Summary
While common obesity accounts for an increasing global health burden, its monogenic forms have taught us underlying mechanisms via more than 20 single-gene disorders. Among these, the most common mechanism is central nervous system dysregulation of food intake and satiety, often accompanied by neurodevelopmental delay (NDD) and autism spectrum disorder. In a family with syndromic obesity, we identified a monoallelic truncating variant in POU3F2 (alias BRN2) encoding a neural transcription factor, which has previously been suggested as a driver of obesity and NDD in individuals with the 6q16.1 deletion. In an international collaboration, we identified ultra-rare truncating and missense variants in another ten individuals sharing autism spectrum disorder, NDD, and adolescent-onset obesity. Affected individuals presented with low-to-normal birth weight and infantile feeding difficulties but developed insulin resistance and hyperphagia during childhood. Except for a variant leading to early truncation of the protein, identified variants showed adequate nuclear translocation but overall disturbed DNA-binding ability and promotor activation. In a cohort with common non-syndromic obesity, we independently observed a negative correlation of POU3F2 gene expression with BMI, suggesting a role beyond monogenic obesity. In summary, we propose deleterious intragenic variants of POU3F2 to cause transcriptional dysregulation associated with hyperphagic obesity of adolescent onset with variable NDD.
Keywords: obesity, neurodevelopmental delay, POU3F2, BRN2, autism, Prader-Willi-like syndrome, hyperphagia, 6q16.1 microdeletion
Graphical abstract

Monogenic forms of obesity have taught us about central nervous system dysregulation of food intake as a disease mechanism. We associate ultra-rare variants in POU3F2, encoding a central nervous system transcription factor, with syndromic obesity and neurodevelopmental delay in 12 individuals. Additionally, we demonstrate variant pathogenicity through in vitro analysis.
Main text
Worldwide obesity prevalence has almost tripled since 1975 and nearly 13% of the global adult population is obese (BMI > 30 kg/m2).1 While common obesity is a polygenic late-onset condition, gene discovery studies yielded more than 20 rare forms of monogenic early-onset obesity over the past two decades.2 Nevertheless, the underlying biology of both polygenic and monogenic obesity is broadly overlapping and rare disease mechanisms have been valuable sources to understand general mechanisms of weight gain and appetite control.2 Interestingly, many monogenic forms provide links to hypothalamic food intake regulation through the leptin-melanocortin pathway. Hereby, adipocytes secrete leptin into the circulation, which in turn is able to mediate satiety through signaling via specialized leptin receptor (LEPR)-expressing neurons within the hypothalamic arcuate nucleus.3,4 Consequently, activation of adjacent melanocortin 4 receptor (MC4R)-expressing neurons in the hypothalamic paraventricular nucleus (PVN) mediate appetite control.2,5 Further upstream, PVN development underlies transcriptional activity of SIM1 (homolog of Drosophila single-minded 1), another critical factor in central nervous energy expenditure. Remarkably, both LEPR (MIM: 614963), MC4R (MIM: 618406)5,6, and SIM17 (MIM: 603128) have been found defective in syndromic, Prader-Willi-like (PWL) obesity, illustrating the importance of this pathway in central nervous feeding behavior. With this study, we propose deleterious single-nucleotide variants (SNVs) in POU3F2 (POU class 3 homeobox 2, alias BRN2 [Brain 2] [MIM: 600494]) to be associated with syndromic hyperphagia-mediated obesity. As a member of the POU-III class of neural transcription factors, POU3F2 regulates hundreds of target genes; among these targets are several genes that have previously been linked to monogenic obesity, such as LEPR, MC4R, BBS7 (Bardet Biedl syndrome 7, [MIM: 615984]8), PCSK1 (proprotein convertase subtilisin/kexon type 1 [MIM: 600955]9), and PHIP (pleckstrin homology domain interacting protein [MIM: 617991]10). Remarkably in 2016, POU3F2 has already been suggested as a candidate when syndromic obesity was associated with monoallelic 6q16.1 deletions encompassing POU3F2 as the most promising driver gene.11 Additionally, a single individual with an SNV in POU3F2 was reported.12 Furthermore, de novo and truncating variants in its paralog POU3F3 have recently been shown to cause a characteristic neurodevelopmental disorder termed Snijders Blok-Fisher syndrome (MIM: 618604),13 rendering POU3F2 a promising candidate for syndromic obesity.
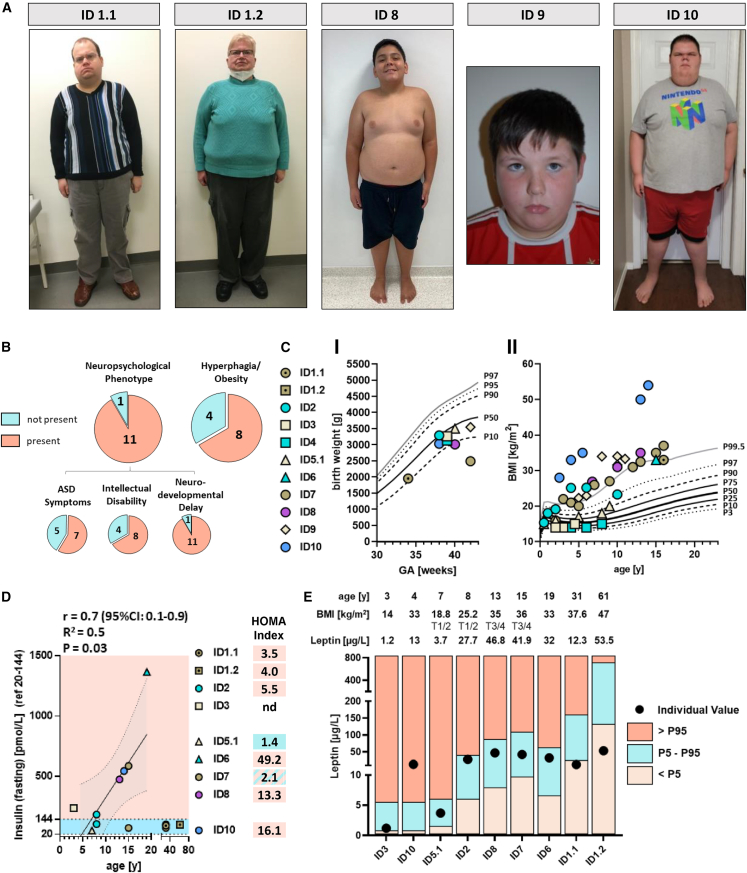
This study was initiated through identification of the following index case: ID 1.1 is a 33-year-old male born from a non-consanguineous union, with congenital anomalies of the kidney and urinary tract (CAKUT), global developmental delay, severe obesity (BMI 43 kg/m2), and autism spectrum disorder (Figure 1A, Table 1). Clinically diagnosed with Prader-Willi syndrome (PWS), this individual was genetically re-evaluated when approaching end-stage kidney disease and kidney transplant waitlisting. Instead of genetic PWS confirmation, however, exome sequencing (ES) surprisingly revealed a rare heterozygous nonsense variant in POU3F2 (c.135C>A [p.Tyr45∗] [GenBank: NM_005604.4]). The respective variant turned out to be inherited from the mother (ID 1.2), a 62-year-old female, who shared severe obesity (BMI 47 kg/m2) and moderate developmental delay but presented with normal kidney function (Figure 1A, Table 1). While birth and infantile weight records appeared normal, hyperphagia with rapid weight gain was first reported by puberty onset.
Figure 1.
Clinical images and metabolic profiles of individuals with ultra-rare POU3F2 variants
(A) Images illustrating obese body stature in the index (ID 1.1) and his mother (ID 1.2) as well as ID 8, ID 9, and ID 10.
(B) Phenotypic spectrum of individuals with POU3F2 variant.
(C) Low-to-normal birth weight is reported in individuals with rare, deleterious POU3F2 variants (<P50) (GA, gestational age) (CI). Longitudinal BMI values showing an increase over time with obesity-onset around puberty (CII).
(D) Fasting insulin values illustrating hyperinsulinemia mostly in adolescents with rare POU3F2 variants. Of note, we removed insulin values from ID 1.1 and ID 1.2 from regression analysis, as both individuals were diagnosed with diabetes mellitus in their twenties.
(E) Serum leptin in individuals with rare POU3F2 variants. While six individuals showed normal values (P5–P95 reference range, adjusted for BMI, Tanner stage, and age), three individuals presented with leptin abnormalities: increased leptin levels in ID 10 (>P95) and diminished leptin values in ID 1.1 and ID 1.2 (<P5) (Leptin ELISA E07, mediagnost). T1/2 and T3/4, Tanner stages 1/2 and 3/4, respectively.
Table 1.
Genotypes and corresponding phenotypes in individuals with ultra-rare POU3F2 variants identified by exome/genome sequencing
| ID 1.1 | ID 1.2 | ID 2 | ID 3 | ID 412 | ID 5.1a | ID 5.2a | ID 6 | ID 7b | ID 8 | ID 9 | ID 10 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | ||||||||||||
| Inheritance | maternal | unknown | de novo | de novo | de novo | paternal | unknown | de novo | de novo | de novo | unknown | de novo |
| Zygosity | het | het | het | het | het | het | het | het | het | het | het | het |
| Coding position | c.135C>A | c.135C>A | c.914A>G | c.41C>T | c.812A>T | c.664C>T | c.664C>T | c.929G>C | c.1249_1252del | c.1064G>T | c.1325T>C | c.1212C>A |
| Protein position | p.Tyr45∗ | p.Tyr45∗ | p.Gln305Arg | p.Ser14Phe | p.Glu271Val | p.Pro222Ser | p.Pro222Ser | p.Arg310Thr | p.Gly417Leufs∗71 | p.Arg355Leu | p.Val442Ala | p.Asn404Lys |
| MAF (gnomAD) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.78 × 10−6 | 0 |
| CADD PHRED | 34.0 | 34.0 | 28.6 | 22.9 | 31.0 | 20.0 | 20.0 | 29.7 | 33 | 32.0 | 28.8 | 29.6 |
| Affecting POU-protein domainsc | yes | yes | yes | no | yes | no | no | yes | no | yes | no | yes |
| Phenotype | ||||||||||||
| Sex | male | female | male | male | female | male | male | male | male | male | male | male |
| Age (current) | 33 years | 62 years | 9 years | 4 years | 10 years | 8 years | >30 years | 20 years | 16 years | 14 years | 11 years | 15 years |
| 1st manifestation | infancy | unknown | postnatal | toddler | infancy | infancy | unknown | infancy | infancy | infancy | infancy | infancy |
| Intellectual disability | yes | no | yes | yes | yes | no | no | no | yes | yes | yes | yes |
| ASD symptoms | yes | no | yes | yes | yes | no | no | no | no | yes | yes | yes |
| Neurodevelopmental delay | yes | no | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Obesity | yes | yes | yes | no | no | no | no | yes | yes | yes | Yes | yes |
| (BMI [kg/m2]) | (43; 29 years) | (47; 60 years) | (25; 6 years) | (14; 4 years) | (15; 8 years) | ND | ND | (33; 15 years) | (36; 15 years) | (35; 13 years) | (34; 10 years) | (54; 14 years) |
| Hyperphagia | yes | yes | yes | no | no | no | no | yes | no | yes | yes | yes |
| Diabetes | yes | yes | no | no | no | no | no | no | no | no | no | no |
| Morphological CNS anomaly | ND | ND | brain MRI normal | ND | brain MRI normal | leuko-encephalopathy | leuko-encephalopathy | brain MRI normal | brain MRI normal | ND | ND | microcephaly |
| Uro-renal anomalies | CAKUT: hydro-nephrosis | no | no | no | no | CAKUT: ureter duplex, dysplastic kidneys | neurogenic bladder | no | no | no | no | no |
The variants reported refer to GenBank: NM_005604.4. Abbreviations: ACMG, American College of Medical Genetics and Genomics; ASD, autism spectrum disorder; CADD, Combined Annotation Dependent Depletion (v1.4); CAKUT, congenital anomalies of the kidney and urinary tract; CNS, central nervous system; het, heterozygous; MAF, minor allele frequency; MRI, magnetic resonance imaging; ND, no data.
Additional variant in COL4A2 (GenBank: NM_001846.3): c.315+1G>C, splice site analysis confirmed pathogenicity by exon skipping.
Additional variant in KDM3B (GenBank: NM_016604.3): c.5191G>A (p.Glu1731Lys).14
According to https://www.uniprot.org/uniprot/P20265; gnomAD v2.1.1 (http://gnomad.broadinstitute.org/).
By using matchmaking initiatives such as GeneMatcher15 and the Genomics England 100,000 Genomes Project (GEL) rare disease database (supplemental methods), we identified another ten individuals from nine families with similar phenotypes and novel (not previously reported in gnomAD and/or ClinVar databases) POU3F2 variants, predicted deleterious by in-silico assessment (Table 1). Phenotypic and genotypic presentation is summarized in the following. ID 2 is an overweight 9-year-old male who was diagnosed with intellectual disability, autism spectrum disorder, and NDD. Genetically, a de novo POU3F2 missense variant affecting the POU-specific domain (c.914A>G [p.Gln305Arg] [GenBank: NM_005604.4]) was found by trio ES (Table 1, Figure 2A). ID 3 is a 4-year-old toddler who presented with intellectual disability, autism, and NDD. This individual was found to carry another de novo POU3F2 missense variant predicted to affect an evolutionary conserved Serine-residue at the N terminus of the encoded protein (c.41C>T [p.Ser14Phe] [GenBank: NM_005604.4]) (Table 1, Figure 2A). In contrast, ID 4 is a 10-year-old, normal-weight female whose clinical presentation (NDD and autism) had previously been reported as potentially associated with a de novo missense change in the POU-specific domain (c.812A>T [p.Glu271Val] [GenBank: NM_005604.4]) (Table 1, Figure 2A).12 Furthermore, by scrutinizing our institutional databank for NDD phenotypes (at Leipzig University, Germany), we identified an individual with familial leukoencephalopathy, CAKUT, and NDD (ID 5). Both the 8-year-old son (ID 5.1) and his father (ID 5.2) were found to carry a rare missense variant within POU3F2 (c.664C>T [p.Pro222Ser] [GenBank: NM_005604.4]) (Table 1, Figure 2A). In this family, however, a monoallelic COL4A2 splice site variant (c.315+1G>C [GenBank: NM_001846.3]) in both affected individuals deserves mentioning, as COL4A2 has previously been linked to autosomal-dominant brain small vessel disease 2 (MIM: 614483).16 For the remaining five male individuals, aged 11 (ID 9), 14 (ID 8), 15 (ID 10), 16 (ID 7), and 20 years (ID 6) (Figure 1A), we saw the full phenotypic presentation of NDD, intellectual disability, autism, and severe hyperphagic obesity (BMI > 35 kg/m2) (Table 1). All five individuals harbored ultra-rare POU3F2 de novo variants, three of which affected one of the two DNA-binding POU domains (c.929G>C [p.Arg310Thr] [GenBank: NM_005604.4] in ID 6; c.1064G>T [p.Arg355Leu] in ID 8; and c.1212C>A [p.Asn404Lys] in ID 10) (Table 1, Figure 2A). In contrast, in ID 7 the detected variant was a monoallelic frameshift with C-terminal truncation downstream of both POU domains (c.1249_1252del [p.Gly417Leufs∗71] [GenBank: NM_005604.4]) (Table 114, Figure 2A).
Figure 2.
POU3F2 variants upon in silico and in vitro analysis
(A) 2D protein structure with identified variants (NLS, nuclear localization signal) and functional domains (based on https://www.uniprot.org/uniprot/P20265) (missense variants, unshaded arrows; truncating variants, striped arrows). Expression plasmids used in in vitro experiments encode an N-terminal YFP (yellow fluorescent protein) and lack the poly-glycine stretch.
(B) 3D protein model indicating amino acid residues that are mutated in the identified individuals (purple, nuclear localization signal).
(C) Assessment of nuclear translocation and aberrant intracellular localization by immunofluorescence microscopy in transiently transfected HeLa cells (yellow, YFP-POU3F2 variants; cyan, nuclei; magenta, nuclear lamina stained with anti-LMNA antibody).
(D) Assessment of DNA-binding activity and promotor activation of POU3F2 variants by dual-luciferase assay; striped bars indicate truncating variants (n ≥ 3; data shown as mean ± SEM). The variant p.Gly417∗ was used as a surrogate for p.Gly417Leufs∗71 (ID 7).
Lastly, for unbiased replication of associated phenotypes and to estimate the frequency of deleterious POU3F2-associated obesity, we searched 71,682 germline genomes of participants from the 100,000 Genomes Project (v16 GRCh38 n = 63363 and v16 GRCh37 n = 8,319) for additional instances of monoallelic POU3F2 alteration. Filtering criteria included truncation or exceedingly rare missense variants with a minor allele frequency < 0.001 and a CADD_PHRED score > 20.17 The only individual with a matching genotype-phenotype combination was ID 9, carrying an ultra-rare missense change at an evolutionary-conserved residue at the C terminus (c.1325T>C [p.Val442Ala] [GenBank: NM_005604.4]) (Table 1, Figure 2A). Of note, methodologically all variants were detected by trio exome sequencing, except for the use of single genome sequencing in ID 9 with the unavailability of parental samples. With the exemption of the previously published p.Glu271Val variant (rs1769989432)12 and p.Val442Ala (rs754333102), a variant with gnomAD entry (allele frequency [AF] = 9.78 × 10−6), these POU3F2 variants were absent from population databases and clinical databases (including gnomAD, ExAc, ClinVar, HGMD Professional Version 2022.2) and showed CADD_PHRED scores (v1.4) greater than 20 (Table 1).17 Similar to loss-of-function variants in its co-expressed ortholog POU3F3, almost all of these POU3F2 variants constituted putatively non-inherited de novo variants (eight out of ten). In contrast to POU3F3-associated phenotypes however, we did not see a specific facial dysmorphic pattern as part of the shared clinical spectrum (Table S1).
In an international collaboration, we teamed up for deep-characterization of all study probands by longitudinal data collection and prospective metabolic analyses after written informed consent (ethics votes from Universities of Leipzig, Barcelona, Philadelphia, Poitiers, Dijon, Bordeaux, and Seattle). Thereby, we observed a pattern of NDD characterized by neonatal hypotonia, early feeding difficulties, speech and motor delay, learning disabilities, and incompletely penetrant intellectual disability (Tables S2 and S3). On the metabolic side, we observed low-to-normal birth weight and normal infantile weight gain but hyperphagia, overweight, and incremental obesity for the individuals who had reached puberty and beyond (Figures 1B, 1C, and S1). Commonly, hyperinsulinemia and insulin resistance (documented by homeostatic model assessment for insulin resistance, HOMA-index > 2) preceded obesity in absence of overt diabetes mellitus (Figure 1D, Table S4). Of note, the only two adult individuals from the index family (ID 1.1 and ID 1.2) developed type 2 diabetes mellitus (T2DM) during their twenties, leaving the possibility that T2DM is yet to develop in younger individuals. Interestingly, impaired insulin sensitivity also plays a causal role in hyperphagia.18
As POU3F2 is known to act downstream of SIM1 and interacts with the leptin and melanocortin pathway, we performed further endocrine assessment by measuring leptin, adiponectin, oxytocin, arginin-vasopressin, fasting insulin/glucose (C-peptide), and cortisol levels in study probands upon availability (Table S4). While leptin levels were mostly within age-, BMI-, and sex-adjusted reference ranges, we found leptin to be elevated in ID 10. In contrast, both individuals from the index family showed intermittently decreased leptin values (ID 1.1/ID 1.2), suggesting a different mechanism upon early POU3F2 truncation (p.Tyr45∗) (Figure 1E). Of note, diminished leptin levels have successfully been approached by therapeutic metreleptin substitution in individuals with rare genetic LEP-deficiency (MIM: 614962), leading to substantial weight loss and improvement of central nervous insulin sensitivity.18 In contrast, oxytocin and cortisol levels presented unaltered in all available serum samples obtained from individuals with a deleterious POU3F2 variation. With the exception of ID 5.1/ID 5.2, who presented with leukoencephalopathy in the presence of the additional COL4A2-truncating variant, morphological anomalies of the cerebrum, notably the pituitary gland, were excluded by magnetic resonance imaging (MRI) whenever available (ID 2, ID 4, ID 6, ID 7).
The transcription factor POU3F2 contains 443 amino acids with a molecular weight of 46.9 kDa (Figures 2A and 2B). By using a three-dimensional protein model from the AlphaFold database,19,20 we further investigated potential functional impact of the identified variants, especially the eight non-truncating alterations. Thereby, we were able to confirm affection of one of the DNA-binding domains, POU-homeo or POU-specific, in the majority of individuals (five out of eight) (Figures 2A and 2B). In the remaining three individuals (p.Ser14Phe, p.Pro222Ser, p.Val442Ala), affected amino acid residues are both orthologously and paralogously well conserved but lie within less characterized protein regions (Figures 2A, 2B, and S2). Interestingly, we found p.Arg355Leu (ID 8) to be located within the linker region between both POU domain structures, which is predicted as the nuclear localization signal (NLS; amino acids 351 to 361) (Figures 2A and 2B).
To further evaluate variant pathogenicity, we generated wild-type and variant plasmids encoding N-terminally YFP-tagged POU3F2 for in vitro overexpression in immortalized cell lines (HeLa) (supplemental methods). To ensure consistency following mutagenesis, we worked with a modified wild-type construct lacking the region encoding the N-terminal poly-glycine stretch at amino acid residues 68 to 88 (delGly WT; Figure 2A). Subsequent analysis of intracellular POU3F2 localization by immunofluorescent live cell imaging demonstrated normal nuclear translocation for all variants except the early truncation p.Tyr45∗, which showed cytosolic retention (Figure 2C). Additionally, overexpression of the missense variant p.Arg310Thr affecting the POU-specific domain presented with adequate translocation but intranuclear aggregates (Figure 2C). Although p.Arg355Leu (ID 8) is predicted to locate directly to the NLS, nuclear translocation was unaltered in our overexpression system (Figure 2C).
To test the hypothesis of impaired DNA-binding ability, we transfected wild-type (WT) and variant constructs in HEK293T cells to investigate POU3F2 binding to the known intronic FOXP2-binding site by use of luciferase assays (NanoGlo Dual-Luciferase) (Figure 2D; supplemental methods). Analogous to previously published protocols on POU3F3 transcriptional activity assessment,13 the construct encoding the YFP-POU3F2 fusion protein was transfected together with a Firefly luciferase construct containing the conserved FOXP2-binding site as well as a Nano-luciferase construct, providing a normalization control. While both WT constructs were able to markedly increase luciferase levels compared to the negative control (luciferase plasmids only) (Figure 2D; Table S5), eight out of ten POU3F2 variants showed significantly reduced promotor activation compared to the delGly WT (Figure 2D; Table S5). As expected, two POU-domain changes (p.Gln305Arg, ID 2; p.Arg310Thr, ID 6) and the early truncation p.Tyr45∗ (index family) were found among the constructs with most severe impairment in transcriptional activation (Figure 2D; Table S5). To our surprise, however, the N-terminal variant p.Ser14Phe (ID 3) also yielded strikingly reduced luciferase levels, consistent with complete or partial loss of DNA-binding ability. Another four variants (p.Pro222Ser, ID 5.1/ID 5.2; p.Asn404Lys, ID 10; p.Gly417∗ [as surrogate for Gly417Leufs∗71, ID 7]; and p.Val442Ala, ID 9) showed partial transactivation capacity that was significantly lower than that of the WT (Figure 2D; Table S5). On the contrary, upon transfection of the plasmid encoding the NLS variant p.Arg355Leu (ID 8), we observed the opposite effect with increased relative luciferase levels, pointing to an enhanced promotor activation. Lastly, no significant difference compared to the WT was seen in p.Gly271Val (ID 4). Interestingly, pathogenic variants of paralogous POU3F3 residues corresponding to POU3F2 p.Arg310Thr, p.Asn404Lys, and p.Arg407Leu were previously identified in POU3F3 cases and showed a comparable impact on FOXP2 promotor activation.13
Therefore, these functional studies provide evidence for loss of function in eight and gain of function in one obesity-associated POU3F2 variant (Figure 2D; Table S5), whereas the missense variant identified in ID 4 (p.Gly271Val) did not impact POU3F2-promotor activation, leaving the possibility for further pathomechanisms.
In order to investigate the role of POU3F2 in common, non-syndromic obesity, we scrutinized exome-sequencing data from cohorts of obese individuals (n = 1,480) and corresponding controls from the UK10K project (supplemental methods); inclusion criteria detailed in supplemental information). As a result, we identified seven individuals with five distinct heterozygous variants with a minor allele frequency (MAF) of <0.01% according to gnomAD and absence in the ethnically matched UK10K control dataset (n = 1,854). Among these was a heterozygous cytosine deletion variant (c.1241del [p.Pro414Leufs∗75] [GenBank: NM_005604.4]) predicted to introduce a frameshift affecting the C terminus directly adjacent to the POU homeodomain (Figure 2A). We also found three other alleles with predicted moderate impact (Table S6) that were not present in the control dataset.
To assess potential mechanisms of POU3F2 in obesity, we next examined POU3F2 expression in visceral (VIS) or subcutaneous (SC) fat from individuals with common obesity. Thus, we employed a large dataset comprising paired samples of omental visceral and abdominal subcutaneous tissues from 1,553 individuals of the Leipzig Obesity Biobank (LOBB). Upon quantitative RNA sequencing from bulk tissue, POU3F2 gene expression was significantly (adj. p = 0.02) less abundant in visceral fat obtained from obese individuals (n = 1,470) when compared to visceral fat from lean controls (n = 83) (Figure S3A). In contrast, we did not find differences concerning subcutaneous POU3F2 expression between lean and obese individuals. When correlating POU3F2 gene expression with additional specific metabolic and anthropometric parameters, we obtained significantly negative (p < 0.05) correlations with serum adiponectin levels (VIS: n = 114, adj. p = 0.02, ρSpearman = −0.22 [Figure S3B]; SC: n = 114, adj. p = 0.02, ρSpearman = −0.21 [Figure S3C]). Correlation analyses of visceral POU3F2 gene expression with body fat (n = 694, ρSpearman = −0.01, p = 0.009), body weight (n = 1,522, ρSpearman = −0.06, p = 0.03), and BMI (VIS: n = 1,553, ρSpearman = −0.06, p = 0.03) also yielded significant results (Figure S3B). Lastly, in subcutaneous fat, POU3F2 gene expression correlated significantly (Figure S3C) with HOMA-indices (n = 444, ρSpearman = 0.14, p = 0.004) and fasting plasma insulin (FPI) (n = 486, ρSpearman = 0.12, p = 0.009).
In this work, we identify rare, monoallelic POU3F2 variants as an etiology of syndromic PWL obesity, similar to the phenotypes that have been reported in individuals with the 6q16.1 microdeletion,11,21,22 encompassing POU3F2 as one out of nine coding candidate genes (POU3F2, FBXL4, FAXC, COQ3, PNISR, USP45, TSTD3, CCNC, PRDM13). In a systematic comparison, we found substantial phenotypic overlap between individuals with intragenic POU3F2 variants and individuals with the microdeletion syndrome. Overlap comprised both NDD traits and metabolic traits and was greatest in terms of speech delay, learning disabilities, neonatal hypotonia, and hyperphagic obesity (Table 2). In contrast, infantile feeding difficulties and low birth weight were less commonly reported in individuals with the 6q16.1 microdeletion as compared to those with an intragenic POU3F2 variant (Table 2).11,21,22
Table 2.
Phenotypic comparison of individuals with a 6q16.1 microdeletion versus intragenic POU3F2 variant
| Category | Trait/phenotype | 6q16.1 deletions (A)11 | POU3F2 intragenic rare variants (B) | Ratio (B/A) |
|---|---|---|---|---|
| NDD traits | Infantile feeding difficulties (yes/total) (%) | 2/9 (22%) | 5/9 (56%) | 2.5 |
| Sex (male/female) (% male) | 6/4 (60%) | 9/2 (82%) | 1.4 | |
| Speech delay (yes/total) (%) | 10/10 (100%) | 10/11 (91%) | 0.9 | |
| Learning disabilities (yes/total) (%) | 10/10 (100%) | 10/11 (91%) | 0.9 | |
| Neonatal hypotonia (yes/total) (%) | 8/10 (80%) | 6/9 (67%) | 0.8 | |
| Motor delay (yes/total) (%) | 9/10 (90%) | 6/11 (55%) | 0.6 | |
| Metabolic traits | Birth weight ≤ 50th percentile (yes/total) (%) | 5/9 (56%) | 9/9 (100%) | 1.8 |
| Hyperphagia (yes/total) (%) | 9/10 (90%) | 7/11 (64%) | 0.7 | |
| Obesity (yes/total) (%) | 9/9 (100%) | 8/11 (73%) | 0.7 |
Traits per category in order of comparative ratio (B/A). Of note, while a theoretical ratio of 1 would constitute 100% phenotypic overlap, ratios < 1 indicate higher trait frequency in individuals with 6q16.1 deletions as compared to intragenic variants and ratios > 1 denote trait predominance in individuals with intragenic variants as compared to microdeletions. Most phenotypic overlap can be found for the following traits: speech delay (ratio 0.9), learning disabilities (ratio 0.9), neonatal hypotonia (ratio 0.8), and hyperphagic obesity (ratio 0.7). Less phenotypic overlap can be observed for infantile feeding difficulties (ratio 2.5), low birth weight (ratio 1.8), male predominance (ratio 1.4), and motor delay (ratio 0.6). NDD, neurodevelopmental delay.
Beyond the syndromic presentation, we aimed to address the question of whether variants in POU3F2 are associated with common obesity. From common obesity cohort analyses (UK10K, LOBB), we propose risk conferring POU3F2 germline variants and reduced abundance of POU3F2 expression to also play a role in development of non-syndromic obesity. Independently, rare missense variants in POU3F2 associated with female BMI (p value 2 × 10−5, nominal significance) in gene burden tests with UK Biobank exome data from 394,841 individuals of European descent.23
The intron-less POU3F2 gene shows high genetic constraint (observed/expected metric for loss-of-function variants [pLoF] 0.08 [confidence interval (CI): 0.03–0.38]; probability of being loss-of-function intolerant [pLI] 0.92 in gnomAD). This supports the notion of haploinsufficiency as the prevailing disease mechanism, consistent with reported haploinsufficiency in individuals with monoallelic 6q16.1 deletions11 and deleterious POU3F3 variants.13
The hypothesis that loss-of-function POU3F2 variants predispose to syndromic obesity is consistent with published data on haploinsufficiency of SIM1, the POU3F2-regulator associated with monogenic obesity.7 However, for one variant (p.Arg355Leu) we found a potential gain-of-function mechanism upon in vitro analysis, which was in line with previous studies on the paralogous residue in POU3F3.13 As POU3F2 gene expression was generally low in visceral fat and even more reduced upon obesity, we suppose POU3F2 transcriptional activity to act in the central nervous system directly, regulating behavioral processes, and eating behavior is one of these. Data on its hypothalamic target genes comprising several known players of appetite control, such as LEPR, MC4R, PCSK1, and PHIP, strongly support this hypothesis.24 Interestingly, among the four class III POU genes (POU3F1 [MIM: 602479], POU3F2, POU3F3 [MIM: 602480], and POU3F4 [MIM: 300039]), only POU3F2 alterations seem to associate with weight gain and insulin resistance, potentially because of its specific expression at the hypothalamic PVN (Allen Human Brain Atlas).11
In future studies, it will be important to delineate the syndromic nature of organ involvement beyond NDD and central obesity in more detail. Typical facial features from individuals with pathogenic POU3F3 variants, such as cupped and low-set ears, open-mouth appearance, and orbital fullness, were not systematically present among the families from our study (Table S1). In this regard, reports on additional individuals will help clarify whether CAKUT (ID 1.1 and ID 5.1) is variably associated.
Importantly, deciphering molecular effects of POU3F2 deficiency will be key to characterizing targets for therapeutic intervention. Conflicting data on the role of leptin, illustrated by the wide spectrum of normal, increased, and decreased levels in our study, limit metreleptin substitution as a reasonable treatment option. However, successful clinical trials on setmelanotide, a MC4R agonist, in individuals with LEPR deficiency25,26 and MC4R deficiency27 recently created promising therapeutic alternatives in the field of severe hyperphagic obesity. Novel pharmacological treatment options become even more important when considering that bariatric surgery appears to be less effective in hyperphagic disorders.28
In summary, we identified 12 individuals from ten unrelated families with likely deleterious intragenic variants in POU3F2, primarily displaying PWL features with NDD and adolescent-onset hyperphagic obesity. Observed phenotypes resemble what has been reported in individuals with the 6q16.1 microdeletion,11,21,22 thereby strongly supporting the previously postulated candidate status of POU3F2 as the driving gene in this nine-gene deletion syndrome.
Acknowledgments
We thank all individuals and their families for participating in this study. We also kindly appreciate the donation of POU3F2 luciferase constructs by Prof. S.E. Fisher. The graphical abstract was created with BioRender.com. J. Halbritter receives funding from the German Research Foundation (DFG, HA 6908/3-1, HA 6908/4-1, HA 6908/7-1, and HA 6908/8-1). R.S. receives funding from the Else Kröner-Fresenius-Stiftung (2019_A96). M.B. received grant funding from the DFG (German Research Foundation), project number 209933838 – SFB 1052 (project B1), and by Deutsches Zentrum für Diabetesforschung (DZD, grant: 82DZD00601). L.D. is funded by MRC DiMEN and the Northern Counties Kidney Research Fund. R.N. is funded by the Barbour Foundation and the Northern Counties Kidney Research Fund. J.A.S. is funded by Kidney Research UK and the Northern Counties Kidney Research Fund. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. The 100,000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100,000 Genomes Project is funded by the National Institutes of Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by participants and their families and collected by the National Health Service as part of their care and support.
Author contributions
R.S. and J. Halbritter designed the study, analyzed the data, prepared the figures, and wrote the manuscript. R.S., W.J., C.H, L. Pöschla, and E.H. performed experiments and analyzed in vitro data. M.B. and C.W. contributed the LOBB fat tissue expression data. A.G., W.S., H.D., and F.N. performed RNA sequencing and pre-processing of the LOBB samples. A.H. performed the bioinformatics analyses of the LOBB sequencing data. M.I.V.P., L.D., J.M, E.B., K.R., J. Hoefele, K.P., R.B., P.C., E.W., M.P., R.N., G.L.G., J.M., A.-L.B., L. Perrin, I.M., M.F., V.M., D.L., S.A.P., M.G., B.P., and J.A.S. provided genotypic and phenotypic information. H.H. contributed UK Biobank analysis. J.A.S. additionally contributed UK10K sequencing data.
Declaration of interests
The authors declare no competing interests.
Published: May 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.04.010.
Web resources
Allen Human Brain Atlas, https://human.brain-map.org
AlphaFold, https://alphafold.ebi.ac.uk/
ClustalW, https://www.genome.jp/tools-bin/clustalw
cNLS Mapper, http://nls-mapper.iab.keio.ac.jp
Decipher, https://decipher.sanger.ac.uk
Genebass (UK biobank), http://genebass.org
Genomics England 100,000 Genomes Project, https://www.genomicsengland.co.uk/
Human Protein Atlas, https://www.proteinatlas.org/
MetaDome, https://stuart.radboudumc.nl/metadome
OMIM, https://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2
UniProt, https://www.uniprot.org
Supplemental information
Data and code availability
Genetics data and human RNA-seq data from the LOBB supporting the current study have not been deposited in a public repository because of consent restriction but are available from the corresponding author on request. Primary data from the 100,000 Genomes Project, which are held in a secure research environment, are available to registered users. Please see https://www.genomicsengland.co.uk/about-gecip/for-gecip-members/data-and-data-access for information.
References
- 1.Abarca-Gómez L., Abdeen Z.A., Hamid Z.A., Abu-Rmeileh N.M., Acosta-Cazares B., Acuin C., Adams R.J., Aekplakorn W., Afsana K., Aguilar-Salinas C.A., et al. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128·9 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loos R.J.F., Yeo G.S.H. The genetics of obesity: from discovery to biology. Nat. Rev. Genet. 2022;23:120–133. doi: 10.1038/s41576-021-00414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clément K., Vaisse C., Lahlou N., Cabrol S., Pelloux V., Cassuto D., Gourmelen M., Dina C., Chambaz J., Lacorte J.-M., et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 4.Friedman J.M., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 5.Vaisse C., Clement K., Guy-Grand B., Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat. Genet. 1998;20:113–114. doi: 10.1038/2407. [DOI] [PubMed] [Google Scholar]
- 6.Yeo G.S., Farooqi I.S., Aminian S., Halsall D.J., Stanhope R.G., O'Rahilly S. A frameshift mutation in MC4R associated with dominantly inherited human obesity. Nat. Genet. 1998;20:111–112. doi: 10.1038/2404. [DOI] [PubMed] [Google Scholar]
- 7.Bonnefond A., Raimondo A., Stutzmann F., Ghoussaini M., Ramachandrappa S., Bersten D.C., Durand E., Vatin V., Balkau B., Lantieri O., et al. Loss-of-function mutations in SIM1 contribute to obesity and Prader-Willi-like features. J. Clin. Invest. 2013;123:3037–3041. doi: 10.1172/JCI68035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badano J.L., Ansley S.J., Leitch C.C., Lewis R.A., Lupski J.R., Katsanis N. Identification of a Novel Bardet-Biedl Syndrome Protein, BBS7, That Shares Structural Features with BBS1 and BBS2. Am. J. Hum. Genet. 2003;72:650–658. doi: 10.1086/368204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson R.S., Creemers J.W., Ohagi S., Raffin-Sanson M.-L., Sanders L., Montague C.T., Hutton J.C., O'Rahilly S. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat. Genet. 1997;16:303–306. doi: 10.1038/ng0797-303. [DOI] [PubMed] [Google Scholar]
- 10.Webster E., Cho M.T., Alexander N., Desai S., Naidu S., Bekheirnia M.R., Lewis A., Retterer K., Juusola J., Chung W.K. De novo PHIP-predicted deleterious variants are associated with developmental delay, intellectual disability, obesity, and dysmorphic features. Cold Spring Harb. Mol. Case Stud. 2016;2:a001172. doi: 10.1101/mcs.a001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasher P.R., Schertz K.E., Thomas M., Jackson A., Annunziata S., Ballesta-Martinez M.J., Campeau P.M., Clayton P.E., Eaton J.L., Granata T., et al. Small 6q16.1 Deletions Encompassing POU3F2 Cause Susceptibility to Obesity and Variable Developmental Delay with Intellectual Disability. Am. J. Hum. Genet. 2016;98:363–372. doi: 10.1016/j.ajhg.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Westphal D.S., Riedhammer K.M., Kovacs-Nagy R., Meitinger T., Hoefele J., Wagner M. A De Novo Missense Variant in POU3F2 Identified in a Child with Global Developmental Delay. Neuropediatrics. 2018;49:401–404. doi: 10.1055/s-0038-1669926. [DOI] [PubMed] [Google Scholar]
- 13.Snijders Blok L., Kleefstra T., Venselaar H., Maas S., Kroes H.Y., Lachmeijer A.M.A., van Gassen K.L.I., Firth H.V., Tomkins S., Bodek S., et al. De Novo Variants Disturbing the Transactivation Capacity of POU3F3 Cause a Characteristic Neurodevelopmental Disorder. Am. J. Hum. Genet. 2019;105:403–412. doi: 10.1016/j.ajhg.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diets I.J., van der Donk R., Baltrunaite K., Waanders E., Reijnders M.R.F., Dingemans A.J.M., Pfundt R., Vulto-van Silfhout A.T., Wiel L., Gilissen C., et al. De Novo and Inherited Pathogenic Variants in KDM3B Cause Intellectual Disability, Short Stature, and Facial Dysmorphism. Am. J. Hum. Genet. 2019;104:758–766. doi: 10.1016/j.ajhg.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoneda Y., Haginoya K., Arai H., Yamaoka S., Tsurusaki Y., Doi H., Miyake N., Yokochi K., Osaka H., Kato M., et al. De Novo and Inherited Mutations in COL4A2, Encoding the Type IV Collagen α2 Chain Cause Porencephaly. Am. J. Hum. Genet. 2012;90:86–90. doi: 10.1016/j.ajhg.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank-Podlech S., von Schnurbein J., Veit R., Heni M., Machann J., Heinze J.M., Kullmann S., Manzoor J., Mahmood S., Häring H.U., et al. Leptin Replacement Reestablishes Brain Insulin Action in the Hypothalamus in Congenital Leptin Deficiency. Diabetes Care. 2018;41:907–910. doi: 10.2337/dc17-1867. [DOI] [PubMed] [Google Scholar]
- 19.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Angelo C.S., Varela M.C., de Castro C.I.E., Otto P.A., Perez A.B.A., Lourenço C.M., Kim C.A., Bertola D.R., Kok F., Garcia-Alonso L., Koiffmann C.P. Chromosomal microarray analysis in the genetic evaluation of 279 patients with syndromic obesity. Mol. Cytogenet. 2018;11:14. doi: 10.1186/s13039-018-0363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumi K., Housam R., Kapadia C., Stallings V.A., Medne L., Shaikh T.H., Kublaoui B.M., Zackai E.H., Grimberg A. Endocrine phenotype of 6q16.1-q21 deletion involving SIM1 and Prader-Willi syndrome-like features. Am. J. Med. Genet. 2013;161A:3137–3143. doi: 10.1002/ajmg.a.36149. [DOI] [PubMed] [Google Scholar]
- 23.Karczewski K.J., Solomonson M., Chao K.R., Goodrich J.K., Tiao G., Lu W., Riley-Gillis B.M., Tsai E.A., Kim H.I., Zheng X., et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genom. 2022;2:100168. doi: 10.1016/j.xgen.2022.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouillard A.D., Gundersen G.W., Fernandez N.F., Wang Z., Monteiro C.D., McDermott M.G., Ma’ayan A. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clément K., Biebermann H., Farooqi I.S., van der Ploeg L., Wolters B., Poitou C., Puder L., Fiedorek F., Gottesdiener K., Kleinau G., et al. MC4R agonism promotes durable weight loss in patients with leptin receptor deficiency. Nat. Med. 2018;24:551–555. doi: 10.1038/s41591-018-0015-9. [DOI] [PubMed] [Google Scholar]
- 26.Clément K., van den Akker E., Argente J., Bahm A., Chung W.K., Connors H., Waele K. de, Farooqi I.S., Gonneau-Lejeune J., Gordon G., et al. Efficacy and safety of setmelanotide, an MC4R agonist, in individuals with severe obesity due to LEPR or POMC deficiency: single-arm, open-label, multicentre, phase 3 trials. The lancet. Diabetes & endocrinology. 2020;8:960–970. doi: 10.1016/S2213-8587(20)30364-8. [DOI] [PubMed] [Google Scholar]
- 27.Collet T.-H., Dubern B., Mokrosinski J., Connors H., Keogh J.M., Mendes de Oliveira E., Henning E., Poitou-Bernert C., Oppert J.-M., Tounian P., et al. Evaluation of a melanocortin-4 receptor (MC4R) agonist (Setmelanotide) in MC4R deficiency. Mol. Metab. 2017;6:1321–1329. doi: 10.1016/j.molmet.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gantz M.G., Driscoll D.J., Miller J.L., Duis J.B., Butler M.G., Gourash L., Forster J., Scheimann A.O. Critical review of bariatric surgical outcomes in patients with Prader-Willi syndrome and other hyperphagic disorders. Obesity. 2022;30:973–981. doi: 10.1002/oby.23385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genetics data and human RNA-seq data from the LOBB supporting the current study have not been deposited in a public repository because of consent restriction but are available from the corresponding author on request. Primary data from the 100,000 Genomes Project, which are held in a secure research environment, are available to registered users. Please see https://www.genomicsengland.co.uk/about-gecip/for-gecip-members/data-and-data-access for information.