fig. 3. Heroin intake with short-term withdrawal strongly activates transcriptional programs in the BLA and vHPC associated with ECM processes and inflammatory cytokines.
(A) Heatmaps with top row displaying all DEGs in the H24 condition ranked from lowest to highest log2 FC, and log2 FC of the corresponding gene in other experimental conditions [heroin 24 hours (H24), saline-heroin (SH), heroin-saline (HS), and heroin-heroin (HH)] in subsequent rows. (B) Results of GO analysis showing enrichment of H24 DEGs in various biological processes. In the BLA, vHPC, and VTA, top enriched terms relate to ECM functions. (C) Upstream regulator analysis revealed numerous molecular regulators of gene expression that are activated (yellow) or suppressed (blue) across multiple brain regions.
GO analysis across brain regions of the H24 condition identified enrichment of numerous biological processes in the BLA, vHPC, and VTA, which predominantly related to the ECM (Fig. 3B and table S2). These results indicate that heroin self-administration initiates a multiregion shift in ECM function, consistent with recent suggestions from human postmortem studies (15). Heroin self-administration also caused broad shifts in upstream regulator activation, which were identified across multiple brain regions (Fig. 3C and table S3). The BLA and VTA showed the strongest enrichment of upstream regulators, many of which overlapped between the two regions (63 of 88). However, many of these upstream regulators showed opposite predicted activation or inhibition between the BLA and VTA, with the majority activated in BLA but inhibited in VTA. Upstream regulators that showed enrichment across four brain regions, including tumor necrosis factor–α, signal transducer and activator of transcription 5B (STAT5B), STAT3, and interferon-α, were largely activated throughout the brain. These upstream regulators are associated with cytokine-mediated transcriptional regulation, suggesting coordinated transcriptional responses throughout the brain in mediating cytokine function. Cytokines work in partnership with the ECM to maintain structural components of the extracellular milieu and support neural and glial function (16, 17). Thus, these results also point to heroin’s ability to disrupt ECM maintenance and cytokine function as a potential brain-wide driver of OUD.
Segregated transcriptional remodeling across the reward circuitry supports drug-specific versus context-specific influences in relapse
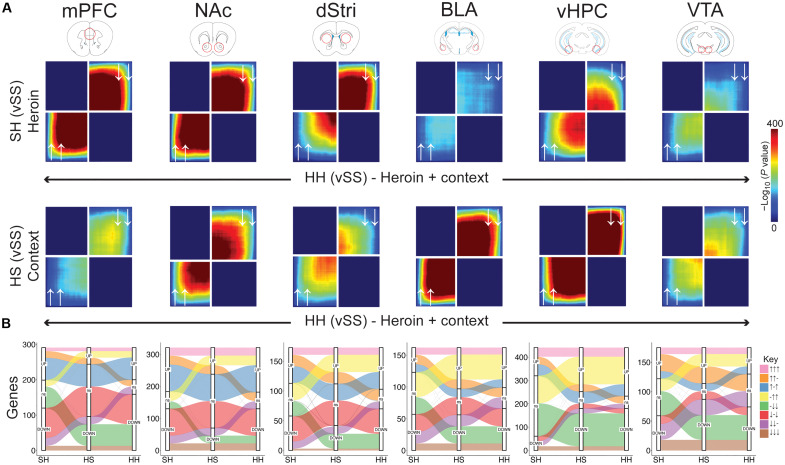
A primary goal for this study was to examine transcriptional mechanisms of relapse following protracted abstinence from heroin intake. Thus, we next focused our analysis on mice that underwent 30 days of forced abstinence from IVSA. Using a threshold-free approach, we first asked how transcriptomic remodeling varied across initial heroin exposure (SH), context-induced drug-seeking (HS), and heroin-primed drug-seeking (HH) as a function of experimental condition. We took the approach that SH and HS conditions contain separable components of the HH condition: SH may capture transcriptional regulation mediated specifically by an acute heroin exposure, whereas HS captures transcriptional regulation mediated specifically by context-dependent memory-related drug-seeking. Rank-rank hypergeometric overlap (RRHO) plots (Fig. 4A) demonstrate that all brain regions show either concordance or no concordance in their transcriptional response across experimental groups—no anticorrelations were apparent. However, we noted that particular brain regions played unique roles in coordinating transcriptional responses to drug experiences. The mPFC, NAc, and dStri showed the strongest transcriptional overlap between SH and HH conditions, suggesting that transcriptional remodeling within these regions may be strongly mediated by direct actions of heroin. These effects may indicate that repeated exposure to heroin causes iterative engagement of this transcriptional signature, priming neurobiological changes induced in relapse. On the other hand, the BLA and vHPC showed exceptionally strong coordination across HS and HH conditions, implicating a role for transcriptional remodeling supporting drug-seeking specifically in these brain regions. These findings are consistent with prior literature implicating BLA and vHPC in cue- and context-induced drug-seeking (18, 19) and extend this idea to molecular reprogramming in these brain regions.
Fig. 4. Distinct regional contributions to heroin-priming and context-priming transcriptional regulation associated with relapse.
(A) Rank-rank hypergeometric overlap plots showing coordinated gene expression between SH (saline-heroin) and HH (heroin-heroin) conditions (top row) or HS (heroin-saline) and HH conditions (bottom row) across brain regions. White arrows represent the direction of gene expression changes across the two groups being compared (i.e., lower left quadrant: genes up-regulated in both groups; upper right quadrant: genes down-regulated in both groups). (B) Alluvial plots showing clusters of genes that exhibit a particular pattern of gene expression through SH, HS, and HH conditions (UP, up-regulated; ns, not significant; DOWN, down-regulated). Plots are stratified for each experimental condition (x axis) to show the number of genes (y axis) corresponding to each pattern, as represented by color-coding of each alluvium [see Key, right, showing changes across SH, HS, and HH conditions as UP (↑), ns (−), or DOWN (↓)].
Our experimental design enables profiling of transcriptomic dynamics across what could be considered increasing exposure or severity in OUD: from the first-ever experience (SH), to abstinence from chronic intake (HS), and lastly to relapse (HH). To examine how sets of genes flow through these heroin experiences, we used a pattern analysis approach to group genes based on their expression pattern of being either significantly up-regulated, unchanged, or significantly down-regulated across SH, HS, and HH conditions. Alluvial plots in Fig. 4B (table S4) demonstrate that many clusters of genes take unique paths through heroin exposure conditions and that the magnitude of genes following particular paths shifts across brain regions. All regions have several genes that show consistent and persistent up- or down-regulation across each heroin exposure condition. Notably, very few genes show patterns of inversion of expression, i.e., genes that are up in one condition are typically not down in another condition. Consistent with RRHO analyses, genes in the mPFC, NAc, and dStri that are up- or down-regulated in SH are quiescent in HS, but reactivated again in HH upon re-exposure to heroin. Additionally,, consistent with RRHO analyses, many genes in the vHPC and BLA that are unaffected by an initial exposure to heroin (SH) exhibit consistent up- or down-regulation in HS and HH conditions. Together, these findings suggest that certain groups of genes in the mPFC and NAc, and potentially the dStri, are heavily influenced by the direct effects of heroin, which may become primed or desensitized under relapse-like conditions. On the other hand, genes in the BLA and vHPC are primarily induced after prolonged withdrawal from heroin consumption and may be involved in processes signaling contextual memory associated with drug experience to drive drug-seeking.
Gene priming and desensitization by a distant history of heroin intake
We next examined how heroin exposure and drug-seeking caused shifts in gene priming and desensitization throughout the reward circuitry. Union heatmaps seeded to the HH condition (fig. S2) illustrate that gene expression changes induced by SH, HS, and HH were largely similar in direction across all brain regions. However, hierarchical clustering of these results revealed unique signatures of heroin exposure for each of the brain regions studied (Fig. 4). Notably, this approach found that SH and HH conditions generally show major differences in their patterns of gene expression, thus demonstrating how a distant (30 days) history of volitional heroin consumption changes the pattern of gene induction produced by acute exposure to heroin. We also noted that context-induced drug-seeking (HS) engaged unique gene sets in multiple brain regions, with the exception of the vHPC, which stood out as having a high degree of overlap with the HH condition. These results are consistent with findings from RRHO analysis between HS and HH conditions, further demonstrating a key role for vHPC as a regulator of relapse-related molecular processes.
Many gene clusters were found to enrich for particular GO processes spanning various domains of biological function (Fig. 5 and table S5). The most apparent reward circuit-wide changes were in processes related to ECM maintenance, identified as various subdomains in ECM biology including collagen, keratan sulfate, and general changes to the ECM. Gene clusters that were most enriched for DEGs in one particular condition tended to also enrich for certain biological processes. In the mPFC, genes specifically down-regulated in HH (cluster 6) enrich for keratan sulfate and sulfur transport, which, by contrast with SH, suggests that repeated heroin exposure primes genes related to these processes. In the dStri, genes predominantly up-regulated in SH (cluster 1) enrich for processes related to neurotransmission, which, by contrast with HH, may imply that repeated exposure to heroin diminishes or refines the heroin-induced transcriptional response in this domain. In the VTA, genes down-regulated specifically in HS (cluster 3) enrich for collagen-related processes. In the vHPC, clusters of genes that are up-regulated specifically in HH enrich for processes related to ECM and vasculature maintenance, while clusters down-regulated in HH enrich for processes related to neurotransmission and ion transport. Considering that the ECM is crucial for supporting synaptogenesis, learning, and memory (20), the observation of a balanced up-regulation of ECM functions with down-regulation of synaptic transmission-related processes may imply an ECM-dependent mechanism of neuroplasticity within the vHPC to support relapse. Together, these results suggest that heroin engages unique gene priming mechanisms affecting broad neurobiological domains to promote drug-seeking and relapse.
Fig. 5. Unique patterns of transcriptional regulation across 30-day withdrawal groups implicate unique biological domains affected by heroin exposure history.
Heatmaps show log2 FC for genes across SH (saline-heroin), HS (heroin-saline), and HH (heroin-heroin) conditions. Genes are organized by hierarchical clustering across experimental conditions, with dendrograms cut manually to emphasize clusters. GO analysis reveals enrichment of biological processes for particular clusters of genes showing distinct patterns across heroin exposure conditions (GO term enrichment: ≥4 genes, P < 0.01).
A history of heroin intake shifts upstream regulator control over transcription in a region-dependent manner
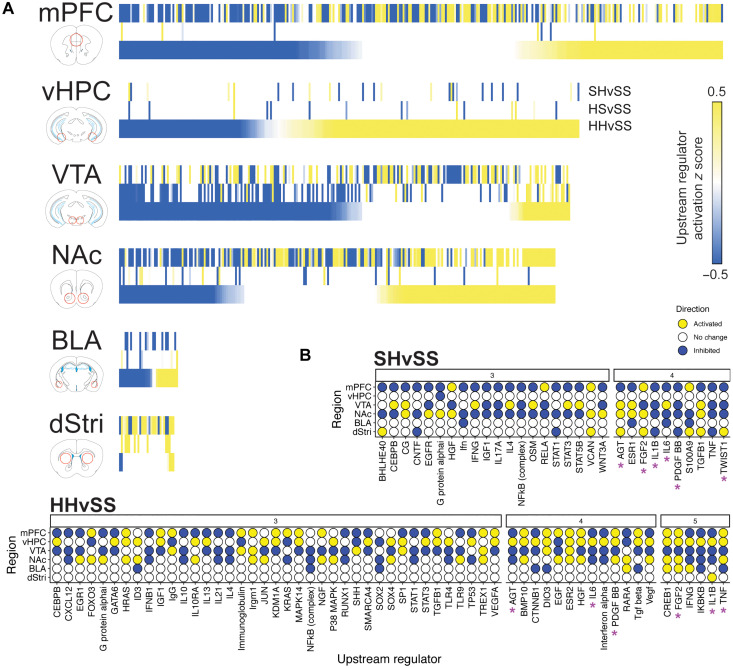
Having identified broad changes to gene expression across heroin exposure conditions, we investigated whether these changes are patterned by particular transcriptional regulatory systems. Using ingenuity pathway analysis (IPA), we deduced whether upstream regulators were significantly activated or inhibited in SH, HS, and HH conditions in the brain regions studied. Many upstream regulators were found to be engaged in a condition-dependent manner across mPFC, NAc, vHPC, and VTA, with fewer total hits for the dStri and BLA (Fig. 6A and table S3). In general, the HS condition elicited comparably few upstream regulators across each brain region, with the exception of the VTA. Union heatmaps showing predicted upstream regulators ranked by activation score in HH (Fig. 6A) demonstrate a clear region-specific pattern of activation and inhibition by heroin exposure conditions. Notably, many upstream regulators were unique to the SH condition, again suggesting that the initial transcriptional response to heroin becomes consolidated with repeated exposure. In the mPFC and NAc, strong overlap was observed between SH and HH in upstream regulators predicted to be inhibited or activated. These findings are consistent with transcriptome-wide analyses above, which demonstrate a heroin-specific response in mPFC and NAc, implicating a critical role for these upstream regulators in mediating drug-priming effects in relapse. On the other hand, the vHPC showed little enrichment of upstream regulators between SH or HS conditions but strong engagement of many upstream regulators in HH. This contrasts the general patterns observed at the DEG level, where overlap is prominent with the HS condition, indicating that transcriptional control within the vHPC is unique to relapse-like conditions. An entirely different pattern was observed in the VTA, where upstream regulators were enriched across all three conditions, but concordance is most evident for HS and HH.
Fig. 6. Upstream regulators coordinate distinct components of relapse-related processes in a region-specific manner.
(A) Heatmaps showing upstream regulators predicted to be inhibited (blue) or activated (yellow) across SH (saline-heroin), HS (heroin-saline), and HH (heroin-heroin) conditions, ranked by activation z score seeded to HH. White indicates upstream regulator not present in experimental condition. (B) Upstream regulators identified to exhibit coordinated engagement across three or more brain regions in SH and HH conditions (none identified in HS). Asterisks signify upstream regulators identified to be engaged in ≥4 brain regions in both SH and HH conditions.
We next examined whether heroin exposure conditions drove coordinated changes to upstream regulators. While the HS condition did not elicit significant upstream regulator activation or inhibition across multiple brain regions, numerous instances were observed for SH and HH. Figure 6B shows upstream regulators engaged across three or more brain regions for SH and HH, respectively. Many upstream regulators were activated or inhibited across up to four brain regions in SH and five brain regions in HH, suggesting a strong, brain-wide coordination of transcriptional responses to heroin regardless of intake history. We noted that between SH and HH, the pattern of activation or inhibition of these multiregion upstream regulators shifts for particular brain regions. For example, the mPFC and NAc shift from mostly inhibited to mostly activated upstream regulators, the vHPC gains numerous upstream regulators, and the dStri loses nearly all upstream regulator engagement. Furthermore, several multiregion upstream regulators were common to both SH and HH, including AGT, FGF2, IL1B, IL6, PDGF-BB, and TNF. Many upstream regulators showing multiregion overlap are commonly inhibited in the mPFC in both SH and HH conditions, implicating common transcriptional responses induced acutely by heroin regardless of a history of heroin intake. Several transcription factors that have previously been implicated in drug addiction, including CREB1 (cAMP response element–binding protein 1), JUN (Jun proto-oncogene, an obligatory binding partner for ∆FOSB), and early growth response 1 (EGR1), are identified in this unbiased dataset as significant upstream regulators of heroin-induced transcriptional regulation (21).
Linking gene expression with addiction-relevant behavioral phenotypes
Thus far, we examined how independent experimental groups reflective of unique OUD conditions influenced transcriptional regulation in brain reward regions. To extend this analysis, we examined how gene expression could predict addiction-relevant behavioral phenotypes and harness individual variability in drug-taking and drug-seeking. Using exploratory factor analysis, we collapsed multidimensional behavioral data collected from animals in all experimental conditions into latent variables (factors) that captured variability across behavioral measures (summarized in Fig. 7A and table S6). Three factors captured the most variability in the dataset and could be considered components of behavioral phenotypes relevant to promoting drug-taking and drug-seeking. Factor 1 was associated with measures related to discrimination of active and inactive lever pressing during self-administration and drug-seeking, and was thus considered a measure of “selectivity” in responding. Factor 2 was associated with total heroin consumed and was thus considered a measure of “intake”. Factor 3 was associated with the number of responses made, particularly during time-outs between periods of drug availability and was thus considered a measure of response “vigor.” To create an individual score for each animal, these factor values were combined into a composite “addiction index” (AI) score [fig. S3A and (13)]. This AI score scaled across factors in that higher levels of each factor contribute to a higher AI score and vice versa (fig. S3B). AI scores also scaled with raw behavioral measures, such that the higher AI scores were generally associated with the higher levels of responding, more lever discrimination, higher time-out responding, and higher drug-seeking (fig. S3C). This approach thus captured aspects of addiction-relevant behavioral phenotypes in a single score for individual animals.
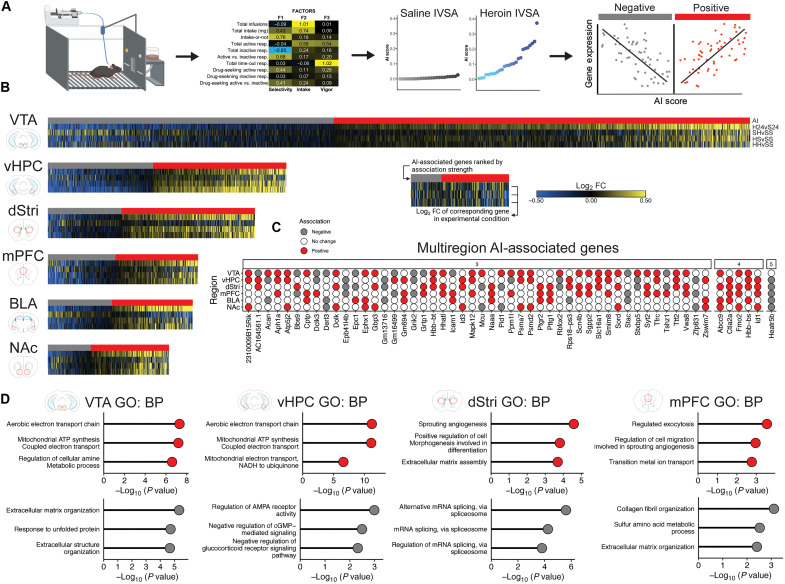
Fig. 7. Genes associated with addiction-relevant behavior enrich for processes related to energetic utilization and extracellular matrix function across multiple regions.
(A) Approach to identify genes associated with addiction-relevant behavioral outcomes of IVSA. Factor analysis was used to collapse all behavioral variables collected during IVSA into three primary latent variables related to selectivity of responding for heroin, total heroin intake, and response vigor. Factors were then combined to generate a single latent variable, termed the AI, for each animal individually, and these scores were higher in animals that self-administered heroin (middle right, blue symbols, shaded by relative AI score) compared to saline (middle right, gray symbols). Linear regression was then used to identify genes in each brain region that were significantly positively or negatively associated with the AI (slope > 20%, nominal P < 0.05). (B) Heatmaps showing AI-associated genes (top row; positive in red, negative in gray) ranked by most negative to most positive, and log2 FC of corresponding gene from DEG analysis from all experimental conditions [subsequent rows; down-regulated in blue, up-regulated in yellow; heroin 24 hours (H24), saline-heroin (SH), heroin-saline (HS), and heroin-heroin (HH)] across all brain regions. (C) Genes identified to be positively (red) or negatively (gray) associated with the AI across ≥3 brain regions, showing strong coordinated patterns throughout the reward circuit. (D) GO analysis on positively associated (red, top graphs) and negatively associated (gray, bottom graphs) AI genes identifies enrichment of biological processes related to extracellular matrix function and metabolic activity across multiple brain regions.
We then used linear regression to link gene expression with AI scores. Notably, because this approach restricts analysis to genes associated with behavior rather than strictly differential expression relative to a control condition, we were now able to examine transcriptional regulation across all primary experimental conditions (H24, SH, HS, and HH). Heatmaps in Fig. 7B (table S7) show numerous genes identified as being positively associated with AI (red; higher AI = higher expression) or negatively associated with AI (gray; higher AI = lower expression) and their respective expression values across each experimental condition in blue (down-regulated versus control) or yellow (up-regulated versus control). In all drug-taking conditions (H24, HS, and HH), positively associated genes are typically up-regulated, while negatively associated genes are typically down-regulated. This suggests that heroin intake drives expression of a set of addiction-relevant genes, which are maintained as primed or desensitized throughout abstinence and reengaged upon drug-seeking. Certain regions also showed differing degrees of coordinated activation or inhibition of addiction-related genes in a condition-specific manner. For example, ongoing intake (H24) engages many AI genes in VTA, BLA, and NAc, which contrasts with relapse-like conditions (HH) causing the strongest engagement of AI genes in the vHPC. Acute exposure (SH), on the other hand, causes a largely distinct pattern of AI-associated gene expression compared to those conditions with a history of heroin intake. However, the ability of SH to engage these genes, rather than having no effect on them, is somewhat unexpected. This suggests that, regardless of a history of heroin intake, initial exposure to heroin engages addiction-relevant gene expression systems throughout the brain reward circuit, which become refined and potentially amplified with increasing exposure.
Threshold-free RRHO analysis found that AI-associated genes were largely independent across brain regions but identified a notable coupling of positive-AI genes between dStri and vHPC, as well as broad concordance between VTA and mPFC (fig. S4). We also identified 54 AI-associated genes that spanned at least three brain regions, which were mostly consistent in being either positively or negatively correlated with the AI (Fig. 7C). Five genes (Hbb-bs, Ctla2a, Id1, Fmo2, and Abcc9) stood out as being positively associated across four brain regions, and one gene (Heatr5b) was negatively associated across five regions. Hbb-bs was also identified as being up-regulated across multiple brain regions in H24 (Fig. 2). In addition, we noted more positive (121) compared to negative (48) AI genes when examining overlaps across brain regions, despite the largely balanced number of positive and negative AI genes identified for each region (Fig. 7B). This may imply a reward circuit-wide coordination of gene expression that promotes addiction-like behavior. GO analysis of AI-associated genes revealed a strong enrichment of positive-AI genes in biological processes related to energy utilization and mitochondrial function (Fig. 7D and table S7). These analyses also identified biological processes related to ECM function enriched in positive-AI genes in the dStri and negative-AI genes in the mPFC and VTA, further implicating ECM dysfunction in OUD-related processes.
Convergence with human RNA-seq and GWAS data reveals conserved addiction signatures and gene targets for treating OUD
We next integrated our heroin IVSA RNA-seq datasets with genome-wide assays from OUD patients to identify commonly regulated genes that may promote addiction (Fig. 8A). First, we compared how gene expression across heroin IVSA conditions in mice compared with human OUD for mPFC and NAc using a recent RNA-seq dataset from postmortem dorsolateral PFC and NAc of OUD patients (15). Patients in these studies had ongoing OUD and died following opioid overdose. Thus, we determined the H24 and HH conditions, which reflect ongoing heroin intake and re-exposure with heroin onboard, respectively, to be the most relevant comparison conditions to identify convergent patterns of gene expression induced by OUD. Pearson correlation analysis in the PFC did not identify a significant association between OUD and H24 (R = 0.029, not significant) but found a slight negative correlation between OUD and HH (R = −0.054, P < 0.01). On the other hand, in the NAc, we found a robust positive association between OUD and H24 (R = 0.216, P < 0.0001) and a negative correlation between OUD and HH (R = −0.199, P < 0.0001). These results are largely consistent with RRHO analyses comparing human and mouse PFC and NAc across all experimental conditions (fig. S5). We then merged significant DEGs from OUD patients and mouse experimental conditions to generate union heatmaps presented in Fig. 8B (table S8). Consistent with correlation analysis, we observed a notable overlap of DEGs between human OUD and H24 conditions in the NAc, while the HH condition showed primarily opposite regulation to the human OUD condition. In contrast, less DEG overlap was observed between mouse and human datasets for the PFC. Using GO analysis, we then examined biological process enrichment in genes identified in significantly correlated conditions. Concordant genes between human and mouse H24 conditions from the NAc enriched for biological processes related to nervous system development, dendrite morphogenesis, protein processing, and lipid transport (all P < 0.001). On the other hand, discordant genes between human and mouse HH conditions in the NAc enriched for lysosome function, protein phosphorylation, transcriptional regulation, and ion transport (all P < 0.001), while in the PFC, these genes enriched for mitogen-activated protein kinase and extracellular signal–regulated kinase signaling, cell adhesion, and RNA processing (all P < 0.001).
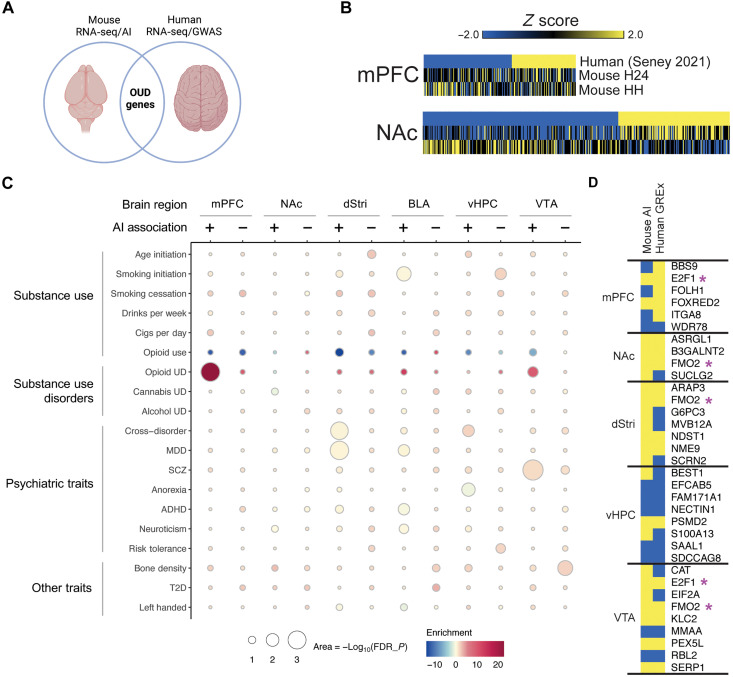
Fig. 8. Convergent molecular drivers of OUD identified through integrated analysis of mouse and human genome-wide data.
(A) Results from the present work were contrasted with available RNA-seq data from postmortem tissue of humans with OUD and with GWAS. (B) Union heatmaps showing gene expression relationships between human OUD patients [top row, down-regulated in blue, up-regulated in yellow; Seney et al. (15)] and two relevant experimental conditions, H24 (heroin 24 hours) and HH (heroin-heroin), for mPFC and NAc. Gene lists were generated by combining common significantly regulated genes (z score > |0.2|, P < 0.05) between human and H24 and human and HH. (C) Association scores between AI genes and GWAS traits related to substance use, substance use disorders, psychiatric traits, and other traits. Columns show brain region, subdivided by negative or positive AI association. (D) Comparison of AI genes and tissue-specific GREx analysis. Overlap of genes identified as significantly (P < 0.05) positively (yellow) or negatively (blue) associated with AI and GREx. Purple asterisks indicate genes that show overlap across multiple brain regions.
While genes identified in association with the AI reflect transcriptional correlates of an addiction-related behavioral phenotype in mice, GWAS identifies genetic variants that associate with human behavioral phenotypes and disorder status at a population scale—results that can be leveraged to identify genomic regions associated with phenotypic risk. Because genetic variation predates lifetime exposures (e.g., drug and stress), GWAS may provide strong causal directionality in the development of addiction. Thus, integration of AI genes and GWAS hits could highlight key gene targets that serve as risk genes for addiction. We first asked whether region-specific AI-associated genes contribute to risk for substance use or other psychiatric phenotypes using partitioned heritability linkage disequilibrium score regression (LDSC) (22). Genes positively and negatively associated with the AI in each brain region were tested for enrichment in single-nucleotide polymorphism (SNP) heritability for each trait using GWAS summary statistics (Fig. 8C and table S9). Overall, we noted much higher enrichment of AI-associated genes in opioid-related conditions compared to other substance use and psychiatric conditions across all brain regions examined. Interestingly, we observed an opposite pattern of enrichment for opioid use GWAS. We also found particularly strong enrichment for genes up-regulated with increased AI (positive association) in the mPFC and genes down-regulated with increased AI (negative association) in the VTA for OUD GWAS. This suggests that AI-associated transcriptional signatures, especially those in mPFC and VTA, are likely to capture causal mechanisms of OUD. Overall, AI gene enrichment was specific to OUD risk as opposed to other substance use and psychiatric disorders. This finding confirms the validity of using animal models of drug intake to investigate mechanisms of addiction and suggests that the molecular signatures captured in this study are primarily unique to OUD.
We then asked whether genes could be identified, which are shared between AI and OUD risk. We translated OUD GWAS [OUD cases versus healthy opioid-exposed controls; (23)] from the single-nucleotide variant level to the gene level using transcriptomic imputation. Specifically, we used Summary-PrediXcan (24) to impute brain region–specific genetically regulated gene expression (GREx) associations for OUD risk using the Genotype-Tissue Expression (GTEx) models (25). These OUD GREx associations (OUD-GREx) reflect baseline transcriptional differences associated with a predisposition for OUD at the functionally translatable level of gene expression for comparison to our AI-associated genes. We found 31 genes that were significant (P < 0.05) in both OUD-GREx and AI associations with brain region specificity—many of which were regulated in the same direction (Fig. 8D). Two genes, FMO2 and E2F1, were identified to be significantly positively associated with OUD-GREx and AI in multiple brain regions. FMO2 (flavin containing dimethylaniline monoxygenase 2) is involved in oxidative metabolism of xenobiotics, which includes heroin (26). Little is known about FMO2 or its related gene partners in addiction, making it an interesting potential brain-wide therapeutic target to blunt drug intake. On the other hand, E2F1 (E2F transcription factor 1) is part of a well-defined family of transcription factors involved in addiction-relevant behavior (27–29). E2F1 has been implicated in coordinating transcriptional responses to fentanyl abstinence (30) and abstinence from morphine self-administration (31) within the NAc. Thus, these genes and others identified represent strong candidates as key players in transcriptional mechanisms driving of OUD vulnerability.
DISCUSSION
These studies combined heroin self-administration in mice, RNA-seq in six brain reward regions, and advanced bioinformatic approaches to generate an atlas of transcriptional regulation relevant to OUD. We identified numerous transcriptomic changes across crucial reward processing centers (mPFC, NAc, dStri, BLA, vHPC, and VTA) driven by initial heroin exposure (SH), ongoing intake (H24), context-induced drug-seeking (HS), and relapse-like conditions (HH). This comprehensive, unbiased assessment of transcriptome-wide changes spanning multiple brain regions and exposure conditions enabled deep characterization of heroin-induced molecular dysfunctions that may promote drug-taking and prime relapse. Integration of addiction-relevant behavioral outcomes to create an AI with transcriptomic data revealed unique roles for brain regions, regulatory factors, and biological processes contributing to heroin intake. Furthermore, comparisons of mouse RNA-seq with that from OUD patients and GWAS results narrow in on key molecular targets with high therapeutic potential for OUD. These results serve as a resource that can be leveraged to develop innumerable avenues of future research into molecular mechanisms and treatment of OUD.
Throughout these studies, we noted that distinct brain regions play partly separable roles in coordinating molecular reprogramming by heroin exposure and heroin-associated contexts. Transcriptome-wide analysis using RRHO identified a strong coordination of gene expression between SH and HH conditions for the mPFC, NAc, and dStri, which was maintained at the level of DEGs. We also noted that SH and HH drive highly overlapping activation or inhibition of upstream regulator function in both mPFC and NAc, which was essentially absent for HS. The SH and HH conditions are similar in that heroin is on board before euthanasia. Coordination of transcriptional responses between these conditions implies a key role for these brain regions in orchestrating the effects of heroin. In addition, genes primed across these two conditions may reflect key molecular mechanisms that undergo cyclical activation or suppression with repeated exposure to heroin and drive consolidation of drug-relevant motivational processes. On the other hand, we found that the BLA and vHPC exhibited a strong coordination of transcriptomic overlap between HS and HH, with particularly strong DEG overlap between conditions for vHPC. These regions did not show much upstream regulator engagement for HS, which could suggest that gene expression related to context-induced drug-seeking is more tightly controlled by fewer upstream regulators, which contrasts with heroin re-exposure. Notably, AI-associated genes in the vHPC exhibit increasing engagement across H24, HS, and HH conditions indicative of a consolidation of gene priming with increasing exposure and relapse conditions. Together, these findings imply a separable compartmentalization of transcriptional regulation induced by heroin exposure itself versus learned associations between heroin experience and heroin exposure context. Cyclical activation of these gene networks may prime or desensitize genes through withdrawal periods, promoting vulnerability to relapse upon re-exposure to drug or drug-associated contexts.
Heroin altered numerous transcriptional regulatory patterns throughout the reward circuitry, but the molecular profile of the VTA stood out as unique. Transcriptome-wide analysis in the VTA identified no obvious bias for gene regulation induced by heroin exposure or context-induced drug-seeking. Compared to other brain regions, overall DEG expression in the VTA was moderate for both up- and down-regulated genes across experimental conditions. However, we found that acute heroin exposure (SH), ongoing intake (H24), context-induced drug-seeking (HS), and relapse-like conditions (HH) all caused a high degree of activation or inhibition of upstream regulator activity. Transcriptomic overlap from RRHO and pattern analysis found little bias between heroin exposure or context-induced seeking but high degree of overlap in upstream regulators engaged by HS and HH. These results suggest that, although heroin does not cause striking DEG activation in the VTA, engagement of these DEGs may be controlled by multiple regulatory mechanisms. In contrast to the moderate level of DEG expression in the VTA, we found this region to have by far the most AI-associated genes. AI genes select for those that may have predictive power in promoting drug-taking and drug-seeking, suggesting that transcriptional changes within the VTA may directly promote ongoing drug-taking. This idea is consistent with findings integrating AI genes with human GWAS results that identified a strong relationship between VTA AI genes and OUD. Furthermore, genes positively associated with the AI were enriched for processes related to metabolic functions and energy utilization. These results are consistent with previous work on the VTA using cocaine, which identified distinct cocaine-induced transcriptional responses depending on exposure paradigm, but a convergent role for disruptions to energetic functions in mediating cocaine effects (32). These results suggest that molecular changes within the VTA may represent a pan-addiction target for disruption of drug intake and potentially drug-seeking.
One major theme uncovered by these studies was heroin-induced dysfunction in processes supporting ECM function throughout the reward circuit, which has been hypothesized to be a key component of substance use disorders (33–36). The ECM of the brain is a complex milieu of proteins, proteoglycans, and polysaccharides that support various processes including cell-cell adhesion, blood–brain barrier integrity, and synaptic plasticity (20). Here, we identified many genes and biological processes either directly or indirectly related to ECM remodeling, spanning collagen biology, chondroitin and keratan sulfate metabolism, and angiogenesis-related processes. Changes in collagen biology were particularly evident for conditions of ongoing heroin intake (H24). Many genes that were found to be differentially regulated across multiple brain regions enriched for biological processes related to ECM functions. These changes were most notable in conditions of ongoing heroin intake and genes associated with drug-taking (AI-associated genes). Considering that the AI is heavily weighted to prior behavior (i.e., before withdrawal), these results may indicate that the strongest ECM changes are state dependent and more specific to ongoing opioid use. These findings are consistent with previous reports that suggest that opioids induce various changes to ECM biology [reviewed in (35)], specifically, recent reports from human OUD patient that identified a key role for heroin-induced changes to ECM biology in promoting opioid use (15). Although ECM changes were most apparent from the AI and H24 conditions in this study, it is plausible that the processes affected by this remodeling may drive functional changes supporting relapse (37). Thus, our findings confirm heroin-induced changes to ECM biology as a key driver of OUD and extend this idea to largely cover the entire reward circuit.
The broad molecular reprogramming by heroin observed in these studies may be directly useful to inform existing knowledge of human OUD and deconstruct relevant patterns of gene expression that promote drug-taking. One approach we took was to examine how our mouse RNA-seq results directly related to recently developed human OUD RNA-seq datasets from the PFC and NAc. In the PFC, we did not observe notable overlap between mouse and human conditions, but this may relate to any number of caveats, the most obvious of which relates to anatomical ambiguity between rodent and human PFC (38), and anatomical isolation of dorsolateral PFC from humans versus infralimbic and prelimbic mPFC from mice. On the other hand, we found strong overlap with the H24 condition in the NAc, but largely opposite regulation in the HH condition when compared to human NAc. The ability of ongoing use to drive gene expression in one direction but invert expression after re-exposure suggests that these genes undergo cyclical patterns of activation and inhibition through periods of abstinence and relapse that affect particular biological processes. We have recently noted a similar pattern of inversion of gene expression with re-exposure after abstinence when comparing RNA-seq results from the NAc across cocaine IVSA in mice and postmortem tissue from humans with cocaine use disorder (39). Together, these studies solidify a crucial role for molecular reprogramming in the NAc to promote ongoing addiction-relevant behavior across drugs and species and suggest that relevant gene expression networks undergo cyclical patterns of restructuring across abstinence and re-exposure conditions. Direct comparisons of gene regulatory networks engaged between periods of drug exposure and abstinence represents a crucial next step in identifying molecular drivers of addiction.
The AI developed in these studies enabled us to identify genes that may have predictive power in their ability to influence patterns of behavior relevant to addiction. This approach is similar to GWAS in humans, which identify risk genes for addiction development. By integrating these two approaches, we identified positive enrichment of AI genes with OUD GWAS hits across all brain regions examined, indicating the utility of this approach in identifying addiction-relevant genes from rodent models. The strongest enrichment was observed for the mPFC and VTA, indicating that these regions may be particularly susceptible to heroin-induced transcriptional regulation to promote addiction. Notably, enrichment of AI genes was largely specific to opioid conditions, further emphasizing that this dimensionality reduction approach effectively refined our analysis to opioid-relevant processes. To identify predictive OUD candidate genes throughout the reward circuitry with this AI approach, we used transcriptomic imputation to convert OUD GWAS to gene-level variants, which revealed brain region–specific GREx associations for OUD risk. This approach revealed 31 genes that were significantly different in both AI and GREx across five brain regions, many of which were regulated in parallel. Two genes, FMO2 and E2F1, were identified to be affected in multiple brain regions, and were consistently positively associated with addiction-relevant outcomes in both AI and GREx across all regions. E2F family transcription factors have recently been implicated in cocaine addiction in rodent models, further highlighting their likely importance in addiction mechanisms (13, 27, 28). Thus, these genes may hold exceptionally high promise as potential therapeutic targets for treating OUD.
The current work uses heroin self-administration in male mice to model OUD and relapse-related processes. Relapse can be modeled in rodents through a number of permutations to experimental design (40). Here, we specifically used a 30-day forced abstinence condition to emulate long-term withdrawal consistent with a time period known to incubate drug craving responses (41). Furthermore, our behavioral readout of relapse-like behavior was under conditions of context-induced heroin-seeking or a combination of context-induced heroin-seeking with experimenter-administered heroin. Future studies could refine this work using variations of our approach, including response extinction procedures, cue-induced drug-seeking, or IVSA following withdrawal. An additional caveat to these studies is the exclusive use of male mice. Because of the complex experimental design and number of animals required, we chose to use males specifically for practical reasons, including our ability to perform heroin self-administration in male mice and to restrict analysis to one sex, thereby reducing variability in molecular analyses. However, sex-specific molecular reprogramming induced by drugs of abuse including opioids are well documented (42–44). It will be crucial to investigate similar transcriptional changes using sex as a biological variable to uncover common as well as unique molecular drivers of OUD.
Together, the present studies identified numerous genes, regulatory systems, and biological processes affected by heroin exposure, intake, seeking, and relapse. Throughout the brain reward circuitry, we uncover networked and compartmentalized transcriptional changes that may drive drug-taking and drug-seeking. Dimensionality reduction approaches to integrate transcriptomic findings and behavioral outcomes refined key addiction-relevant gene expression signatures that promote overlapping functional changes across brain reward regions. Furthermore, we show that these mouse data are directly applicable to the human condition and can be leveraged to isolate convergent targets relevant to OUD. These results thus present a broad landscape of heroin-induced molecular reprogramming relevant to OUD that can be mined to uncover new mechanisms of addiction and potentially inform novel treatment strategies.
MATERIALS AND METHODS
Animals
Adult male C57BL/6J mice (9 to 10 weeks old at the beginning of experimentation) were housed on a 12-hour reverse light-dark cycle (lights on at 19:00) and maintained on ad libitum food and water access, except during pretraining wherein mice were food-restricted as described. Mice were group-housed until catheterization, after which point they were single-housed for the duration of the study. All experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Mount Sinai (protocol number 08-0465).
Heroin self-administration procedure
Pretraining
Mice were maintained on mild, overnight food restriction and first trained to lever press for a food reward in operant boxes (Med Associates, Fairfax, VT). Two levers were presented to mice at the beginning of the session and responding on the active lever led to the delivery of a chocolate pellet (Bio-Serv, Flemington, NJ) according to a fixed-ratio 1 (FR1) schedule of reinforcement. Responding on the inactive lever had no consequence. Sessions lasted for 1 hour or until 30 rewards had been earned. Mice were trained until acquisition criteria for lever pressing had been met (30 pellets earned in two consecutive sessions; three to four sessions required on average). Following acquisition, mice were returned to ad libitum food access for the remainder of the study.
Jugular vein catheterization
Following acquisition of responding for food, mice (N = 72) maintained under inhaled isoflurane anesthesia (2%) received surgery to implant a preconstructed intravenous catheter (Strategic Applications Incorporated, Lake Villa, IL; SBD-05C) in the right jugular vein. Catheter tubing (0.013-inch inner diameter) was threaded subcutaneously over the right shoulder and inserted 1 cm into the jugular vein. The catheter cannula exited the skin from the animal’s mid-back. Ketoprofen (5 mg/kg) was injected subcutaneously upon completion of surgical procedures as postoperative analgesia. Following surgery, mice were single-housed for the duration of the study and given 3 to 4 days of recovery. During recovery, catheters were flushed once daily with 0.03 ml of heparinized saline (30 U) containing ampicillin antibiotic (5 mg/ml) to forestall infection.
Heroin self-administration
In 15 daily 4-hour sessions, responding on the active lever, which previously delivered food, now delivered an intravenous infusion of heroin (0.05 mg/kg per infusion, 24.5-μl infusion volume) on an FR1 schedule of reinforcement. Infusions were signaled by the illumination of a cue light located above the active lever and were followed by a 20-s time-out, during which the cue light remained illuminated and levers remained extended. During time-out, responses on the active lever were recorded but had no consequence. Throughout testing, responses on the inactive lever were recorded but had no consequence. Catheters were flushed with heparinized saline (30 U) before and after self-administration sessions. For the first two sessions, the maximum number of infusions was capped at 60 to minimize overdosing. Thereafter, maximum infusions were capped at 100. Following the 15th self-administration session, saline and heroin self-administering animals were separated into two withdrawal conditions: 24-hour withdrawal (S24 or H24) or extended 30-day homecage withdrawal (i.e., forced abstinence). In the extended withdrawal condition, 30 days after the last self-administration session, mice were further subdivided into two groups receiving a challenge injection of either saline (SS or HS) or heroin subcutaneously at a dose of 1 mg/kg (SH and HH) immediately followed by a drug-seeking test. Mice in all four groups were balanced for baseline heroin intake. Here, mice were placed back into their original self-administration chambers with levers extended for 2 hours. During this test, lever pressing is recorded but does not lead to drug delivery or cue light illumination, thus measuring anticipatory responses for heroin. Animals were run in two cohorts to ensure similar ages at the time of euthanasia between the 24-hour and 30-day withdrawal groups; while the first cohort was undergoing 30-day withdrawal, the second cohort was being tested. Twenty-one mice were removed from the study due to catheter patency loss, poor behavioral outcomes, or death unrelated to behavioral procedures.
RNA-seq and differential expression analysis
Immediately after the drug-seeking test, mice were removed from operant boxes and killed via cervical dislocation. Brains were rapidly extracted on ice, sectioned into 1-mm-thick coronal slices using a brain matrix, and brain punches of regions of interest were collected and flash-frozen on dry ice. Unilateral midline punches were taken from mPFC (12 gauge), and bilateral punches were taken from NAc (14 gauge), dStri (14 gauge), BLA (15 gauge), vHPC (16 gauge), and VTA (16 gauge). One BLA sample was lost during tissue collection.
Total RNA was isolated and purified using the RNeasy Micro Kit (Qiagen, Hilden, Germany). All RNA samples used for RNA-seq had 260/280 values >1.6 (NanoDrop) and RNA integrity number values >8.1 (Tapestation). Library preparation and sequencing were performed by Genewiz/Azenta (Chelmsford, MA). Using a minimum of 200 ng of RNA, sequencing libraries were generated for each sample individually with an Illumina TruSeq Gold kit with ribosomal RNA depletion. Sequencing was conducted on an Illumina Hi-Seq machine with a 2 × 150–base pair paired-end read configuration. All samples (305 in total) were multiplexed and run concurrently to produce 40 million paired-end reads per sample. Raw sequencing reads underwent adapter trimming and were mapped to mm10 (https://ftp.ensembl.org/pub/release-90/fasta/mus_musculus/dna/) using HISAT2. Duplicates were removed using samtools rmdup. Counts of reads mapping to genes were obtained using htseq-count against Ensembl v90 annotation. Quality control was performed using FastQC. Four VTA samples did not meet quality control criteria or were depleted for dopaminergic gene expression (V-261, V-258, V-279, and V-280) and were excluded before differential expression analysis. Normalized reads were filtered to include only the top third of the genome, yielding a minimum base mean of ~12, and a total list of ~17,800 genes. Unless otherwise specified, significance for DEGs was set at 30% expression fold change (FC ± 30%) and P < 0.05. This threshold has been used extensively by our laboratory and others to identify broad patterns of gene expression changes across experimental conditions. Differential expression was performed on this filtered gene list in R version 4.0.2 using the DESeq2 package version 1.28.1 (45), with built-in independent filtering disabled. For differential expression analysis, experimental conditions were always compared to their respective control group (i.e., H24 versus S24 and SH/HS/HH versus SS). DEG lists are available with associated raw data on the Gene Expression Omnibus (GSE228031).
Biotype and cell-type enrichment
For gene biotype analysis, gene lists were filtered across all conditions into five biotype groupings according to the Ensembl database: long noncoding RNA (lncRNA) (combined antisense, bidirectional promoter lncRNA, sense intronic, and processed transcripts), small noncoding RNA (microRNA, miscellaneous RNA, small nuclear RNA, and small nucleolar RNA), pseudogene (processed pseudogene, unprocessed pseudogene, and transcribed processed pseudogene), to be experimentally confirmed, and protein coding. For cell-type enrichment, DEG lists were merged with a comprehensive single-cell RNA-seq database (46), which identified between 850 and 1050 cell type–specific genes for astrocytes, endothelial cells, microglia, neurons, oligodendrocytes, and oligodendrocyte precursor cells in brain.
GO analysis
EnrichR (47) was used to query the 2021 GO:BP database of the Gene Ontology Consortium (48). Criteria for presentation of GO terms are as described. Specific terms presented throughout results are summarized from output lists containing redundancies.
Upstream regulator analysis
Predicted upstream regulators were identified using Ingenuity Pathway Analysis (Qiagen, Fredrick, MD). Genes included in analyses met a criterion of 30% fold change and P < 0.05. Only upstream regulators considered “molecules” (genes and proteins) were examined. Upstream regulators considered for analyses met a Benjamini-Hochberg–corrected P value of P < 0.05 and had a predicted activation score of >0 or inhibition score of <0. Union heatmaps of upstream regulators for each brain region were generated by creating a reference list of all upstream regulators present in each condition and merging each condition with this reference list.
Rank-rank hypergeometric overlap
RRHO provides the ability to compare gene expression profiles of two conditions in a threshold-free manner to identify the degree and significance of overlap (49). RRHOs were generated to compare transcriptomic overlap across DEG lists for two distinct experimental conditions or brain regions (e.g., SH × HH compares DEG lists from SH versus SS against DEG lists from HH versus SS). RRHO plots were generated using the RRHO2 package with default settings (github.com/RRHO2/RRHO2).
Generation of the addiction index
The R package “psych” was used to conduct factor analysis to reduce the dimensions of the behavioral variables in the dataset. All animals with the exception of one outlier (ID no. 23) were included in the analysis. Behavioral measures for IVSA included were infusions, intake, active lever presses, inactive lever presses, time-out responses, and active versus inactive lever presses. For total intake, an additional variable referred to as intake-or-not was included to denote whether the total intake was greater than 0. Behavioral measures included from drug-seeking tests were active lever presses, inactive lever presses, and active versus inactive lever presses. To determine the optimal number of factors, the function “vss” (very simple structure) of the package psych was used. The number of factors was set to 3. To determine the factors, the function “fa” was run with the factoring method set to “minchi.” Factor scores across all three factors for individual animals were estimated using the “Bartlett” method. To compute the AI, factor scores were first transformed to eliminate negative values, which resulted in values ranging from 0 to 1 for each factor. The product of the transformed factors was calculated for each animal to obtain the AI. Voom Limma was used to determine the genes associated with factors (50). Factors 1, 2, and 3 along with the AI were included as a covariate to the Voom Limma regression model to obtain genes associated with the factors and AI.
Pattern analysis: Alluvial plots
To generate alluvial plots, genes were categorized for significance (P < 0.05) in each experimental condition, and patterns of common regulation across conditions were mapped using the ggalluvial package in R, version 0.12.3.
Comparisons with human RNA-seq OUD datasets
Human RNA-seq data from the dorsolateral PFC and NAc were accessed from (15). To compare heroin IVSA gene expression associations with human OUD gene expression associations, genes identified as being significantly up- or down-regulated at P < 0.05 were merged. Union heatmaps were generated across human, mouse H24, and mouse HH conditions to identify convergence or divergence across these conditions. Pearson correlations were calculated to quantify strength of correlation between transcriptional signatures.
Partitioned heritability LDSC
Partitioned heritability LDSC is a modified version of LDSC regression that partitions the heritability of a trait by a set of annotated single SNPs (22). It then tests for enrichment of the annotated SNPs in the heritability of the trait using GWAS summary statistics. We defined gene sets of genes positively and negatively associated with addiction index and annotated SNPs [HapMap3 MAF > 5% SNPs; (51)] within 100 kb of the transcribed region of each gene to derive annotation-specific LD scores using 1000 Genomes EUR ancestry as reference (52). We also derived background annotation-specific LD scores for all SNPs and for all genes captured in our RNA-seq datasets across brain regions. We then partition heritability of six substance use traits, three substance use disorder traits, seven psychiatric traits, and three nonpsychiatric or substance use traits using publicly available GWAS summary statistics with partitioned LD score regression conditioning on background LD scores. For negative control traits, we used left handedness, bone mineral density, and type 2 diabetes (T2D) as nonpsychiatric, nonsubstance use, brain-related (left handedness), and non–brain-related traits (bone density and T2D). Enrichment was calculated as proportion of heritability normalized by proportion of SNPs annotated, and P values were derived from significance testing procedures as implemented in ldsc software and previously described (22).
Transcriptomic imputation of OUD GWAS
Transcriptomic imputation methods use previously calculated associations between genetic variation and gene expression data to impute GREx given an independent dataset of genotype array or genome sequencing data. Summary imputation methods translate GWAS associations for a trait from the SNP-level to tissue-specific gene expression–level associations using summary statistics. We used Summary-PrediXcan (24) to impute gene-level associations of human OUD risk (23) from a GWAS comparing opioid dependence cases to opioid-exposed controls. We used brain region–specific elastic net predictor models of 13 brain regions and whole blood from GTEx data (24, 25, 53) to compute OUD-GREx associations for each brain region. Z scores for OUD-GREx were compared to the AI association z scores to identify overlapping gene expression signatures. GWAS data were accessed from (23, 54–66) and table S9.
Statistical analyses
All behavioral data were plotted as mean ± SEM, and statistical analysis was performed using two-way ANOVAs or unpaired t tests, as described. Morpheus (https://software.broadinstitute.org/morpheus) was used to make heatmaps and perform hierarchical clustering analysis with default settings (one minus Pearson correlation and average linkage method). Gene expression data are plotted on the basis of log2 FC and P value thresholds as indicated throughout. Data visualization was performed using the tidyverse package, version 1.3.1.
Acknowledgments
Funding: This work was supported by funding from the National Science and Research Council of Canada (Postdoctoral Fellowship to C.J.B.) and the NIH (P01DA047233 to E.J.N.).
Author contributions: Conceptualization: C.J.B. and E.J.N. Methodology: E.M.H. and L.M.H. Investigation: C.J.B., R.F., A.M-T., F.J.M.-R., A.G., E.M.P., A.T-B., A.M.C., and P.J.H. Visualization: C.J.B., E.M.H., and A.R. Formal analysis: C.J.B., E.M.H., A.R., and M.E. Resources: Y.L.H. Supervision: D.M.W., L.M.H., Y.L.H., L.S., and E.J.N. Writing: C.J.B. and E.J.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All RNA-seq data reported in this study are deposited publicly in the Gene Expression Omnibus (GSE228031).
Supplementary Materials
This PDF file includes:
Figs. S1 to S5
Legends for supplementary files S1 to S9
Other Supplementary Material for this manuscript includes the following:
Supplementary files S1 to S9
REFERENCES AND NOTES
- 1.F. B. Ahmad, J. A. Cisewski, L. M. Rossen, P. Sutton, Provisional Drug Overdose Death Counts (National Center for Health Statistics, 2023).
- 2.H. Hedegaard, A. M. Miniño, M. R. Spencer, M. Warner, Drug Overdose Deaths in the United States, 1999–2020, NCHS Data Brief, no. 428 (National Center for Health Statistics, Hyattsville, MD, 2021).
- 3.G. Bart, Maintenance medication for opiate addiction: The foundation of recovery. J. Addict. Dis. 31, 207–225 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.B. P. Smyth, J. Barry, E. Keenan, K. Ducray, Lapse and relapse following inpatient treatment of opiate dependence. Ir. Med. J. 103, 176–179 (2010). [PubMed] [Google Scholar]
- 5.C. P. O'Brien, Research advances in the understanding and treatment of addiction. Am. J. Addict. 12, S36–S47 (2003). [PubMed] [Google Scholar]
- 6.G. F. Koob, N. D. Volkow, Neurocircuitry of addiction. Neuropsychopharmacology 35, 217–238 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.T. E. Robinson, K. C. Berridge, The neural basis of drug craving: An incentive-sensitization theory of addiction. Brain Res. Brain Res. Rev. 18, 247–291 (1993). [DOI] [PubMed] [Google Scholar]
- 8.S. E. Hyman, R. C. Malenka, E. J. Nestler, Neural mechanisms of addiction: The role of reward-related learning and memory. Annu. Rev. Neurosci. 29, 565–598 (2006). [DOI] [PubMed] [Google Scholar]
- 9.C. J. Browne, A. Godino, M. Salery, E. J. Nestler, Epigenetic mechanisms of opioid addiction. Biol. Psychiatry 87, 22–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E. H. Jacobs, S. Spijker, C. W. Verhoog, K. Kamprath, T. J. Vries, A. B. Smit, A. N. M. Schoffelmeer, Active heroin administration induces specific genomic responses in the nucleus accumbens shell. FASEB J. 16, 1961–1963 (2002). [DOI] [PubMed] [Google Scholar]
- 11.C. L. Pickens, M. Airavaara, F. Theberge, S. Fanous, B. T. Hope, Y. Shaham, Neurobiology of the incubation of drug craving. Trends Neurosci. 34, 411–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.U. Shalev, M. Morales, B. Hope, J. Yap, Y. Shaham, Time-dependent changes in extinction behavior and stress-induced reinstatement of drug seeking following withdrawal from heroin in rats. Psychopharmacology (Berl) 156, 98–107 (2001). [DOI] [PubMed] [Google Scholar]
- 13.D. M. Walker, H. M. Cates, Y. H. E. Loh, I. Purushothaman, A. Ramakrishnan, K. M. Cahill, C. K. Lardner, A. Godino, H. G. Kronman, J. Rabkin, Z. S. Lorsch, P. Mews, M. A. Doyle, J. Feng, B. Labonté, J. W. Koo, R. C. Bagot, R. W. Logan, M. L. Seney, E. S. Calipari, L. Shen, E. J. Nestler, Cocaine self-administration alters transcriptome-wide responses in the brain's reward circuitry. Biol. Psychiatry 84, 867–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.W. Davies, A. Isles, R. Smith, D. Karunadasa, D. Burrmann, T. Humby, O. Ojarikre, C. Biggin, D. Skuse, P. Burgoyne, L. Wilkinson, Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat. Genet. 37, 625–629 (2005). [DOI] [PubMed] [Google Scholar]
- 15.M. L. Seney, S. M. Kim, J. R. Glausier, M. A. Hildebrand, X. Xue, W. Zong, J. Wang, M. A. Shelton, B. D. N. Phan, C. Srinivasan, A. R. Pfenning, G. C. Tseng, D. A. Lewis, Z. Freyberg, R. W. Logan, Transcriptional alterations in dorsolateral prefrontal cortex and nucleus accumbens implicate neuroinflammation and synaptic remodeling in opioid use disorder. Biol. Psychiatry 90, 550–562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Schönherr, H. J. Hausser, Extracellular matrix and cytokines: A functional unit. Dev. Immunol. 7, 89–101 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Wiranowska, A. Plaas, Cytokines and extracellular matrix remodeling in the central nervous system. Neuroimmune Biol. 6, 167–197 (2008). [Google Scholar]
- 18.R. E. See, R. A. Fuchs, C. C. Ledford, J. McLaughlin, Drug addiction, relapse, and the amygdala. Ann. N. Y. Acad. Sci. 985, 294–307 (2003). [DOI] [PubMed] [Google Scholar]
- 19.J. M. Bossert, A. L. Stern, Role of ventral subiculum in context-induced reinstatement of heroin seeking in rats. Addict. Biol. 19, 338–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Dityatev, M. Schachner, P. Sonderegger, The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat. Rev. Neurosci. 11, 735–746 (2010). [DOI] [PubMed] [Google Scholar]
- 21.C. D. Teague, E. J. Nestler, Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol. Psychiatry 27, 687–709 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.H. K. Finucane, B. Bulik-Sullivan, A. Gusev, G. Trynka, Y. Reshef, P.-R. Loh, V. Anttila, H. Xu, C. Zang, K. Farh, S. Ripke, F. R. Day; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium, S. Purcell, E. Stahl, S. Lindstrom, J. R. B. Perry, Y. Okada, S. Raychaudhuri, M. J. Daly, N. Patterson, B. M. Neale, A. L. Price, Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. Polimanti, R. K. Walters, E. C. Johnson, J. N. McClintick, A. E. Adkins, D. E. Adkins, S. A. Bacanu, L. J. Bierut, T. B. Bigdeli, S. Brown, K. K. Bucholz, W. E. Copeland, E. J. Costello, L. Degenhardt, L. A. Farrer, T. M. Foroud, L. Fox, A. M. Goate, R. Grucza, L. M. Hack, D. B. Hancock, S. M. Hartz, A. C. Heath, J. K. Hewitt, C. J. Hopfer, E. O. Johnson, K. S. Kendler, H. R. Kranzler, K. Krauter, D. Lai, P. A. F. Madden, N. G. Martin, H. H. Maes, E. C. Nelson, R. E. Peterson, B. Porjesz, B. P. Riley, N. Saccone, M. Stallings, T. L. Wall, B. T. Webb, L. Wetherill; Psychiatric Genomics Consortium Substance Use Disorders Workgroup, H. J. Edenberg, A. Agrawal, J. Gelernter, Leveraging genome-wide data to investigate differences between opioid use versus opioid dependence in 41,176 individuals from the Psychiatric Genomics Consortium. Mol. Psychiatry 25, 1673–1687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. N. Barbeira, S. P. Dickinson, R. Bonazzola, J. Zheng, H. E. Wheeler, J. M. Torres, E. S. Torstenson, K. P. Shah, T. Garcia, T. L. Edwards, E. A. Stahl, L. M. Huckins; GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group, F. Aguet, K. G. Ardlie, B. B. Cummings, E. T. Gelfand, G. Getz, K. Hadley, R. E. Handsaker, K. H. Huang, S. Kashin, K. J. Karczewski, M. Lek, X. Li, D. G. MacArthur, J. L. Nedzel, D. T. Nguyen, M. S. Noble, A. V. Segrè, C. A. Trowbridge, T. Tukiainen; Statistical Methods groups—Analysis Working Group, N. S. Abell, B. Balliu, R. Barshir, O. Basha, A. Battle, G. K. Bogu, A. Brown, C. D. Brown, S. E. Castel, L. S. Chen, C. Chiang, D. F. Conrad, F. N. Damani, J. R. Davis, O. Delaneau, E. T. Dermitzakis, B. E. Engelhardt, E. Eskin, P. G. Ferreira, L. Frésard, E. R. Gamazon, D. Garrido-Martín, A. D. H. Gewirtz, G. Gliner, M. J. Gloudemans, R. Guigo, I. M. Hall, B. Han, Y. He, F. Hormozdiari, C. Howald, B. Jo, E. Y. Kang, Y. Kim, S. Kim-Hellmuth, T. Lappalainen, G. Li, X. Li, B. Liu, S. Mangul, M. I. McCarthy, I. C. McDowell, P. Mohammadi, J. Monlong, S. B. Montgomery, M. Muñoz-Aguirre, A. W. Ndungu, A. B. Nobel, M. Oliva, H. Ongen, J. J. Palowitch, N. Panousis, P. Papasaikas, Y. S. Park, P. Parsana, A. J. Payne, C. B. Peterson, J. Quan, F. Reverter, C. Sabatti, A. Saha, M. Sammeth, A. J. Scott, A. A. Shabalin, R. Sodaei, M. Stephens, B. E. Stranger, B. J. Strober, J. H. Sul, E. K. Tsang, S. Urbut, M. van de Bunt, G. Wang, X. Wen, F. A. Wright, H. S. Xi, E. Yeger-Lotem, Z. Zappala, J. B. Zaugg, Y. H. Zhou; Enhancing GTEx (eGTEx) groups, J. M. Akey, D. Bates, J. Chan, L. S. Chen, M. Claussnitzer, K. Demanelis, M. Diegel, J. A. Doherty, A. P. Feinberg, M. S. Fernando, J. Halow, K. D. Hansen, E. Haugen, P. F. Hickey, L. Hou, F. Jasmine, R. Jian, L. Jiang, A. Johnson, R. Kaul, M. Kellis, M. G. Kibriya, K. Lee, J. B. Li, Q. Li, X. Li, J. Lin, S. Lin, S. Linder, C. Linke, Y. Liu, M. T. Maurano, B. Molinie, S. B. Montgomery, J. Nelson, F. J. Neri, M. Oliva, Y. Park, B. L. Pierce, N. J. Rinaldi, L. F. Rizzardi, R. Sandstrom, A. Skol, K. S. Smith, M. P. Snyder, J. Stamatoyannopoulos, B. E. Stranger, H. Tang, E. K. Tsang, L. Wang, M. Wang, N. van Wittenberghe, F. Wu, R. Zhang; NIH Common Fund, C. R. Nierras; NIH/NCI, P. A. Branton, L. J. Carithers, P. Guan, H. M. Moore, A. Rao, J. B. Vaught; NIH/NHGrI, S. E. Gould, N. C. Lockart, C. Martin, J. P. Struewing, S. Volpi; NIH/NIMH, A. M. Addington, S. E. Koester; NIH/NIDA, A. R. Little; Biospecimen Collection Source Site—NDrI, L. E. Brigham, R. Hasz, M. Hunter, C. Johns, M. Johnson, G. Kopen, W. F. Leinweber, J. T. Lonsdale, A. McDonald, B. Mestichelli, K. Myer, B. Roe, M. Salvatore, S. Shad, J. A. Thomas, G. Walters, M. Washington, J. Wheeler; Biospecimen Collection Source Site—rPCI, J. Bridge, B. A. Foster, B. M. Gillard, E. Karasik, R. Kumar, M. Miklos, M. T. Moser; Biospecimen Core resource—VArI, S. D. Jewell, R. G. Montroy, D. C. Rohrer, D. R. Valley; Brain Bank repository—University of Miami Brain Endowment Bank, D. A. Davis, D. C. Mash; Leidos Biomedical—Project Management, A. H. Undale, A. M. Smith, D. E. Tabor, N. V. Roche, J. A. McLean, N. Vatanian, K. L. Robinson, L. Sobin, M. E. Barcus, K. M. Valentino, L. Qi, S. Hunter, P. Hariharan, S. Singh, K. S. Um, T. Matose, M. M. Tomaszewski; ELSI Study, L. K. Barker, M. Mosavel, L. A. Siminoff, H. M. Traino; Genome Browser Data Integration & Visualization—EBI, P. Flicek, T. Juettemann, M. Ruffier, D. Sheppard, K. Taylor, S. J. Trevanion, D. R. Zerbino; Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz, B. Craft, M. Goldman, M. Haeussler, W. J. Kent, C. M. Lee, B. Paten, K. R. Rosenbloom, J. Vivian, J. Zhu, D. L. Nicolae, N. J. Cox, H. K. Im, Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825–1820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GTEx Consortium , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. K. Krueger, D. E. Williams, Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 106, 357–387 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.H. M. Cates, R. C. Bagot, E. A. Heller, I. Purushothaman, C. K. Lardner, D. M. Walker, C. J. Peña, R. L. Neve, L. Shen, E. J. Nestler, A novel role for E2F3b in regulating cocaine action in the prefrontal cortex. Neuropsychopharmacology 44, 776–784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.H. M. Cates, E. A. Heller, C. K. Lardner, I. Purushothaman, C. J. Peña, D. M. Walker, M. E. Cahill, R. L. Neve, L. Shen, R. C. Bagot, E. J. Nestler, Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol. Psychiatry 84, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Feng, M. Wilkinson, X. Liu, I. Purushothaman, D. Ferguson, V. Vialou, I. Maze, N. Shao, P. Kennedy, J. W. Koo, C. Dias, B. Laitman, V. Stockman, Q. LaPlant, M. E. Cahill, E. J. Nestler, L. Shen, Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15, R65–R18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. E. Fox, A. B. Wulff, D. Franco, E. Y. Choi, C. A. Calarco, M. Engeln, M. D. Turner, R. Chandra, V. M. Rhodes, S. M. Thompson, S. A. Ament, M. K. Lobo, Adaptations in nucleus accumbens neuron subtypes mediate negative affective behaviors in fentanyl abstinence. Biol. Psychiatry 93, 489–501 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.H. L. Mayberry, C. C. Bavley, R. Karbalaei, D. R. Peterson, A. R. Bongiovanni, A. S. Ellis, S. H. Downey, A. B. Toussaint, M. E. Wimmer, Transcriptomics in the nucleus accumbens shell reveal sex- and reinforcer-specific signatures associated with morphine and sucrose craving. Neuropsychopharmacology 47, 1764–1775 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R. R. Campbell, S. Chen, J. H. Beardwood, A. J. López, L. V. Pham, A. M. Keiser, J. E. Childs, D. P. Matheos, V. Swarup, P. Baldi, M. A. Wood, Cocaine induces paradigm-specific changes to the transcriptome within the ventral tegmental area. Neuropsychopharmacology 46, 1768–1779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. C. W. Smith, M. D. Scofield, P. W. Kalivas, The tetrapartite synapse: Extracellular matrix remodeling contributes to corticoaccumbens plasticity underlying drug addiction. Brain Res. 1628, 29–39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Ferrer-Ferrer, A. Dityatev, Shaping synapses by the neural extracellular matrix. Front. Neuroanat. 12, 40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. H. Ray, B. R. Williams, M. K. Kuppe, C. D. Bryant, R. W. Logan, A glitch in the matrix: The role of extracellular matrix remodeling in opioid use disorder. Front. Integr. Neurosci. 16, 899637 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Kruyer, V. C. Chioma, P. W. Kalivas, The opioid-addicted tetrapartite synapse. Biol. Psychiatry 87, 34–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.V. C. Chioma, A. Kruyer, A. C. Bobadilla, A. Angelis, Z. Ellison, R. Hodebourg, M. D. Scofield, P. W. Kalivas, Heroin seeking and extinction from seeking activate matrix metalloproteinases at synapses on distinct subpopulations of accumbens cells. Biol. Psychiatry 89, 947–958 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Laubach, L. M. Amarante, K. Swanson, S. R. White, What, if anything, is rodent rrefrontal cortex? eNeuro 5, ENEURO.0315-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.P. Mews, A. M. Cunningham, J. Scarpa, A. Ramakrishnan, E. M. Hicks, S. Bolnick, S. Garamszegi, L. Shen, D. C. Mash, E. J. Nestler, Convergent abnormalities in striatal gene networks in human cocaine use disorder and mouse cocaine administration models. Sci. Adv. 9, eadd8946 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Venniro, M. L. Banks, M. Heilig, D. H. Epstein, Y. Shaham, Improving translation of animal models of addiction and relapse by reverse translation. Nat. Rev. Neurosci. 21, 625–643 (2020). [DOI] [PubMed] [Google Scholar]
- 41.J. W. Grimm, B. T. Hope, R. A. Wise, Y. Shaham, Neuroadaptation. Incubation of cocaine craving after withdrawal. Nature 412, 141–142 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. J. López, A. R. Johnson, T. J. Euston, R. Wilson, S. O. Nolan, L. J. Brady, K. C. Thibeault, S. J. Kelly, V. Kondev, P. Melugin, M. G. Kutlu, E. Chuang, T. K. T. Lam, D. D. Kiraly, E. S. Calipari, Cocaine self-administration induces sex-dependent protein expression in the nucleus accumbens. Commun. Biol. 4, 883–813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E. A. Townsend, R. K. Kim, H. L. Robinson, S. A. Marsh, M. L. Banks, P. J. Hamilton, Opioid withdrawal produces sex-specific effects on fentanyl-versus-food choice and mesolimbic transcription. Biol. Psychiatry Glob. Open Sci. 1, 112–122 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. M. Walker, X. Zhou, A. M. Cunningham, A. P. Lipschultz, A. Ramakrishnan, H. M. Cates, R. C. Bagot, L. Shen, B. Zhang, E. J. Nestler, Sex-specific transcriptional changes in response to adolescent social stress in the brain's reward circuitry. Biol. Psychiatry 91, 118–128 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. I. Love, W. Huber, S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.A. T. McKenzie, M. Wang, M. E. Hauberg, J. F. Fullard, A. Kozlenkov, A. Keenan, Y. L. Hurd, S. Dracheva, P. Casaccia, P. Roussos, B. Zhang, Brain cell type specific gene expression and co-expression network architectures. Sci. Rep. 8, 8868–8819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Z. Xie, A. Bailey, M. V. Kuleshov, D. J. B. Clarke, J. E. Evangelista, S. L. Jenkins, A. Lachmann, M. L. Wojciechowicz, E. Kropiwnicki, K. M. Jagodnik, M. Jeon, A. Ma’ayan, Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gene Ontology Consortium , The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.S. B. Plaisier, R. Taschereau, J. A. Wong, T. G. Graeber, Rank-rank hypergeometric overlap: Identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. W. Law, Y. Chen, W. Shi, G. K. Smyth, voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.International HapMap 3 Consortium , Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.1000 Genomes Project Consortium , An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.E. R. Gamazon, H. E. Wheeler, K. P. Shah, S. V. Mozaffari, K. Aquino-Michaels, R. J. Carroll, A. E. Eyler, J. C. Denny; GTEx Consortium, D. L. Nicolae, N. J. Cox, H. K. Im, A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Liu, Y. Jiang, R. Wedow, Y. Li, D. M. Brazel, F. Chen, G. Datta, J. Davila-Velderrain, D. M. Guire, C. Tian, X. Zhan; 23andMe Research Team; HUNT All-In Psychiatry, H. Choquet, A. R. Docherty, J. D. Faul, J. R. Foerster, L. G. Fritsche, M. E. Gabrielsen, S. D. Gordon, J. Haessler, J.-J. Hottenga, H. Huang, S.-K. Jang, P. R. Jansen, Y. Ling, R. Mägi, N. Matoba, G. M. Mahon, A. Mulas, V. Orrù, T. Palviainen, A. Pandit, G. W. Reginsson, A. H. Skogholt, J. A. Smith, A. E. Taylor, C. Turman, G. Willemsen, H. Young, K. A. Young, G. J. M. Zajac, W. Zhao, W. Zhou, G. Bjornsdottir, J. D. Boardman, M. Boehnke, D. I. Boomsma, C. Chen, F. Cucca, G. E. Davies, C. B. Eaton, M. A. Ehringer, T. Esko, E. Fiorillo, N. A. Gillespie, D. F. Gudbjartsson, T. Haller, K. M. Harris, A. C. Heath, J. K. Hewitt, I. B. Hickie, J. E. Hokanson, C. J. Hopfer, D. J. Hunter, W. G. Iacono, E. O. Johnson, Y. Kamatani, S. L. R. Kardia, M. C. Keller, M. Kellis, C. Kooperberg, P. Kraft, K. S. Krauter, M. Laakso, P. A. Lind, A. Loukola, S. M. Lutz, P. A. F. Madden, N. G. Martin, M. M. Gue, M. B. Mc Queen, S. E. Medland, A. Metspalu, K. L. Mohlke, J. B. Nielsen, Y. Okada, U. Peters, T. J. C. Polderman, D. Posthuma, A. P. Reiner, J. P. Rice, E. Rimm, R. J. Rose, V. Runarsdottir, M. C. Stallings, A. Staňáková, H. Stefansson, K. K. Thai, H. A. Tindle, T. Tyrfingsson, T. L. Wall, D. R. Weir, C. Weisner, J. B. Whitfield, B. S. Winsvold, J. Yin, L. Zuccolo, L. J. Bierut, K. Hveem, J. J. Lee, M. R. Munafò, N. L. Saccone, C. J. Willer, M. C. Cornelis, S. P. David, D. A. Hinds, E. Jorgenson, J. Kaprio, J. A. Stitzel, K. Stefansson, T. E. Thorgeirsson, G. Abecasis, D. J. Liu, S. Vrieze, Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.E. C. Johnson, D. Demontis, T. E. Thorgeirsson, R. K. Walters, R. Polimanti, A. S. Hatoum, S. Sanchez-Roige, S. E. Paul, F. R. Wendt, T. K. Clarke, D. Lai, G. W. Reginsson, H. Zhou, J. He, D. A. A. Baranger, D. F. Gudbjartsson, R. Wedow, D. E. Adkins, A. E. Adkins, J. Alexander, S. A. Bacanu, T. B. Bigdeli, J. Boden, S. A. Brown, K. K. Bucholz, J. Bybjerg-Grauholm, R. P. Corley, L. Degenhardt, D. M. Dick, B. W. Domingue, L. Fox, A. M. Goate, S. D. Gordon, L. M. Hack, D. B. Hancock, S. M. Hartz, I. B. Hickie, D. M. Hougaard, K. Krauter, P. A. Lind, J. N. McClintick, M. B. McQueen, J. L. Meyers, G. W. Montgomery, O. Mors, P. B. Mortensen, M. Nordentoft, J. F. Pearson, R. E. Peterson, M. D. Reynolds, J. P. Rice, V. Runarsdottir, N. L. Saccone, R. Sherva, J. L. Silberg, R. E. Tarter, T. Tyrfingsson, T. L. Wall, B. T. Webb, T. Werge, L. Wetherill, M. J. Wright, S. Zellers, M. J. Adams, L. J. Bierut, J. D. Boardman, W. E. Copeland, L. A. Farrer, T. M. Foroud, N. A. Gillespie, R. A. Grucza, K. M. Harris, A. C. Heath, V. Hesselbrock, J. K. Hewitt, C. J. Hopfer, J. Horwood, W. G. Iacono, E. O. Johnson, K. S. Kendler, M. A. Kennedy, H. R. Kranzler, P. A. F. Madden, H. H. Maes, B. S. Maher, N. G. Martin, M. McGue, A. M. McIntosh, S. E. Medland, E. C. Nelson, B. Porjesz, B. P. Riley, M. C. Stallings, M. M. Vanyukov, S. Vrieze, L. K. Davis, R. Bogdan, J. Gelernter, H. J. Edenberg, K. Stefansson, A. D. Børglum, A. Agrawal, R. Walters, R. Polimanti, E. Johnson, J. McClintick, A. Hatoum, J. He, F. Wendt, H. Zhou, M. Adams, A. Adkins, F. Aliev, S. A. Bacanu, A. Batzler, S. Bertelsen, J. Biernacka, T. Bigdeli, L. S. Chen, T. K. Clarke, Y. L. Chou, F. Degenhardt, A. Docherty, A. Edwards, P. Fontanillas, J. Foo, L. Fox, J. Frank, I. Giegling, S. Gordon, L. Hack, A. Hartmann, S. Hartz, S. Heilmann-Heimbach, S. Herms, C. Hodgkinson, P. Hoffman, J. Hottenga, M. Kennedy, M. Alanne-Kinnunen, B. Konte, J. Lahti, M. Lahti-Pulkkinen, D. Lai, L. Ligthart, A. Loukola, B. Maher, H. Mbarek, A. McIntosh, M. McQueen, J. Meyers, Y. Milaneschi, T. Palviainen, J. Pearson, R. Peterson, S. Ripatti, E. Ryu, N. Saccone, J. Salvatore, S. Sanchez-Roige, M. Schwandt, R. Sherva, F. Streit, J. Strohmaier, N. Thomas, J. C. Wang, B. Webb, R. Wedow, L. Wetherill, A. Wills, J. Boardman, D. Chen, D. S. Choi, W. Copeland, R. Culverhouse, N. Dahmen, L. Degenhardt, B. Domingue, S. Elson, M. Frye, W. Gäbel, C. Hayward, M. Ising, M. Keyes, F. Kiefer, J. Kramer, S. Kuperman, S. Lucae, M. Lynskey, W. Maier, K. Mann, S. Männistö, B. Müller-Myhsok, A. Murray, J. Nurnberger, A. Palotie, U. Preuss, K. Räikkönen, M. Reynolds, M. Ridinger, N. Scherbaum, M. Schuckit, M. Soyka, J. Treutlein, S. Witt, N. Wodarz, P. Zill, D. Adkins, J. Boden, D. Boomsma, L. Bierut, S. Brown, K. Bucholz, S. Cichon, E. J. Costello, H. de Wit, N. Diazgranados, D. Dick, J. Eriksson, L. Farrer, T. Foroud, N. Gillespie, A. Goate, D. Goldman, R. Grucza, D. Hancock, K. M. Harris, A. Heath, V. Hesselbrock, J. Hewitt, C. Hopfer, J. Horwood, W. Iacono, E. Johnson, J. Kaprio, V. Karpyak, K. Kendler, H. Kranzler, K. Krauter, P. Lichtenstein, P. Lind, M. McGue, J. MacKillop, P. Madden, H. Maes, P. Magnusson, N. Martin, S. Medland, G. Montgomery, E. Nelson, M. Nöthen, A. Palmer, N. Pederson, B. Penninx, B. Porjesz, J. Rice, M. Rietschel, B. Riley, R. Rose, D. Rujescu, P. H. Shen, J. Silberg, M. Stallings, R. Tarter, M. Vanyukov, S. Vrieze, T. Wall, J. Whitfield, H. Zhao, B. Neale, J. Gelernter, H. Edenberg, A. Agrawal, A large-scale genome-wide association study meta-analysis of cannabis use disorder. Lancet Psychiatry 7, 1032–1045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Sanchez-Roige, A. A. Palmer, P. Fontanillas, S. L. Elson; 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, M. J. Adams, D. M. Howard, H. J. Edenberg, G. Davies, R. C. Crist, I. J. Deary, A. M. McIntosh, T. K. Clarke, Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. Am. J. Psychiatry 176, 107–118 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D. Demontis, R. K. Walters, J. Martin, M. Mattheisen, T. D. Als, E. Agerbo, G. Baldursson, R. Belliveau, J. Bybjerg-Grauholm, M. Bækvad-Hansen, F. Cerrato, K. Chambert, C. Churchhouse, A. Dumont, N. Eriksson, M. Gandal, J. I. Goldstein, K. L. Grasby, J. Grove, O. O. Gudmundsson, C. S. Hansen, M. E. Hauberg, M. V. Hollegaard, D. P. Howrigan, H. Huang, J. B. Maller, A. R. Martin, N. G. Martin, J. Moran, J. Pallesen, D. S. Palmer, C. B. Pedersen, M. G. Pedersen, T. Poterba, J. B. Poulsen, S. Ripke, E. B. Robinson, F. K. Satterstrom, H. Stefansson, C. Stevens, P. Turley, G. B. Walters, H. Won, M. J. Wright; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team, O. A. Andreassen, P. Asherson, C. L. Burton, D. I. Boomsma, B. Cormand, S. Dalsgaard, B. Franke, J. Gelernter, D. Geschwind, H. Hakonarson, J. Haavik, H. R. Kranzler, J. Kuntsi, K. Langley, K.-P. Lesch, C. Middeldorp, A. Reif, L. A. Rohde, P. Roussos, R. Schachar, P. Sklar, E. J. S. Sonuga-Barke, P. F. Sullivan, A. Thapar, J. Y. Tung, I. D. Waldman, S. E. Medland, K. Stefansson, M. Nordentoft, D. M. Hougaard, T. Werge, O. Mors, P. B. Mortensen, M. J. Daly, S. V. Faraone, A. D. Børglum, B. M. Neale, Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.L. Duncan, Z. Yilmaz, H. Gaspar, R. Walters, J. Goldstein, V. Anttila, B. Bulik-Sullivan, S. Ripke; Eating Disorders Working Group of the Psychiatric Genomics Consortium, L. Thornton, A. Hinney, M. Daly, P. F. Sullivan, E. Zeggini, G. Breen, C. M. Bulik, L. Duncan, Z. Yilmaz, H. Gaspar, R. Walters, J. Goldstein, V. Anttila, B. Bulik-Sullivan, S. Ripke, R. Adan, L. Alfredsson, T. Ando, O. Andreassen, H. Aschauer, J. Baker, J. Barrett, V. Bencko, A. Bergen, W. Berrettini, A. Birgegård, C. Boni, V. B. Perica, H. Brandt, R. Burghardt, L. Carlberg, M. Cassina, C. Cesta, S. Cichon, M. Clementi, S. Cohen-Woods, J. Coleman, R. Cone, P. Courtet, S. Crawford, S. Crow, J. Crowley, U. Danner, O. Davis, M. de Zwaan, G. Dedoussis, D. Degortes, J. DeSocio, D. Dick, D. Dikeos, C. Dina, B. Ding, M. Dmitrzak-Weglarz, E. Docampo, K. Egberts, S. Ehrlich, G. Escaramís, T. Esko, T. Espeseth, X. Estivill, A. Favaro, F. Fernández-Aranda, M. Fichter, C. Finan, K. Fischer, J. Floyd, M. Föcker, L. Foretova, M. Forzan, C. Fox, C. Franklin, V. Gaborieau, S. Gallinger, G. Gambaro, I. Giegling, F. Gonidakis, P. Gorwood, M. Gratacos, S. Guillaume, Y. Guo, H. Hakonarson, K. Halmi, R. Harrison, K. Hatzikotoulas, J. Hauser, J. Hebebrand, S. Helder, J. Hendriks, S. Herms, B. Herpertz-Dahlmann, W. Herzog, C. Hilliard, L. Huckins, J. Hudson, J. Huemer, H. Imgart, H. Inoko, S. Jall, S. Jamain, V. Janout, S. Jiménez-Murcia, C. Johnson, J. Jordan, A. Julià, A. Juréus, G. Kalsi, A. Kaplan, J. Kaprio, L. Karhunen, A. Karwautz, M. Kas, W. Kaye, M. Kennedy, J. Kennedy, A. Keski-Rahkonen, K. Kiezebrink, Y. R. Kim, L. Klareskog, K. Klump, G. P. Knudsen, B. Koeleman, D. Koubek, M. la Via, M. Landén, S. le Hellard, M. Leboyer, R. Levitan, D. Li, P. Lichtenstein, L. Lilenfeld, J. Lissowska, A. Lundervold, P. Magistretti, M. Maj, K. Mannik, S. Marsal, D. Kaminska, N. Martin, M. Mattingsdal, S. McDevitt, P. McGuffin, E. Merl, A. Metspalu, I. Meulenbelt, N. Micali, J. Mitchell, K. Mitchell, P. Monteleone, A. M. Monteleone, G. Montgomery, P. Mortensen, M. Munn-Chernoff, T. Müller, B. Nacmias, M. Navratilova, I. Nilsson, C. Norring, I. Ntalla, R. Ophoff, J. O’Toole, A. Palotie, J. Pantel, H. Papezova, R. Parker, D. Pinto, R. Rabionet, A. Raevuori, A. Rajewski, N. Ramoz, N. W. Rayner, T. Reichborn-Kjennerud, V. Ricca, S. Ripatti, F. Ritschel, M. Roberts, A. Rotondo, D. Rujescu, F. Rybakowski, P. Santonastaso, A. Scherag, S. Scherer, U. Schmidt, N. Schork, A. Schosser, L. Scott, J. Seitz, L. Slachtova, R. Sladek, P. E. Slagboom, M. S. O. ’t Landt, A. Slopien, T. Smith, N. Soranzo, S. Sorbi, L. Southam, V. Steen, E. Strengman, M. Strober, J. Szatkiewicz, N. Szeszenia-Dabrowska, I. Tachmazidou, E. Tenconi, A. Tortorella, F. Tozzi, J. Treasure, M. Tschöp, A. Tsitsika, K. Tziouvas, A. van Elburg, E. van Furth, T. Wade, G. Wagner, E. Walton, H. Watson, H. E. Wichmann, E. Widen, D. B. Woodside, J. Yanovski, S. Yao, S. Zerwas, S. Zipfel, L. Thornton, A. Hinney, M. Daly, P. F. Sullivan, E. Zeggini, G. Breen, C. M. Bulik, Significant locus and metabolic genetic correlations revealed in genome-wide association study of anorexia nervosa. Am. J. Psychiatry 174, 850–858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schizophrenia Working Group of the Psychiatric Genomics Consortium , Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D. M. Howard, M. J. Adams, T. K. Clarke, J. D. Hafferty, J. Gibson, M. Shirali, J. R. I. Coleman, S. P. Hagenaars, J. Ward, E. M. Wigmore, C. Alloza, X. Shen, M. C. Barbu, E. Y. Xu, H. C. Whalley, R. E. Marioni, D. J. Porteous, G. Davies, I. J. Deary, G. Hemani, K. Berger, H. Teismann, R. Rawal, V. Arolt, B. T. Baune, U. Dannlowski, K. Domschke, C. Tian, D. A. Hinds; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, M. Trzaskowski, E. M. Byrne, S. Ripke, D. J. Smith, P. F. Sullivan, N. R. Wray, G. Breen, C. M. Lewis, A. M. McIntosh, Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cross-Disorder Group of the Psychiatric Genomics Consortium , Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482.e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. Okbay, B. M. L. Baselmans, J.-E. De Neve, P. Turley, M. G. Nivard, M. A. Fontana, S. F. W. Meddens, R. K. Linnér, C. A. Rietveld, J. Derringer, J. Gratten, J. J. Lee, J. Z. Liu, R. de Vlaming, T. S. Ahluwalia, J. Buchwald, A. Cavadino, A. C. Frazier-Wood, N. A. Furlotte, V. Garfield, M. H. Geisel, J. R. Gonzalez, S. Haitjema, R. Karlsson, S. W. van der Laan, K.-H. Ladwig, J. Lahti, S. J. van der Lee, P. A. Lind, T. Liu, L. Matteson, E. Mihailov, M. B. Miller, C. C. Minica, I. M. Nolte, D. Mook-Kanamori, P. J. van der Most, C. Oldmeadow, Y. Qian, O. Raitakari, R. Rawal, A. Realo, R. Rueedi, B. Schmidt, A. V. Smith, E. Stergiakouli, T. Tanaka, K. Taylor, G. Thorleifsson, J. Wedenoja, J. Wellmann, H.-J. Westra, S. M. Willems, W. Zhao; LifeLines Cohort Study, N. Amin, A. Bakshi, S. Bergmann, G. Bjornsdottir, P. A. Boyle, S. Cherney, S. R. Cox, G. Davies, O. S. P. Davis, J. Ding, N. Direk, P. Eibich, R. T. Emeny, G. Fatemifar, J. D. Faul, L. Ferrucci, A. J. Forstner, C. Gieger, R. Gupta, T. B. Harris, J. M. Harris, E. G. Holliday, J.-J. Hottenga, P. L. De Jager, M. A. Kaakinen, E. Kajantie, V. Karhunen, I. Kolcic, M. Kumari, L. J. Launer, L. Franke, R. Li-Gao, D. C. Liewald, M. Koini, A. Loukola, P. Marques-Vidal, G. W. Montgomery, M. A. Mosing, L. Paternoster, A. Pattie, K. E. Petrovic, L. Pulkki-Råback, L. Quaye, K. Räikkönen, I. Rudan, R. J. Scott, J. A. Smith, A. R. Sutin, M. Trzaskowski, A. E. Vinkhuyzen, L. Yu, D. Zabaneh, J. R. Attia, D. A. Bennett, K. Berger, L. Bertram, D. I. Boomsma, H. Snieder, S.-C. Chang, F. Cucca, I. J. Deary, C. M. van Duijn, J. G. Eriksson, U. Bültmann, E. J. C. de Geus, P. J. F. Groenen, V. Gudnason, T. Hansen, C. A. Hartman, C. M. A. Haworth, C. Hayward, A. C. Heath, D. A. Hinds, E. Hyppönen, W. G. Iacono, M.-R. Järvelin, K.-H. Jöckel, J. Kaprio, S. L. R. Kardia, L. Keltikangas-Järvinen, P. Kraft, L. D. Kubzansky, T. Lehtimäki, P. K. E. Magnusson, N. G. Martin, M. M. Gue, A. Metspalu, M. Mills, R. de Mutsert, A. J. Oldehinkel, G. Pasterkamp, N. L. Pedersen, R. Plomin, O. Polasek, C. Power, S. S. Rich, F. R. Rosendaal, H. M. den Ruijter, D. Schlessinger, H. Schmidt, R. Svento, R. Schmidt, B. Z. Alizadeh, T. I. A. Sørensen, T. D. Spector, J. M. Starr, K. Stefansson, A. Steptoe, A. Terracciano, U. Thorsteinsdottir, A. R. Thurik, N. J. Timpson, H. Tiemeier, A. G. Uitterlinden, P. Vollenweider, G. G. Wagner, D. R. Weir, J. Yang, D. C. Conley, G. D. Smith, A. Hofman, M. Johannesson, D. I. Laibson, S. E. Medland, M. N. Meyer, J. K. Pickrell, T. Esko, R. F. Krueger, J. P. Beauchamp, P. D. Koellinger, D. J. Benjamin, M. Bartels, D. Cesarini, Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R. Karlsson Linnér, P. Biroli, E. Kong, S. F. W. Meddens, R. Wedow, M. A. Fontana, M. Lebreton, S. P. Tino, A. Abdellaoui, A. R. Hammerschlag, M. G. Nivard, A. Okbay, C. A. Rietveld, P. N. Timshel, M. Trzaskowski, R. de Vlaming, C. L. Zünd, Y. Bao, L. Buzdugan, A. H. Caplin, C.-Y. Chen, P. Eibich, P. Fontanillas, J. R. Gonzalez, P. K. Joshi, V. Karhunen, A. Kleinman, R. Z. Levin, C. M. Lill, G. A. Meddens, G. Muntané, S. Sanchez-Roige, F. J. van Rooij, E. Taskesen, Y. Wu, F. Zhang; 23and Me Research Team; eQTLgen Consortium; International Cannabis Consortium; Social Science Genetic Association Consortium, A. Auton, J. D. Boardman, D. W. Clark, A. Conlin, C. C. Dolan, U. Fischbacher, P. J. F. Groenen, K. M. Harris, G. Hasler, A. Hofman, M. A. Ikram, S. Jain, R. Karlsson, R. C. Kessler, M. Kooyman, J. M. Killop, M. Männikkö, C. Morcillo-Suarez, M. B. McQueen, K. M. Schmidt, M. C. Smart, M. Sutter, A. R. Thurik, A. G. Uitterlinden, J. White, H. de Wit, J. Yang, L. Bertram, D. I. Boomsma, T. Esko, E. Fehr, D. A. Hinds, M. Johannesson, M. Kumari, D. Laibson, P. K. E. Magnusson, M. N. Meyer, A. Navarro, A. A. Palmer, T. H. Pers, D. Posthuma, D. Schunk, M. B. Stein, R. Svento, H. Tiemeier, P. R. H. J. Timmers, P. Turley, R. J. Ursano, G. G. Wagner, J. F. Wilson, J. Gratten, J. J. Lee, D. Cesarini, D. J. Benjamin, P. D. Koellinger, J. P. Beauchamp, Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat. Genet. 51, 245–257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.C. Sudlow, J. Gallacher, N. Allen, V. Beral, P. Burton, J. Danesh, P. Downey, P. Elliott, J. Green, M. Landray, B. Liu, P. Matthews, G. Ong, J. Pell, A. Silman, A. Young, T. Sprosen, T. Peakman, R. Collins, UK Biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLOS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.G. Cuellar-Partida, J. Y. Tung, N. Eriksson, E. Albrecht, F. Aliev, O. A. Andreassen, I. Barroso, J. S. Beckmann, M. P. Boks, D. I. Boomsma, H. A. Boyd, M. M. B. Breteler, H. Campbell, D. I. Chasman, L. F. Cherkas, G. Davies, E. J. C. de Geus, I. J. Deary, P. Deloukas, D. M. Dick, D. L. Duffy, J. G. Eriksson, T. Esko, B. Feenstra, F. Geller, C. Gieger, I. Giegling, S. D. Gordon, J. Han, T. F. Hansen, A. M. Hartmann, C. Hayward, K. Heikkilä, A. A. Hicks, J. N. Hirschhorn, J. J. Hottenga, J. E. Huffman, L. D. Hwang, M. A. Ikram, J. Kaprio, J. P. Kemp, K. T. Khaw, N. Klopp, B. Konte, Z. Kutalik, J. Lahti, X. Li, R. J. F. Loos, M. Luciano, S. H. Magnusson, M. Mangino, P. Marques-Vidal, N. G. Martin, W. McArdle, M. McCarthy, C. Medina-Gomez, M. Melbye, S. A. Melville, A. Metspalu, L. Milani, V. Mooser, M. Nelis, D. R. Nyholt, K. S. O'Connell, R. A. Ophoff, C. Palmer, A. Palotie, T. Palviainen, G. Pare, L. Paternoster, L. Peltonen, B. W. J. H. Penninx, O. Polasek, P. P. Pramstaller, I. Prokopenko, K. Raikkonen, S. Ripatti, F. Rivadeneira, I. Rudan, D. Rujescu, J. H. Smit, G. D. Smith, J. W. Smoller, N. Soranzo, T. D. Spector, B. S. Pourcain, J. M. Starr, H. Stefánsson, S. Steinberg, M. Teder-Laving, G. Thorleifsson, K. Stefánsson, N. J. Timpson, A. G. Uitterlinden, C. van Duijn, F. van Rooij, J. M. Vink, P. Vollenweider, E. Vuoksimaa, G. Waeber, N. J. Wareham, N. Warrington, D. Waterworth, T. Werge, H. E. Wichmann, E. Widen, G. Willemsen, A. F. Wright, M. J. Wright, M. Xu, J. H. Zhao, P. Kraft, D. A. Hinds, C. M. Lindgren, R. Mägi, B. M. Neale, D. M. Evans, S. E. Medland, Genome-wide association study identifies 48 common genetic variants associated with handedness. Nat. Hum. Behav. 5, 59–70 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.J. P. Kemp, J. A. Morris, C. Medina-Gomez, V. Forgetta, N. M. Warrington, S. E. Youlten, J. Zheng, C. L. Gregson, E. Grundberg, K. Trajanoska, J. G. Logan, A. S. Pollard, P. C. Sparkes, E. J. Ghirardello, R. Allen, V. D. Leitch, N. C. Butterfield, D. Komla-Ebri, A. T. Adoum, K. F. Curry, J. K. White, F. Kussy, K. M. Greenlaw, C. Xu, N. C. Harvey, C. Cooper, D. J. Adams, C. M. T. Greenwood, M. T. Maurano, S. Kaptoge, F. Rivadeneira, J. H. Tobias, P. I. Croucher, C. L. Ackert-Bicknell, J. H. D. Bassett, G. R. Williams, J. B. Richards, D. M. Evans, Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat. Genet. 49, 1468–1475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S5
Legends for supplementary files S1 to S9
Supplementary files S1 to S9