Summary
De novo variants are a leading cause of neurodevelopmental disorders (NDDs), but because every monogenic NDD is different and usually extremely rare, it remains a major challenge to understand the complete phenotype and genotype spectrum of any morbid gene. According to OMIM, heterozygous variants in KDM6B cause “neurodevelopmental disorder with coarse facies and mild distal skeletal abnormalities.” Here, by examining the molecular and clinical spectrum of 85 reported individuals with mostly de novo (likely) pathogenic KDM6B variants, we demonstrate that this description is inaccurate and potentially misleading. Cognitive deficits are seen consistently in all individuals, but the overall phenotype is highly variable. Notably, coarse facies and distal skeletal anomalies, as defined by OMIM, are rare in this expanded cohort while other features are unexpectedly common (e.g., hypotonia, psychosis, etc.). Using 3D protein structure analysis and an innovative dual Drosophila gain-of-function assay, we demonstrated a disruptive effect of 11 missense/in-frame indels located in or near the enzymatic JmJC or Zn-containing domain of KDM6B. Consistent with the role of KDM6B in human cognition, we demonstrated a role for the Drosophila KDM6B ortholog in memory and behavior. Taken together, we accurately define the broad clinical spectrum of the KDM6B-related NDD, introduce an innovative functional testing paradigm for the assessment of KDM6B variants, and demonstrate a conserved role for KDM6B in cognition and behavior. Our study demonstrates the critical importance of international collaboration, sharing of clinical data, and rigorous functional analysis of genetic variants to ensure correct disease diagnosis for rare disorders.
Keywords: neurodevelopmental disorders, COMPASS, Mendelian disorders, missense variants, de novo variants, Drosophila, KDM6B
Clinical and molecular analysis of the KDM6B-related neurodevelopmental disorder highlights inaccuracies in its current OMIM definition and demonstrates the critical importance of international collaboration, sharing of clinical data, and rigorous functional analysis of genetic variants for an unbiased, accurate, and comprehensive definition of rare genetic disorders.
Introduction
The development of the brain is a complex process requiring precise control of gene expression by epigenetic regulators,1 including proteins involved in enzymatic modification of histone tails, ATP-dependent chromatin remodeling, and DNA methylation. Dysfunction of epigenetic regulators frequently results in neurodevelopmental disorders (NDDs).2 Pathogenic variants in genes encoding epigenetic regulators, including histone methylases and demethylases, are a common cause of monogenic NDDs.3,4
The complex of proteins associated with Set1 (COMPASS) and COMPASS-like complexes are important components of the epigenetic machinery.5 The COMPASS complexes are highly conserved among species, including Drosophila and yeast, and their main function is to promote gene expression by methylating histone H3 on lysine 4 (H3K4) with enzymes containing a SET domain and demethylating histone H3 on lysine 27 (H3K27) through the enzymatic activity of the KDM6A and KDM6B demethylases.5
KDM6A and KDM6B demethylate di- and trimethylated H3K27 through the catalytic activity of the iron-containing jumonji C (JmJC) domain, which is common to different histone demethylases.6 KDM6B can act independently or as a component of a COMPASS-like complex.7 KDM6B can also influence transcription independent of its enzymatic activity, although the non-demethylase function of KDM6B is poorly understood.8 KDM6B dysfunction has also been implicated in various disorders, including cancer, immunologic, and developmental disorders.9
Recently, Stolerman et al. reported a cohort (n = 12) of individuals with de novo KDM6B (MIM: 611577) variants, suggesting that haploinsufficiency of KDM6B may result in a novel syndromic NDD with multisystem involvement.10 However, current knowledge regarding the molecular and clinical spectrum of the KDM6B-related NDD is limited and the function of KDM6B in neurons remains undefined. OMIM currently classifies this KDM6B-related NDD as “neurodevelopmental disorder with coarse facies and mild distal skeletal abnormalities” (MIM: 618505). Here, we further characterized the clinical and molecular spectrum of this disorder on the basis of a large cohort (n = 85) of individuals with (likely) pathogenic KDM6B variants. In addition, we developed Drosophila models to assess the impact of identified KDM6B variants and to examine the role of KDM6B in regulating cognitive function and behavior. Our results elucidate a more complete clinical and molecular spectrum for the KDM6B-related NDD and indicate an urgent need to reassess the current OMIM description for KDM6B. These findings highlight the challenges in defining rare NDDs in general.
Material and methods
Cohort recruitment
We have collected genetic and clinical data from 85 individuals with rare heterozygous (mostly de novo) variants in KDM6B. The variants were annotated with the GRCh37 reference and GenBank: NM_001080424.2/ENST00000254846.9 transcript. The individuals were recruited from the Radboudumc in-house diagnostic laboratory, international collaborators, individuals registered in GeneMatcher,11 and individuals included in various research cohorts, such as the Simons Simplex Collection (SSC),12 Deciphering Developmental Delay (DDD),13 100,000 Genome Project,14 Pediatric Cardiac Genomics Consortium (PCGC),15 Autism Sequencing Consortium (ASC),16,17 and MSSNG.18 The variants were identified by performing exome or genome sequencing in diagnostic or research settings with standard laboratory methods.19,20,21,22,23,24,25,26,27 For individuals identified through the DDD Study, a complementary analysis project (CAP #83) was approved that filtered for variants in chromatin remodeling genes throughout the entire cohort. This list was then filtered for rare de novo variants with damaging in silico predictions. Panel-agnostic re-analysis of locally unsolved cases from the DDD Study (CAP #147) was also performed as previously described.4,28 For the 100,000 Genome Project, tiered variants from the third September 2020 data release were accessed/filtered via LabKey. Variants were filtered for de novo inheritance and clinicians contacted through the AirLock.
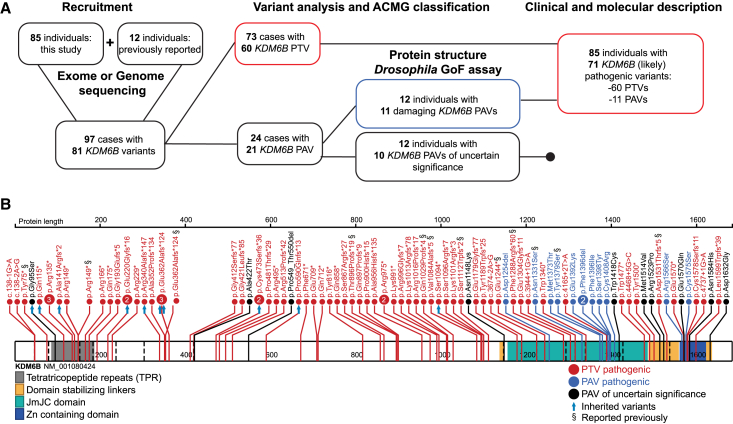
After collecting all evidence, we re-interpreted all identified variants according to ACMG variant classification guidelines29: variants in 73 individuals were classified as (likely) pathogenic, but variants in 12 individuals were classified as variants of uncertain significance (VUSs) as a result of limited or controversial evidence. Clinical features of only individuals with (likely) pathogenic variants were further analyzed. An overview of the study design is shown in Figure 1A.
Figure 1.
Overview of the study design and the identified KDM6B variants
(A) Schematic illustration of the study design.
(B) KDM6B variants, and their positions, identified in independent families. JmJC, Jumonji C domain; Zn, zinc; PTVs, protein-truncating (nonsense, frameshift, canonical splice) variants; PAVs, protein altering (missense and in-frame indel) variants.
Detailed descriptions of 73 individuals with (likely) pathogenic KDM6B variants, their molecular findings, and corresponding study type (clinical or research) are provided in Table S1. In most individuals (52/73), variants occur de novo, but nine truncating variants were inherited from a mildly affected or unaffected parent; for 12 individuals, the inheritance was unknown. To provide a more precise description of different clinical feature frequencies, we aggregated our individual data with the previously published 12 individuals (resulting in cohort of 85 individuals with [likely] pathogenic KDM6B variants). We corrected for incomplete data across individuals when calculating total feature frequency and we did not include one individual (#17) with a pathogenic KDM6B variant in combination with a pathogenic HNRNPU (MIM: 602869) variant in the clinical feature frequency calculations (Table 1) to minimize possible effects from additional genetic variants. Detailed clinical and molecular descriptions of 12 individuals with KDM6B VUSs are provided in Table S2. All variants identified in this study were deposited to the ClinVar database (ClinVar accession numbers: SCV002570417–SCV002570487).
Table 1.
Main clinical features among individuals with (likely) pathogenic KDM6B variants
|
Feature |
PTVs N = 73a |
PAVs N = 12 |
p value (PTVs vs. PAVs) |
Total N = 85a(%) |
|---|---|---|---|---|
| Sex (males/total) | 51/72 | 10/12 | 0.50 | 61/84 (73%) |
| Growth | ||||
| Increased birth weight [>2 SD] | 10/50 | 0/8 | 0.33 | 10/58 (17%) |
| Increased weight [>2 SD] | 9/56 | 0/8 | 0.59 | 9/64 (14%) |
| Tall stature [>2 SD] | 5/58 | 0/8 | 1.0 | 5/66 (8%) |
| Macrocephaly [>2 SD] | 15/57 | 2/8 | 1.0 | 17/65 (26%) |
| At least one feature of overgrowth | 18/59 | 3/10 | 1.0 | 21/69 (30%) |
| Neurodevelopmental and psychiatric issues | ||||
| Language/speech delay | 63/66 | 9/11 | 0.15 | 72/77 (94%) |
| Motor delay | 56/63 | 10/11 | 1.0 | 66/74 (89%) |
| ID or learning problems | 37/56 | 3/8 | 0.14 | 40/64 (63%) |
| ASD | 38/65 | 8/11 | 0.51 | 46/76 (61%) |
| Behavior problems, non-ASD | 40/65 | 4/8 | 0.70 | 44/73 (60%) |
| Psychotic disorders [≥12 years old] | 4/18 | 0/2 | 1.0 | 4/20 (20%) |
| Neurological issues | ||||
| Seizures | 9/62 | 0/7 | 0.58 | 9/69 (13%) |
| Sleep disturbances | 21/59 | 0/7 | 0.09 | 21/66 (32%) |
| Movement disorder/gait disturbances/hypertonia/ataxia | 15/59 | 1/8 | 0.67 | 16/67 (24%) |
| Hypotonia | 36/63 | 4/6 | 1.0 | 40/70 (57%) |
| Gastrointestinal issues | ||||
| Neonatal feeding difficulties or gastroesophageal reflux | 29/58 | 4/7 | 1.0 | 33/65 (51%) |
| Constipation | 10/55 | 1/6 | 1.0 | 11/61 (18%) |
| Congenital anomalies | ||||
| Congenital heart disease | 8/57 | 0/7 | 0.58 | 8/64 (13%) |
| Cleft lip/palate/uvula | 1/60 | 2/7 | 0.03b | 3/67 (4%) |
| Genitourinary abnormalities | 6/55 | 0/7 | 1.0 | 6/62 (10%) |
| Musculoskeletal and limb abnormalities | ||||
| Joint hypermobility | 24/56 | 2/6 | 1.0 | 26/62 (42%) |
| Scoliosis/kyphosis/lordosis | 8/57 | 0/7 | 0.58 | 8/64 (13%) |
| Syndactyly | 4/58 | 2/8 | 0.15 | 6/66 (9%) |
| Short fingers or toes | 6/57 | 0/7 | 1.0 | 6/64 (9%) |
| Broad fingers/fingertips/hands/toes/feet | 12/58 | 1/7 | 1.0 | 13/65 (20%) |
| Sensory issues | ||||
| Myopia/amblyopia | 20/54 | 0/7 | 0.08 | 20/61 (33%) |
| Strabismus | 8/57 | 0/7 | 0.58 | 8/64 (13%) |
| Hearing loss | 1/55 | 0/7 | 1.0 | 1/62 (2%) |
| Recurrent ear infections | 7/54 | 0/5 | 1.0 | 7/59 (12%) |
PTVs, protein-truncating variants; PAVs, protein-altering variants (only [likely] pathogenic variants included); ID, intellectual disability; ASD, autism spectrum disorder; SD, standard deviation.
Single individual with a second pathogenic variant in HNRNPU was not included in the calculations.
Not significant after correction for multiple testing.
Out of 85 individuals with (likely) pathogenic KDM6B variants, 12 had protein-altering variants (PAVs) and 73 had protein-truncating variants (PTVs). We used Fisher’s exact test to compare the frequency of clinical features between the individuals with (likely) pathogenic PAVs and PTVs (Table 1). We used Bonferroni correction to account for multiple testing.
Ethics
This study was approved by the institutional review board “Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen” under number 2011/188. The study participants or their caregivers gave informed consent to participate in the research, of whom 21 consented also to photo publishing (15 individuals with pathogenic KDM6B variants and six with VUSs). Sample data obtained from contributing sites was based on their original ethics protocols referenced in the methods.
Protein structure analysis
The solved three-dimensional (3D) crystal structure of the KDM6B protein was used for analysis of the possible effects of identified variants on the protein. Possible effects of PAVs were predicted on the basis of the wild-type amino acid position and interactions (with other amino acids, other proteins or ligands) and biophysical differences with the mutant amino acid, similarly as described previously.30 The detailed description of the predicted effects is provided in the Table S3. The protein structure used (PDB: 5OY3) contains the C-terminal part of the protein (amino acids [aa] 1141–1643) with the JmJC and Zn-containing domains required for the H3 tail binding and H3K27-specific demethylation together with ligands and co-factors.6 KDM6A JmJC domain structure (PDB: 3AVR) was used for comparison with KDM6B.31 Information from UniProt (ID: O15054)32 (e.g., about disordered regions or modified residues), as well as the AlphaFold33 O15054 ab initio model,34 were used for interpretation of the variants in KDM6B protein regions without solved structure (mostly the N-terminal). The analysis and visualization were performed with YASARA Structure software.35
Variant clustering analysis
Significance of variant clustering was calculated separately for PAVs and PTVs as described before.36 Shortly, we compared geometric mean distance on linear protein structure for the observed variants with randomly permutated variants, performing 1,000,000 permutations and Bonferroni correction for two experiments by using SpatialClustering tool.
Drosophila strains and culture
Flies were reared on standard cornmeal-agar media at 25°C with a 12 h/12 h light/dark cycle in 70% humidity. The mushroom body (MB) driver, R14H06-Gal4 (stock #48667), UAS-Utx-RNAi (UtxRNAi1; stock #34076), UAS-mCherry-RNAi (control 1; stock #35785), ubiquitous driver Act5C-Gal4/CyO (Act-Gal4; stock #4414), and wing-specific driver MS1096-Gal4 (stock #8860) fly lines were obtained from the Bloomington Drosophila Stock Center. A second UAS-Utx-RNAi (UtxRNAi2; stock #37664) and its genetic background control line (control 2; stock #60000) were obtained from Vienna Drosophila Research Center. Control 1 was used as a control because it shares a common genetic background with UtxRNAi1 and expresses a non-targeting double-stranded RNA that controls for any non-specific effects of a RNAi in general. Null mutations in Utx are known to cause lethality in flies.37 In agreement, expression of UtxRNAi lines with a ubiquitous Act-Gal4 driver resulted in lethality, suggesting that the RNAi lines are effective at inducing knockdown.
UAS-KDM6B transgenic flies were generated through Gateway cloning of the reference KDM6B cDNA from KIAA0346 in pENTR3C-KDM6B (a gift from Professor Kristian Helin38) into pGW-HA.attB (GenBank #KC896838) to create pGW-UAS-KDM6B.attB (UAS-KDM6Bref). 19 different PAVs (Table S3) were introduced into pGW-UAS-KDM6B.attB via PCR-based site-directed mutagenesis. The JmJC domain deletion of KDM6B (KDM6BΔJmJC) was completed with the ligation method and primers adapted from Xiang et al.39 We used a similar ligation method to generate domain deletions of the N terminus (KDM6BΔNterm; p.Met1_Pro1100) and the Zn-containing domain (KDM6BΔZndom; p.Tyr1563_Leu1619). All 23 pGW-UAS-KDM6B.attB constructs were validated by Sanger sequencing and inserted into the third chromosome attP2 landing site through phiC31-mediated transgenesis at Genome Prolab (Sherbrooke, QC, Canada).
Drosophila memory, activity, and sleep assays
UtxRNAi and genetic control fly lines were crossed to R14H06-Gal4, and the resulting progeny were analyzed in memory, activity, and sleep assays. Short-term memory (STM) and long-term memory (LTM) were assessed with courtship conditioning in 5-day-old male flies, as previously described.40 Briefly, for each fly pair, a courtship index (CI), which is the proportion of time spent courting over 10 min, was calculated. A minimum of 30 flies were assayed for each genotype in STM and LTM assays. Within each genotype, naive flies were compared to trained flies by Kruskal-Wallis test with uncorrected Dunnett’s test for multiple comparisons. To assess naive courting behavior, we pooled CI from naive short- and long-term experiments for each genotype. UtxRNAi flies were compared to their genetic control by Kruskal-Wallis test with uncorrected Dunnett’s test for multiple comparisons.
Total activity and sleep of flies were monitored as previously described.41 Briefly, a total of 32 flies for each phenotype, males aged 1–4 days, were loaded into activity monitor chambers (Trikinetics, MA, USA). After 1 day of acclimation, we recorded fly locomotion over a 48-h period of 12 h/12 h light/dark cycle and averaged this for each fly to reveal typical 24-h locomotion patterns. Total beam breaks/day were compared between genetic control and corresponding UtxRNAi by t test with two-tailed distribution and unequal variance. A 5-min period of no activity is defined as “sleep.”42,43 We averaged total minutes of sleep over a 48-h period for each fly to reveal typical 24-h sleep patterns and compared these by using t test with two-tailed distributions and unequal variance.
Drosophila MB morphology
To determine whether morphological defects could be responsible for observed memory and behavioral phenotypes, we visualized the structural morphology of the Drosophila MB. UtxRNAi and genetic control fly lines were crossed to R14H06-Gal4 and males and females aged 2–5 days of the resulting progeny were examined for MB morphology. Brains were dissected in PBS, fixed with 4% paraformaldehyde for 45 min at room temperature, and mounted in Vectashield (Vector Laboratories). Brains were imaged with a Zeiss LSM800 confocal microscope at 200× magnification. Confocal stacks were processed with ImageJ software.44 Gross MB morphology was assessed qualitatively and was consistent between at least 10 brains for each genotype.
Drosophila gain-of-function assays
Gain-of-function (GoF) phenotypes were observed upon ectopic expression of UAS-KDM6Bref with the ubiquitous Act-Gal4 driver and the MS1096-Gal4 wing driver. Ubiquitous overexpression of KDM6Bref causes lethality. Act-Gal4/CyO flies were crossed to all UAS-KDM6B variants, and the percentage lethality was calculated by comparison of the number of progeny receiving Act-Gal4 to those receiving the CyO balancer chromosome (% lethality = (1 – # Act-Gal4/# CyO) × 100%). For each ubiquitous UAS-KDM6B variant cross, between 50 and 350 progeny were assessed. Percent lethality of UAS-KDM6B variant transgenes was compared to UAS-KDM6Bref by a chi-square two-sample test for equality of proportions. Expression of UAS-KDM6Bref with MS1096-Gal4 wing driver causes a defect in the formation of the L5 vein in the posterior compartment of the wing. This leads to splitting of L5 at the distal end and results in the appearance of an extra vein protruding into the third posterior cell. A range of 18–35 male flies, aged 2–5 days, were analyzed in wing-specific overexpression for each pGW-UAS-KDM6B.attB variant. Fly wings were mounted in glycerol and imaged with Nikon SMZ800N stereo microscope under 40× magnification. The length of the extra vein was measured and quantified with ImageJ software.44 Statistical comparison of wing vein length was performed via ANOVA with Dunnett’s test for multiple comparisons.
Since KDM6BΔJmJC and pathogenic PAVs cannot induce GoF effects in either assay (i.e., no lethality and no extra vein protrusion), only PAVs that showed effects on both GoF assays were interpreted as functionally disruptive.
Results
The spectrum of identified KDM6B variants
To define the clinical and molecular spectrum of the KDM6B-related NDD, we summarized clinical and genetic information from 85 individuals presenting with an NDD with rare heterozygous (mostly de novo) variants in KDM6B. When combined with the previously reported 12 individuals,10 the total cohort of 97 individuals included information about 81 unique variants in KDM6B (Figure 1). The vast majority of variants are present in a single individual, and only seven variants (three nonsense, three frameshifts, and one in-frame indel) are recurrent in two or three unrelated individuals (as de novo and/or inherited). The variants included 60 PTVs (32 frameshift, 21 nonsense, and seven canonical splicing variants) and 21 PAVs, including 18 missense variants and three in-frame indels. On the basis of ACMG guidelines, all PTVs (n = 60) were classified as likely pathogenic (Figure 1). For PAVs (n = 21), classification was refined with protein structure data and functional analysis in Drosophila models and 11 were classified as (likely) pathogenic and ten as VUSs (see below).
Predicted effect of PAVs on 3D protein structure
KDM6B has two known functional domains, the JmJC domain (aa 1157−1485), which is required for the enzymatic activity of the protein (demethylating H3K27me3/2), and a Zn-containing domain (aa 1563−1620). The Zn-containing domain is structurally similar to GATA-type zinc fingers, but unlike zinc fingers, it is responsible for the specific binding to the H3 tail.6 The interaction between the Zn domain and H3 ensures the specificity of KDM6B to demethylate H3K27. Adjacent to the Zn-containing and JmJC domains, there are two linker regions (aa 1490−1558 and aa 1623−1635) that interact with and are predicted to stabilize the domains.6 While PTVs are scattered throughout the gene (including the last and penultimate exons) (corrected p value = 1 for clustering), 18 of 21 identified PAVs significantly cluster at the C terminus of the protein, in or near the JmJC and Zn-containing domains (Figure 1B) (corrected p value = 2.0 × 10−6), at positions that are predicted to be intolerant to missense variation (Figure S1). The remaining three PAVs (c.283G>A [p.Gly95Ser], c.1264G>A [p.Ala422Thr], c.1645_1650del [p.Pro549_Thr550del]) are located outside of the defined KDM6B protein structure: c.1264G>A (p.Ala422Thr) and c.1645_1650del (p.Pro549_Thr550del) are located in a disordered (without structure) region of the protein, without known functions, while p.Gly95Ser is located in the tetratricopeptide repeat (TPR) domain (predicted on the basis of sequence with AlphaFold model and amino acid homology with other TPR domains), whose function in KDM6B is currently also unknown.
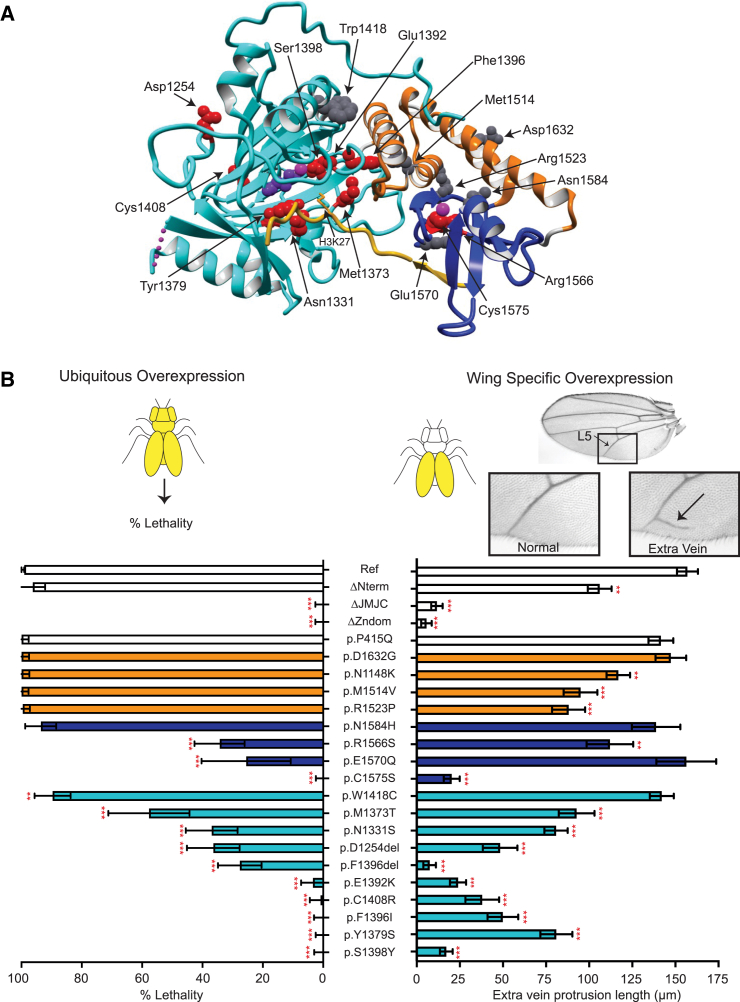
We used available KDM6B and KDM6A protein structures for the C-terminal region spanning amino acids 1147–16406,31 to predict the effect of PAVs on KDM6B function. Of the 21 identified KDM6B PAVs, 18 are located in this region of the protein, allowing for prediction of the effect of each amino acid substitution on protein function. Based on the analysis, 16 of the PAVs are predicted to have a disruptive effect on KDM6B protein structure: nine variants are predicted to disrupt the JmJC domain structure or active site; three PAVs are predicted to disrupt the Zn-containing domain structure or H3 binding; and four stabilizing domain variants were predicted to have a significant impact on the structure of the linker or local structure of the C-terminal region, while the remaining two stabilizing linker variants (c.3444T>G [p.Asn1148Lys] and c.4895A>G [p.Asp1632Gly]) are predicted to have a minimal effect (Figure 2A). Table S3 contains a detailed description of the structural predictions (in addition to ACMG classification) for all 21 KDM6B PAVs.
Figure 2.
Analysis of KDM6B PAVs via protein 3D structure analysis and a dual Drosophila gain-of-function assay
(A) KDM6B fragment (PDB: 5OY3, aa 1157–1639) bound to the H3 tail fragment (aa 17–33). The JmJC domain is shown in cyan with 2-oxoglutaric acid (purple) bound with an Fe ion (magenta), which is necessary for the enzymatic demethylation of H3K27. The Zn-containing domain is shown in blue with Zn ion (magenta). Two out of three JmJC and Zn-containing domain-stabilizing linkers are also visible in the structure (orange). The H3 tail with K27 residue positioned into the active center of the JmJC domain is shown in yellow. Amino acids affected by missense or in-frame indels are shown as balls (pathogenic, red; VUSs, gray), affecting all shown domains as well as binding to H3 tail (see Table S3 for more details on specific variants).
(B) A dual Drosophila gain-of-function assay was used to assess the disruptive potential of KDM6B PAVs. Ubiquitous overexpression (left) of KDM6Bref with the UAS/Gal4 system results in complete lethality. Percent lethality assessed for KDM6BΔNterm, KDM6BΔJmJC, KDM6BΔZndom, KDM6BP415Q as a benign control, and 18 KDM6B variants were compared to KDM6Bref (chi-squared test). n = 50–230 flies for each genotype; data are represented as mean ± 95% confidence interval. Wing-specific overexpression (right) of KDM6Bref in the fly wings results in the formation of an extra vein protruding off the L5 vein. The length of the extra vein was compared to KDM6Bref (Dunnet’s test). n = 18–35 flies for each sample; data are represented as mean ± SEM. PAVs are colored on the basis of the domain (JmJC, cyan; Zn-containing, blue; stabilizing linkers, orange; no domain, white; same as in Figures 1B and 2A). ∗∗p < 0.01, ∗∗∗p < 0.0001.
Experimental testing of KDM6B PAVs with a dual Drosophila GoF assay
We developed a robust dual GoF assay using Drosophila melanogaster to experimentally assess the damaging effect of KDM6B PAVs on protein function. A UAS-KDM6Bref transgene was generated, which revealed that overexpression of human KDM6B in different tissues in flies can induce highly consistent GoF phenotypes. Expression of UAS-KDM6Bref with the ubiquitous Actin-Gal4 driver results in lethality, while expression in the wing with MS1096-Gal4 results in the formation of an extra vein protruding into the third posterior cell (Figure 2B). Overexpression of KDM6B mutants lacking the enzymatic JmJC domain (KDM6BΔJmJC) and/or the Zn-containing domain (KDM6BΔZndom) did not induce lethality or the formation of an extra wing vein. This demonstrates that these GoF phenotypes can be used to detect loss-of-function related to absence of KDM6B enzymatic activity (mediated by the JmJC domain) and/or histone binding (mediated by the Zn-containing domain). In contrast, expression of a KDM6B construct with a deletion of the entire N-terminal region of the protein (KDM6BΔNterm) was able to induce lethality (similar to KDM6Bref), and extra wing vein formation was only mildly reduced when compared to the KDM6B reference protein. This shows that our GoF assay can robustly assess KDM6B functionality related to the C-terminal portion of the protein including the JmJC domain and the Zn-containing domain, while the role of the KDM6B N-terminal region is either dispensable or cannot be reliably assessed with this assay.
We used our dual GoF assay to assess the functional effects of all 18 identified KDM6B PAVs that were present in the C-terminal region of the protein. One benign variant, c.1244C>A (p.Pro415Gln) (KDM6BP415Q), was used as a negative control based on a frequent occurrence in the gnomAD database (allele frequency: 3.62 × 10−5).45 Expression of KDM6BP415Q induced lethality and extra wing vein protrusions similar to KDM6Bref, indicating a functional protein as expected (Figure 2B). Of the 18 PAVs tested, eight of the nine variants located in the JmJC domain failed to induce the GoF phenotypes or showed significantly reduced magnitude of the phenotypes compared to the controls, indicating a clear loss of KDM6B function associated with these variants. Variants located in the Zn-containing domain and the domain stabilizing linkers showed less consistent loss-of-function phenotypes. Variants in the Zn-containing domain showed diverse effects from complete loss of function (c.4724G>C [p.Cys1575Ser]) to reduced function (c.4696C>A [p.Arg1566Ser] and c.4708G>C [p.Glu1570Gln]) or no loss of function (c.4750A>C [p.Asn1584His]). The p.Glu1570Gln variant shows only effects on lethality but not on the wing vein, suggesting a different (or mild) effect. Variants in the domain-stabilizing linkers showed little effect on KDM6B function, and only the wing phenotypes were moderately reduced for some alleles. Importantly, the only JmJC domain variant that did not show loss of KDM6B function (c.4254G>T [p.Trp1418Cys]) is predicted to disrupt a interactions with a linker region and not the JmJC catalytic site (Figure 2A and Table S3), which is consistent with the GoF assay results. These data highlight the sensitivity of the JmJC and Zn-containing domains to missense variants and suggest that the stabilizing function of the linkers is more tolerant to missense variation in general.
Taken together, these data provide evidence for 11 KDM6B PAVs (found in 12 individuals) to be classified as (likely) pathogenic, on the basis of the ACMG guidelines. The remaining ten PAVs are classified as VUSs, including the untested three PAVs located in the KDM6B N-terminal region and six alleles that did not show reduced functionality in the dual GoF assay. Details of the ACMG classification for all PAVs are provided in Table S3.
The Drosophila KDM6B ortholog Utx is required in neurons for normal cognition and behavior
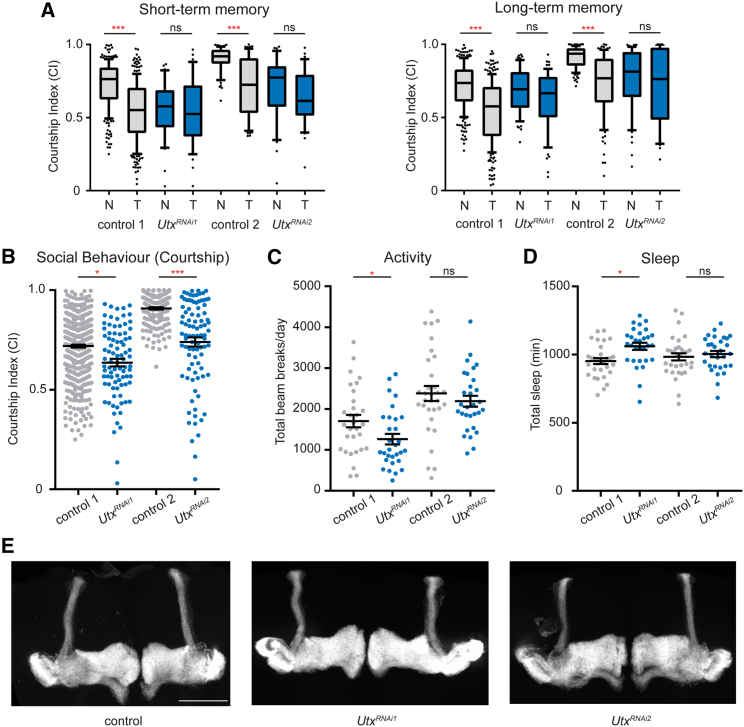
We aimed to understand the potential fundamental role of KDM6B in the brain by using a Drosophila loss-of-function model. Drosophila has a single ortholog of KDM6A and KDM6B, Utx, which is ubiquitously expressed in the fly brain. Germline loss of Utx is lethal,37 and so we used RNAi knockdown to deplete Utx in Drosophila memory neurons of the MB. MB-specific RNAi knockdown was achieved with the R14H06-Gal4 driver line, which is highly specific for post-mitotic MB neurons in the adult and larval fly brain.46 Utx MB knockdown flies were assessed for memory, courtship behavior, activity, and sleep. Two unique Utx RNAi lines both caused defects in STM and LTM and overall courtship behavior (Figures 3A and 3B). In addition, overall daily activity and sleep were affected modestly; one of two RNAi lines showed significant increase in sleep and a significant reduction in activity (Figures 3C and 3D).
Figure 3.
The Drosophila KDM6B ortholog, Utx, is required in neurons for normal memory and behavior
(A) Short term (STM) and long term (LTM) courtship memory was assessed upon MB-specific expression (R14H06-Gal4) of two independent Utx RNAi lines (UtxRNAi1 and UtxRNAi2) and their genetic controls (control 1 and control 2). Boxplots show the distribution of courtship indices (CIs) for naive (N) and trained (T) male flies aged 5 days. Memory was observed when a significant reduction in CI occurred between naive and trained conditions of the same genotype (Kruskal Wallis test). All controls show a significant reduction in courtship in trained vs. naive groups, while Utx RNAi knockdown flies did not. At least 30 flies were tested per condition.
(B) Naive courting behavior was pooled from short- and long-term memory assays and compared between MB-specific Utx RNAi knockdown flies and their genetic controls. At least 60 male flies aged 5 days were tested.
(C and D) MB-specific Utx RNAi knockdown caused reduced daily activity (C) and increased sleep (D) compared to genetic controls, but these differences were only significant for UtxRNAi1 (t test). n = 32 flies for each genotype.
(E) No morphological defects were found following MB-specific knockdown of Utx compared to their genetic controls. MB morphology was consistent in at least ten brains for each genotype. Scale bar represents 50 μm. Data are represented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.0001.
The Drosophila MB undergoes extensive post-mitotic morphological remodeling during fly development, and disruption of these morphogenic processes could underly the observed memory and behavior defects. However, confocal imaging of Utx RNAi knockdown flies showed normal morphology (Figure 3E), suggesting that Utx-dependent memory and behavior defects are not caused by disrupted MB morphogenesis. These findings reflect the broad behavioral effects of the KDM6B haploinsufficiency described in our cohort, confirming a role for this protein family in regulating cognition.
The clinical spectrum of the KDM6B-related NDD
In total, the genetic and functional analyses presented in this study confirmed the likely pathogenic effect of 71 different KDM6B variants identified in 85 individuals (Figure 1A). For 64/85 (75%) of these individuals, de novo occurrence of the variant was demonstrated, but nine of the individuals inherited the variant (five maternal, four paternal) from a mildly affected (developmental delay [DD], learning problems, autism spectrum disorder [ASD]) or clinically unaffected parent. All variants, their classifications, and their location within the protein are shown in Figure 1B.
For comprehensive characterization of the KDM6B-related NDD clinical spectrum, we assessed clinical information only from 85 individuals with KDM6B variants that we classified as (likely) pathogenic: 73 described in this study and 12 published previously.10 For 16/73 newly identified individuals recruited from large cohort research studies, only limited clinical data were available. Proportional statistics along with detailed clinical information for the individuals with the (likely) pathogenic variants are provided in Tables 1 and S1, respectively. To focus on the KDM6B phenotype, we did not include one of 73 individuals in the calculation of the clinical features because of an additional pathogenic variant in HNRNPU. As the effects of the identified VUSs are currently unknown, the clinical characteristics of these individuals are provided separately (Table S2) and were not included in the clinical feature description or frequency calculations.
Neurodevelopmental abnormalities were present in all individuals with likely pathogenic KDM6B variants. Developmental delay (speech-language, motor, or global) was the most common feature, present in all except two individuals. However, at age ≥5 years, when more objective developmental parameters can be assessed, neurodevelopmental problems were less prevalent. Most individuals had intellectual disability (ID), ASD, or both. The level of ID was mostly mild and was present in 63% of individuals. Importantly, severe ID was reported in only two individuals, and one of the individuals had a second diagnosis due to a pathogenic HNRNPU variant (c.970A>G [p.Arg324Gly] [GenBank: NM_031844.2]).47 ASD was reported in 61% of individuals, and other behavioral problems were reported commonly (60%). A psychotic disorder was present in 20% (4/20) of individuals ≥ 12 years of age.
A significant proportion of the individuals showed various neurological abnormalities, including hypotonia (57%), sleep disturbances (32%), seizures (13%), and movement disorders (24%), including gait abnormalities, dystonia-like movement, spasticity, and hypertonia with toe walking. In several individuals, these movement disorders were the main presenting feature and reason for performing genetic testing. Movement disorders resolved over time in two individuals, while one individual required treatment with botulinum toxin injections because of spasticity. Similarly, in several individuals, severe hypotonia was the main presenting feature, leading to muscle biopsy and/or muscle disorder gene panel sequencing to exclude primary myopathies.
Approximately one-third (30%) of individuals with likely pathogenic KDM6B variants displayed features of postnatal overgrowth; tall stature was reported in 8% of individuals and macrocephaly in 26%. Increased weight was reported in 14% of individuals, and 16% (10/63) of individuals had increased birth weight, of whom 7/10 showed overgrowth features later in life. None of the individuals had short stature and the majority have normal growth parameters.
Gastrointestinal issues were common and sometimes severe with a significant impact on the individuals' care. Neonatal feeding difficulties or gastroesophageal reflux was present in half (51%). For several individuals, severe neonatal feeding difficulties required nasogastric tube feeding or even resulted in admission to neonatal intensive care. Constipation, often chronic, was reported in 18% of individuals and, in some, was the major health concern requiring active treatment and regular follow-ups.
Congenital anomalies of different organ systems were also seen in this cohort. Congenital heart disease affected 13% of individuals. Other congenital abnormalities included cleft lip and/or palate, affecting 4%, and congenital genitourinary system anomalies, observed in 10% of individuals. Musculoskeletal system and limb abnormalities were relatively common but mild and variable; cutaneous (II, III, and sometimes IV toe) syndactyly was reported in 9% of individuals, spine curvature abnormalities in 13%, short fingers and/or toes present in 9%, and broad fingers and hands or broad toes and feet in 20%.
Dysmorphic facial features were noted for most of the individuals (Figure 4) and included anteverted nares with depressed nasal bridge, deep-set eyes with down-slanting and narrow palpebral fissures, and prominent forehead. Additionally, some individuals presented with flat face, synophrys, and overfolded helices. Coarse facial features were uncommon and were reported only in four individuals from this study.
Figure 4.
Photos of individuals with identified KDM6B variants
Individuals with (likely) pathogenic variants are shown in the red box above, and those with a VUS are shown in the gray box.
KDM6B-related clinical features described by OMIM (neurodevelopmental disorder with coarse facies and mild distal skeletal abnormalities [MIM: 618505]) were found to be rare in our large cohort with only 4% incidence of coarse facies and 9% incidence of very mild distal skeletal abnormalities identified. Overall, the phenotype of the affected individuals was extremely variable, without clear genotype-phenotype correlation for PAVs versus PTVs (Table 1), ranging from isolated developmental delay or neuropsychiatric problems with normal IQ to severe NDD associated with severe ID and/or multiple affected organ systems.
Discussion
In this study, we describe the molecular and clinical spectrum of the KDM6B-related NDD by using a large cohort of individuals possessing heterozygous PTVs and PAVs in KDM6B. Analyses of KDM6B 3D protein structure combined with an innovative dual GoF assay in Drosophila proved effective for classifying KDM6B PAVs. Pathogenic variants in KDM6B result in loss of one allele, most likely reducing the enzymatic demethylation function of the protein. Our large cohort analysis redefines the KDM6B-related clinical spectrum, which includes ID, ASD, facial dysmorphisms, macrocephaly, various neurological and gastrointestinal problems, congenital anomalies, and a relatively high prevalence of psychotic disorders among adult individuals. The neurodevelopmental phenotype observed in our cohort was recapitulated in a Drosophila neuronal knockdown model, confirming a conserved role of this family of histone demethylases in cognition and behavior and providing a system to further elucidate the underlying molecular mechanisms.
Analysis of KDM6B protein structure largely explains the effect of the identified PAVs on protein function. The C-terminal region of the protein contains a JmJC and Zn-containing domain in addition to domain-stabilizing linker regions, which are required for normal catalytic activity.6 According to ACMG variant interpretation guidelines,29 pathogenic moderate (PM1) criteria can be applied for variants located in a functional domain and/or mutational hot-spot. Not surprisingly, pathogenic de novo KDM6B PAVs significantly cluster at the KDM6B C-terminal region (hot-spot), disrupting the catalytic JmJC domain and the Zn-containing domain, which is required for interaction with histones. Therefore, variants located in these regions could result in loss of H3K27 demethylase activity. To test this, we developed a GoF overexpression assay that can detect loss of KDM6B function associated with its JmJC and Zn-containing domains. Experimental testing of identified PAVs using this Drosophila dual GoF assay confirmed the detrimental effects on KDM6B for PAVs occurring in the JmJC domain. However, variable effects were observed for PAVs in the Zn-containing domain and the predicted domain-stabilizing linkers. Our assay identified a strong loss of function for the p.Cys1575Ser variant, which directly effects a cysteine in the Zn-containing domain that directly interacts with the Zn ion. In contrast, moderate loss of function was observed for PAVs predicted to stabilize loops in the Zn-containing domain (p.Arg1566Ser and p.Glu1570Gln), while no loss of function was observed for a PAV present on the surface of the Zn-containing domain that interacts with the domain-stabilizing linkers. These results suggest that core amino acids of the domain are more important for protein function than those playing a role in domain stabilization. Indeed, for all four PAVs located in the domain stabilizing linkers, we observed only a minimal (if any) effect on KDM6B function. These results raise doubts about the pathogenicity of these variants, which we classify as VUSs. One limitation of our dual GoF assay is that it might be not sensitive to defects in the N-terminal region of the protein as we did not have any clearly pathogenic variant to validate the assay and the N-terminal deletion showed effect only a mild effect on the wing vein phenotype. Since this region of the protein is disordered, it was not possible to classify PAVs near the N terminus of KDM6B. On one hand, N-terminal variants are expected to be benign because this region is mostly disordered, its deletion did affect lethality in our Drosophila GoF assay, it does not have a known tertiary structure (except for a short predicted TPR domain), and it is predicted to be mostly tolerant to missense variants on a populational level (Figures 2 and S1).48 However, we cannot exclude that this region is important for the KDM6B non-enzymatic activity (e.g., binding to other proteins) and the variants may act by a different mechanism.
Truncating variants in the last and penultimate exons are usually interpreted with caution, as they can escape nonsense-mediated decay (NMD), resulting in translation of a truncated protein. In this study, three individuals with likely pathogenic truncating variants predicted to escape NMD were identified, which were predicted to result in a protein lacking the critical (Zn-containing) domain49 and predicted to result in functional loss. Supporting this, our GoF assay results show loss of KDM6B function due to deletion of the Zn-containing domain (Figure 2B).
Individuals with pathogenic KDM6B variants display a wide spectrum of symptoms with variable expressivity. Developmental delay was present in almost all individuals and was the most common clinical feature. In total, most reported individuals (∼90%) had ID, ASD, or both. This highlights that cognitive deficits are the main consistent clinical feature resulting from KDM6B pathogenic variants. Other frequent clinical features included behavioral and psychiatric problems, features of overgrowth, neonatal feeding difficulties, constipation, hypotonia, or movement disorders (spasticity, hypertonia, or ataxia). These features occurred in ∼20%–60% of individuals and showed variable expressivity. Additionally, most of the individuals also presented with some facial dysmorphism, but most of the dysmorphic features are mild and variable among the individuals and the condition does not have a recognizable facial gestalt. Considering highly variable expressivity, it is not surprising that 9/85 (11%) of individuals had a pathogenic variant inherited from a mildly affected or clinically unaffected parent. Taken together, this condition is unlikely to be recognized on the basis of clinical and dysmorphic features alone. This is similar to other conditions recently described where the phenotype including facial dysmorphism is very broad, requiring additional evidence to prove causality.41,50,51
Psychotic disorders were reported in four individuals in our study. While it may seem to be a rare feature, it corresponds to 20% of individuals older than 12 years, an age threshold used because psychotic disorders rarely manifest before that age.52 For the same reason, its true frequency may be underestimated, since only a minority (20/85) of the reported individuals are ≥12 years. These findings are in line with other monogenic NDDs that manifest with DD/ID in childhood and psychotic disorders in adolescence/adulthood, e.g., SETD1A (MIM: 611052),41 KMT2C (MIM: 606833),53 SRCAP (MIM: 611421),50 and EHMT1 (MIM: 607001).54 Additionally, de novo and rare KDM6B variants were recently reported to be associated with schizophrenia at false discovery rate < 5%, especially among cases with developmental delay,52 thus confirming findings from our individual-based cohort. Psychotic disorders are not only complicated to identify and diagnose in such individuals but they can also have therapeutic implications, as has been recently shown for Kleefstra syndrome, caused by EHMT1 haploinsufficiency.54 Therefore, being aware of such risk is critical for accurately diagnosing and providing appropriate treatment and care for these individuals.
There was no genotype-phenotype correlation observed that could explain the variable expressivity of clinical phenotypes. Even though PAVs had different effect sizes in the Drosophila GoF assays, individuals with different KDM6B variant types (PAVs or PTVs) did not display clear differences in the phenotype. Therefore, we hypothesized that the main differences are most likely explained by other genetic or environmental factors. For example, we have observed a significant sex bias in the cohort: ∼3/4 of the affected individuals were males. While gnomAD,45 which does not contain individuals with severe pediatric disorders, is depleted of protein-truncating variants (probability of loss-of-function intolerace [pLI] = 1, loss of function observed over expected upper bound fraction [LOEUF] = 0.14), 13 individuals with high quality truncating variants are present with no sex bias (six females and seven males). These data suggest that the previously described55,56 female protective effect is also at play for the KDM6B pathogenic variants. It is likely that in addition to genetic background, rare variants in other NDD genes also contribute to this variability,57,58 as we have observed for one individual with a severe phenotype with two pathogenic variants (in KDM6B and HNRNPU) (Table S2).
Recently, KDM6B was independently described to be significantly enriched with de novo variants among ∼31,000 individuals with NDDs59 and ∼11,000 individuals with ASD.16 These observations prove the causality of pathogenic heterozygous KDM6B variants in the development of different NDDs and suggest that they are a common NDD cause. Interestingly, in the study by Satterstrom et al., KDM6B is categorized as “ASD predominant” by virtue of having a higher frequency of disruptive de novo variants in ASD-ascertained probands than in NDD-ascertained probands.16 However, in our cohort, collected from various sources (including both studies described above), developmental delay is present in almost all individuals, while ASD is present in about two-thirds of the individuals. This shows the importance of gathering detailed clinical data to evaluate the findings of meta-analyses conducted by large consortia.
Based on the initial description of 12 individuals with KDM6B variants by Stolerman et al.,10 the disorder has been named a “neurodevelopmental disorder with coarse facies and mild distal skeletal abnormalities” by OMIM (MIM: 618505). However, after analyzing this large cohort of individuals with pathogenic KDM6B variants, we see that coarse facies, as well as mild distal skeletal abnormalities, are rare and not typical. As it currently stands, such designation could be misleading to professional and patient communities and therefore urgent redefinition is required. On the basis of the wide array of symptoms caused by pathogenic KDM6B variants in our cohort, we propose that the name “KDM6B-related NDD” would better describe this condition.
While the literature on the role of KDM6B in development is extensive, most studies are limited to cell culture models and often KDM6B haploinsufficiency is used as the control condition in these cell culture studies.60,61 This suggests that KDM6B haploinsufficiency has a limited effect on cell development and differentiation. This is consistent with the low penetrance of developmental/morphological phenotypes observed in our cohort (Tables 1 and S1). Our analysis of Utx in Drosophila memory shows that Utx is required for the normal function of adult memory neurons, post development (Figure 3). Consistent with this, postnatal knockout of Kdm6b in excitatory neurons of mice impairs learning and memory through regulation of dendritic spine formation in the adult mouse brain.62 Interestingly, a recent study identified autistic-like behavioral deficits in Kdm6b haploinsufficient mice,63 which seem to replicate some autistic and cognitive features seen on our cohort. Taken together these functional studies suggest that KDM6B-mediated demethylation of H3K27 may have an evolutionarily conserved role in adult brain function, which could underly the primary cognitive deficits observed in our cohort.
There are some limitations in this study. First, the cohort has ascertainment bias toward individuals who were genetically tested for NDDs and most likely underrepresents mildly affected individuals. There is evidence that some individuals are mildly affected; for example, 13 individuals with KDM6B PTVs are present in gnomAD, that excluded cases with severe pediatric disorders. Additionally, for the majority of the parents with KDM6B pathogenic variants, an NDD phenotype (such as speech delay, learning problems, ASD) was reported, but they were not deeply phenotyped and could not be included in the study. However, they most likely represent a milder spectrum of the condition. For better representation of the phenotypic spectrum of the KDM6B-related NDD and to reduce bias, we recruited a large cohort of individuals from various sources, both diagnostic testing and research cohorts focusing on various phenotypes. Next, even though we used a dual GoF assay, which allowed us to accurately evaluate the effects of multiple PAVs, the assay might be not sensitive to loss of the N-terminal region function, but the role and functions of this region are unknown. Lastly, Utx is the only H3K27 demethylase present in flies, with strong homology to both KDM6B and KDM6A. Behavioral results using a Utx knockdown model are, therefore, relevant to both KDM6A and KDM6B. Disorders caused by mutations in these two genes are both characterized by cognitive deficits, however, there are differences in other clinical phenotypes. As an example, KDM6A (MIM: 300128) pathogenic variants result in Kabuki syndrome type 2 (MIM: 300867), with specific facial features and short stature/microcephaly, while KDM6B-related NDD is associated with overgrowth.64 It is unclear whether the different clinical effect of KDM6A and KDM6B is due to different molecular functions of the two proteins or different expression during human development. There is evidence that KDM6A and KDM6B have some redundancy but also have unique roles and differing expression patterns have been observed for the two genes. A recent study found that in the adult mouse brain Kdm6b is specifically expressed in neurons, while Kdm6a is expressed in neurons and different types of glia.62 While the expression patterns of the two are not well studied in humans, the detailed analysis of mouse expression suggests that clinical differences (for example high prevalence of ASD in our cohort but not in individuals with KDM6A pathogenic variants) may arise from differing expression patterns. It will be interesting to compare future functional analysis of mouse Kdm6a, Kdm6b, and fly Utx in the context of cognitive function to understand the true level of redundancy and evolutionary conservation in the brain.
Our study demonstrates the critical importance of international collaboration, sharing of genomic data, and rigorous functional analysis of genetic variants for an unbiased, accurate, and comprehensive definition of rare genetic disorders. Clinically, the KDM6B-related NDD is a neurodevelopmental disorder characterized by cognitive defects with broad clinical features of variable expressivity, requiring a molecular diagnosis.
Data and code availability
All data are available in the main text or the supplemental materials. The accession numbers for all identified variants and their classification are submitted to ClinVar: SCV002570417–SCV002570487.
Acknowledgments
Funding for this work was provided by an Aspasia grant of the Dutch Research Council (015.014.036 to T.K.), the Netherlands Organisation for Health Research and Development (91718310 to T.K.), NSERC PGSD grant to T.E.J., and a Canadian Institutes of Health Research Project grant (PJT 469689) to J.M.K. Additional funding sources are listed in the supplemental information. We thank the Simons Simplex Collection (SSC),12 Deciphering Developmental Delay (DDD),13 the 100,000 Genome Project,14 the Pediatric Cardiac Genomics Consortium (PCGC),15 the Autism Sequencing Consortium (ASC),16,17 the Autism Speaks MSSNG project,18 the Biobank of the Laboratory of Human Genetics, IRCCS Istituto G. Gaslini, the 2025 French Genomic Medicine Initiative, and GeneDx for providing cases, samples, and/or molecular diagnostic data. We also thank the Bloomington Drosophila Stock Center and the Vienna Drosophila Resource Center for providing fly stocks and Anastasia Mereshchuk for technical assistance.
Author contributions
Conceptualization: D.R., T.E.J., J.M.K., L.E.L.M.V., H.G.B., T.K. Methodology: D.R., T.E.J., J.M.K., T.K. Investigation: all authors. Formal analysis: D.R., T.E.J. Visualization: D.R., T.E.J., V.I. Supervision: L.E.L.M.V., H.G.B., J.M.K., T.K., D.T. Writing—original draft: D.R., T.E.J., J.M.K., T.K. Writing—review & editing: all authors.
Declaration of interests
S.W.S. is a scientific consultant of Population Bio and the King Abdullaziz University, and Athena Diagnostics has licensed intellectual property from his work held by the Hospital for Sick Children, Toronto.
Published: May 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.04.008.
Contributor Information
Jamie M. Kramer, Email: jkramer@dal.ca.
Tjitske Kleefstra, Email: t.kleefstra@erasmusmc.nl.
Supplemental information
References
- 1.Gerrard D.T., Berry A.A., Jennings R.E., Birket M.J., Zarrineh P., Garstang M.G., Withey S.L., Short P., Jiménez-Gancedo S., Firbas P.N., et al. Dynamic changes in the epigenomic landscape regulate human organogenesis and link to developmental disorders. Nat. Commun. 2020;11:3920. doi: 10.1038/s41467-020-17305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciptasari U., van Bokhoven H. The phenomenal epigenome in neurodevelopmental disorders. Hum. Mol. Genet. 2020;29:R42–R50. doi: 10.1093/hmg/ddaa175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 4.Faundes V., Newman W.G., Bernardini L., Canham N., Clayton-Smith J., Dallapiccola B., Davies S.J., Demos M.K., Goldman A., Gill H., et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2018;102:175–187. doi: 10.1016/j.ajhg.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cenik B.K., Shilatifard A. COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 2021;22:38–58. doi: 10.1038/s41576-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 6.Jones S.E., Olsen L., Gajhede M. Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity. Biochemistry. 2018;57:585–592. doi: 10.1021/acs.biochem.7b01152. [DOI] [PubMed] [Google Scholar]
- 7.De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Meng Y., Li H., Liu C., Zheng L., Shen B. Jumonji domain-containing protein family: the functions beyond lysine demethylation. J. Mol. Cell Biol. 2018;10:371–373. doi: 10.1093/jmcb/mjy010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X., Liu L., Yuan X., Wei Y., Wei X. JMJD3 in the regulation of human diseases. Protein Cell. 2019;10:864–882. doi: 10.1007/s13238-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolerman E.S., Francisco E., Stallworth J.L., Jones J.R., Monaghan K.G., Keller-Ramey J., Person R., Wentzensen I.M., McWalter K., Keren B., et al. Genetic variants in the KDM6B gene are associated with neurodevelopmental delays and dysmorphic features. Am. J. Med. Genet. 2019;179:1276–1286. doi: 10.1002/ajmg.a.61173. [DOI] [PubMed] [Google Scholar]
- 11.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.100000 Genomes Project Pilot Investigators. Smedley D., Smith K.R., Martin A., Thomas E.A., McDonagh E.M., Cipriani V., Ellingford J.M., Arno G., Tucci A., et al. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care - Preliminary Report. N. Engl. J. Med. 2021;385:1868–1880. doi: 10.1056/NEJMoa2035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin S.C., Homsy J., Zaidi S., Lu Q., Morton S., DePalma S.R., Zeng X., Qi H., Chang W., Sierant M.C., et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat. Genet. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L., et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buxbaum J.D., Daly M.J., Devlin B., Lehner T., Roeder K., State M.W., Autism Sequencing Consortium The autism sequencing consortium: large-scale, high-throughput sequencing in autism spectrum disorders. Neuron. 2012;76:1052–1056. doi: 10.1016/j.neuron.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C Yuen R.K., Merico D., Bookman M., L Howe J., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z., et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K., et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 20.Guillen Sacoto M.J., Tchasovnikarova I.A., Torti E., Forster C., Andrew E.H., Anselm I., Baranano K.W., Briere L.C., Cohen J.S., Craigen W.J., et al. De Novo Variants in the ATPase Module of MORC2 Cause a Neurodevelopmental Disorder with Growth Retardation and Variable Craniofacial Dysmorphism. Am. J. Hum. Genet. 2020;107:352–363. doi: 10.1016/j.ajhg.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunet T., Jech R., Brugger M., Kovacs R., Alhaddad B., Leszinski G., Riedhammer K.M., Westphal D.S., Mahle I., Mayerhanser K., et al. De novo variants in neurodevelopmental disorders-experiences from a tertiary care center. Clin. Genet. 2021;100:14–28. doi: 10.1111/cge.13946. [DOI] [PubMed] [Google Scholar]
- 22.Lecoquierre F., Bonnevalle A., Chadie A., Gayet C., Dumant-Forest C., Renaux-Petel M., Leca J.B., Hazelzet T., Brasseur-Daudruy M., Louillet F., et al. Confirmation and further delineation of the SMG9-deficiency syndrome, a rare and severe developmental disorder. Am. J. Med. Genet. 2019;179:2257–2262. doi: 10.1002/ajmg.a.61317. [DOI] [PubMed] [Google Scholar]
- 23.Husson T., Lecoquierre F., Cassinari K., Charbonnier C., Quenez O., Goldenberg A., Guerrot A.M., Richard A.C., Drouin-Garraud V., Brehin A.C., et al. Rare genetic susceptibility variants assessment in autism spectrum disorder: detection rate and practical use. Transl. Psychiatry. 2020;10:77. doi: 10.1038/s41398-020-0760-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falb R.J., Müller A.J., Klein W., Grimmel M., Grasshoff U., Spranger S., Stöbe P., Gauck D., Kuechler A., Dikow N., et al. Bi-allelic loss-of-function variants in KIF21A cause severe fetal akinesia with arthrogryposis multiplex. J. Med. Genet. 2021;60:48–56. doi: 10.1136/jmedgenet-2021-108064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pezzani L., Marchetti D., Cereda A., Caffi L.G., Manara O., Mamoli D., Pezzoli L., Lincesso A.R., Perego L., Pellicioli I., et al. Atypical presentation of pediatric BRAF RASopathy with acute encephalopathy. Am. J. Med. Genet. 2018;176:2867–2871. doi: 10.1002/ajmg.a.40635. [DOI] [PubMed] [Google Scholar]
- 26.Hertz J.M., Svenningsen P., Dimke H., Engelund M.B., Nørgaard H., Hansen A., Marcussen N., Thiesson H.C., Bergmann C., Larsen M.J. Detection of DZIP1L mutations by whole-exome sequencing in consanguineous families with polycystic kidney disease. Pediatr. Nephrol. 2022;37:2657–2665. doi: 10.1007/s00467-022-05441-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schobers G., Schieving J.H., Yntema H.G., Pennings M., Pfundt R., Derks R., Hofste T., de Wijs I., Wieskamp N., van den Heuvel S., et al. Reanalysis of exome negative patients with rare disease: a pragmatic workflow for diagnostic applications. Genome Med. 2022;14:66. doi: 10.1186/s13073-022-01069-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson A., Banka S., Stewart H., Genomics England Research Consortium. Robinson H., Lovell S., Clayton-Smith J. Recurrent KCNT2 missense variants affecting p.Arg190 result in a recognizable phenotype. Am. J. Med. Genet. 2021;185:3083–3091. doi: 10.1002/ajmg.a.62370. [DOI] [PubMed] [Google Scholar]
- 29.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijders Blok L., Verseput J., Rots D., Venselaar H., Innes A.M., Stumpel C., Õunap K., Reinson K., Seaby E.G., McKee S., et al. A clustering of heterozygous missense variants in the crucial chromatin modifier WDR5 defines a new neurodevelopmental disorder. HGG Adv. 2023;4:100157. doi: 10.1016/j.xhgg.2022.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengoku T., Yokoyama S. Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 2011;25:2266–2277. doi: 10.1101/gad.172296.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:439–444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krieger E., Vriend G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics. 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L., Gilissen C. Spatial Clustering of de Novo Missense Mutations Identifies Candidate Neurodevelopmental Disorder-Associated Genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copur Ö., Müller J. The histone H3-K27 demethylase Utx regulates HOX gene expression in Drosophila in a temporally restricted manner. Development. 2013;140:3478–3485. doi: 10.1242/dev.097204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agger K., Cloos P.A.C., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 39.Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C.D. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 40.Siegel R.W., Hall J.C. Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl. Acad. Sci. USA. 1979;76:3430–3434. doi: 10.1073/pnas.76.7.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kummeling J., Stremmelaar D.E., Raun N., Reijnders M.R.F., Willemsen M.H., Ruiterkamp-Versteeg M., Schepens M., Man C.C.O., Gilissen C., Cho M.T., et al. Characterization of SETD1A haploinsufficiency in humans and Drosophila defines a novel neurodevelopmental syndrome. Mol. Psychiatry. 2021;26:2013–2024. doi: 10.1038/s41380-020-0725-5. [DOI] [PubMed] [Google Scholar]
- 42.Shaw P.J., Cirelli C., Greenspan R.J., Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 43.Huber R., Hill S.L., Holladay C., Biesiadecki M., Tononi G., Cirelli C. Sleep homeostasis in Drosophila melanogaster. Sleep. 2004;27:628–639. doi: 10.1093/sleep/27.4.628. [DOI] [PubMed] [Google Scholar]
- 44.Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chubak M.C., Nixon K.C.J., Stone M.H., Raun N., Rice S.L., Sarikahya M., Jones S.G., Lyons T.A., Jakub T.E., Mainland R.L.M., et al. Individual components of the SWI/SNF chromatin remodelling complex have distinct roles in memory neurons of the Drosophila mushroom body. Dis. Model. Mech. 2019;12:dmm037325. doi: 10.1242/dmm.037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bramswig N.C., Lüdecke H.J., Hamdan F.F., Altmüller J., Beleggia F., Elcioglu N.H., Freyer C., Gerkes E.H., Demirkol Y.K., Knupp K.G., et al. Heterozygous HNRNPU variants cause early onset epilepsy and severe intellectual disability. Hum. Genet. 2017;136:821–834. doi: 10.1007/s00439-017-1795-6. [DOI] [PubMed] [Google Scholar]
- 48.Wiel L., Baakman C., Gilissen D., Veltman J.A., Vriend G., Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum. Mutat. 2019;40:1030–1038. doi: 10.1002/humu.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abou Tayoun A.N., Pesaran T., DiStefano M.T., Oza A., Rehm H.L., Biesecker L.G., Harrison S.M., ClinGen Sequence Variant Interpretation Working Group ClinGen SVI Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rots D., Chater-Diehl E., Dingemans A.J.M., Goodman S.J., Siu M.T., Cytrynbaum C., Choufani S., Hoang N., Walker S., Awamleh Z., et al. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am. J. Hum. Genet. 2021;108:1053–1068. doi: 10.1016/j.ajhg.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vissers L.E.L., Kalvakuri S., de Boer E., Geuer S., Oud M., van Outersterp I., Kwint M., Witmond M., Kersten S., Polla D.L., et al. De Novo Variants in CNOT1, a Central Component of the CCR4-NOT Complex Involved in Gene Expression and RNA and Protein Stability, Cause Neurodevelopmental Delay. Am. J. Hum. Genet. 2020;107:164–172. doi: 10.1016/j.ajhg.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J.D., Bass N., Bigdeli T.B., Breen G., Bromet E.J., et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–516. doi: 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howrigan D.P., Rose S.A., Samocha K.E., Fromer M., Cerrato F., Chen W.J., Churchhouse C., Chambert K., Chandler S.D., Daly M.J., et al. Exome sequencing in schizophrenia-affected parent-offspring trios reveals risk conferred by protein-coding de novo mutations. Nat. Neurosci. 2020;23:185–193. doi: 10.1038/s41593-019-0564-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vermeulen K., Staal W.G., Janzing J.G., van Bokhoven H., Egger J.I.M., Kleefstra T. Sleep Disturbance as a Precursor of Severe Regression in Kleefstra Syndrome Suggests a Need for Firm and Rapid Pharmacological Treatment. Clin. Neuropharmacol. 2017;40:185–188. doi: 10.1097/wnf.0000000000000226. [DOI] [PubMed] [Google Scholar]
- 55.Jacquemont S., Coe B.P., Hersch M., Duyzend M.H., Krumm N., Bergmann S., Beckmann J.S., Rosenfeld J.A., Eichler E.E. A higher mutational burden in females supports a "female protective model" in neurodevelopmental disorders. Am. J. Hum. Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wigdor E.M., Weiner D.J., Grove J., Fu J.M., Thompson W.K., Carey C.E., Baya N., van der Merwe C., Walters R.K., Satterstrom F.K., et al. The female protective effect against autism spectrum disorder. Cell Genom. 2022;2:100134. doi: 10.1016/j.xgen.2022.100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niemi M.E.K., Martin H.C., Rice D.L., Gallone G., Gordon S., Kelemen M., McAloney K., McRae J., Radford E.J., Yu S., et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature. 2018;562:268–271. doi: 10.1038/s41586-018-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parenti I., Rabaneda L.G., Schoen H., Novarino G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020;43:608–621. doi: 10.1016/j.tins.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 59.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo T., Han X., He J., Feng J., Jing J., Janečková E., Lei J., Ho T.V., Xu J., Chai Y. KDM6B interacts with TFDP1 to activate P53 signaling in regulating mouse palatogenesis. Elife. 2022;11:e74595. doi: 10.7554/eLife.74595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W., Cho H., Lee J.W., Lee S.K. The histone demethylase Kdm6b regulates subtype diversification of mouse spinal motor neurons during development. Nat. Commun. 2022;13:958. doi: 10.1038/s41467-022-28636-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y., Khandelwal N., Liu S., Zhou M., Bao L., Wang J.E., Kumar A., Xing C., Gibson J.R., Wang Y. KDM6B cooperates with Tau and regulates synaptic plasticity and cognition via inducing VGLUT1/2. Mol. Psychiatry. 2022;27:5213–5226. doi: 10.1038/s41380-022-01750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao Y., Aljazi M.B., He J. Kdm6b Haploinsufficiency Causes ASD/ADHD-Like Behavioral Deficits in Mice. Front. Behav. Neurosci. 2022;16:905783. doi: 10.3389/fnbeh.2022.905783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faundes V., Goh S., Akilapa R., Bezuidenhout H., Bjornsson H.T., Bradley L., Brady A.F., Brischoux-Boucher E., Brunner H., Bulk S., et al. Clinical delineation, sex differences, and genotype-phenotype correlation in pathogenic KDM6A variants causing X-linked Kabuki syndrome type 2. Genet. Med. 2021;23:1202–1210. doi: 10.1038/s41436-021-01119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplemental materials. The accession numbers for all identified variants and their classification are submitted to ClinVar: SCV002570417–SCV002570487.