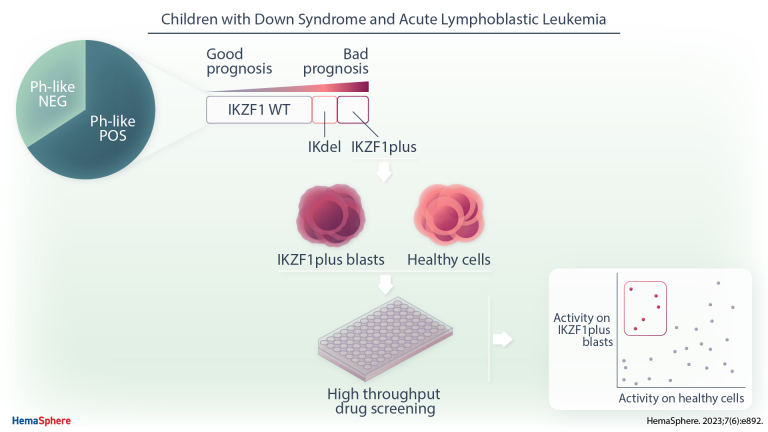
Abstract
Children with Down syndrome have an augmented risk for B-cell acute lymphoblastic leukemia (DS-ALL), which is associated with lower survival than in non-DS-ALL. It is known that cytogenetic abnormalities common in childhood ALL are less frequent in DS-ALL, while other genetic aberrancies (ie, CRLF2 overexpression and IKZF1 deletions) are increased. A possible cause for the lower survival of DS-ALL that we herewith evaluated for the first time was the incidence and prognostic value of the Philadelphia-like (Ph-like) profile and the IKZF1plus pattern. These features have been associated with poor outcome in non-DS ALL and therefore introduced in current therapeutic protocols. Forty-six out of 70 DS-ALL patients treated in Italy from 2000 to 2014 displayed Ph-like signature, mostly characterized by CRLF2 (n = 33) and IKZF1 (n = 16) alterations; only 2 cases were positive for ABL-class or PAX5-fusion genes. Moreover, in an Italian and German joint cohort of 134 DS-ALL patients, we observed 18% patients positive for IKZF1plus feature. Ph-like signature and IKZF1 deletion were associated with poor outcome (cumulative incidence of relapse: 27.7 ± 6.8% versus 13 ± 7%; P = 0.04 and 35.2 ± 8.6% versus 17 ± 3.9%; P = 0.007, respectively), which further worsens when IKZF1 deletion was co-occurring with P2RY8::CRLF2, qualifying for the IKZF1plus definition (13/15 patients had an event of relapse or treatment-related death). Notably, ex vivo drug screening revealed sensitivity of IKZF1plus blasts for drugs active against Ph-like ALL such as Birinapant and histone deacetylase inhibitors. We provided data in a large setting of a rare condition (DS-ALL) supporting that these patients, not associated with other high-risk features, need tailored therapeutic strategies.
INTRODUCTION
Children with Down syndrome have a high risk for acute lymphoblastic leukemia (DS-ALL), which is one of their more frequent causes of death.1 They have an inferior outcome compared with non-DS children with ALL due to both increased chemotherapy-related toxicity and excess of relapses,2 thus demanding tailored therapeutic strategies.
DS-ALL is almost exclusively of B-cell phenotype and exhibits distinct somatic features from non-DS-ALL. Approximately 60% of DS-ALL patients have aberrant expression of CRLF2, caused by the P2RY8::CRLF2 fusion or the IGH::CRLF2 translocation.3,4 Elevated CRLF2 expression in DS-ALL was found to be frequently associated with activating mutations in JAK2 (20%), but neither of these alterations predicted worse outcome.5 Activating mutations affecting other signaling effectors, such as NRAS, KRAS, KIT, FLT3, and PTPN11, are also frequently found in DS-ALL.6 Consistent with the recurrent constitutive activation of these pathways, a recent study on a limited number of DS-ALL revealed the enrichment of the Philadelphia-like (Ph-like) transcription signature, a gene expression profile (GEP) like that observed in Philadelphia chromosome-positive ALL, but lacking the chromosomal aberration.7 In non-DS-ALL, the Ph-like subgroup is characterized by the presence of potentially targetable gene fusions and was reported as associated with poor outcome.8
Copy number variations in cell cycle regulator genes and in transcription factors involved in B-cell development have been described in DS and non-DS-ALL patients.9 In particular, the frequency of IKZF1 deletions in DS-ALL patients was comparable to that of high-risk (HR) non-DS-ALL patients (~30%) and this alteration appeared to be the only one with prognostic significance in DS-ALL based on the Dutch Childhood Oncology Group and UK trials.5
In non-DS patients treated in the Associazione Italiana Ematologia ed Oncologia Pediatrica–Berlin-Frankfurt-Muenster (AIEOP-BFM) ALL2000 trial, the feature IKZF1plus, defined as the co-occurrence of an IKZF1 deletion with deletions in CDKN2A, CDKN2B, PAX5, or PAR1 in the absence of ERG deletion, has been described as a minimal residual disease (MRD)-dependent very poor prognostic profile in childhood BCP-ALL.10
IKZF1plus status has been incorporated in the new stratification strategy and experimental treatment approaches in the ongoing AIEOP-BFM ALL2017 trial (clinicaltrials.gov identifier: 03643276), while patients carrying ABL-class fusion genes, characteristic of the Ph-like signature, are eligible to the current amended EsPhALL2017 protocol (clinicaltrials.gov identifier: NCT03007147). However, no study has yet addressed the incidence and prognostic relevance of these 2 features in DS-ALL patients.
The aim of this study was to evaluate the incidence and the prognostic value of the Ph-like transcription profile with its recurrent fusion events, and of the IKZF1plus feature in a large cohort of children with DS-ALL treated in Italy or Germany in AIEOP-BFM ALL protocols.
METHODS
Patients
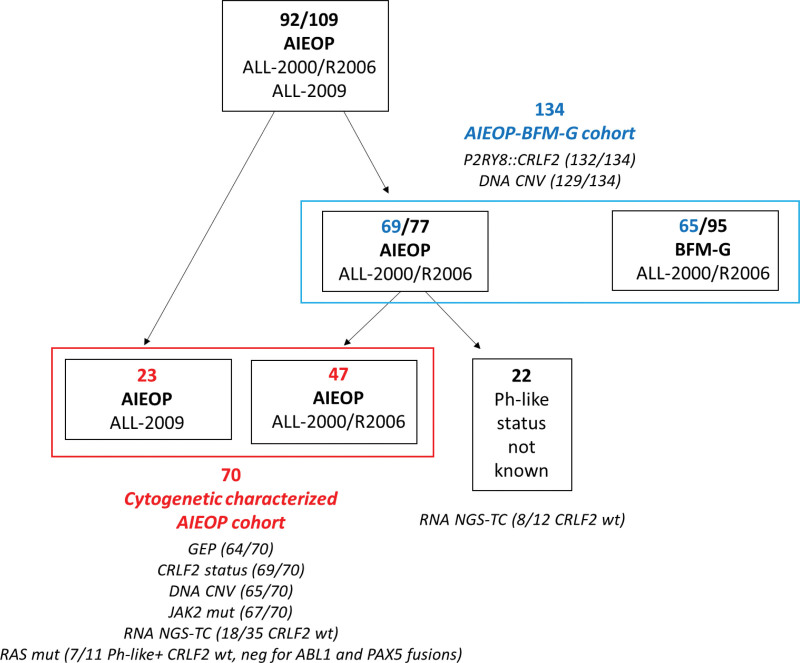
Ninety-two out of 109 DS-ALL patients, consecutively enrolled in AIEOP-BFM protocols in Italian centers from 2000 to 2014, were analyzed in this study, and 17 were excluded due to lack of material. Seventy were evaluated for cytogenetic status and for Ph-like ALL profile (Cytogenetic characterized AIEOP cohort) (Figure 1). Sixty-four of them had RNA available for GEP assay, while 6 DS-ALL, with no leftover RNA, were included in the cohort and classified as ALL ETV6::RUNX1 because positive for the rearrangement by standard reverse transcriptase-polymerase chain reaction (RT-PCR).11
Figure 1.
Schematic representation of the DS-ALL patient cohorts of this study. In the scheme are reported the number of patients and the different therapeutic protocols and centers in which they were enrolled. The patients evaluated for Ph-like ALL profile (Cytogenetic Characterized AIEOP cohort) are highlighted in red and those evaluated for the prognostic relevance of P2RY8::CRLF2 fusion, IKZF1 deletion, and IKZF1plus feature (AIEOP-BFM-G cohort) are highlighted in blue. The molecular analyses performed for each group of patients are indicated. DS-ALL = Down syndrome-acute lymphoblastic leukemia; Ph-like = Philadelphia-like.
No significant differences were observed with respect to event-free survival (EFS) between the 70 analyzed and the 39 not analyzed AIEOP patients diagnosed in the same period of this study (5-y EFS: 64.5% versus 74.4%; P = 0.33; Suppl. Figure S1A).
The prognostic relevance of P2RY8::CRLF2 fusion, IKZF1 deletion, and IKZF1plus feature was analyzed in a larger joint cohort of 134 DS-ALL patients enrolled in the AIEOP-BFM ALL protocols in Italian and German centers. Sixty-nine of these patients were treated in Italian centers from 2000 to 2011 (47 of them overlap with the Cytogenetic characterized AIEOP cohort described earlier) and 65 in German centers from 2000 to 2010 (AIEOP-BFM-G cohort) (Figure 1). They represented almost all (89.6%) patients enrolled in that period in Italy and the 68.4% in Germany. No significant differences were observed with respect to EFS between BFM-G analyzed and not analyzed patients (P = 0.84; Suppl. Figure S1B).
Details of AIEOP-BFM ALL2000 protocol and its subsequent amendment in 2006 have been previously described,12,13 and differences between the 2000 and the 2009 study have been reported.14 The AIEOP-BFM ALL2000 study is registered at https://www.clinicaltrials.gov by BFM as NCT00430118 and by AIEOP as NCT00613457; the AIEOP-BFM ALL2009 study is registered as NCT01117441.
Informed consent to participate in the study was obtained for all patients from their parents or legal guardians.
Gene expression profile
GEP was analyzed by Affymetrix HG-U133 Plus 2.0 arrays for the 64 DS-ALL AIEOP patients (GEO accession number: GSE2000864). Each patient was classified according to the previously published diagnostic classifier (DC) based on the GEP;15–17 a cohort of 289 childhood BCP-ALL non-DS cases at diagnosis enrolled in Italy in the AIEOP-BFM ALL2000/R2006 protocols was also included (GEO accession numbers: GSE79547, GSE13164, GSE13159, and GSE13204).18 Details are given in the supplementary material.
CRLF2 alterations and JAK2 mutations
CRLF2 transcripts levels, P2RY8::CRLF2, IGH::CRLF2, and JAK2 mutations were analyzed as previously described19–21 and briefly recapitulated in the supplementary file.
Copy number variants
DNA copy number variants (CNVs) of 56 key target regions known to have a role in ALL were analyzed by the digital multiplex ligation-dependent probe amplification (digitalMLPA) kit D007 ALL (MRC-Holland, Amsterdam, the Netherlands), following manufacturer’s instructions.22
Statistical analysis
EFS was calculated from the date of diagnosis to the date of the first event. Events considered were resistance, defined as failure to achieve complete remission (absence of physical signs of leukemia or detectable leukemia cells on blood smears, a bone marrow with active hematopoiesis and <5% blasts, and morphologically normal cerebrospinal fluid) by the end of the third high risk block of chemotherapy,12–14 relapse, death, or second malignant neoplasm, whichever occurred first.
EFS curve was estimated according to the Kaplan-Meier method and compared according to the log-rank test. Cumulative incidence of relapse (CIR) at 5 years was estimated by adjusting for competing risks of other events and comparison performed with the Gray test.
Follow-up was updated in January 2014 for AIEOP-BFM ALL 2000 protocol and in February 2019 for AIEOP-BFM ALL 2009 protocol.
RESULTS
Ph-like gene expression signature in AIEOP DS-ALL patients
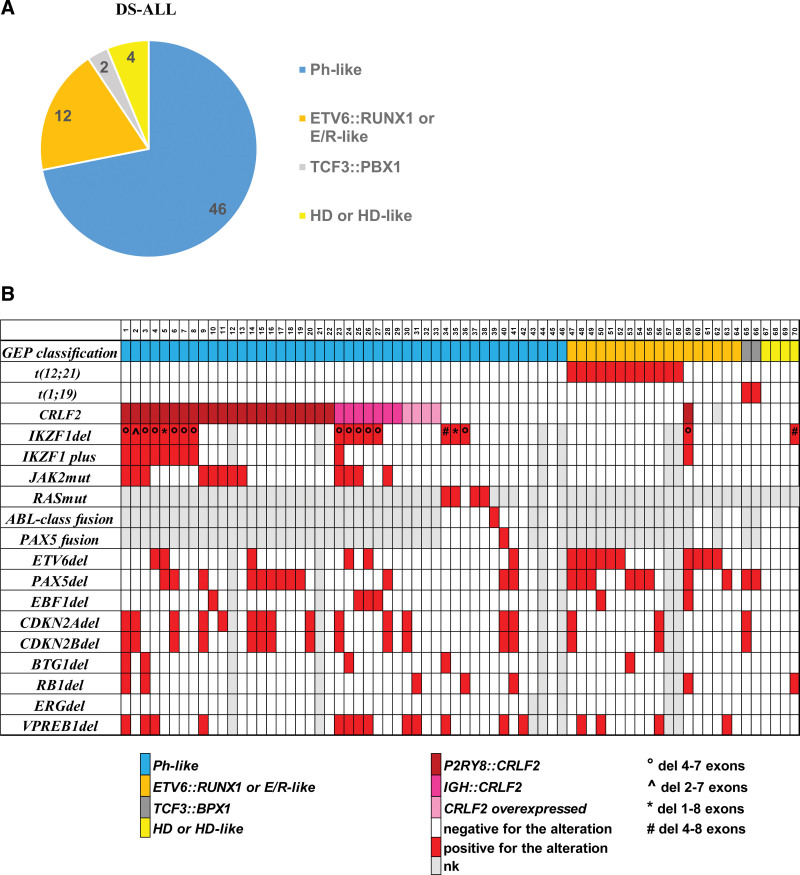
We analyzed 64 DS-ALL patients at diagnosis, treated within AIEOP-BFM protocols in Italian centers from 2000 to 2014 for GEP using PCR, karyotype analysis, and the DC model developed during the Microarray Innovation Leukaemia (MILE) study.15–17 Most of the DS-ALL patients (46/64) were classified as Ph-like, 12 cases as ETV6::RUNX1 or ETV6::RUNX1-like (6/12 carried the t[12;21] translocation), 2 as TCF3::PBX1 (both positive for the t[1;19] translocation), and 4 as ALL with a hyperdiploid/hyperdiploid-like karyotype (all 4 cases had 48 chromosomes) (Figure 2A). The t-SNE analysis, using the top 1000 variable genes of both DS-ALL and no DS-ALL patients, showed that the Down syndrome patients were not characterized by a unique and peculiar expression profile, but shared features with the different BCP-ALL subgroups18 (Suppl. Figure S2).
Figure 2.
Gene expression profile and molecular characterization of AIEOP DS-ALL. The pie chart (A) shows the distribution of the 64 AIEOP DS-ALL patients in the different cytogenetic subgroups based on the GEP analysis. Plot (B) summarizing the GEP and molecular characterization of the DS-ALL in the Cytogenetic Characterized AIEOP cohort. E/R = ETV6::RUNX1; DS-ALL = Down syndrome-acute lymphoblastic leukemia; HD = hyperdiploid; del = deletion; mut = mutation; nk = not known; GEP = gene expression profile.
No DS-ALL patients were characterized by ERG-related signatures23 nor carried KMT2A or MEF2D-rearrangements (Figure 2A and Suppl. Figure S2). These 64 patients, together with 6 additional DS-ALL patients classified as ETV6::RUNX1, were included in the Cytogenetic characterized AIEOP cohort (Figure 1).
Clinical and molecular characteristics of patients positive and negative for the Ph-like feature of this cohort are described in Suppl. Table S1. In particular, the majority of Ph-like positive patients had been enrolled in non-HR therapeutic arms of the protocol (82.6%).
Thirty-three Ph-like patients (71.7% of the Ph-like subgroup) were positive for CRLF2 alterations (22 P2RY8::CRLF2, 7 IgH::CRLF2, and 4 CRLF2 overexpressed with no material to confirm the mechanism). JAK2 mutations were detected in 12 of the 33 CRLF2+ patients (8 P2RY8::CRLF2 and 4 IGH::CRLF2). Sixteen of the 43 Ph-like DS-ALL patients showed IKZF1 deletion (13/16 were also CRLF2+) and 9 of these were classifiable as IKZF1plus, because of the co-occurrence of IKZF1 deletion with other specific deletions, as previously reported.10 Of note, all 9 IKZF1plus patients were positive for CRLF2 alterations.
Conversely, only 1 of the 23 Ph-like negative DS-ALL patients was positive for CRLF2 alterations, 2 for IKZF1 deletions and one of these was IKZF1plus (Suppl. Table S1 and Figure 2B).
RNA next-generation sequencing-targeted capture (NGS-TC) strategy was performed for the detection of fusion genes on 18 of 36 patients negative for CRLF2 alterations for which leftover RNA was available. Notably, we identified 1 patient positive for an ABL-class fusion (RANBP2::ABL1) and 1 positive for a PAX5 fusion (PAX5::FAM219A) previously recognized as Ph-like subgroup (Figure 2B) and visualized in the tSNE in the PAX5t group (Suppl. Figure S2). Although the RANBP2::ABL1 fusion (breakpoints in RANBP2 intron 18–19 and in ABL1 intron 1–2) was already observed in the non-DS Ph-like ALL population,24 the PAX5::FAM219A chimera (breakpoints in PAX5 intron 6–7 and FAM219A intron 1–2) was previously reported only in breast tumor.25
Activating variants in the RAS pathway (NRAS and KRAS variants) were identified in 4 of 7 Ph-like positive patients wild-type (wt) for CRLF2 alterations, ABL and PAX5 fusions by DNA-targeted NGS [2 patients were positive for NRAS pathogenic variants, 2 for KRAS, and 1 of the latter was also positive for a germline BRAF variant of unknown significance (VUS)] (Suppl. Table S2).
Analysis of the DNA CNV in Ph-like versus non-Ph-like DS-ALL patients showed, in addition to the higher incidence of PAR1 deletions and IKZF1 deletions already described earlier, a large number of Ph-like patients carrying deletions in CDKN2A (14/42, 33.3%), CDKN2B (13/42, 31%), BTG1 (4/42, 9.5%), and VPREB1 genes (13/41, 31.7%). In reverse, Ph-like negative DS-ALL patients more frequently carried ETV6 deletion (45.5% versus 14.3%, P = 0.01), consistent with the presence in this group of ETV6::RUNX1 or ETV6::RUNX1-like patients, and a large number of non-Ph-like patients were positive for PAX5 gene deletion (10/22, 45.5%). We did not observe any DS-ALL patients positive for ERG deletion, although subclonal deletions could not be ruled out. Interestingly, the average number of ALL-specific CNVs per patient was higher in the Ph-like versus the non-Ph-like group (2.6 versus 1.7; P = 0.048). (Figure 2B and Suppl. Table S3).
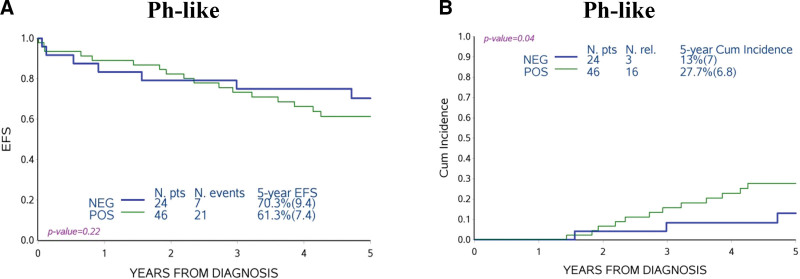
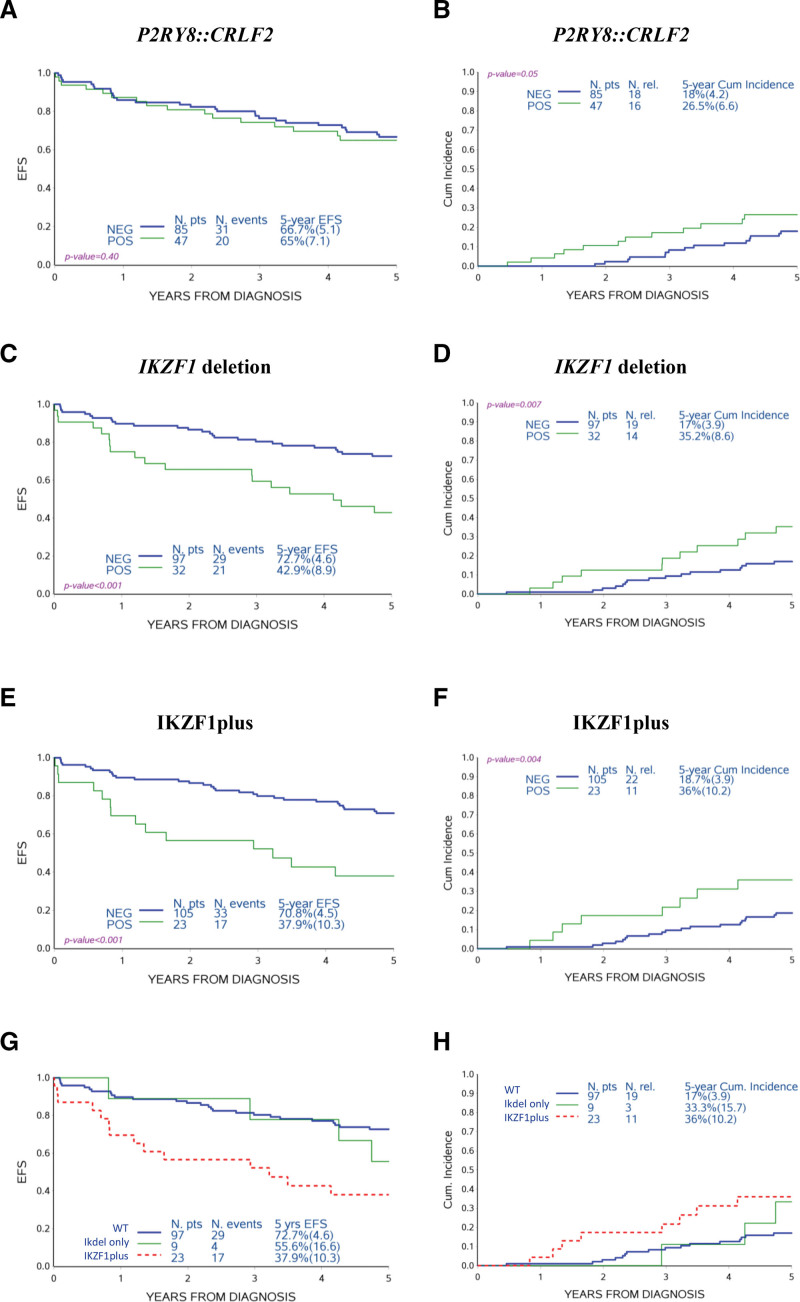
Ph-like DS-ALL patients were not significantly different in EFS compared with Ph-like negative patients, but did have an increased CIR, largely due to late relapses (27.7 ± 6.8% versus 13 ± 7%; P = 0.04) (Figure 3A, B) and a slightly decreased incidence of competing events (deaths and second malignant neoplasms; 11.0 ± 4.6% versus 16.7 ± 7.6%; P = 0.52, data not shown). The negative prognostic impact of the Ph-like feature was more marked in non-HR patients (30.2 ± 7.7% versus 9.7 ± 6.5%; P = 0.02) (Suppl. Figure S3).
Figure 3.
Association of Ph-like profile to treatment outcome. EFS (A) and CIR (B) of DS-ALL Cytogenetic Characterized AIEOP cohort comparing non-Ph-like (NEG) and Ph-like (POS) patients. Ph-like = Philadelphia-like; EFS = event-free survival; CIR = cumulative incidence of relapse; DS-ALL = Down syndrome-acute lymphoblastic leukemia; NEG = negative; POS = positive.
Incidence and prognostic impact of P2RY8-CRLF2, IKZF1 deletion, and IKZF1plus in DS-ALL patients
To evaluate the prognostic impact of P2RY8::CRLF2 fusion, IKZF1 deletion, and IKZF1plus feature, we analyzed 134 DS-ALL patients treated with the same protocol in Italian and German centers from 2000 to 2011 (Suppl. Table S4). Although more patients in the BFM-G study cohort were stratified as standard or HR compared with the Italian cohort, the EFS and CIR curves of the 2 cohorts were overlapping, even when excluding HR patients from the analysis (Suppl. Figure S4).
The incidence of the 3 aberrations was very similar in Italian and German cohorts, and in the joint cohort we observed 35.6% patients positive for P2RY8::CRLF2 fusion, 24.8% for IKZF1 deletion, and 18% for IKZF1plus (Suppl. Table S5). Of note, the majority of patients carrying these alterations had not been identified as being HR (Suppl. Table S6).
Regarding the prognostic impact, the P2RY8::CRLF2 fusion was not associated to a different EFS, despite a tendency for an increased CIR (26.5 ± 6.6% versus 18 ± 4.2%; P = 0.05, Figure 4A, B). Instead, IKZF1 deletion and IKZF1plus were associated with an inferior EFS than the respective negative cases for the alteration taken into consideration in the analysis (42.9 ± 8.9% versus 72.7 ± 4.6%; P < 0.001 and 37.9 ± 10.3% versus 70.8 ± 4.5%; P < 0.001, respectively), due not only to a higher treatment-related mortality (TRM) (21.9 ± 7.3% versus 10.3 ± 3.1%; P = 0.08 and 26.1 ± 9.2% versus 10.5 ± 3.0%; P = 0.038, data not shown), but also mainly to a higher CIR (35.2 ± 8.6% versus 17 ± 3.9%; P = 0.007 and 36 ± 10.2% versus 18.7 ± 3.9%; P = 0.004, respectively, Figure 4C–F). The strong prognostic relevance was maintained when HR patients were excluded (CIR IKZF1 deletion positive non-HR patients: 34.4 ± 9.3% versus 15.6 ± 3.8%; P = 0.006 and CIR IKZF1plus positive non-HR patients: 36.4 ± 11% versus 16.8 ± 3.8%; P = 0.002; Suppl. Figure S5).
Figure 4.
Association of P2RY8::CRLF2 fusion, IKZF1 deletion, and IKZF1plus feature to treatment outcome. EFS and CIR of DS-ALL AIEOP-BFM-G cohort comparing patients positive (POS) and negative (NEG) for the indicated molecular alterations (A)–(F). The comparator group was the respective negative cases for the alteration taken into consideration in the analysis: for (A) and (B) cases negative for P2RY8::CRLF2 fusion, for (C) and (D) cases negative for IKZF1 deletion, and for (E) and (F) cases negative for IKZF1plus feature. EFS and CIR comparing patients negative for IKZF1 deletions (wt), positive for IKZF1 deletions but without the other co-occurring CNV that define the IKZF1plus feature (IKdel only) and IKZF1plus (G) and (H). EFS = event-free survival; CIR = Cumulative incidence of relapse; CNV = copy number variant; DS-ALL = Down syndrome-acute lymphoblastic leukemia; wt= wild type; IKdel only = IKZF1 deleted patients.
Although numbers are quite small, it can be noted that excluding IKZF1plus patients from the analysis of IKZF1 deleted patients (IKdel only), the IKZF1plus characteristic was the alteration with the most negative prognosis (Figure 4G, H). In particular, in the subgroup of IKZF1plus patients positive for the P2RY8::CRLF2 fusion, the incidence of adverse events (relapses or treatment-related deaths) was very high (13/15; Suppl. Figure S6).
In light of the observed poor prognosis of the IKZF1 alterations, we investigated whether the high CIR described earlier in AIEOP Ph-like DS-ALL patients was the direct consequence of a high incidence of IKZF1 alterations in this subgroup. As shown in Suppl. Figure S7, despite the exclusion of IKZF1 deleted cases, a tendency for an increased CIR was still observed in the Ph-like positive group, although the low number of patients examined does not allow it to reach a statistical significance (22.6 ± 8.1% versus 10.7 ± 7.2%; P = 0.11).
Moreover, to verify in this enlarged cohort whether other DS-ALL patients carried any fusion gene, we analyzed 8 of the 12 Italian CRLF2 negative patients with no GEP data available (and therefore not included in the previous cohort) by NGS-TC strategy. We identified 1 patient positive for the PAX5::ETV6 fusion (joining PAX5 exon 5 to ETV6 exon 3) and 1 patient positive for ETV6::NTRK3 fusion (joining ETV6 exon 5 to NTRK3 exon 15), 2 rearrangements already observed in non-DS Ph-like ALL population.26,27 These 2 patients, as well as the other 2 carrying PAX5 or kinase fusion genes described earlier, were negative for IKZF1 deletion and remained in continuous complete remission.
Gene expression and drug response profile of IKZF1plus DS-ALL
Considering samples for which GEP was available, we identified 175 differentially expressed genes among the 3 subgroups (IKZF1 wt versus IKdel only; IKZF1 wt versus IKZF1plus; and IKZF1plus versus IKdel only) (Suppl. Figure S8A-D). In particular, the greatest difference in gene expression was observed between patients IKdel only compared with wt, while very few genes were statistically differentially expressed [false discovery rate (FDR) < 0.05] among IKZF1plus and IKZF1 wt patients (Suppl. Figure S8A-D).
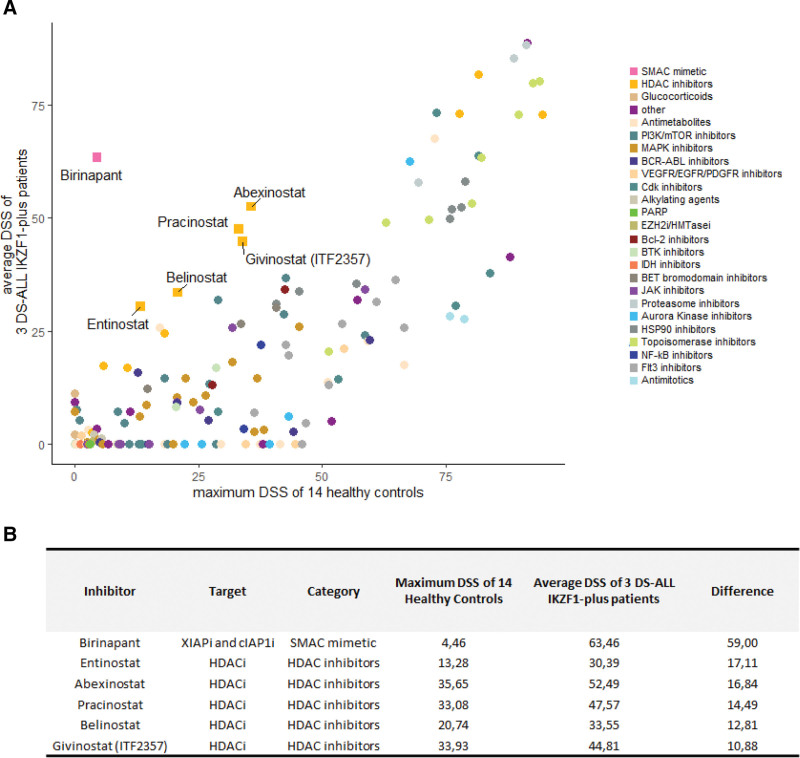
Moreover, we performed an extended ex vivo drug screening with 174 drugs or inhibitors (Suppl. Table S7) in early to late clinical trials on blasts of 3 IKZF1plus DS-ALL patients and on 14 different controls (5 B-cell lymphoblastoid cell lines, 3 peripheral blood mononuclear cells (PBMCs), 3 T cells, and 3 CD34+ cells, all derived from healthy donors). Compounds known to be effective in Ph-like patients such as Birinapant, a SMAC mimetic,28 and histone deacetylase (HDAC) inhibitors29 show the highest efficacy on IKZF1plus blasts in comparison to healthy controls (average Drug Sensitivity Score [DSS]30 of blasts >20 and difference between the average DSS of the patients and the maximum DSS of healthy controls >10; Figure 5).
Figure 5.
Drug response profile of IKZF1plus DS-ALL vs healthy controls. Scatter plot of the average DSS of 3 IKZF1plus DS-ALL patients vs the maximum DSS of 14 different controls (5 B-cell lymphoblastoid cell lines, 3 PBMCs, 3 T cells, and 3 CD34+ cells, all derived from healthy donors). The drugs are colored according to their drug class (A). The drugs with the average DSS of blasts >20 and with a difference from the maximum DSS of healthy controls of at least 10 are highlighted and listed in the table (B). DSS = drug sensitivity scores; DS-ALL = Down syndrome-acute lymphoblastic leukemia; PBMC= peripheral blood mononuclear cell.
Genomic aberrations at relapse
Twenty paired diagnostic and relapse specimens of DS-ALL AIEOP patients were analyzed for P2RY8::CRLF2 fusion, JAK2 mutations, and IKZF1 deletion. One out of 9 P2RY8::CRLF2-positive patients at diagnosis lost the rearrangement at relapse and a de novo fusion was not detected. JAK2 mutations were lost in 1 of 5 patients at relapse, remained stable in 2 of 5 cases, and in 2 cases was different between diagnosis and relapse. IKZF1 deletion was acquired at relapse in 1 of 8 patients, remained stable in 5 cases, and was larger in 2 cases (Suppl. Table S8).
DISCUSSION
Although many studies have reported a worse clinical outcome for DS-ALL and have shown that this disease is genetically different from non-DS-ALL, DS-ALL children are generally cured without specific therapeutic interventions. The only modification of the standard protocol is a reduction in the chemotherapy dose due to higher incidence of chemotherapy-related toxicity.2
Currently, Italian and German DS-ALL children are enrolled in the collaborative prospective AIEOP-BFM ALL2017 trial together with non-DS children. In this treatment protocol, several risk factors in addition to the MRD level have been used in the stratification strategy and in the experimental treatment approaches, including the presence of the IKZF1plus feature and fusion genes characteristic of the Ph-like condition.
Taking the advantage of a large cohort of pediatric DS-ALL patients diagnosed over 10 years, this is the first study that evaluated the incidence and the prognostic impact of the IKZF1plus feature and the effect of the Ph-like profile on clinical outcome, as well as looking for Ph-related fusion genes, other than CRLF2, in this subgroup of patients.
By molecular and GEP analyses, we confirmed that DS-ALL is a heterogeneous disease, characterized by many genetic profiles, similar to those observed in non-DS-ALL, but with a different distribution.4,7 As expected, we found that the majority of DS-ALL patients displayed a Ph-like ALL gene expression signature, mostly characterized by CRLF2 and IKZF1 alterations. In accordance with the high incidence of these 2 alterations and their frequent co-occurrence in DS-ALL, 20% of Ph-like patients were classifiable as IKZF1plus.
Unexpectedly, a single IKZF1plus patient of our cohort showed a profile like patients positive for the ETV6::RUNX1 fusion, although negative for the t(12;21) translocation. We cannot exclude the presence of a noncanonical fusion of ETV6::RUNX1 with loss of ETV6 exon 531 not detectable with the standard RT-PCR used for diagnostic purposes.11
In general, we observed a higher average number of ALL-specific CNVs per patient in the Ph-like group. Further whole genome CNV analysis is needed to establish whether this subgroup is indeed characterized by increased genetic instability.
Importantly, overall CIR analyses (also after excluding HR patients) showed that the Ph-like feature had a negative prognostic impact also in DS-ALL. Considering its very high incidence (65.7%), the Ph-like feature could represent the main risk factor for this subgroup. It is also noteworthy that most of the Ph-like cases were not identified at diagnosis as having a HR of relapse (82.6%).
Analyzing patients without CRLF2 overexpression, we identified fusion genes characteristic of Ph-like non-DS-ALL involving PAX5 or kinases (ABL1 and NTRK3) in 2 Ph-like patients and in 2 patients for whom no GEP data were available. To the best of our knowledge, these fusions were not reported in DS-ALL yet. Notably, these patients remain in continuous complete remission; however, due to the small number, the prognostic value cannot be assessed.
Moreover, we identified activating mutations in the RAS signaling pathway in 4 of 7 Ph-like positive patients, wt for CRLF2 and fusion genes.
The large joint Italian and German study cohort allowed us to observe that the IKZF1plus feature in DS-ALL was 3 times more frequent than in non-DS-ALL (18% versus 6%).10 With regard to the prognostic impact, in accordance with previous reports,2,5 we observed that although the P2RY8::CRLF2 fusion was not significantly associated to a different outcome, IKZF1 deletion was associated with an inferior EFS and with a higher CIR. Importantly, although it is not possible to draw solid conclusions based on the analyses of small subgroups of patients, this study suggests that the IKZF1plus characteristic may be the alteration associated with the most negative prognosis, especially when determined by the combination of IKZF1 deletion and the P2RY8::CRLF2 fusion.
In a recent article, Michels et al32 addressed the question whether the increased risk of relapse in DS-ALL patients was exclusively due to a higher incidence of adverse genetic markers in this subgroup or was intrinsic to DS. The authors concluded that DS itself provides an additional risk of relapse in patients with a IKZF1 deletion, but they did not have the opportunity to address the effect of CRLF2 aberrations. Our present study highlights the possibility that the different outcome observed by Michels et al in DS-ALL patients compared with non-DS-ALL patients with IKZF1 deletion may be due to a higher incidence of IKZF1plus patients with the co-occurrence of IKZF1 and CRLF2 gene alterations in the first group. Matched case-control studies that consider this combination of aberrations would be necessary to clarify this issue.
The drug screening analysis showed that the blasts of IKZF1plus DS-ALL patients are particularly sensitive to drugs that do not act directly in restoring the activity of the transcription factor, but have been described to be effective in Ph-like cases, such as Birinapant28 and drugs belonging to the class of HDAC inhibitors such as Givinostat.29
Eventually, we had the opportunity to analyze 20 paired diagnostic and relapse samples of AIEOP DS-ALL patients. Interestingly, as we previously described in non-DS-ALL,19 no de novo P2RY8::CRLF2 fusion was detected at relapse, while 1 case positive at diagnosis lost the fusion at relapse. Moreover, as already reported in a collaborative study,33 we observed that clones with JAK2 mutations are often unstable in DS-ALL. This observation could be an indirect clue that these are not primary lesions. On the contrary, we found that IKZF1 deletion was acquired at relapse in 1 case and was larger in 2 cases, suggesting that this alteration could play an important role in the blast cells at each stage of the disease. However, a higher number of paired samples is required to confirm these observations.
In the current AIEOP-BFM ALL2017 protocol, non-DS and DS-ALL IKZF1plus patients with any MRD positivity after induction treatment are allocated in the HR therapeutic arm and randomized for innovative therapies. All HR DS-ALL patients receive Blinatumomab instead of 2 of 3 intensive and high-toxic HR blocks. The poor outcome of this patient group in the study presented here, with a high incidence of relapses but also a high TRM, confirms the correctness of this procedure. In our cohort, only 3 IKZF1plus DS-ALL patients were MRD negative (of which 2 remain in continuous complete remission, while 1 deceased); this low number does not allow us to draw conclusions on the relevance of IKZF1plus feature on the outcome of MRD-SR DS-ALL.
In summary, we reported here in a large cohort of DS-ALL that only very few cases were positive for gene fusions involving PAX5 or kinases. The IKZF1plus feature in DS-ALL was 3 times more frequent than in non-DS-ALL. Ph-like feature and IKZF1 deletions were associated with poor outcome, with the risk of relapse further increased for IKZF1plus patients, in keeping with the current concept for treatment risk stratification criteria for ALL children, regardless of the DS status.10 These alterations characterize subgroups of DS-ALL patients, who for the most part do not have other HR features, who need tailored therapeutic strategies. Despite ex vivo drug screening revealed sensitivity of IKZF1plus blasts for drugs known to be active against Ph-like leukemia samples, further studies are necessary to identify the best therapeutic plan for these fragile patients.
AUTHOR CONTRIBUTIONS
CP, SB, GF, CS, MG, and MB performed the experiments. DS and MGV collected the trial data and performed the statistical analyses. AO, KS, S Bhatia, and AB developed and analyzed drug screening. BB, LLN, VC, and AB provided the data of the AIEOP patients. SJ, M Zaliova, M Stanulla, AM, M Zimmermann, M Schrappe, and G Cario provided the data of the BFM-G patients. CP, SB, G Kronnie, and G Cazzaniga designed the study and analyzed the data. All the authors contributed in writing the article.
DISCLOSURES
The authors have no conflicts of interest to disclose.
SOURCES OF FUNDING
This work was supported by the Italian Ministry of Health, grant Ricerca Finalizzata-Giovani Ricercatori (GR-2016-02364753 to CP, GF, MB), Fondazione Cariparo (17/07_1FCR to SB and 20/12FCR to SB, BB), Italian Association for Cancer Research (AIRC) (grants IG2017 no. 20564 to AB and IG2015 no. 17593 to GiC), Transcan2 (ID 189 to AB). S Bhatia acknowledges the financial support from Gesellschaft für KinderKrebsForschung e.V. This project has received funding from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 813091 (ARCH, age-related changes in hematopoiesis).
Supplementary Material
Footnotes
CP and SB have contributed equally to this work.
Supplemental digital content is available for this article.
REFERENCES
- 1.Ravindranath Y. Down syndrome and leukemia: new insights into the epidemiology, pathogenesis, and treatment. Pediatr Blood Cancer. 2005;44:1–7. [DOI] [PubMed] [Google Scholar]
- 2.Buitenkamp TD, Izraeli S, Zimmermann M, et al. Acute lymphoblastic leukemia in children with Down syndrome: a retrospective analysis from the Ponte di Legno study group. Blood. 2014;123:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullighan CG, Collins-Underwood JR, Phillips LA, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009;41:1243–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–1017. [DOI] [PubMed] [Google Scholar]
- 5.Buitenkamp TD, Pieters R, Gallimore NE, et al. Outcome in children with Down’s syndrome and acute lymphoblastic leukemia: role of IKZF1 deletions and CRLF2 aberrations. Leukemia. 2012;26:2204–2211. [DOI] [PubMed] [Google Scholar]
- 6.Nikolaev SI, Garieri M, Santoni F, et al. Frequent cases of RAS-mutated Down syndrome acute lymphoblastic leukaemia lack JAK2 mutations. Nat Commun. 2014;5:4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kubota Y, Uryu K, Ito T, et al. Integrated genetic and epigenetic analysis revealed heterogeneity of acute lymphoblastic leukemia in Down syndrome. Cancer Sci. 2019;110:3358–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shiraz P, Payne KJ, Muffly L. The current genomic and molecular landscape of Philadelphia-like acute lymphoblastic leukemia. Int J Mol Sci . 2020;21:2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laurent AP, Kotecha RS, Malinge S. Gain of chromosome 21 in hematological malignancies: lessons from studying leukemia in children with Down syndrome. Leukemia. 2020;34:1984–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanulla M, Dagdan E, Zaliova M, et al. IKZF1(plus) defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-cell precursor acute lymphoblastic leukemia. J Clin Oncol. 2018;36:1240–1249. [DOI] [PubMed] [Google Scholar]
- 11.van Dongen JJ, Macintyre EA, Gabert JA, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. [DOI] [PubMed] [Google Scholar]
- 12.Conter V, Bartram CR, Valsecchi MG, et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood. 2010;115:3206–3214. [DOI] [PubMed] [Google Scholar]
- 13.Schrappe M, Valsecchi MG, Bartram CR, et al. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood. 2011;118:2077–2084. [DOI] [PubMed] [Google Scholar]
- 14.Cario G, Leoni V, Conter V, et al. Relapses and treatment-related events contributed equally to poor prognosis in children with ABL-class fusion positive B-cell acute lymphoblastic leukemia treated according to AIEOP-BFM protocols. Haematologica. 2020;105:1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills KI, Kohlmann A, Williams PM, et al. Microarray-based classifiers and prognosis models identify subgroups with distinct clinical outcomes and high risk of AML transformation of myelodysplastic syndrome. Blood. 2009;114:1063–1072. [DOI] [PubMed] [Google Scholar]
- 16.Haferlach T, Kohlmann A, Wieczorek L, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haferlach T, Kohlmann A, Schnittger S, et al. Global approach to the diagnosis of leukemia using gene expression profiling. Blood. 2005;106:1189–1198. [DOI] [PubMed] [Google Scholar]
- 18.Fazio G, Bresolin S, Silvestri D, et al. PAX5 fusion genes are frequent in poor risk childhood acute lymphoblastic leukaemia and can be targeted with BIBF1120. EBioMedicine. 2022;83:104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmi C, Vendramini E, Silvestri D, et al. Poor prognosis for P2RY8-CRLF2 fusion but not for CRLF2 over-expression in children with intermediate risk B-cell precursor acute lymphoblastic leukemia. Leukemia. 2012;26:2245–2253. [DOI] [PubMed] [Google Scholar]
- 20.Bugarin C, Sarno J, Palmi C, et al. Fine tuning of surface CRLF2 expression and its associated signaling profile in childhood B-cell precursor acute lymphoblastic leukemia. Haematologica. 2015;100:e229–e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palmi C, Savino AM, Silvestri D, et al. CRLF2 over-expression is a poor prognostic marker in children with high risk T-cell acute lymphoblastic leukemia. Oncotarget. 2016;7:59260–59272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benard-Slagter A, Zondervan I, de Groot K, et al. Digital multiplex ligation-dependent probe amplification for detection of key copy number alterations in T- and B-cell lymphoblastic leukemia. J Mol Diagn. 2017;19:659–672. [DOI] [PubMed] [Google Scholar]
- 23.Vendramini E, Giordan M, Giarin E, et al. High expression of miR-125b-2 and SNORD116 noncoding RNA clusters characterize ERG-related B cell precursor acute lymphoblastic leukemia. Oncotarget. 2017;8:42398–42413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alaei-Mahabadi B, Bhadury J, Karlsson JW, et al. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc Natl Acad Sci U S A. 2016;113:13768–13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazzaniga G, Daniotti M, Tosi S, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–4670. [PubMed] [Google Scholar]
- 27.Roberts KG, Reshmi SC, Harvey RC, et al. Genomic and outcome analyses of Ph-like ALL in NCI standard-risk patients: a report from the Children’s Oncology Group. Blood. 2018;132:815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond J, Robbins A, Evans K, et al. Acute Sensitivity of Ph-like acute lymphoblastic leukemia to the SMAC-mimetic birinapant. Cancer Res. 2016;76:4579–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savino AM, Sarno J, Trentin L, et al. The histone deacetylase inhibitor givinostat (ITF2357) exhibits potent anti-tumor activity against CRLF2-rearranged BCP-ALL. Leukemia. 2017;31:2365–2375. [DOI] [PubMed] [Google Scholar]
- 30.Yadav B, Pemovska T, Szwajda A, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep. 2014;4:5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaliova M, Meyer C, Cario G, et al. TEL/AML1-positive patients lacking TEL exon 5 resemble canonical TEL/AML1 cases. Pediatr Blood Cancer. 2011;56:217–225. [DOI] [PubMed] [Google Scholar]
- 32.Michels N, Boer JM, Enshaei A, et al. Minimal residual disease, long-term outcome, and IKZF1 deletions in children and adolescents with Down syndrome and acute lymphocytic leukaemia: a matched cohort study. Lancet Haematol. 2021;8:e700–e710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartzman O, Savino AM, Gombert M, et al. Suppressors and activators of JAK-STAT signaling at diagnosis and relapse of acute lymphoblastic leukemia in Down syndrome. Proc Natl Acad Sci U S A. 2017;114:E4030–E4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.