Abstract
Introduction
Small cell lung cancer (SCLC) is a subtype of lung cancer with high malignancy and poor prognosis. Rapid acquisition of chemoresistance is one of the main reasons leading to clinical treatment failure of SCLC. Studies have indicated that circRNAs participate in multiple processes of tumor progression, including chemoresistance. However, the molecular mechanisms of circRNAs driving the chemoresistance of SCLC are not well specified.
Methods
The differentially expressed circRNAs were screened by transcriptome sequencing of chemoresistant and chemosensitive SCLC cells. The EVs of SCLC cells were isolated and identified by ultracentrifugation, Western blotting, transmission electron microscopy, nanoparticle tracking analysis and EVs uptake assays. The expression levels of circSH3PXD2A in serum and EVs of SCLC patients and healthy individuals were detected by qRT‒PCR. The characteristics of circSH3PXD2A were detected by Sanger sequencing, RNase R assay, nuclear-cytoplasmic fraction assay, and fluorescence in situ hybridization assay. The mechanisms of circSH3PXD2A inhibiting SCLC progression were studied by bioinformatics analysis, chemoresistance assay, proliferation assay, apoptosis assay, transwell assay, pull-down assay, luciferase reporting assay, and mouse xenograft assay.
Results
It was identified that the circSH3PXD2A was a prominently downregulated circRNA in chemoresistant SCLC cells. The expression level of circSH3PXD2A in EVs of SCLC patients was negatively associated with chemoresistance, and the combination of EVs-derived circSH3PXD2A and serum ProGRP (Progastrin-releasing peptide) levels had better indications for DDP-resistant SCLC patients. CircSH3PXD2A inhibited the chemoresistance, proliferation, migration, and invasion of SCLC cells through miR-375-3p/YAP1 axis in vivo and in vitro. SCLC cells cocultured with EVs secreted by circSH3PXD2A-overexpressing cells exhibited decreased chemoresistance and cell proliferation.
Conclusion
Our results manifest that EVs-derived circSH3PXD2A inhibits the chemoresistance of SCLC through miR-375-3p/YAP1 axis. Moreover, EVs-derived circSH3PXD2A may serve as a predictive biomarker for DDP-resistant SCLC patients.
Keywords: circSH3PXD2A, extracellular vesicles, miR-375-3p, YAP1, chemoresistance, small cell lung cancer
Introduction
Lung cancer has the highest incidence and mortality rate worldwide. In 2018, there were approximately 2.1 million new lung cancer cases and 1.8 million lung cancer deaths.1 Lung cancer can be generally categorized into two subtypes: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC).2 The proportion of SCLC is approximately 15%,3 which has a high degree of malignancy and rapid proliferation and tends to metastasize in the early stage.4,5 Due to the nature of high metastasis rate of SCLC, most patients have already developed distant metastases at the time of treatment and have therefore missed the opportunity for surgical treatment. Because of that, chemotherapy is the preferred treatment for SCLC. The widely accepted first-line therapeutic method for SCLC is chemotherapy-based comprehensive treatment. The standard chemotherapy regimen is a combined chemotherapy regimen containing platinum.5,6 Despite the fact that patients with SCLC have a high response rate to first-line therapy, the 5-year overall survival (OS) rate is merely 6% due to the rapid progression of SCLC and the development of chemoresistance.7 Therefore, one of the core topics of SCLC research is to systematically study the mechanism of chemoresistance in SCLC and develop effective molecular diagnostic methods to guide clinical chemotherapy more accurately.
CircRNAs were first discovered in RNA viruses in 19768 and were once thought to be splicing noise in organisms. In recent years, circRNAs have become a research focus in the wake of the advancement of technologies and methods such as high-throughput sequencing and bioinformatics.9 Multiple studies have manifested that circRNAs may adjust and control gene expression at multiple levels, including regulations of transcription of parental genes, homologous linear RNA cleavage and RNA-binding protein function, sponges of microRNAs, and polypeptide coding.10 A growing number of studies have shown that circRNAs participate in various physiological and pathological processes, especially in cancer. Dysregulated circRNAs participate in multiple developmental processes of malignant tumors, including tumorigenesis, growth, metastasis, apoptosis, angiogenesis, and chemoresistance.11,12 Recent studies have shown that circRNAs can positively or negatively modulate the resistance of a variety of tumors to conventional chemotherapy or immunotherapy,12,13 suggesting that circRNAs may be a new and promising biomarker for the prevention, diagnosis, and treatment of tumors, which are of great importance in scientific research or clinical fields.
Extracellular vesicles (EVs) are small nanovesicles with a diameter of approximately 30–150 nm14 that contain multiple components such as proteins, nucleic acids, lipids, miRNAs, lncRNAs, and circRNAs.15 EVs can create favorable conditions for cancer progression and metastasis by transporting their contents to recipient cells and modulating the interaction between cancer cells and surrounding cells.16 In recent years, studies have shown that EVs derived from drug-resistant tumor cells may transmit drug resistance to chemosensitive cells by delivering proteins, miRNAs, or circRNAs, thus increasing the chemoresistance of tumor cells. Meanwhile, EVs in fluids can be used as indicators to evaluate the response of patients to drug efficacy in view of their easy accessibility.17–19 However, the potential mechanism of EVs-derived circRNAs in chemoresistance is still unknown in SCLC.
Considering the important role of circRNAs in the progression of tumorigenesis and development, and the fact that few studies on the mechanism of circRNAs in the progression of SCLC, especially in chemoresistance, it is necessary to study circRNAs involved in chemoresistance of SCLC. By analyzing the transcriptome sequencing results of chemoresistant and chemosensitive SCLC cells, we find significant differences in the expression of cricSH3PXD2A between the two cell lines. CircSH3PXDA2 is a novel circRNA that has not yet been studied. The role of circSH3PXD2A in tumorigenesis and development remains unclear. Our study shows that circSH3PXD2A is downregulated in chemoresistant SCLC cells and is negatively correlated with chemoresistance in SCLC patients. Mechanistically, we find that EVs-derived circSH3PXD2A inhibits the chemoresistance of SCLC through sponging miR-375-3p to promote the expression of YAP1. Our findings provide new insights into the mechanisms of SCLC chemoresistance.
Materials and Methods
Cell Lines and Reagents
The SCLC cell lines SBC-3, H446, H69, H82 and SHP77 were purchased from ATCC, and DMS273 was purchased from AcceGen Biotech. Six human SCLC cell lines were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 incubator.20 Selleck Chemicals (Shanghai, China) provided the YAP1 inhibitor (CA3). Antibodies against YAP1 and GAPDH were obtained from Abways Technology (Shanghai, China) and antibodies against CD63 and TSG101 were obtained from Cell Signaling Technology (United States).
Clinical Samples
From July 2021 to June 2022, we collected peripheral serum samples and clinical case data from 30 SCLC patients at the First Affiliated Hospital of University of Science and Technology of China. They were all treated with cisplatin chemotherapy, with clinical disease progression as a marker of resistance. Serum samples of SCLC patients were collected at the beginning of cisplatin treatment and at the time of clinical progression. In addition, the peripheral blood serum of 30 healthy individuals in our hospital was collected as the control group. The inclusion criteria for healthy controls were no previous history of malignant tumors, normal tumor markers, and normal liver and kidney functions. The study was approved by the Ethics Committee of West District of the First Affiliated Hospital of University of Science and Technology of China (2020-kl-120). All subjects signed informed consent forms, and the information of the patients is shown in Table 1.
Table 1.
Clinical Associations Between circSH3PXD2A and Clinicopathological Variables in SCLC Patients
| Variable | Serum circSH3PXD2A | P value | EVs-Derived circSH3PXD2A | P value | ||
|---|---|---|---|---|---|---|
| Low (n=15) | High (n=15) | Low (n=15) | High (n=15) | |||
| Age (years) | ||||||
| <63.0 | 8 | 6 | 0.464 | 7 | 7 | 1.000 |
| ≥ 63.0 | 7 | 9 | 8 | 8 | ||
| Gender | ||||||
| Male | 10 | 11 | 0.690 | 10 | 11 | 0.690 |
| Female | 5 | 4 | 5 | 4 | ||
| Smoking history | ||||||
| Yes | 2 | 5 | 0.195 | 3 | 4 | 0.666 |
| No | 13 | 10 | 12 | 11 | ||
| Time to development of chemoresistance (months) | ||||||
| <7.0 | 4 | 10 | 0.067 | 7 | 7 | 1.000 |
| ≥ 7.0 | 11 | 5 | 8 | 8 | ||
| Clinical stages | ||||||
| LD | 1 | 3 | 0.283 | 1 | 3 | 0.283 |
| ED | 14 | 12 | 14 | 12 | ||
| Lymph node metastasis | ||||||
| Yes | 14 | 12 | 0.283 | 14 | 12 | 0.283 |
| No | 1 | 3 | 1 | 3 | ||
| Distant metastasis | ||||||
| Yes | 12 | 9 | 0.232 | 13 | 8 | 0.046 |
| No | 3 | 6 | 2 | 7 | ||
Abbreviations: LD, limited disease; ED, extensive disease.
Tumor Marker Detection
Peripheral venous blood samples were collected at the beginning of cisplatin treatment and at the time of clinical progression and centrifuged at 3000 × g for 10 minutes to separate serum. Tumor markers were detected by Roche electrochemiluminescence immunoanalyzer E601 (Roche, Germany). All operations were conducted strictly following the reagent and instrument instructions.
Cell Transfection
The circSH3PXD2A overexpression lentivirus was supplied by HANBIO (Shanghai, China). The circSH3PXD2A siRNA, miR-375-3p mimic or inhibitor and their negative controls were purchased from RiboBio (Guangzhou, China). The sequences of circSH3PXD2A are shown in Table S1. All operations were conducted following the reagent instructions.
Chemoresistance Assay
Small cell lung cancer cells (5000 cells per well) were seeded in 96-well plates and then treated with diluted drugs for 24 hours after cell adhesion. Cell survival was measured by CCK-8 according to the reagent instructions. The 50% inhibitory concentration (IC50) (the concentration of drug required for 50% of cells to be killed) was calculated. Once the IC50 values of SCLC cell lines to different drugs were determined, the IC50 dose of each drug was used to treat SCLC cells for subsequent chemoresistance experiments, and CCK-8 was used to detect the viability of the cells to determine the chemoresistance of the cells.
Cell Proliferation Assay
Small cell lung cancer cells (3000 cells per well) were seeded in 96-well plates. Ten microliters of CCK-8 reagent was added to the 96-well plate at different points in time. Then, the 96-well plates with cells were incubated in the incubator at 37°C for 2 h. The absorbance was detected at 450 nm.
Transwell Assay
The transwell chamber (Corning Costar) coated without (cell migration) or with (cell invasion) Matrigel was used in this assay. Briefly, digested and resuspended H446 cells were plated in the upper chamber, RPMI-1640 medium containing 20% FBS was added to the bottom of the chamber. After 48 hours of cultivation, the chamber was swabbed with cotton swabs and then fixed and stained. After observation with a microscope, the cell number that migrated and invaded to the bottom of the chamber was counted.
Quantitative Real-Time PCR
Total RNA of SCLC cells was isolated with TRIzol (TIANGEN) and reverse transcribed by HiScript II qRT Supermix (Vazyme). qPCR was conducted to quantify RNA expression levels. The RNA expression levels were analyzed by the 2−ΔΔCt method. The internal control for mRNA was GAPDH, and that for miRNA was U6. The primers for U6, miR-370-3p and miR-375-3p were designed and synthesized by RiboBio (Guangzhou, China). All primer sequences are shown in Table S1.
Cell Apoptosis Assay
All operations were conducted in accordance with detection kit (Bestbio) instructions. Briefly, SCLC cells were digested and then collected after washing twice with PBS. The collected cells were incubated with Annexin V-Alexa Fluor 660/Annexin V-FITC and PI after treatment with DDP for 48 hours. The cell apoptosis rate was detected by flow cytometry.
RNase R Treatment
Total RNA of SCLC cells was isolated with TRIzol reagent (TIANGEN) and then treated without or with RNase R (Geneseed Biotech). After incubation at 37°C for 10 min, qRT‒PCR was performed to analyze the expression of circSH3PXD2A and SH3PXD2A. GAPDH was selected as the internal reference.
Western Blot Assay
The proteins of SCLC cells were extracted with protein lysis buffer. The extracted proteins were subjected to SDS-PAGE electrophoresis and membrane transfer. The membrane that contains the transferred protein was then blocked with 5% skim milk powder at room temperature for 2 hours, and the blocked membrane was incubated with the primary antibody and the secondary antibody, respectively. Finally, the protein expression level was detected by chemiluminescence detection kit (ECL, Thermo).
Nuclear‑Cytoplasmic Fraction Assay
An RNA Subcellular Isolation Kit (Active Motif) and TRIZOL (TIANGEN) were used to separate the RNA from the nucleus and cytoplasm of SCLC cells. Then, qRT‒PCR was conducted to determine the ratio of nuclear and cytoplasm distribution. The internal control for cytoplasmic and nuclear was GAPDH and U6 respectively.
Fluorescence in situ Hybridization Assay
Cy3-labeled circSH3PXD2A FISH probes were purchased from RiboBio (Guangzhou, China). The localization of circSH3PXD2A in SCLC cells was analyzed by the fluorescent in situ hybridization kit (RiboBio, China). All operations were strictly performed in accordance with the reagent instructions.
Biotin‑coupled Probe Pull‑down Assay
Bio-miR-375-3p-WT (biotin-labeled miR-375-3p wild-type) or bio-miR-375-3p-Mut (biotin-labeled miR-375-3p mutant type) as well as bio-miR-NC (a negative control) were transfected into cells using the riboFECT CP Transfection Kit (Ribobio). After 48 hours, the transfected cell lysates were incubated with Dynabeads MyOneTM Streptavidin C1 (Invitrogen). After that, the bead-bound RNA was washed down and isolated by TRIzol (TIANGEN). The enrichment of circSH3PXD2A or YAP1 was analyzed by qRT‒PCR.
Luciferase Reporter Assay
Wild-type (WT) or mutant (Mut) circSH3PXD2A or YAP1 was synthesized and then subcloned into the pmiR-RB-ReportTM vector. MiR-375-3p mimics or miR-NC were cotransfected with WT-circSH3PXD2A, Mut-circSH3PXD2A, WT-YAP1 or Mut-YAP1 into cells. After 48 hours, the Dual-Luciferase Reporter Assay System (Promega) was used for analyzing the firefly and Renilla luciferase activities.
EVs Purification and Identification
The ultracentrifugation method was used to isolate EVs from the serum of patients with SCLC and the cell culture medium of SCLC cells. The separated serum or the medium with cell debris removed was centrifuged at 16,000 × g at 4°C for 45 min. Then, the supernatant was transferred to a new ultracentrifuge tube and ultracentrifuged at 100,000 × g at 4°C for 1 hour. After ultracentrifugation, the supernatant was discarded and the pellet was resuspended in PBS and filtrated with a 0.22 μm filter. Finally, the EVs were centrifuged at 100,000 × g at 4°C for 1 hour and dissolved in PBS. The protein concentration of EVs was determined by BCA protein assay kit. Protein markers of EVs, such as CD63 and TSG101, were used to examine the EVs. The extracted EVs were detected through Western blotting, nanoparticle tracking analysis (NTA) and transmission electron microscopy (TEM) as previously mentioned.21,22
EVs Uptake Experiment
The red fluorescent dye PKH26 (Sigma‒Aldrich) was used to stain EVs. The extracted EVs (10 μg) were dissolved and mixed with RPMI-1640 medium (serum-free) and 4 μL of PKH26. The mixture of EVs and PKH26 was incubated and centrifuged. The red fluorescent labeled EVs were then dissolved in PBS and cocultured with recipient cells for 12–24 hours. After DAPI staining, the cells containing red fluorescently labeled EVs were analyzed using a fluorescence microscope (Olympus).
Animal Experiments
To assess the function of circSH3PXD2A in the chemoresistance of SCLC. A total of 5×106 SHP77 vector and SHP77 circSH3PXD2A-oe cells were subcutaneously injected into BALB/c nude mice (4–6 weeks old). When the tumor size reached 100 mm3, the mice were randomly divided into four groups (SHP77 vector + PBS, SHP77 vector +DDP, SHP77 circSH3PXD2A-oe + PBS, SHP77 circSH3PXD2A-oe + DDP) and intraperitoneally injected with DDP (3 mg/kg) or PBS. Tumor volume was measured every three days with a caliper. After 4 weeks, the mice were sacrificed, and tumor samples were collected for qRT‒PCR, Western blotting, and histological examination.
To analyze the influence of EVs-derived circSH3PXD2A on the chemoresistance of SCLC. H446 cells (5x106) were subcutaneously injected into BALB/c nude mice (4–6 weeks). When the diameter of the tumor reached 6 mm, the mice were randomly divided into four groups (H446 vector-EVs + PBS, H446 vector-EVs + DDP, H446 circSH3PXD2A-oe-EVs + PBS, H446 circSH3PXD2A-oe-EVs + DDP) and were intratumorally injected with 50 μg of EVs or PBS and intraperitoneally injected with DDP. Tumor volume was measured every three days with a caliper.
Bioinformatics Analysis of SCLC Samples from GEO Platform
Clinical sample datasets of SCLC, GSE60052,23 GSE149507,24 and GSE1037,25 were enrolled in this study. The expression data and clinical characteristics were downloaded from the Gene Expression Omnibus (GEO) Platform (http://www.ncbi.nlm.nih.gov/geo). The GSE60052 dataset contained RNA-sequencing data of 7 normal lung tissue and 79 SCLC tumor tissues. The GSE149507 dataset contained microarray data of 18 pairs of SCLC tumor and adjacent lung tissues. The GSE1037 dataset contained microarray data of 19 normal lung tissues, 15 SCLC tumor tissues, and other types of lung cancer tissues. The mRNA expression values were normalized, log2-transformed using R (version 4.1.3), and then analyzed using GraphPad Prism software.
Statistical Analysis
GraphPad Prism 7 software (San Diego, CA, United States) and SPSS 25.0 (IBM Corp., Armonk, NY, United States) were applied for all statistical analyses. Two-tailed Student’s t-test and analysis of variance (ANOVA) were used for pairwise comparisons or multivariate analysis. The Mann‒Whitney U-test was used for data analysis of SCLC patients and healthy individuals. The receiver operator characteristic curve (ROC) was used to obtain the area under the curve (AUC) for each tumor marker. Each experiment was repeated at least three times, and all the data in our study were presented as the means ± standard deviations (SD). P < 0.05 was regarded to be statistically significant.
Results
CircSH3PXD2A Was Downregulated in Chemoresistant SCLC Cells and Negatively Correlated with Chemoresistance in SCLC Patients
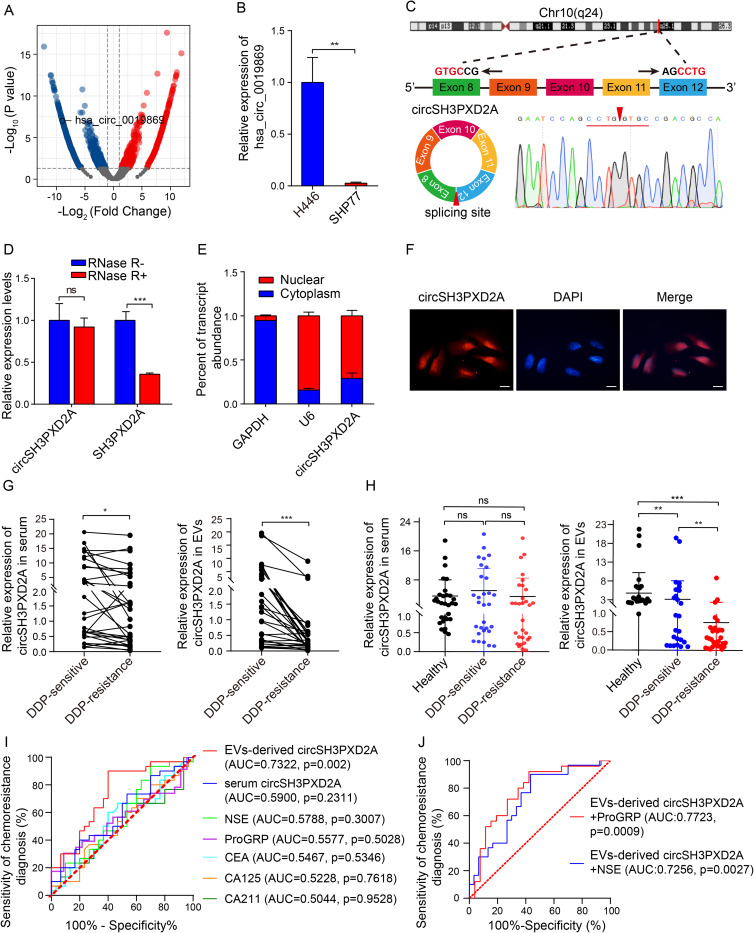
To identify candidate circRNAs related to chemotherapeutic sensitivity in SCLC, we first examined the chemosensitivity of six SCLC cell lines, including H446, SHP77, SBC-3, DMS273, H82, and H69. The IC50 (the drug dose required for 50% of cells killed) of first-line chemotherapy drugs of SCLC, including cis-platinum (DDP), etoposide (VP-16), and Adriamycin (ADM), was assayed by CCK-8 assay in these cell lines (Figure S1). According to the calculation of the relative IC50 and chemoresistance index (the numerical value of the total relative IC50 over the number of drugs), a chemoresistant cell line (SHP77) and a chemosensitive cell line (H446) were determined. The transcriptomes of these two cell lines were then sequenced. There were 1181 differentially expressed circRNAs between two cell lines. Among them, hsa_circ_0019869 was downregulated in the chemoresistant SHP77 cell line (Figure 1A), which was subsequently verified through qRT-PCR (Figure 1B). Hsa_circ_0019869 (chr10:105,371,338–105,420,872), also termed circSH3PXD2A, was derived from the SH3PXD2A gene and generated by cyclization of exons 8–12 (752 bp). The back-spliced site of circSH3PXD2A was confirmed by qRT-PCR based on divergent primers and Sanger sequencing (Figure 1C). RNase R treatment was then used to validate circSH3PXD2A. The linear SH3PXD2A was degraded, while the level of circSH3PXD2A remained almost unchanged after treatment (Figure 1D). Nuclear‑cytoplasmic fractionation assays and FISH assays showed that circSH3PXD2A was predominantly present in nuclear sections (Figure 1E and Figure 1F).
Figure 1.
CircSH3PXD2A was downregulated in chemoresistant SCLC cells and negatively correlated with chemoresistance. (A) Volcano plot for differentially expressed circRNAs between chemosensitive H446 and chemoresistant SHP77 cells. (B) qRT-PCR analysis of hsa_circ_0019869 levels in H446 and SHP77 cells. (C) Schematic diagram of circSH3PXD2A and its back-splicing site. The red arrow indicates the back-splicing site. (D) The expression levels of circSH3PXD2A and its host gene SH3PXD2A were determined by qRT‒PCR after treatment with RNase R or without RNase R. (E and F) Nuclear‑cytoplasmic fraction assay (E) and FISH assay (F) were employed to confirm the distribution of circSH3PXD2A. Scale bar, 10 μm (G) The circSH3PXD2A levels in serum (left) and EVs (right) in SCLC patients before and after cisplatin resistance (n=30). (H) qRT-qPCR analysis of the serum (left) and EVs-derived (right) circSH3PXD2A expression levels of DDP-resistant patients (n=30), DDP-sensitive patients (n=30) and normal controls (n=30). (I) Comparison of the diagnostic efficiency of EVs-derived and serum circSH3PXD2A and serum tumor markers in patients with SCLC before and after DDP resistance. (J) The diagnostic efficiency of serum tumor markers combined with EVs-derived circSH3PXD2A in patients with SCLC before and after DDP resistance. *p < 0.05 **p < 0.01, ***p<0.001.
Abbreviations: ns, no significance; DDP, cis-platinum.
To explore the clinical value of circSH3PXD2A in SCLC patients, we collected clinical case data and peripheral samples from 30 SCLC patients at the beginning of cisplatin treatment (DDP-sensitive) and at the time of the first clinical progression (DDP-resistance) respectively, and we analyzed the level of serum circSH3PXD2A. The results showed that serum circSH3PXD2A levels decreased in SCLC patients after DDP resistance. CircRNAs can be transmitted between tumor cells through EVs to regulate the occurrence and development of tumors.18,26–28 Serum EVs were also successfully isolated and identified by Western blotting, NTA, and TEM in our previous studies.22 Based on that, we studied the expression level of circSH3PXD2A in SCLC patients-derived EVs. The circSH3PXD2A levels in EVs decreased significantly after DDP resistance, and the expression difference of circSH3PXD2A in EVs was larger than that in serum (Figure 1G). We also found that EVs-derived circSH3PXD2A levels in DDP-resistance SCLC patients were much lower than in DDP-sensitive SCLC patients and healthy people. Unlike EVs-derived circSH3PXD2A, serum circSH3PXD2A showed no significant difference among the three groups (Figure 1H). After analyzing the clinical association between circSH3PXD2A and clinicopathological variables in SCLC patients, we found that DDP-resistant patients with low EVs-derived circSH3PXD2A levels had more distant metastasis (Table 1). By comparing the diagnostic efficiency of EVs-derived circSH3PXD2A, serum circSH3PXD2A and clinical diagnostic markers for lung cancer including CEA, CA125, CA211, NSE, and ProGRP, we found that EVs-derived circSH3PXD2A was superior to other clinical diagnostic markers in predicting chemoresistance to DDP in SCLC patients. Moreover, the combination of EVs-derived circSH3PXD2A and serum ProGRP had a better predictive performance for DDP-resistant SCLC patients (Figure 1I and Figure 1J, Table S2).
CircSH3PXD2A Inhibits Chemoresistance of SCLC
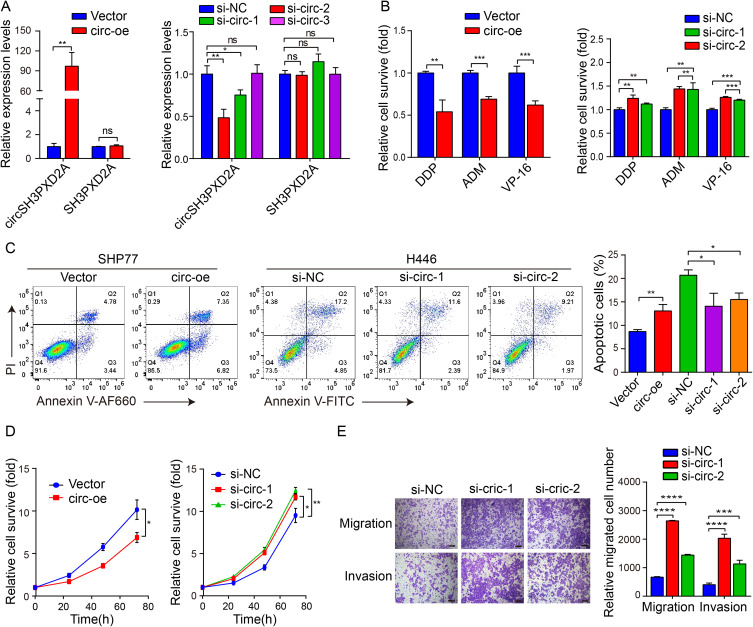
As the level of circSH3PXD2A was negatively associated with chemoresistance in SCLC, we wanted to examine whether circSH3PXD2A could inhibit chemoresistance of SCLC. SHP77 and H446 cells were transfected with circSH3PXD2A vector and siRNAs that targeted the cyclization sites of circSH3PXD2A respectively. CircSH3PXD2A vector transfection effectively increased the levels of circSH3PXD2A but not linear SH3PXD2A in SHP77 cells, and circSH3PXD2A siRNAs transfection significantly decreased the levels of circSH3PXD2A but not linear SH3PXD2A in H446 cells (Figure 2A). The circSH3PXD2A overexpression in SHP77 significantly reduced the ratio of surviving cells treated with DDP, ADM, and VP-16, and increased the ratio of cell apoptosis, while knockdown of circSH3PXD2A resulted in the opposite results (Figure 2B and Figure 2C). Also, a significant increase in cell proliferation, migration, and invasion of circSH3PXD2A knockdown H446 cells was observed when compared to the control. Additionally, circSH3PXD2A overexpression promoted cell proliferation in SHP77 cells (Figure 2D and Figure 2E). Due to the fact that SHP77 cells are semiadherent and semisuspended in nature, the migration and invasion capacities of the cells are weak.Therefore, we were unable to perform invasion and migration experiments in these cells. To conclude, our results show that circSH3PXD2A inhibits the chemoresistance, proliferation, migration, and invasion of SCLC cells.
Figure 2.
CircSH3PXD2A inhibits chemoresistance of SCLC. (A) The expression levels of circSH3PXD2A and SH3PXD2A were detected after transfection with circSH3PXD2A lentivirus or siRNA in SCLC cells. (B) Chemoresistance assays were performed to analyze the chemoresistance of SCLC cells to the chemotherapeutic drugs DDP, ADM, and VP-16. (C) Flow cytometric analyses for apoptosis in SCLC cells transfected with lentivirus or siRNA after DDP treatment. (D) CCK-8 assay was employed to analyze the effect of circSH3PXD2A on SCLC cell proliferation. (E) After transfected with circSH3PXD2A siRNA or negative control, the migration and invasion capacities of H446 cells were detected through transwell assays. Scale bar, 100 μm. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Abbreviations: ns, no significance; OE, overexpression; si, siRNA.
CircSH3PXD2A Acts as a Sponge for miR-375-3p to Suppress Chemoresistance in SCLC
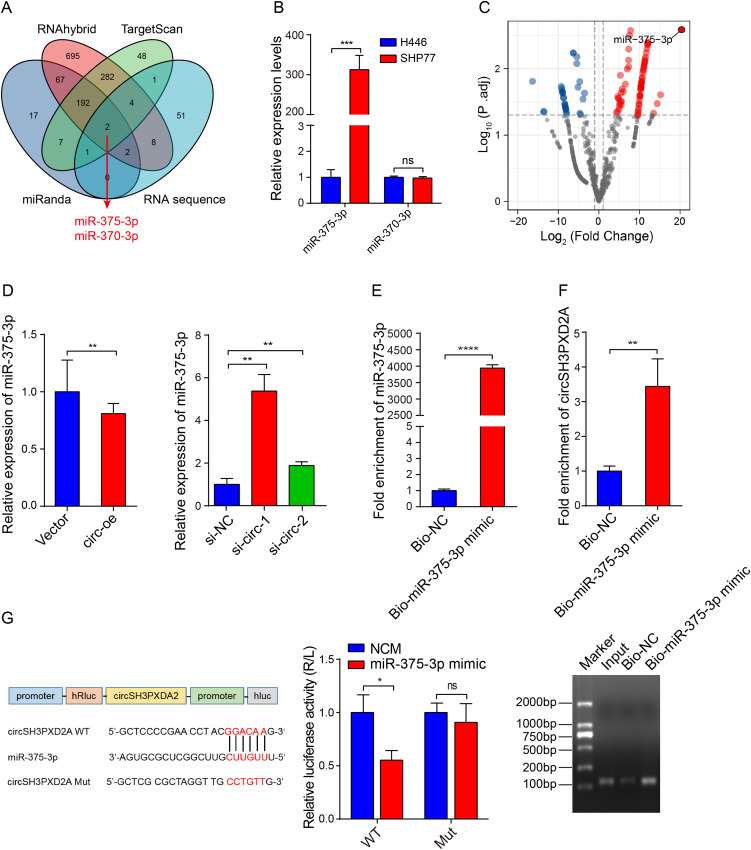
Increasing number of studies have shown that circRNAs could function by sponging miRNAs. The potential miRNAs binding to circSH3PXD2A were predicted through miRanda, RNAhybrid, and TargetScan. After intersection with our miRNA sequencing data, we found that miR-375-3p and miR-370-3p might be miRNAs interacting with circSH3PXD2A (Figure 3A). The results of qRT-PCR showed that the miR-375-3p expression level of SHP77 (chemoresistant SCLC cell line) was higher than that of H446 (chemosensitive SCLC cell line), but there was no significant difference in the expression of miR-370-3p between them (Figure 3B). By analyzing the sequencing data of chemoresistant and chemosensitive SCLC cells, we found that miR-375-3p had the most significant difference (Figure 3C). This result is consistent with our earlier research that miR-375 is dramatically increased in the plasma of chemoresistant SCLC patients.29 Further research revealed that the expression of miR-375-3p was reduced in circSH3PXD2A-upregulated cells, whereas its expression was increased in circSH3PXD2A-downregulated cells (Figure 3D). Biotin‑coupled probe pull‑down assay using biotin‑miR-375-3p revealed that circSH3PXD2A was enriched in the biotin-coupled group, indicating that it could bind with miR-375-3p (Figure 3E and Figure 3F). Furthermore, the binding site of circSH3PXD2A and miR-375-3p was shown, and the dual-luciferase reporter assay showed that miR-375-3p mimics reduced the luciferase activity of circSH3PXD2A wild-type rather than mutant-type (Figure 3G). In conclusion, these results manifest that miR-375-3p is upregulated in chemoresistant SCLC cells, and circSH3PXD2A could interact with miR-375-3p.
Figure 3.
Interaction between circSH3PXD2A and miR-375-3p. (A) Venn diagram showing circSH3PXD2A-related miRNAs screened by the intersection of miRanda, RNAhybrid, TargetScan, and circRNA sequence data. (B) Expression levels of candidate circSH3PXD2A-related miRNAs (miR-375-3p and miR-370-3p) in H446 and SHP77 cells. (C) Volcano plot for differentially expressed miRNAs in H446 and SHP77 cells. (D) The expression level of miR-375-3p was detected in SCLC cells transfected with circSH3PXD2A overexpression lentivirus or circSH3PXD2A siRNA. (E and F) Biotin-coupled miR-375-3p mimic was used to pull down circSH3PXD2A, and the expression level of circSH3PXD2A was analyzed through qRT‒PCR and agarose gel electrophoresis. (G) The sequences of circSH3PXD2A wild type and mutant type, and the detection of luciferase activities of circSH3PXD2A in H446 cells cotransfected with miR-375-3p mimic or negative controls. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Abbreviation: ns, no significance.
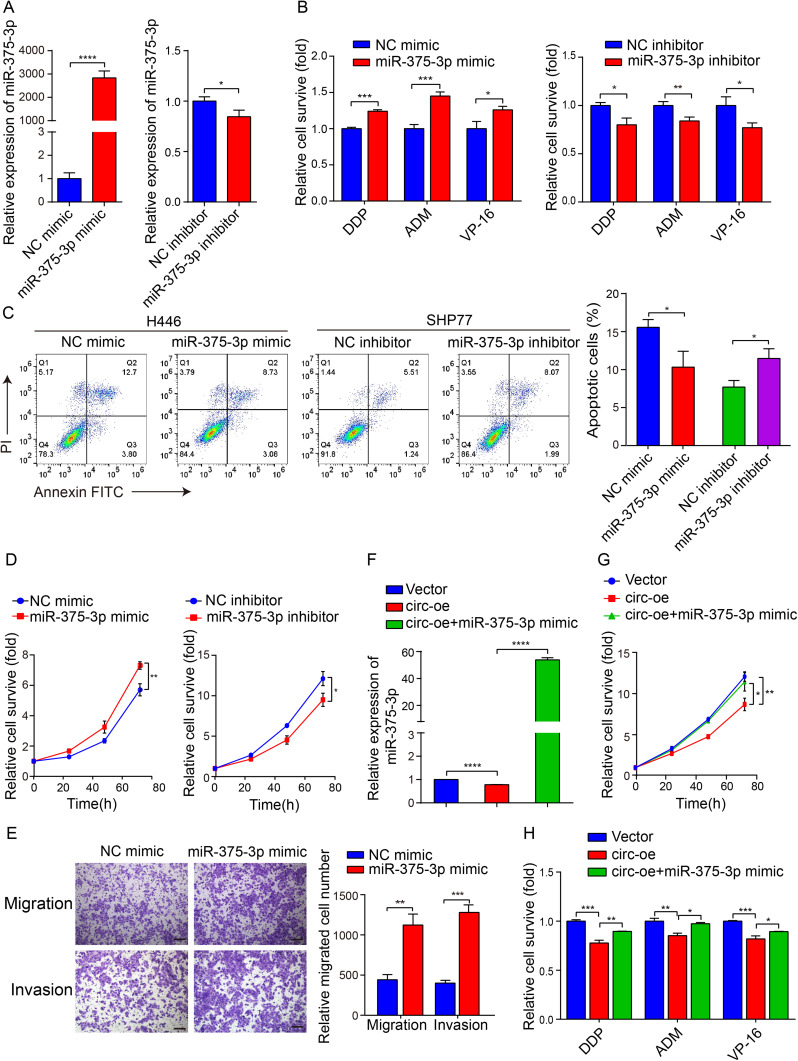
We then tried to confirm that circSH3PXD2A functions by inhibiting miR-375-3p in SCLC cells. To explore the functional role of miR-375-3p, miR-375-3p mimic or inhibitor were used to alter the expression level of miR-375-3p in SCLC cells, later detected by qRT-PCR (Figure 4A). The chemoresistance, proliferation, migration and invasion of SCLC cells were enhanced when transfected with miR-375-3p mimic. Conversely, the chemoresistance and proliferation of SCLC cells were weakened when transfected with miR-375-3p inhibitor (Figure 4B–E). Moreover, cotransfection of circSH3PXD2A and miR-375-3p mimic in SCLC cells revealed that miR-375-3p counteracted tumor suppressive effects of circSH3PXD2A, including proliferation and chemoresistance (Figure 4F–H). Our data suggests that circSH3PXD2A decreases the chemoresistance, proliferation, migration, and invasion of SCLC cells through miR-375-3p.
Figure 4.
CircSH3PXD2A acts as a sponge for miR-375-3p to suppress chemoresistance in SCLC. (A) The transfection efficiency of miR-375-3p mimic (left) or inhibitor (right) was determined by qRT‒PCR. (B) After transfected with miR-375-3p mimic (left) or inhibitor (left), the chemoresistance of SCLC cells to the chemotherapeutic drugs DDP, ADM and VP-16 was detected by chemoresistance assay. (C) The effect of miR-375-3p on the apoptosis of SCLC cells was analyzed by flow cytometry. (D) The proliferation rate of SCLC cells was detected by CCK-8 assay. (E) Transwell assays were applied to analyze the influence of miR-375-3p on the migration and invasion of SCLC cells. Scale bar, 100 μm. (F) Comparison of the miR-375-3p levels in SHP77 cells transfected with vector, circ-oe, and circ-oe+miR-375-3p mimic. (G and H) The cell proliferation ability (G) and chemoresistance (H) of SCLC cells were detected after cotransfection of circSH3PXD2A and miR-375-3p mimic. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Direct Regulation of SCLC Chemoresistance Through circSH3PXD2A/miR-375-3p/YAP1 Axis in vitro and in vivo
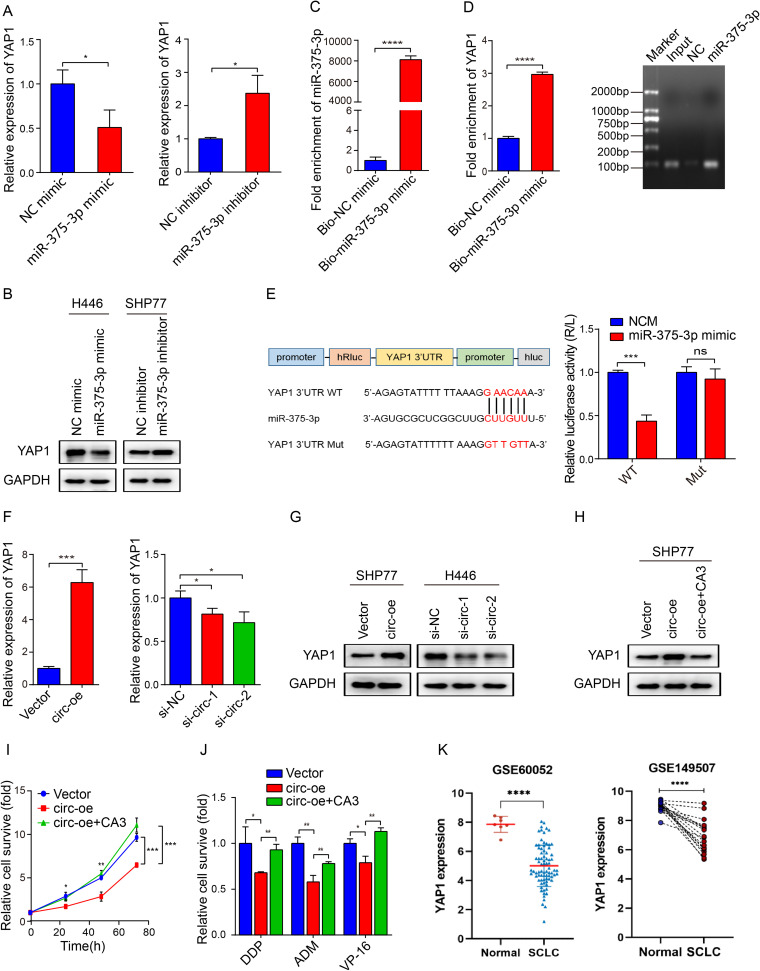
To investigate the interplay mechanism by which circSH3PXD2A/miR-375-3p axis works, we screened the targets of miR-375-3p using the PicTar, TargetScan, and mRNA sequence data, and found that YAP1 was a candidate target of miR-375-3p (Figure S2A). After detecting the mRNA expression of YAP1 and two other reported miR-375-3p downstream target genes, SLC7A11 and LDHB, we found that the mRNA expression level of YAP1, rather than the other two genes, was dramatically changed with the alteration of miR-375-3p level (Figure 5A and Figure S2B). Western blot analysis also showed that miR-375-3p could inhibit the protein expression level of YAP1 (Figure 5B). Moreover, the expression level of YAP1 decreased with increasing chemoresistance in six SCLC cell lines (Figures S1 and S2C). To further validate the association between miR-375-3p and YAP1, biotin-labeled miR-375-3p mimic was used to conduct a pull-down assay. The results indicated that the enrichment of YAP1 was higher in the biotin‑labeled miR-375-3p group than in the negative control (Figure 5C and Figure 5D). Then, the YAP1 3’UTR (wild-type or mutant) was cotransfected with miR-375-3p mimic into H446 cells to verify the mutual effect between YAP1 and miR-375-3p. The results showed that the miR-375-3p mimic reduced the luciferase activity of YAP1 3’UTR wild-type but not the mutant-type (Figure 5E). Furthermore, we also found overexpression of circSH3PXD2A elevated YAP1 expression, and knockdown of circSH3PXD2A reduced its expression (Figure 5F and Figure 5G).
Figure 5.
Direct regulation of SCLC chemoresistance through circSH3PXD2A/miR-375-3p/YAP1 axis in vitro. (A) The mRNA expression levels of YAP1 in SCLC cells transfected with miR-375-3p mimic (left) or inhibitor (right). (B) Western blotting was conducted to analyze the effect of miR-375-3p on the expression level of YAP1. (C and D) Pull-down assay revealed the enrichment of YAP1 by biotin-labeled miR-375-3p mimic. (E) Schematic diagram showing the site at which miR-375-3p binds to the 3’UTR of YAP1, and a luciferase reporter assay suggested the interaction of YAP1 with miR-375-3p. (F and G) Determination of the mRNA (F) and protein (G) expression levels of YAP1. (H) The protein expression level of SCLC cells lentivirally transfected with vector, circSH3PXD2A or circSH3PXD2A combined with CA3 (1 μM) was detected with Western blotting. (I and J) Effects of YAP1 on the proliferation (I) and chemoresistance (J) of SCLC cells. (K) Comparison of YAP1 levels in SCLC patients and healthy people. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Abbreviation: ns, no significance.
To find out whether YAP1 is critical for the action of circSH3PXD2A in SCLC, we inhibited YAP1 expression through a YAP1 inhibitor named CA3. Inhibition of YAP1 expression with CA3 (1 μM) significantly reversed circSH3PXD2A-mediated effects, including chemoresistance and proliferation ability on SHP77 cells (Figure 5H–J and Figure S2D). Moreover, we compared YAP1 expression levels in SCLC and normal tissues from three data sets (GSE60052, GSE149507 and GSE1037), and found that YAP1 was downregulated in SCLC patients (Figure 5K and Figure S2E).
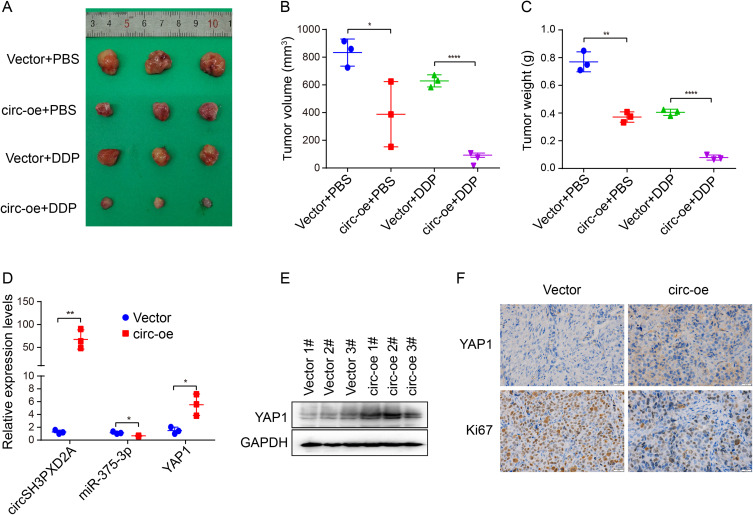
We also studied the influence of circSH3PXD2A on the chemoresistance of SCLC in vivo. We subcutaneously injected SHP77 vector and SHP77 circ-oe cells into the flanks of BALB/c nude mice and intraperitoneally injected DDP or PBS. Compared with the vector group, xenograft tumor size and weight were smaller in the circ-oe group. The circ-oe group combined with DDP showed the lowest tumor volume and weight among the four groups (Figure 6A–C). qRT‒PCR or Western blotting was employed to analyze the expression levels of circSH3PXD2A, miR-375-3p, and YAP1. The results manifested that the expression of circSH3PXD2A and YAP1 increased while miR-375-3p decreased in the circSH3PXD2A overexpression group, which coincided with the in vitro results (Figure 6D and Figure 6E). IHC staining showed that YAP1 expression increased and Ki67, which is a universal cell proliferation marker, decreased in the circSH3PXD2A overexpression group (Figure 6F). These results show that circSH3PXD2A inhibits the chemoresistance of SCLC through the miR-375-3p/YAP1 axis in vivo.
Figure 6.
Effects of the circSH3PXD2A/miR-375-3p/YAP1 axis on the chemoresistance of SCLC in vivo. (A) Representative graphs of xenograft tumors in balb/c mice that were subcutaneously injected with vector or circ-oe cells and treated with DDP or PBS. (B and C) Analysis of tumor size (B) and tumor weight (C) from balb/c mice. (D and E) The mRNA expression levels of circSH3PXD2A, miR-375-3p, and YAP1 (D) and the protein level of YAP1 (E) in xenograft tumors were analyzed through qRT-PCR and Western blotting, respectively. (F) IHC detection of YAP1 and Ki67 protein levels in vector and circSH3PXD2A-oe transfected tumors. Scale bar, 20 μm. *p<0.05, **p<0.01, ****p<0.0001.
Effect of EVs-Delivered circSH3PXD2A on Chemoresistance of SCLC in vitro and in vivo
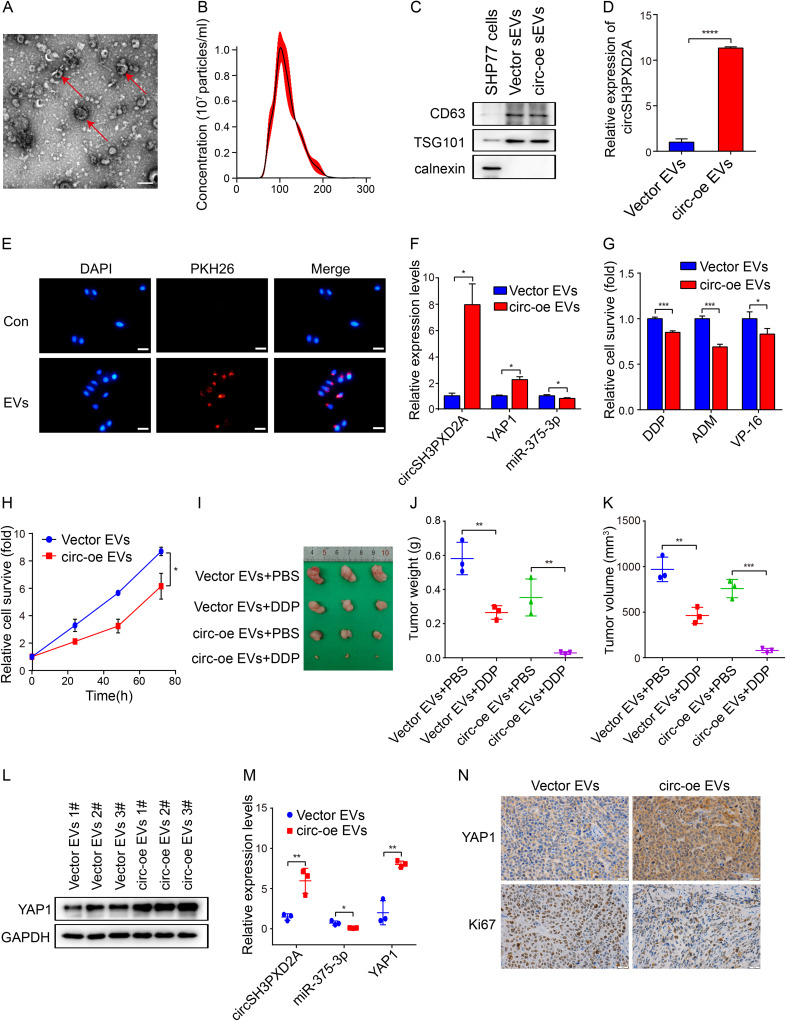
Numerous studies have shown that circRNAs can be transported to recipient cells to affect the biological function of recipient cells through EVs and to participate in tumorigenesis and development, including tumor chemoresistance.15,30,31 We wonder whether circSH3PXD2A inhibits SCLC chemoresistance through EVs. We collected and characterized the EVs from the cell culture medium of SCLC cell lines. Transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) showed that the isolated EVs had a round morphology and were 50–150 nm in diameter (Figure 7A and Figure 7B). Moreover, Western blotting showed that CD63 and TSG101, protein markers of EVs, were enriched in EVs, and the negative protein marker, calnexin was not found in EVs (Figure 7C). These results indicated that EVs were successfully collected. Then, qRT‒PCR analysis of RNA in these isolated EVs revealed that circSH3PXD2A levels were increased in the EVs of circSH3PXD2A overexpressed cells compared with the EVs of control cells (Figure 7D). To test whether the EVs could be taken by SCLC cells, the red fluorescent dye PKH26 labeled EVs isolated from SCLC cells were cocultured with H446 cells. As shown in Figure 7E, these EVs were absorbed and internalized by the H446 cells. Furthermore, after coculture with circSH3PXD2A-oe EVs, the expression of circSH3PXD2A and YAP1 was upregulated, while miR-375-3p was downregulated (Figure 7F). Functionally, the chemoresistance and proliferation of the cells cocultured with circSH3PXD2A-oe EVs were impaired (Figure 7G and Figure 7H).
Figure 7.
Effect of EVs-delivered circSH3PXD2A on the chemoresistance of SCLC in vitro and in vivo. (A) Transmission electron microscopy showed the morphology of EVs. Scale bar, 200 nm. (B) Nanoparticle tracking analysis determined EVs size. (C) Western blotting showing the EVs protein markers CD63 and TSG101 were enriched in EVs secreted by SCLC cells. Calnexin was used as a negative control. (D) The expression level of circSH3PXD2A in vector EVs and circ-oe EVs.(E) EVs uptake assay revealed that PKH26-labeled EVs were absorbed by SCLC cells. Scale bar, 100 μm. (F) Determination of the mRNA expression levels of circSH3PXD2A, miR-375-3p and YAP1 in H446 cells after coculture with vector EVs or circ-oe EVs. (G and H) Chemoresistance (G) and cell proliferation ability (H) were analyzed in H446 cells treated with vector EVs or circ-oe EVs. (I) Representative images of tumors from balb/c mice. (J and K) Comparison of tumor weight (J) and tumor weight (K) of sacrificed mice. (L and M) Analysis of the protein levels of YAP1 (L) and the mRNA levels of circSH3PXD2A, miR-375-3p and YAP1 (M). (N) IHC detection of YAP1 and Ki67 protein levels in tumors from balb/c mice. Scale bar, 20 μm. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Xenotransplantation experiments were then performed to investigate the role of EVs-derived circSH3PXD2A in vivo. H446 cells were subcutaneously injected into BALB/c nude mice to form tumors. These tumors were intratumorally injected with vector EVs or circSH3PXD2A-oe-EVs, and then intraperitoneally injected DDP or PBS one day later. Tumor size and weight measurement suggested that DDP inhibited tumor growth and circSH3PXD2A-oe-EVs strengthened this inhibitory effect (Figure 7I–K). qRT‒PCR or Western blotting manifested that circSH3PXD2A and YAP1 expression was elevated in the circ-oe-EVs group and negatively related to miR-375-3p (Figure 7L and Figure 7M). The expression of YAP1 and Ki67 was analyzed by IHC.Our results revealed that YAP1 was increased and Ki67 was decreased in the circ-oe-EVs group (Figure 7N). In conclusion, our data manifests that EVs mediate the intercellular delivery of circSH3PXD2A and suppress chemoresistance through miR-375-3p/YAP1 in SCLC.
Discussion
Rapid acquisition of chemoresistance and lack of effective follow-up treatment are the main reasons leading to failure of clinical diagnosis and treatment in SCLC. For the past few years, multiple studies have demonstrated that dysregulated circRNAs participate in multiple processes in cancer, such as chemoresistance and metastasis. Li et al found that circ_0000098 facilitates doxorubicin resistance in hepatocellular carcinoma through the miR-383/MCUR1 axis.32 Wei et al revealed that circPARD3 inhibits autophagy and promotes laryngeal squamous cell carcinoma chemoresistance by targeting miR-145-5p.12 In colorectal cancer, circ_001680 enhances resistance to irinotecan by upregulating BMI1, and circEXOC6B decreases chemoresistance to 5-fluorouracil through binding to RRAGB.13,33 However, there are limited number of studies on the connection between circRNAs and chemoresistance in SCLC. In our study, we found that circSH3PXD2A was abnormally downregulated in chemoresistant SCLC cells and that upregulation of cricSH3PXD2A inhibited chemoresistance, proliferation, invasion and migration of SCLC cells. The current clinical diagnostic markers of SCLC mainly include CEA, CA125, CA211, NSE and ProGRP. In our study, we found that EVs-derived circSH3PXD2A combined with serum ProGRP had a better diagnostic indication on DDP-resistant SCLC. This finding may provide a new method for the diagnosis and prediction of SCLC with DDP resistance.
Since circRNAs take part in the development of tumors by sponging miRNAs,34 we screened miRNAs interacting with circSH3PXD2A by bioinformatics and miRNA sequencing data and found that circSH3PXD2A interacted with miR-375-3p. In vitro and in vivo assays manifested that circSH3PXD2A inhibits the chemoresistance of SCLC through miR-375-3p. In our previous research, we found that miR-375 was dramatically increased in the plasma of chemoresistant SCLC patients.29 Mao et al found that exosomal miR-375-3p derived from SCLC cells targets claudin-1 of vascular endothelial cells to disrupt the vascular barrier and promote metastasis in SCLC.35 Our study further validated the positive regulatory role of miR-375-3p in the development of SCLC.
MiRNAs are involved in tumorigenesis and development by suppressing the expression of target genes.13 To obtain more specific knowledge of the mechanism by which circSH3PXD2A influences the chemoresistance of SCLC, we analyzed the downstream target genes of miR-375-3p and found that YAP1 was its target gene. Yes-associated protein 1 (YAP1) has been reported to play diverse functions in cancers. In SCLC, YAP1, ASCL1, NEUROD1, and POU2F3 were used as markers for subtype classification of SCLC.36 Several studies have shown that YAP1 functions as an oncogene to accelerate cancer cell survival, metastasis, tumorigenesis, and chemoresistance.37–40 However, there are also some studies suggesting that YAP1 has a tumor-inhibiting effect in tumors. Studies have revealed that YAP1 is lessened in esophageal squamous cell carcinoma and medullary thyroid carcinoma.41–43 Downregulated expression of YAP1 accelerates resistance to chemotherapy and targeted drugs in FLT3-ITD+ AML cells and docetaxel resistance in prostate cancer.44,45 In our study, we confirmed that miR-375-3p targets YAP1. Our further research showed that circSH3PXD2A promotes YAP1 expression by sponging miR-375-3p and suppresses the chemoresistance of SCLC in vitro and in vivo. Additionally, inhibition of YAP1 expression promotes chemoresistance and cell proliferation in SCLC. Our results are in support of the research by Nishikawa et al, who found that upregulation of YAP1 inhibits the proliferative capacity of SCLC cells.46 However, a recent research revealed that activation of YAP1 promotes the transition of the neuroendocrine to non-neuroendocrine fate of SCLC and increases acquired resistance in mouse SCLC cells, which is inconsistent with our conclusion.47 We hold the opinion that primary and acquired drug resistance may have contributed to the different results. Taken together, our results indicate that circSH3PXD2A suppresses chemoresistance through the miR-375-3p/YAP1 axis in SCLC.
Multiple studies have manifested that EVs derived from cancer cells act as messengers for cell communication and participate in tumorigenesis, tumor survival, metastasis, chemoresistance, and radiosensitivity in various cancers.30,48 Zang et al revealed that exosomal circ_0000337 derived from CDDP-resistant cells could increase chemoresistance, cell survival, and metastasis in CDDP-sensitive cells through the miR-337-3p/JAK2 axis in esophageal cancer cells.30 Ding et al found that TMZ-resistant cell-derived exosomal circ_0072083 could increase drug resistance of TMZ-sensitive cells in glioma.18 In our study, EVs secreted by circSH3PXD2A-overexpressing cells inhibit the chemoresistance and proliferation of SCLC in vitro and in vivo. Additionally, circSH3PXD2A-overexpressing cell-derived EVs downregulate miR-375-3p expression and upregulate YAP1 expression in SCLC. These data indicate that EVs mediate intercellular delivery of circSH3PXD2A and suppress chemoresistance through the miR-375-3p/YAP1 axis in SCLC.
Conclusions
In summary, our results demonstrated the important role of EVs-derived circSH3PXD2A in regulating the chemoresistance of SCLC cells. EVs-derived circSH3PXD2A inhibits the chemoresistance of SCLC cells by upregulating YAP1 through sponge adsorption of miR-375-3p. Furthermore, we found that EVs-derived circSH3PXD2A combined with serum ProGRP had a better indication for SCLC patients with DDP-resistance. Our study underlined a previously unknown function of circSH3PXD2A in SCLC and the potential of circSH3PXD2A as a prognostic biomarker for SCLC patients with DDP-resistance.
Acknowledgments
We thank the researchers who provided the public data set used in this study.
Funding Statement
This study was supported by the Natural Science Foundation of Anhui Province (No. 2008085MH288 and 2208085MH249), the Major program of Education Department of Anhui Province (No. 2022AH040183), the Youth Fund of Anhui Cancer Hospital (No. 2020YJQN003) and the Fundamental Research Funds for the Central Universities (No. WK9110000188).
Abbreviations
SCLC, Small cell lung cancer; NSCLC, Non-small cell lung cancer; DDP, Cisplatinum; ADM, Adriamycin; VP-16, Etoposide; 3’ UTR, 3’-untranslated region; DAPI, 4’,6-Diamidino-2-phenylindole; qRT‒PCR, Quantitative real-time PCR; IHC, Immunohistochemical; FISH, Fluorescence in situ hybridization; NTA, Nanoparticle tracking analysis; TEM, Transmission electron microscopy; CEA, Carcino-embryonic antigen; CA125, Carbohydrate antigen 125; CA211, Cytokeratins 21-1; NSE, Neuronspecific enolase; ProGRP, Pro-gastrin-releasing peptide.
Data Sharing Statement
The NGS data for this study (GSE193854) can be found on the National Center for Biotechnology Information website.
Ethics Approval and Informed Consent
This study was received and approved by The Institutional Biomedical Ethics Committee and The Institutional Animal Care and Use Committee of The First Affiliated Hospital of University of Science and Technology of China (2021-N (A) −158). All animal experiments were conducted in accordance with the Guideline for Ethical Review of Experimental Animal Welfare (GB/T 35892-2018) to ensure the welfare of experimental animals.
Consent for Publication
We obtained consent to publish this paper from all the participants of this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Davidson MR, Gazdar AF, Clarke BE. The pivotal role of pathology in the management of lung cancer. J Thorac Dis. 2013;5(4):S463–S478. doi: 10.3978/j.issn.2072-1439.2013.08.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24(28):4539–4544. doi: 10.1200/JCO.2005.04.4859 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez E, Lilenbaum RC. Small cell lung cancer: past, present, and future. Curr Oncol Rep. 2010;12(5):327–334. doi: 10.1007/s11912-010-0120-5 [DOI] [PubMed] [Google Scholar]
- 5.Bunn PA, Minna JD, Augustyn A, et al. Small cell lung cancer: can recent advances in biology and molecular biology be translated into improved outcomes? J Thorac Oncol. 2016;11(4):453–474. doi: 10.1016/j.jtho.2016.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietanza M, Byers L, Minna J, et al. Small cell lung cancer: will recent progress lead to improved outcomes? Clin Cancer Res. 2015;21(10):2244–2255. doi: 10.1158/1078-0432.CCR-14-2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Tang J, Sun T, et al. Survival changes in patients with small cell lung cancer and disparities between different sexes, socioeconomic statuses and ages. Sci Rep. 2017;7(1):1–13. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanger HL, Klotz G, Riesner D, et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristensen L, Hansen T, Venø M, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J, Meng J, Zhu L, et al. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol Cancer. 2020;19(1):66. doi: 10.1186/s12943-020-01189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers. Onco Targets Ther. 2020;13:2067. doi: 10.2147/OTT.S233672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W, Guo H, Niu M, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer. 2020;19(1):166. doi: 10.1186/s12943-020-01279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jian X, He H, Zhu J, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19(1):20. doi: 10.1186/s12943-020-1134-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Guo T, Gao X, et al. Exosome-derived noncoding RNAs in gastric cancer: functions and clinical applications. Mol Cancer. 2021;20(1):99. doi: 10.1186/s12943-021-01396-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi: 10.1186/s12943-019-1041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Zhang G, Liu C, et al. The potential role of exosomal circRNAs in the tumor microenvironment: insights into cancer diagnosis and therapy. Theranostics. 2022;12(1):87–104. doi: 10.7150/thno.64096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challagundla K, Wise P, Neviani P, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. JNCI. 2015;107(7). doi: 10.1093/jnci/djv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding C, Yi X, Chen X, et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021;40(1):164. doi: 10.1186/s13046-021-01942-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding C, Yi X, Wu X, et al. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 20.Hua Y, Dai C, He Q, et al. Autoantibody panel on small extracellular vesicles for the early detection of lung cancer. Clin Immunol. 2022;245(109175):109175. doi: 10.1016/j.clim.2022.109175 [DOI] [PubMed] [Google Scholar]
- 21.Li M, Shan W, Hua Y, et al. Exosomal miR-92b-3p promotes chemoresistance of small cell lung cancer through the PTEN/AKT pathway. Front Cell Dev Biol. 2021;9:661602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dou X, Hua Y, Chen Z, et al. Extracellular vesicles containing PD-L1 contribute to CD8+ T-cell immune suppression and predict poor outcomes in small cell lung cancer. Clin Exp Immunol. 2022;207(3):307–317. doi: 10.1093/cei/uxac006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang L, Huang J, Higgs BW, et al. Genomic landscape survey identifies SRSF1 as a key oncodriver in small cell lung cancer. PLoS Genet. 2016;12(4):e1005895. doi: 10.1371/journal.pgen.1005895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai L, Liu H, Huang F, et al. Cell-autonomous immune gene expression is repressed in pulmonary neuroendocrine cells and small cell lung cancer. Commun Biol. 2021;4(1):314. doi: 10.1038/s42003-021-01842-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones MH, Virtanen C, Honjoh D, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet. 2004;363(9411):775–781. doi: 10.1016/S0140-6736(04)15693-6 [DOI] [PubMed] [Google Scholar]
- 26.Yang C, Wu S, Mou Z, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30(3):1054–1070. doi: 10.1016/j.ymthe.2022.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong W, Xue M, Jiang J, et al. Circular RNA circ-CPA4/ let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res. 2020;39(1):149. doi: 10.1186/s13046-020-01648-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Shan W, Hong B, et al. Circulating miR-92b and miR-375 for monitoring the chemoresistance and prognosis of small cell lung cancer. Sci Rep. 2020;10(1):12705. doi: 10.1038/s41598-020-69615-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang R, Qiu X, Song Y, et al. Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p. Front Cell Dev Biol. 2021;9(673237). doi: 10.3389/fcell.2021.673237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi: 10.1002/1878-0261.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Wu A, Chen L, et al. Hsa_circ_0000098 is a novel therapeutic target that promotes hepatocellular carcinoma development and resistance to doxorubicin. J Exp Clin Cancer Res. 2022;41(1):267. doi: 10.1186/s13046-022-02482-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Wang J, Lin W, et al. circEXOC6B interacting with RRAGB, an mTORC1 activator, inhibits the progression of colorectal cancer by antagonizing the HIF1A-RRAGB-mTORC1 positive feedback loop. Mol Cancer. 2022;21(1):135. doi: 10.1186/s12943-022-01600-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen T, Jensen T, Clausen B, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 35.Mao S, Zheng S, Lu Z, et al. Exosomal miR-375-3p breaks vascular barrier and promotes small cell lung cancer metastasis by targeting claudin-1. Transl Lung Cancer Res. 2021;10(7):3155–3172. doi: 10.21037/tlcr-21-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baine M, Hsieh M, Lai W, et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol. 2020;15(12):1823–1835. doi: 10.1016/j.jtho.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu G, Huang K, Jie Z, et al. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol Cancer. 2018;17(1):170. doi: 10.1186/s12943-018-0917-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan H, Li H, Li P, et al. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J Exp Clin Cancer Res. 2018;37(1):237. doi: 10.1186/s13046-018-0910-4 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Chen M, Wu L, Tu J, et al. miR-590-5p suppresses hepatocellular carcinoma chemoresistance by targeting YAP1 expression. EBioMedicine. 2018;35(142):142–154. doi: 10.1016/j.ebiom.2018.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Liu T, Li Y, et al. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 2018;37(18):2422–2431. doi: 10.1038/s41388-018-0134-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson J, Duncavage E, Tamburrino A, et al. Overexpression of miR-10a and miR-375 and downregulation of YAP1 in medullary thyroid carcinoma. Exp Mol Pathol. 2013;95(1):62–67. doi: 10.1016/j.yexmp.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galuppini F, Bertazza L, Barollo S, et al. MiR-375 and YAP1 expression profiling in medullary thyroid carcinoma and their correlation with clinical-pathological features and outcome. Virchows Arch. 2017;471(5):651–658. doi: 10.1007/s00428-017-2227-7 [DOI] [PubMed] [Google Scholar]
- 43.Kuo YZ, Kang YR, Chang WL, et al. YAP1 acts as a negative regulator of pro-tumor TAZ expression in esophageal squamous cell carcinoma. Cell Onco. 2022;45:893–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng P, Zhang J, Zhang J, et al. Deacetylation of YAP1 promotes the resistance to chemo- and targeted therapy in FLT3-ITD AML cells. Front Cell Dev Biol. 2022;10(842214). doi: 10.3389/fcell.2022.842214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Lieberman R, Pan J, et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol Cancer. 2016;15(1):70. doi: 10.1186/s12943-016-0556-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishikawa E, Osada H, Okazaki Y, et al. miR-375 is activated by ASH1 and inhibits YAP1 in a lineage-dependent manner in lung cancer. Cancer Res. 2011;71(19):6165–6173. doi: 10.1158/0008-5472.CAN-11-1020 [DOI] [PubMed] [Google Scholar]
- 47.Wu Q, Guo J, Liu Y, et al. YAP drives fate conversion and chemoresistance of small cell lung cancer. Sci Adv. 2021;7(40):eabg1850. doi: 10.1126/sciadv.abg1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu X, Shi Y, Dong M, et al. Exosomal transfer of tumor-associated macrophage-derived hsa_circ_0001610 reduces radiosensitivity in endometrial cancer. Cell Death Dis. 2021;12(9):818. doi: 10.1038/s41419-021-04087-8 [DOI] [PMC free article] [PubMed] [Google Scholar]