Abstract
Ethylene-Insensitive3 (EIN3) is a transcription factor that works in the ethylene signaling pathway in Arabidopsis. We isolated a tobacco cDNA encoding an EIN3 homolog as a sequence-specific DNA-binding protein. The encoded protein TEIL (tobacco EIN3-like) shares 60% identity in amino acid sequence with EIN3. The DNA-binding domain was localized in the N-terminal half, which shows 92% identity in amino acid sequence with the corresponding region of EIN3, suggesting a conserved function in DNA-binding specificity. TEIL was indeed functionally similar to EIN3 because, like EIN3-overexpressing plants, transgenic Arabidopsis seedlings overexpressing TEIL cDNA exhibited constitutive triple response phenotypes. Random binding site selection analysis revealed that the consensus binding sequence for TEIL is AYGWAYCT, where Y and W represent A or C and A or T, respectively. A reporter plasmid containing the TEIL binding sites showed a 7- to 10-fold higher activation relative to that containing a mutated TEIL-binding sequence in tobacco protoplasts. A further 2- to 3-fold increase in activation was observed when a plasmid for TEIL overproduction was co-transfected, indicating that TEIL is a transcriptional activator. Moreover, nuclear extracts from ethylene-treated leaves showed an increase in DNA-binding activity specific to the TEIL-binding sequence, despite the level of the transcripts being unchanged. These observations suggest that TEIL functions as a transcription activator with a relatively redundant DNA-binding specificity, and its function may be regulated at least in part by modulation of the DNA-binding activity through ethylene signaling.
INTRODUCTION
The gaseous plant hormone ethylene affects many processes in plant growth and development, being involved in the regulation of seed germination, cell elongation, root hair formation, Rhizobium infection, fruit ripening, senescence and abscission (1). As another aspect of ethylene action, Abeles (2) proposed ‘stress ethylene’, which mediates tolerance to environmental and biological stresses. Some stresses, such as mechanical injury or pathogen attack, accelerate ethylene evolution (3), and the increased ethylene induces stress-related gene expression, probably thereby conferring resistance to some pathogens and stresses (4,5).
These effects are controlled by both ethylene biosynthesis and perception followed by successive signaling to putative transcription factors that regulate ethylene-responsive genes. Considerable findings delineating ethylene signaling have been obtained from a genetic approach with Arabidopsis mutants defective in response to ethylene (6–8). A number of ethylene-insensitive or constitutive ethylene response mutants in Arabidopsis have been isolated, based on morphological changes in etiolated seedlings treated with ethylene known as the ‘triple response’, an exaggeration of apical hook curvature, inhibition of hypocotyl and root elongation and radial swelling of the hypocotyl and root. Of these mutants, several genes have been cloned to date.
All of the four mutant alleles (etr1-1–etr1-4) of the ETR1 gene confer ethylene insensitivity and are dominant to the wild-type allele. The ETR1 gene product contains a sequence with a striking similarity to response regulators of bacterial two-component systems (9). ETR1-expressing yeast have been shown to bind ethylene, suggesting that ETR1 functions as an ethylene receptor in plants (10). It has recently been demonstrated that ethylene receptor proteins, including ETR1, negatively regulate ethylene responses and ethylene inhibits the signaling activities of these ethylene receptors (11).
In contrast to etr1 mutants, mutations at the CTR1 locus lead to a constitutive ethylene response, indicating that CTR1 is a negative regulator of ethylene signal transduction. The CTR1 gene is epistatic to ETR1 and encodes a protein with homology to Raf kinase (12). Recently, it has been found that CTR1 directly interacts with ETR1 (13).
The third gene, Ethylene-Insensitive3 (EIN3), cloned by Chao et al. (14), is genetically epistatic to ETR1 and CTR1 and, at present, located at the most downstream position of the genetic pathway. As well as etr1, ein3 mutants show a loss-of-function phenotype for ethylene responses represented by the triple response and are impaired in expression of some ethylene-regulated genes encoding a chitinase and a glutathione S-transferase (14,15). The protein encoded by the EIN3 gene has shown no significant homology to other amino acid sequences in the databases.
Chao et al. have isolated three other cDNAs encoding EIN3-related products, EIL1, EIL2 and EIL3. EILs have significant homology to the N-terminal half of EIN3 and appear to be functionally similar to each other because expression of these cDNAs (i.e. EIL1 and EIL2) can complement the eil3 mutation as well as EIN3 (14). It has been found that EIN3 and EIL1 were localized exclusively in the nucleus when they were expressed as a fusion protein with the GUS reporter in Arabidopsis protoplasts, suggesting that these proteins are transcription factors (14). More recently, Solano et al. (16) have reported that EIN3 is indeed a transcription factor, whose primary target is Ethylene-Response-Factor1 (ERF1), an early ethylene-responsive gene that encodes a GCC-box-binding protein. The EIN3/EIL proteins specifically bind to a palindromic repeat in the ERF promoter. This finding indicates a transcriptional cascade in ethylene signaling with EIN3 acting upstream of ERF1, which regulates late ethylene-responsive genes that contain a GCC-box in their promoters.
In tobacco, many ethylene-regulated genes, including the pathogenesis-related (PR) protein and GCC-box-binding protein (EREBPs) genes, have been isolated and their regulatory mechanisms studied extensively by molecular and biochemical approaches (17–24). Tobacco plants are a useful material, especially for biochemical analyses, which are often limited to Arabidopsis because of its small size. In these respects, isolation and characterization of tobacco EIN3 homologs would help progress in the study of ethylene signaling in plants. In addition, determination of the consensus DNA-binding sequence, which is not clear for EIN3 protein, would help to define ethylene-responsive elements in their target promoters. In this study, we report the isolation of an EIN3 homolog from tobacco, designated TEIL, and its consensus DNA-binding sequence. Moreover, we demonstrate that the consensus sequence binds proteins in nuclear extracts from ethylene-treated leaves but not from untreated ones.
MATERIALS AND METHODS
Plasmid constructions
To construct a reporter gene for the yeast one-hybrid screening, a fragment containing four copies of a ps1 sequence, which corresponds to a proximal site in the tobacco PR1a promoter (25), was generated by annealing and ligating oligonucleotides 5′-TCTAGAAGTACATGAATAATCACCGTGAAATCTTCAT-3′ and 5′-TCTAGATGAAGATTTC-ACGGTGATTATTCATGTACTT-3′, and was cloned in the blunt-ended SacII site of the CYC1–HIS3 construct (26). A fragment corresponding to (ps1)4–CYC1–HIS3 was excised with SacI, treated with T4 polymerase and then digested with EcoRI and cloned into the EcoRI and blunt-ended AatII sites of a yeast integration vector (26). The resulting reporter construct, YIHps1, is an integration vector harboring the HIS3 reporter gene directed by four copies of ps1. A control construct, YIH, containing the CYC1–HIS3 gene was generated as described previously (26).
For a thioredoxin (Trx) fusion, Trx–TEIL, a product amplified with the primer set NT (5′-GGATCCAGAATTCGGGCCTGCCAAAATGATG-3′) and CT (5′-CTACACAAGCTTAGAATTCTGATACCAAACCGGAACATC-3′) was digested with BamHI and HindIII and inserted into the corresponding sites of pET32b (Novagen, Madison, WI) to generate pET-TEIL. In a similar way, N-terminal deletion constructs were generated by inserting a PCR-amplified fragment into pET32a. pET-ΔC1, pET-ΔC2 and pET-ΔC3 constructs were generated by deleting SacI–HindIII, XhoI and SacII–HindIII fragments from pET-TEIL, respectively.
For construction of reporter plasmids used in the transient gene expression assay, a four-copy sequence of obs1, which is an optimal binding sequence for TEIL, was prepared by ligation of annealed products of oligonucleotide obs1, 5′-TC-GAGAACGATGTACCTGGTCGTATTG-3′ and 5′-TCGAC-AATACGACCAGGTACATCGTTC-3′. This fragment was directly cloned into a XhoI site, located upstream of a cauliflower mosaic virus (CaMV) 35S minimal promoter–GUS construct [35S(–54)–GUS] that contained a region extending to position –54 upstream of the transcription start site of the 35S promoter, the uidA gene of Escherichia coli encoding β-glucuronidase (GUS) and the nopaline synthase (NOS) terminator, to generate a obs1–GUS reporter plasmid. In the same way, an obsm2–GUS reporter plasmid was generated as a control using oligonucleotides obsm2. Effector plasmid 35S–TEIL was generated by cloning the SpeI–XhoI TEIL cDNA fragment containing the complete ORF into the XbaI and XhoI sites of a plant expression vector pCEP2, which consisted of a modified CaMV 35S promoter containing two repeat 35S enhancer regions and the NOS terminator. In a similar way, 35S–NPTII used as a control effector plasmid was generated by cloning a fragment encoding neomycin phosphotransferase II into the pCEP2 vector.
A vector, pBCEP5, which contained the CaMV 35S promoter fused with the TMV Ω 5′-untranslated sequence and NOS terminator, was used for Arabidopsis transformation. The SpeI–XhoI TEIL cDNA fragment was inserted into the corresponding sites of pBCEP5, generating pBCEP5-TEIL.
Yeast one-hybrid screening
The yeast one-hybrid screening and yeast manipulations, including culture and transformation, were performed as described previously (26). A plasmid library with pGAD424 vector (Clontech, Palo Alto, CA) was prepared from random primed cDNA which was synthesized from poly(A)+ RNA of mature tobacco leaves (Nicotiana tabacum cv Samsun NN) treated with 1 mM salicylic acid using a cDNA synthesis kit (Pharmacia). Of approximately 5 × 106 Leu+ transformants, one clone that activated the YIHps1 reporter gene but not the YIH reporter was sequenced using a dye terminator cycle sequencing kit (Applied Biosystems) and a 373A DNA sequencer (Applied Biosystems).
The full-length cDNA for TEIL was assembled by adding 486 bp obtained by 5′-RACE cloning as a template of the pGAD cDNA library and using a primer set (5′-TAATACCACTACAATGGATG-3′ and 5′-CTACACGGATCCGCATTATCAGCTTGGTAC-3′). The amplified product was digested with EcoRI and BstEII and inserted into the corresponding sites of the initially isolated TEIL cDNA, which had been subcloned into the EcoRI and SalI sites of pBluescript SK (Stratagene, La Jolla, CA). The sequence integrity was confirmed by sequencing of two independent amplified products.
Manipulation with Arabidopsis
Arabidopsis ecotype Wassilewskija was transformed with Agrobacterium tumefaciens strain LBA4404 containing the pBCEP5-TEIL or pBCEP5 plasmid by the in planta vacuum infiltration method (27). Kanamycin-resistant T1 plants were selected by plating seeds on medium containing a half strength of the MS salt, 1% sucrose and 50 µg/ml kanamycin, and transferred to soil. T2 seeds harvested from individual T1 plants were screened for the triple response phenotype, as described by Guzman and Ecker (28).
Production of Trx fusion proteins
pET-TEIL and its deletion mutant constructs were introduced into E.coli (ADA494[DE3]). The E.coli was cultured in Terrific broth until the OD600 reached ~0.8. Thereafter IPTG was added to a final concentration of 1.0 mM and the broth was further incubated for 8 h at 25°C. The collected cells were lysed and the insoluble fraction obtained after several washing steps was solubilized and purified with a nickel chelate matrix under denaturing conditions according to the manufacturer’s instructions (Novagen). Renaturation of the denatured protein was achieved by reducing the concentration of guanidine–HCl in the washing buffer.
Preparation of nuclear extracts
Nuclear extracts from ethylene-treated or untreated tobacco leaves were prepared as described (29), except for the use of nuclear isolation buffers containing 20 mM sodium pyrophosphate and 10 mM sodium fluoride. Ethylene treatment was performed by placing detached mature leaves or whole plants for 8 h in a closed chamber containing 20 p.p.m. ethylene.
Electrophoretic mobility shift assays
DNA probes were generated by annealing oligonucleotides spanning the regions of interest and by filling in the single-strand overhangs with [α-32P]dCTP using the Klenow enzyme. Sequences of double-stranded oligonucleotides for obs1 and ps1, the optimal binding sequence of TEIL and the proximal site of the tobacco PR1a promoter (25), respectively, are represented in Figure 4. The G-box oligonucleotides are 5′-TCGACG-CCACGTGGCCC-3′ and 5′-TCGAGGGCCACGTGGCG-3′. Oligonucleotide IIa, containing site IIa of the rice proliferating cell nuclear antigen promoter, was as described previously (26).
Figure 4.
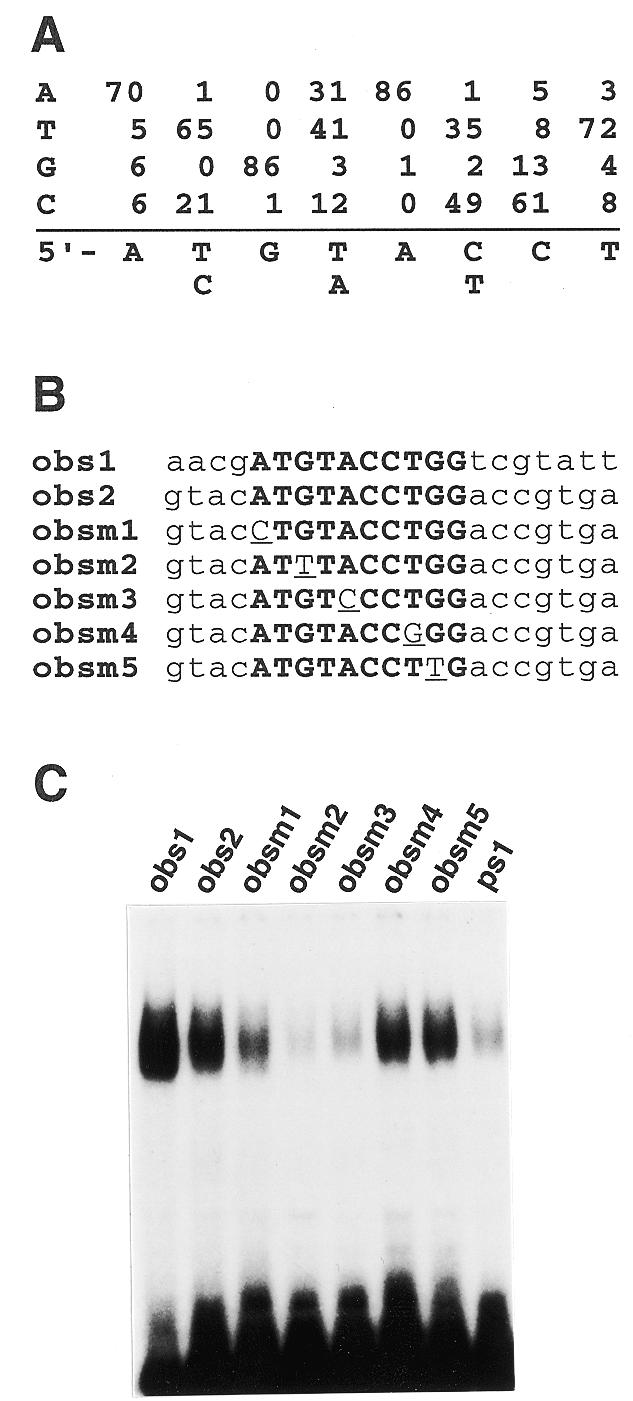
Optimization of the binding sequence of TEIL protein. (A) A consensus DNA-binding sequence of TEIL. The consensus binding sequence was determined by a random binding site selection method. A total of 87 binding sites of TEIL were sequenced and the consensus sequence was deduced from their aligned sequences. (B) Mutational analysis of the optimized TEIL-binding sequence. The sequences of obs1, with the highest binding affinity, and its mutants are represented. Bold letters represent the consensus sequence of TEIL. Lower case represents the flanking sequence in obs1 to be exchanged in obs2. Underlining indicates mutated nucleotides. (C) EMSA with the mutant oligonucleotide probes. Oligonucleotides with the sequences shown in (B) were 32P-labeled and incubated with the recombinant Trx–TEIL protein.
The binding reaction proceeded in a volume of 10 µl which contained 50 fmol of the oligonucleotide probe, 0.2–4.0 µg poly(dI·dC), 1× binding buffer (20 mM HEPES–KOH, pH 7.8, 80 mM KCl, 1 mM EDTA, 1 mM DTT, 0.05% BSA and 10% glycerol) and 50–100 ng of recombinant protein or 2 µg of nuclear extract proteins. The mixtures were incubated for 30 min at room temperature and loaded on native 5% polyacrylamide gels. Electrophoresis was conducted at 4 V/cm for 40 min with × TBE buffer at room temperature. Gels were dried and autoradiographed using an intensifying screen.
Random binding site selection
Double-stranded oligonucleotide BS18N, which contained random 18mer sequences flanked by BamHI, EcoRI, SalI and XhoI sites, was prepared by annealing oligonucleotides bs-18N [5′-CGCGAATTCGGATCCAAGC(N)18CGTTGTCGACTC-GAGTCGA-3′] and rs-1 (5′-TCGACTCGAGTCGACAACG-3′), followed by primer extension with Klenow fragment. The recombinant Trx–TEIL protein (250 ng) was incubated with 1.5 µg of BS18N in 50 µl of 1× EMSA binding buffer containing 0.05% BSA and 1.0 µg of poly(dI·dC). The DNA–protein complex was separated by polyacrylamide gel electrophoresis and the DNA of the complex was eluted from the gel and dissolved in 20 µl of TE. The recovered DNA was amplified by PCR with the primer set rs-1 and fb-1 (5′-CGCGAATTCGGATCCAAGC-3′). The PCR amplification was carried out in 30 µl of a cocktail containing 10 mM Tris–HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.1% Triton, 100 µM each dNTP, 0.2 µM each primer, 2 µl DNA template and 1 U Taq DNA polymerase (Toyobo, Japan) at 95°C for 30 s, 55°C for 30 s and 70°C for 60 s for 20 cycles. The PCR product (30 µl) was extracted with phenol/chloroform and ether, and 10 µl was subjected to a subsequent round of selection. For the third and fourth rounds of selection, the amount of template DNA and the number of PCR cycles were reduced to 2–5 µl and 15, respectively, to avoid generation of concatenated high molecular weight DNA. The DNA from the fourth round of selection was amplified by PCR (27 cycles), digested with BamHI and SalI, purified on a 20% polyacrylamide gel and cloned into a pGEM3Z(+) vector (Promega) for sequencing.
Transient gene expression assay
Transfection of plasmids into tobacco mesophyll protoplasts for the transient gene expression assay was performed as follows. Fully expanded leaves from young plants were sterilized with a 2% CaClO solution and then digested with an enzyme solution [1% cellulase onozuka R10 (Yakult, Japan), 0.02% pectolyase Y-23 (Kikkoman, Japan), 5 mM CaCl2 and 0.5 M mannitol, at a final pH of 7.2] at 28°C for 2 h. Protoplasts were collected and washed with 0.5 M mannitol twice and then resuspended in 0.5 M mannitol supplemented with 5 mM HEPES–KOH, pH 6.5, at a density of 6 × 105 cells/ml. A 0.5 ml aliquot of the protoplast suspension was mixed with 5 µg of reporter plasmid and 5 µg of effector plasmid, and a single rectangular pulse with a field strength of 450 V/cm and a pulse length of 30 ms was delivered to the mixture on ice. The protoplasts were cultured in medium (30) containing 0.2 M sucrose and 0.2 M mannitol at 28°C for 48 h in the dark. GUS activity was determined with 4-methylumbelliferyl-d-glucuronide (31).
RESULTS
TEIL is a tobacco homolog of EIN3
We have previously found that a proximal site (ps1) in the tobacco PR1a promoter is bound by nuclear factors present in healthy leaves of Nicotiana tabacum cv Samsun NN but not in the interspecific hybrid of Nicotiana glutinosa × Nicotiana debneyi, which constitutively produces the acidic PR1 protein (25). We isolated a cDNA clone that shows specific binding to the ps1 sequence using the yeast one-hybrid system. The cDNA in vector pGAD424 activated a HIS3 reporter gene downstream of a ps1 site, (ps1)4–CYC1–HIS3, but not a ps1-less reporter gene, CYC1–HIS3.
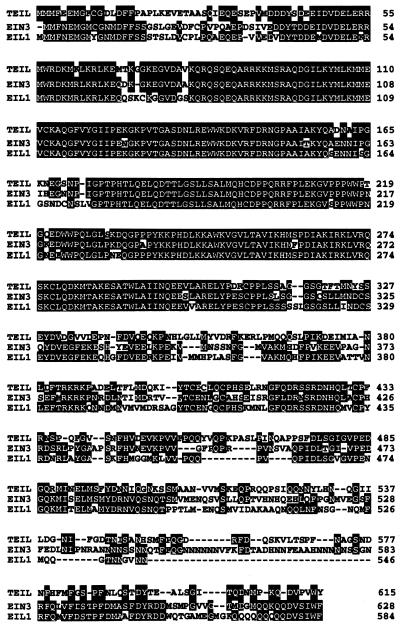
Sequence analysis revealed that an ORF of the cDNA in pGAD424 was in-frame with the GAL4 activation domain and encoded 534 amino acid residues. The extended N-terminal region isolated by a PCR-based cloning strategy encoded 81 amino acid residues and, thus, the full-length cDNA encoded 615 amino acid residues. In the deduced amino acid sequence, there was no motif characteristic of a DNA-binding protein, such as bZIP, zinc finger or basic helix–loop–helix. We, however, found that this protein shared significant homology with Ethylene-Insensitive3 (EIN3) when we used the deduced amino acid sequence as a query for the SWISS-PROT database (Fig. 1).
Figure 1.
Amino acid sequence comparisons between the TEIL, EIN3 and EIL1 polypeptides. White characters with black boxes indicate amino acids identical in all three proteins. The nucleotide sequence of the TEIL cDNA has been submitted to DDBJ under accession no. AB015855.
EIN3 is encoded by an Arabidopsis gene responsible for an ethylene-insensitive mutant, ein3, and acts downstream of ETR1, CTR1 and EIN2 in the ethylene signaling pathway (14). The deduced protein, designated TEIL (tobacco EIN3-like), shares 60% identity with the overall EIN3 amino acid sequence and 92% with the N-terminal halves that extend from residues 80 to 300 in these amino acid sequences (Fig. 1). The TEIL protein is also highly homologous to EILs from Arabidopsis (58–35% identity), which have been isolated as proteins related to EIN3 (14), with EIL1 being the most closely related to TEIL (Fig. 1) and EIL2 and EIL3 the most distantly related. In the degree of overall similarity and the presence of a basic domain IV, which is characteristic of these proteins except for EIL2, it seems that TEIL is closest to EIN3 or EIL1 among the Arabidopsis EIN3 family. However we failed to determine which of the proteins, EIN3 or EIL1, would function as an ortholog for TEIL.
Phenotypes of transgenic Arabidopsis seedlings overexpressing the TEIL cDNA
Seedlings of EIN3- or EIL1-overexpressing Arabidopsis have been shown to exhibit a constitutive triple response phenotype even in the absence of ethylene (14). To determine whether TEIL is functionally similar to EIN3 or EIL1, we introduced the TEIL cDNA under control of the strong and constitutive promoter CaMV 35S into Arabidopsis. Two independent transformed lines were produced, and expression of the transgene was confirmed by northern blot analysis (data not shown). Four-day-old etiolated T2 generation seedlings of these transgenic lines were examined for ethylene response phenotypes in the presence or absence of 1-aminocyclopropane-d-carboxylic acid (ACC), a precursor of ethylene (Fig. 2). Control seedlings, containing only the vector transgenes, showed triple response phenotypes in the presence of ACC but were highly elongated in its absence. TEIL-expressing seedlings showed a triple response under both conditions. These observations indicate that TEIL is functionally similar to EIN3 or EIL1.
Figure 2.
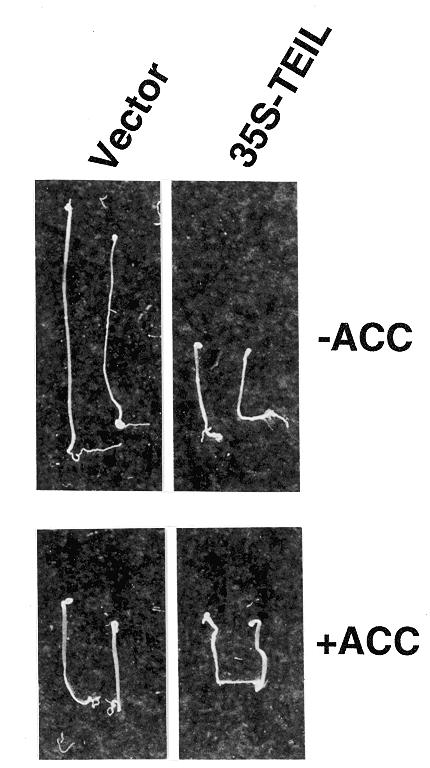
Overexpression of TEIL cDNA in Arabidopsis plants. The left and right panels show transgenic Arabidopsis seedlings containing vector DNA as a control and the TEIL cDNA under control of CaMV 35S promoter, respectively. The seedlings were germinated in the dark in the presence (ACC) or absence (air) of 10 µM ACC for 4 days.
The DNA-binding domain is localized in the N-terminal region
To delineate the region responsible for the DNA binding of TEIL, several deletion mutants of TEIL were generated and tested for DNA-binding ability. Recombinant thioredoxin (Trx) fusions with full-length TEIL and the N- or C-terminal deletion mutants were produced and purified on an affinity column (Fig. 3A) and then subjected to electrophoretic mobility shift assay (EMSA) with an obs1 probe DNA which exhibits strong affinity for TEIL binding, which will be described later. Because some recombinant proteins, especially those containing the N-terminal region, were hardly expressed in E.coli, the quantities of these recombinant proteins in E.coli extracts were estimated by western blot analysis with S protein (data not shown). Trx-ΔN1, a mutant whose N-terminal region extending from residue 1 to 81 of the TEIL amino acid sequence was deleted, retained the ability to bind the probe DNA, although its affinity was reduced to ~30% relative to the full-length (WT) protein (Fig. 3B). Further deletion of the N-terminal region to residues 93, 132 and 160, corresponding to ΔN2, ΔN3 and ΔN4, resulted in a complete loss of DNA-binding ability. In addition, ΔC1 and ΔC2, mutants whose C-terminal regions extending from residues 413 to 615 and 303 to 615, respectively, were deleted, had 10–15% activities relative to the WT protein. Further deletion to residue 255, corresponding to ΔC3, resulted in a complete loss of activity.
Figure 3.
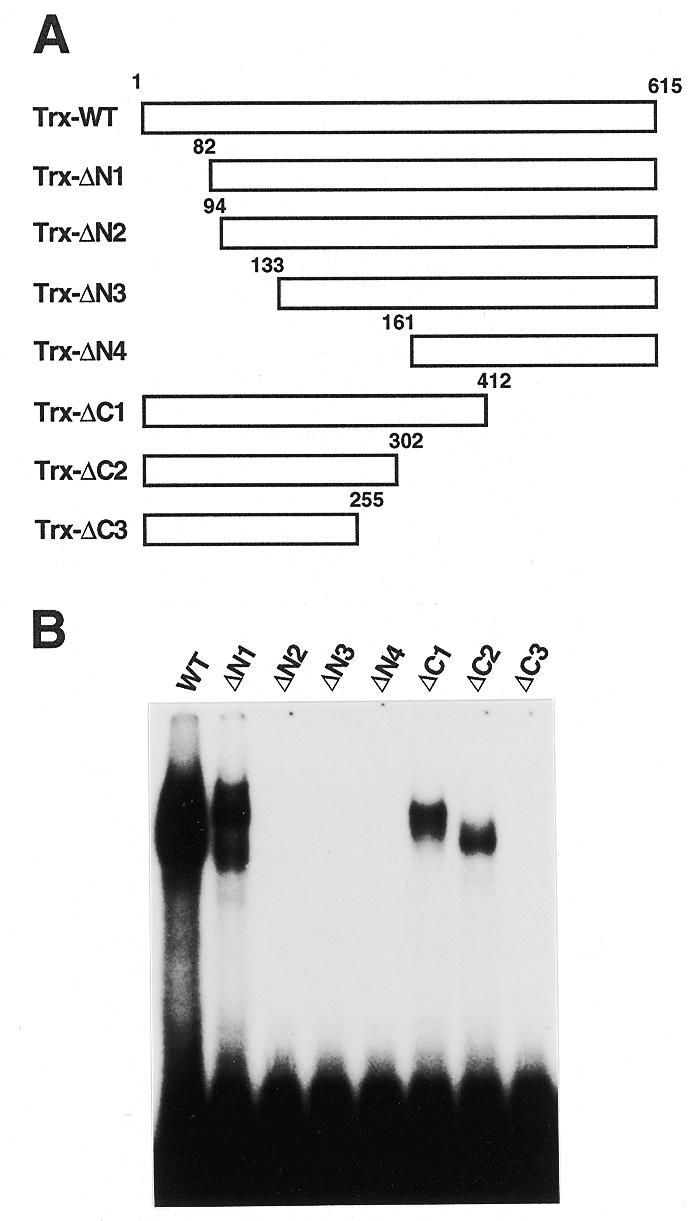
Localization of the DNA-binding domain of TEIL. (A) Schematic representation of deletion mutants of TEIL protein. A series of N-terminal (ΔN1–ΔN4) or C-terminal (ΔC1–ΔC3) deletion mutant proteins and the full-length protein (WT) were produced as Trx fusions. (B) An EMSA with the TEIL deletion mutants. Approximately 50 ng of the Trx protein fused with the TEIL deletion mutants was incubated with the 32P-labeled obs1 probe.
The DNA-binding activity of the C-terminal deletion mutants was also confirmed by in vivo experiments using a yeast one-hybrid assay with the (ps1)4–CYC1–HIS3 reporter. The ΔC3 construct cloned in pGBT9 did not activate the reporter gene whereas the ΔC2 construct did, but only weakly relative to the wild type (data not shown). These results indicate that the region extending from amino acid residue 82 to 302, at least, is necessary for DNA binding and that regions extending further toward the N- and C-termini were required for full DNA-binding activity.
Optimal binding sequence of TEIL
To determine a consensus DNA-binding sequence for TEIL, we performed a random binding site selection analysis with the recombinant fusion protein Trx–TEIL. After four rounds of selection, the selected oligonucleotides were cloned into a plasmid and 87 inserts from independent clones were sequenced. The aligned sequences revealed that the consensus sequence for TEIL is A(T/C)G(A/T)A(C/T)CT (Fig. 4A). This sequence matches 6 bp of a sequence in ps1 (ATGAATAA).
We further determined the relative affinities of 39 fragments randomly selected from the 87 sequences by using them as probe DNAs for EMSA. Although these sequences had a broad range of affinity for TEIL, almost all of them that showed high affinity for TEIL correlated with the consensus sequence (data not shown).
In the consensus sequence A(T/C)G(A/T)A(C/T)CT the underlined nucleotides would appear to be required for strong binding to TEIL, given their high degree of conservation in the selected sequences. To confirm this, we further conducted EMSA analysis using oligonucleotide probes mutated in these nucleotides (Fig. 4B). We used one of the selected sequences with the highest affinity for the probe as an optimal binding sequence and named it obs1. At first, to examine the effect of the sequence flanking the consensus sequence in the obs1 oligonucleotide on TEIL binding, the flanking sequence was exchanged for 11 other nucleotides to generate obs2. The obs2 sequence had a similar activity in binding to TEIL as obs1, indicating little effect of the flanking sequence in obs1 on binding (Fig. 4C, lane 1).
A mutant for the first nucleotide of the consensus sequence with an A→C conversion, obsm1, had about half the affinity of obs2 for binding. Mutation of the third or fifth nucleotide (obsm2 or obsm3) resulted in a significant reduction in TEIL binding, indicating the absolute necessity of these nucleotides for binding. Unexpectedly, mutation of nucleotide 8 by a T→G conversion (obsm4) did not affect binding affinity. Nucleotide 9, which was biased mainly to A or G in the binding selection, also appeared to be unaffected by the mutation (Fig. 4C, lane 7). In addition, the obs1 sequence had a much higher affinity for TEIL than ps1 (Fig. 4C, lane 8). These results suggest that A1 and T8 in the consensus made moderate and small contributions, respectively, to TEIL binding. However, other nucleotide conversions (e.g. a T→A instead of T→G conversion of nucleotide 8) or double mutations (e.g. A→C and T→G at nucleotides 7 and 8, respectively) may give different results from those in this analysis.
Activation of the obs1 reporter gene in tobacco protoplasts and transactivation by TEIL
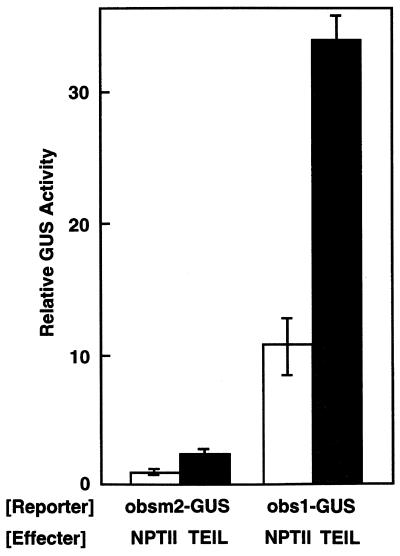
We then examined the action of the TEIL-binding sequence as a transcriptional regulatory element in tobacco mesophyll protoplasts using an obs1 reporter gene (Fig. 5). When the obs1 reporter construct (obs1–GUS), a bacterial β-glucuronidase (GUS) gene fusion with four copies of obs1 located upstream of the CaMV 35S(–54) promoter, was transfected into tobacco mesophyll protoplasts, a 7- to 10-fold increase in the activity of GUS was observed relative to a control reporter construct (obsm2–GUS), which contained four copies of obsm2 instead of obs1 in the obs1–GUS reporter. This observation suggests that an active form of TEIL or a related protein was present in tobacco mesophyll protoplasts and activated the reporter gene through specific binding to the obs1 sequence. Moreover, when both the obs1–GUS reporter and an effector plasmid (35S–TEIL), containing the TEIL cDNA under control of the 35S promoter, were co-transfected into tobacco mesophyll protoplasts, 2- to 3-fold further activation of the reporter gene was observed relative to a control effector construct (35S–NPTII), the neomycin phosphotransferase gene driven by the 35S promoter. In contrast, co-transfection with the obsm2–GUS reporter construct resulted in only low levels of activation of the reporter gene. These results indicate that TEIL has a transcriptional activation function and exerts it through sequence-specific binding to obs1.
Figure 5.
Activation of the tebs reporter gene in tobacco protoplasts and transactivation by TEIL overexpression. Transient expression assays were carried out with tobacco mesophyll protoplasts. The reporter plasmid consisted of a 35S(–54)–GUS gene containing a tetramer of obs1 (obs1–GUS) or a tetramer of obsm2 (obsm2–GUS). The effector plasmid (35S-TEIL) carried the TEIL cDNA under control of the CaMV 35S promoter. A control effector (35S–NPTII) contained the NPTII coding sequence instead of the TEIL cDNA. Five micrograms of the reporter plasmid, either alone or with the effector plasmid, was transfected into the protoplasts by electroporation and the activity of GUS was measured 48 h after culturing.
Enhanced tebs (TEIL-binding sites) binding activities in nuclear extracts from ethylene-treated leaves
The recombinant TEIL protein bound strongly to obs1, a high affinity sequence of TEIL-binding sites (tebs). To confirm that there is binding activity for tebs in tobacco nuclear extracts, we conducted EMSA with a DNA probe containing four copies of obs1. Whereas nuclear extract from healthy tobacco leaves contained little activity binding to the probe, the extract from ethylene-treated leaves had sufficient activity to make a clear shifted band (Fig. 6A). These nuclear extracts contained almost equal activities for binding to the G-box sequence and a site IIa element that interacts with PCFs, nuclear factors bound to promoter elements in the rice proliferating cell nuclear antigen gene (26; Fig. 6A). This result indicates quantitative and qualitative equivalence in ethylene-independent DNA-binding activities of the two extracts. Formation of the obs1–protein complex was abolished by the addition of excess obs1 competitor oligonucleotides (Fig. 6B). On the other hand, excess obsm2 or obsm3 oligonucleotides, which have very weak TEIL-binding activity, had little effect on the competition of DNA binding, indicating the presence of DNA-binding activity specific for tebs in nuclear extracts from ethylene-treated tobacco leaves but not from untreated leaves (Fig. 6B). It is likely that these tebs-binding activities stimulated by ethylene treatment were derived from TEIL or related proteins because their sequence preferences were very similar.
Figure 6.
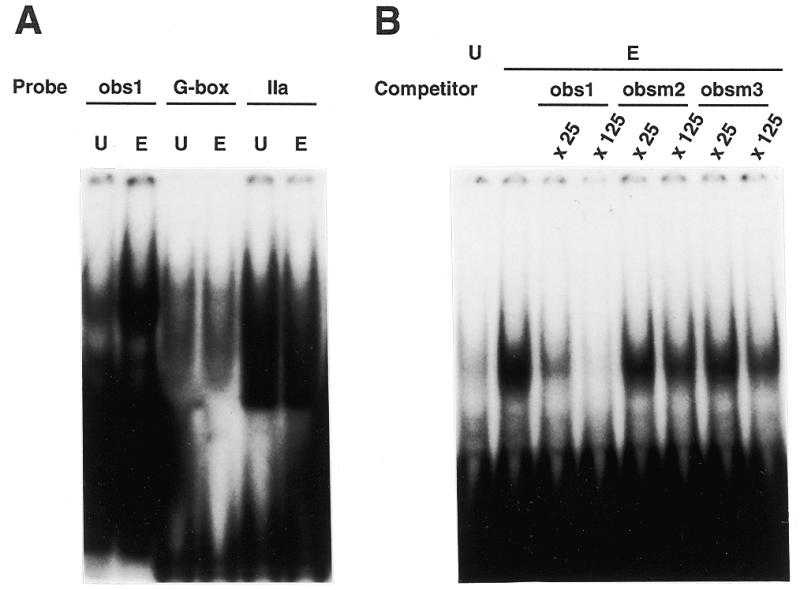
Increased tebs-binding activities in ethylene-treated tobacco leaves. (A) An EMSA for obs1-binding activity in nuclear extracts. Nuclear extracts (2.5 µg) prepared from ethylene-treated (E) or untreated healthy tobacco leaves (U) were mixed with a 32P-labeled DNA probe containing four copies of the obs1 sequence (obs1), the G-box (G-box) or a site IIa element (IIa). (B) Competition binding analysis with double-stranded oligonucleotides. The nuclear extracts from ethylene-treated leaves were mixed with the obs1 probe. Subsequently, a 25- and a 125-fold molar excess of double-stranded oligonucleotides of obs1, obsm2 and obsm3 were added as competitor DNAs to the EMSA reaction.
DISCUSSION
The TEIL protein shares 92% identity with the N-terminal halves, extending from residues 80 to 300, of the EIN3 and EIL1 amino acid sequences. This region is also highly conserved among the EIN3 and EIL proteins, with 60–89% identity, suggesting an essential function of the N-terminal region. The function of the conserved region seems to be primarily DNA binding, as shown in the localization analysis of the DNA-binding domain. The similarity between the region responsible for DNA binding of TEIL and EIN3 or EILs suggests that the DNA sequence recognized by EIN3 or EILs is identical or highly similar to that of TEIL. This is consistent with our observation that transgenic Arabidopsis seedlings overexpressing the TEIL cDNA exhibit constitutive triple response phenotypes, like EIN3- and EIL1-overproducing plants.
The amino acid sequence necessary for DNA binding of TEIL does not contain any known DNA-binding motifs but contains a region rich in α-helical structure as predicted by computer analysis. This helical structure, which also exists in EIN3 and EILs, seems to be involved in DNA binding and protein–protein interactions, including homodimer and heterodimer formation. In addition, two clusters of conserved basic amino acids, corresponding to basic domains I and II of EIN3, are found in the N-terminus of TEIL. Because deletion of the region containing these domains led to complete loss of DNA-binding activity, it is likely that the α-helices and the basic domains constitute a novel DNA-binding domain.
Another noticeable feature of the DNA-binding region of TEIL is that a broad region appears to be required for full DNA-binding activity. Removal of 203 amino acid residues from the C-terminus resulted in a considerable reduction in the binding activity, indicating that almost all of the TEIL protein is required for full DNA-binding activity. There might be a subdomain in the C-terminal half, which acts to support the function of the N-terminal domain. At the C-terminus, however, a transcriptional activation domain has been found to be restricted to within a region of 134 amino acids by a yeast assay (data not shown).
The function of TEIL as a transcriptional activator could be conferred through a signaling factor for ethylene detection. The tebs reporter gene was considerably activated in protoplasts, probably by endogenously activated TEIL or related proteins. In protoplasts, stress-inducible genes are activated by elicitor-like factors released from the cell wall during preparation of the protoplasts (32) or in the cell wall-digesting enzyme material itself and/or by the direct wounding effect, which triggers ethylene evolution (3). In addition, nuclear extracts from ethylene-treated leaves exhibited increased tebs-binding activity derived from TEIL or closely related proteins. TEIL transcripts are not induced by ethylene treatment but are found in all tissues examined, with higher levels in stems, roots and wounded leaves (data not shown). Chao et al. (14) have also observed that the level of EIN3 protein is not changed by ethylene treatment. Thus, it is likely that the increased tebs-binding activity is attributable to ethylene-mediated post-translational activation of an inactive TEIL that is present in healthy tissues. These observations imply that ethylene-mediated gene activation may be controlled, at least in part, by ethylene-modulated stimulation of the DNA-binding activity of TEIL.
Solano et al. (16) have found that EIN3 directly targets the ERF1 gene by binding an element in its promoter. The ERF1 gene, which is rapidly induced by ethylene, codes for a transcription factor that binds a GCC-box, an element commonly found in promoter regions of ethylene-regulated genes. This finding presents a model of ethylene signaling by a transcriptional cascade: EIN3 directs the expression of primary targets such as ERF1, which then regulates secondary targets including basic chitinase and defensin genes. The region essential for EIN3 binding in the ERF1 promoter contains ATGTATAT and ATGCCCCT, partially overlapping in opposite orientation to each other, which are similar to the consensus TEIL-binding sequence. We have observed that this site can also be bound by recombinant TEIL protein, but with considerably less affinity than the TEIL consensus sequence (data not shown). This observation may suggest a small difference in the binding preference between TEIL and EIN3. Moreover, Solano et al. demonstrated that the EIN3-binding site shares similarity with a promoter element required for ethylene responsiveness in the tomato E4 gene. However, we detected little interaction of the E4 element with recombinant TEIL protein, although the E4 element also contains a sequence similar to the consensus TEIL-binding sequence (data not shown).
The TEIL-binding sequence is 8 bp in length, but redundant in nature; nucleotides 2, 4, 6 and 8 in the consensus are not restricted to a single nucleotide. We found sequences that showed good matches (at least six of eight nucleotides) with the consensus TEIL-binding sequence in a large number of promoter regions, including ethylene-regulated genes. For example, there are three tebs as well as two GCC-boxes in the promoter region (–503 to –358) necessary for ethylene-inducible expression of the tobacco basic-type chitinase gene, CHN48 (23). Although it is not clear whether these tebs are functionally relevant, the recombinant TEIL protein indeed binds these sequences with moderate affinity (data not shown). Given the wide prevalence of tebs in plant genomes, tebs in a promoter may function in concert with other regulatory elements.
As another possible target for TEIL/EIN3 besides ERF1, we would note certain kinds of elicitor-responsive genes. An elicitor-responsive region between –168 and –108 of the parsley PR2 promoter (33) contains three potential tebs. Interestingly, this 60 bp region does not contain the sequence (T)TGAC(C), which is necessary for elicitor responsiveness of the parsley PR1, maize PRms and potato PR-10a genes (34–36), and also shows no similarity to elicitor-responsive regions of the promoters of the PAL and 4CL gene families. This suggests the involvement of distinct classes of elicitor-responsive elements and transcription factors in the regulation of these stress-responsive genes. It is plausible that the elicitor-responsiveness of the parsley PR2 gene is mediated by ethylene and conferred by the putative tebs found in the elicitor-responsive region. This would imply that the direct target genes for TEIL and certainly for EIN3 could include elicitor-responsive genes whose promoters do not contain any known elicitor-responsive elements such as the TGAC element.
Although TEIL was initially isolated as a protein that bound to the ps1 site of the tobacco acidic PR-1a promoter, we do not have direct evidence that TEIL is involved in regulation of the PR-1a gene. Acidic type PR genes are activated by salicylic acid-mediated signaling, which is a different pathway from the ethylene and jasmonate signalings that mediate activation of the basic type PR genes. Lawton et al. (37) have observed that salicylic acid-mediated induction of the acidic PR genes is not affected in an etr1 mutant. On the other hand, it has been found that expression of the acidic PR genes is synergistically induced by combined treatment with ethylene and salicylic acid or methyl jasmonate (37,38). Hence, although the binding affinity of TEIL for ps1 is substantially lower than that of the optimal binding sequence, we are not able to exclude the possibility that TEIL affects the regulation of acidic PR gene expression.
One of the findings in this study is that EIN3 and its tobacco homolog TEIL share structural and functional similarities. Given the evolutionarily high degree of conservation between these proteins, the DNA-binding properties of TEIL, which include the DNA-binding specificity and the ethylene-mediated increase in DNA binding, also seem to be highly similar to those of its homologs from other plants, including EIN3. Thus the consensus DNA-binding sequence of TEIL would help to define ethylene-responsive elements in ethylene-regulated genes from a wide variety of plant species. On the other hand, the relatively redundant nature of the DNA-binding specificity indicates a wide prevalence of potential TEIL-binding sites in plant genomes. Further studies will be required to elucidate how TEIL specifically activates a subset of ethylene-regulated genes despite its redundant DNA-binding specificity.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Hideaki Shinshi and Professor Fumihiko Sato for helpful discussions. This work and S.K. were supported by a grant from the Core Research of Science and Technology (CREST), Japan Science and Technology Corporation.
REFERENCES
- 1.Abeles F.B., Morgan,P.W. and Saltveit,M.E.,Jr (1992) Ethylene in Plant Biology, 2nd Edn. Academic Press, New York, NY.
- 2.Abeles F.B. (1973) Ethylene in Plant Biology. Academic Press, New York, NY.
- 3.O’Donnell P.J., Calvert,C., Atzorn,R., Wasternack,C., Leyser,H.M.O. and Bowles,D.J. (1996) Science, 274, 1914–1917. [DOI] [PubMed] [Google Scholar]
- 4.Morgan P.W. and Drew,M.C. (1997) Physiol. Plant, 100, 620–630.
- 5.Knoester M., van Loon,L.C., van den Heuvel,J., Hennig,J. and Bol,J.F. (1998) Proc. Natl Acad. Sci. USA, 95, 1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ecker J.R. (1995) Science, 268, 667–675. [DOI] [PubMed] [Google Scholar]
- 7.Bleecker A.B. and Schaller,G.E. (1996) Plant Physiol., 111, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C. (1996) Trends Biochem. Sci., 21, 129–133. [PubMed] [Google Scholar]
- 9.Chang C., Kwok,S.F., Bleecker,A.B. and Meyerowitz,E.M. (1993) Science, 262, 539–544. [DOI] [PubMed] [Google Scholar]
- 10.Schaller G.E. and Bleecker,A.B. (1995) Science, 270, 1809–1811. [DOI] [PubMed] [Google Scholar]
- 11.Hua J. and Meyerowitz,E.M. (1998) Cell, 94, 261–271. [DOI] [PubMed] [Google Scholar]
- 12.Kieber J.J., Rothenberg,M., Roman,G., Feldmann,K.A. and Ecker,J.R. (1993) Cell, 72, 427–441. [DOI] [PubMed] [Google Scholar]
- 13.Clark K.L., Larsen,P.B., Wang,X. and Chang,C. (1998) Proc. Natl Acad. Sci. USA, 95, 5401–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao Q., Rothenberg,M., Solano,R., Roman,G., Terzaghi,W. and Ecker,J.R. (1997) Cell, 89, 1133–1144. [DOI] [PubMed] [Google Scholar]
- 15.Roman G., Lubarsky,B., Kieber,J.J., Rothenberg,M. and Ecker,J.R. (1995) Genetics, 139, 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solano R., Stepanova,A., Chao,Q. and Echer,J.R. (1998) Genes Dev., 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alonso E., Niebel,F.C., Obregón,P., Gheysen,G., Inzé.D, Van Montagu,M. and Castresana,C. (1995) Plant J., 7, 309–320. [DOI] [PubMed] [Google Scholar]
- 18.Eyal Y., Meller,Y., Lev-Yadun,S. and Fluhr,R. (1993) Plant J., 4, 225–234. [DOI] [PubMed] [Google Scholar]
- 19.Hart C.M., Nagy,F. and Meins,F.,Jr (1993) Plant Mol. Biol., 21, 121–131. [DOI] [PubMed] [Google Scholar]
- 20.Ohme-Takagi M. and Shinshi,H. (1995) Plant Cell, 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghothama K.G., Maggio,A., Narasimhan,M.L., Kononowicz,K.A., Wang,G., D’Urzo,M.P., Hasegawa,P.M. and Bressan,R.A. (1997) Plant Mol. Biol., 34, 393–402. [DOI] [PubMed] [Google Scholar]
- 22.Sato F., Kitajima,S., Koyama,T. and Yamada,Y. (1996) Plant Cell Physiol., 37, 249–255. [DOI] [PubMed] [Google Scholar]
- 23.Shinshi H., Usami,S. and Ohme-Takagi,M. (1995) Plant Mol. Biol., 27, 923–932. [DOI] [PubMed] [Google Scholar]
- 24.van de Rhee M.D., Lemmers,R. and Bol,J.F. (1993) Plant Mol. Biol., 21, 451–461. [DOI] [PubMed] [Google Scholar]
- 25.Hagiwara H., Matsuoka,M., Ohshima,M., Watanabe,M., Hosokawa,D. and Ohashi,Y. (1993) Mol. Gen. Genet., 240, 197–205. [DOI] [PubMed] [Google Scholar]
- 26.Kosugi S. and Ohashi,Y. (1997) Plant Cell, 9, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bechtold N., Ellis,J. and Pelletier,G. (1993) C. R. Acad. Sci. Paris Life Sci., 316, 1194–1199. [Google Scholar]
- 28.Guzman P. and Ecker,J.R. (1990) Plant Cell, 2, 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watson J.C. and Thompson,W.T. (1986) Methods Enzymol., 118, 57–75. [Google Scholar]
- 30.Nagata T. and Takebe,I. (1970) Planta, 92, 301–308. [DOI] [PubMed] [Google Scholar]
- 31.Jefferson R.A., Kavanagh,T.A. and Bevan,M. (1987) EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis K.R. and Hahlbrock,K. (1987) Plant Physiol., 84, 1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van de Löcht U., Meier,I., Hahlbrock,K. and Somssich,I.E. (1990) EMBO J., 9, 2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Després C., Subramaniam,R., Matton,D.P. and Brisson,N. (1995) Plant Cell, 7, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raventós D., Jensen,A.B., Rask,M.-B., Casacuberta,J.M., Mundy,J. and San Segundo,B. (1995) Plant J., 7, 147–155. [DOI] [PubMed] [Google Scholar]
- 36.Rushton P.J., Torres,J.T., Parniske,M., Wernert,P., Hahlbrock,K. and Somssich,I,E. (1996) EMBO J., 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- 37.Lawton K.A., Potter,S.L., Ukness,S. and Ryals,J. (1994) Plant Cell, 6, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y., Chang,P.-F.L., Liu,D., Narasimhan,M.L., Raghothama,K.G., Hasegawa,P.M. and Bressan,R.A. (1994) Plant Cell, 6, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]