Summary
10 years ago, a detailed analysis showed that only 33% of genome-wide association study (GWAS) results included the X chromosome. Multiple recommendations were made to combat such exclusion. Here, we re-surveyed the research landscape to determine whether these earlier recommendations had been translated. Unfortunately, among the genome-wide summary statistics reported in 2021 in the NHGRI-EBI GWAS Catalog, only 25% provided results for the X chromosome and 3% for the Y chromosome, suggesting that the exclusion phenomenon not only persists but has also expanded into an exclusionary problem. Normalizing by physical length of the chromosome, the average number of studies published through November 2022 with genome-wide-significant findings on the X chromosome is ∼1 study/Mb. By contrast, it ranges from ∼6 to ∼16 studies/Mb for chromosomes 4 and 19, respectively. Compared with the autosomal growth rate of ∼0.086 studies/Mb/year over the last decade, studies of the X chromosome grew at less than one-seventh that rate, only ∼0.012 studies/Mb/year. Among the studies that reported significant associations on the X chromosome, we noted extreme heterogeneities in data analysis and reporting of results, suggesting the need for clear guidelines. Unsurprisingly, among the 430 scores sampled from the PolyGenic Score Catalog, 0% contained weights for sex chromosomal SNPs. To overcome the dearth of sex chromosome analyses, we provide five sets of recommendations and future directions. Finally, until the sex chromosomes are included in a whole-genome study, instead of GWASs, we propose such studies would more properly be referred to as “AWASs,” meaning “autosome-wide scans.”
Keywords: GWAS, sex, X chromosome, Y chromosome, quality control, data analysis
A decade ago, paucity of GWAS signals on the X chromosome was noted, in part because only 1/3 of GWASs included the X chromosome. Revisiting this topic, we find that the situation has become even worse: only 1/4 GWASs analyzed the X chromosome.
Introduction
In the 10 years since Wise et al. (2013)1 brought the exclusion of the X chromosome from genome-wide association studies (GWASs) to the attention of the community, little has improved regarding the analysis and reporting of the sex chromosomal variants in GWASs.2,3,4 The X chromosome accounts for ∼5% of the haploid genome and carries ∼800 protein-coding genes. However, to date (November 2022), even after the call for including the X chromosome in GWASs by Wise et al.,1 approximately only 0.5% of associated SNPs in the NHGRI-EBI GWAS Catalog5,6 are on the X chromosome, a 10-fold paucity compared to the autosomes.
The paucity of research on the sex chromosomes includes both the X and Y chromosomes. For the Y chromosome,7 as of November 29, 2022, only nine out of 447,939 associations reported in NHGRI-EBI GWAS Catalog5,6 belong to the Y chromosome. Coverage is scarce on GWAS arrays for the male-only Y chromosome, in part because of repetitive sequences that make variant calling difficult. If Y chromosomal variants are available in the non-pseudo-autosomal region (NPR), they can be analyzed via existing methods. However, there appears to be “a lack of will” to do so.8
The X chromosome presents multiple analytical challenges,9,10,11,12,13,14 including (1) a male has one copy of the X chromosome while a female has two, in contrast to the autosomes; (2) the X chromosome in male germ cells only recombines with the Y chromosome in the pseudo-autosomal regions (PARs) but not in the NPR; (3) in contrast to males, the two copies in female germ cells recombine across the entire X chromosome; (4) the two female copies are also subject to X inactivation (i.e., X chromosome dosage compensation); (5) the X-inactivation status at the population level can be random, skewed, or absent (i.e., X-inactivation escape); and (6) the true X-inactivation status at the individual level cannot be derived from GWAS data alone.
Thus, the existing bioinformatic, statistical, and machine learning methods developed specifically for the autosomes are not suitable for the sex chromosomes. For example, most bioinformatic tools are autosome-centric, meaning that even if the sex chromosomes were included in the pipelines, tool developments were not tailored for the sex chromosomes.11 These include variant calling,15,16 data quality control (QC) prior to imputation17,18 (e.g., cryptic relatedness,19,20 Hardy-Weinberg equilibrium [HWE]21,22), and imputation.15,23,24,25,26,27,28,29,30 Similarly, most association methods are not tailored for the sex chromosomes, including population stratification via principal-component analysis (PCA),31 and the association methodology itself.32,33,34,35,36 Finally, the recent polygenic risk score (PRS)-based disease risk prediction methods37,38,39,40 rarely include the sex chromosomes.
Back in 2013, after examining 743 GWAS papers published between January 2010 and December 2011 and in the NHGRI GWAS Catalog,41 Wise and colleagues noted that only ∼33% GWASs included the X chromosome1; the Y chromosome was not explicitly examined, though it is implicitly involved in the X chromosome through the PARs. Additionally, the authors commented on QC and power concerns, including poorer coverage of the X chromosome in early GWAS arrays and lower genotyping and imputation accuracy as compared to autosomes, as well as X-inactivation-related analytical complexities that may reduce power of an association study. Finally, the authors concluded that “many interesting biological insights could be revealed if we end the exclusion of the X chromosome in future GWAS.”
Thus, 10 years later, we first re-surveyed the research landscape to determine whether the earlier recommendations of including the X (and Y) chromosome(s) in GWASs had been translated into changes in practice. Second, as genotyping and sequencing technologies have also evolved, including imputation panels based on next-generation sequencing data,15,42 we then scanned the literature for emerging issues and insights. Finally, we make new recommendations.
Sex chromosome results in the NHGRI-EBI GWAS and PolyGenic Score (PGS) Catalogs
Lack of X and Y SNP-trait associations in the NHGRI-EBI GWAS Catalog
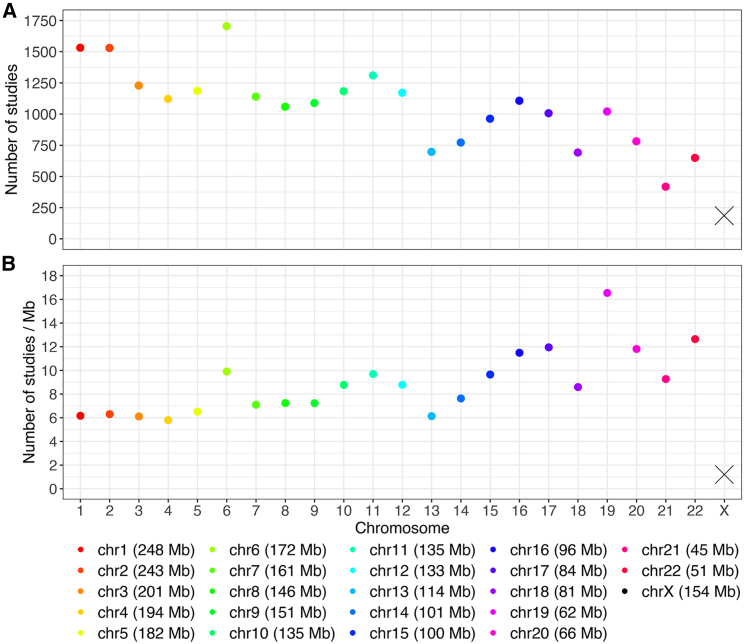
As of Nov 29, 2022, the NHGRI-EBI GWAS Catalog5,6 contained 6,130 published studies, of which 4,208 reported at least one genome-wide-significant association (p value < 5 × 10−8).43 However, only 186 studies (4.4%) had signals on the X chromosome (Figure 1A). In contrast, chromosome 21 had twice the number of signals (418 studies; 9.9%), despite being less than one-third the length (Figure 1A).
Figure 1.
Total number of studies and average number of studies per Mb reporting at least one genome-wide significant finding (p value < 5 × 10−8) stratified by chromosome, from the NHGRI-EBI GWAS Catalog
(A and B) Total number of studies (A) and average number of studies (B). Genetic associations were indexed by unique PubMed IDs up to November 29, 2022. Studies reporting associations with multiple traits were only counted once.
Before investigating how often the X chromosome was analyzed to begin with (in the next section), we first normalized each chromosome by its physical length (Figure 1B). It is clear that signal densities vary across the autosomes. However, the most striking feature is the continued paucity of signals on the X chromosome since 2010–2011.1
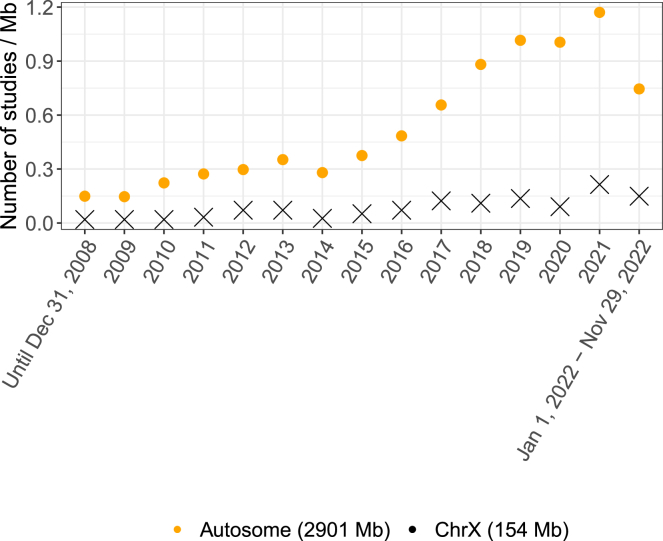
To investigate whether the 2013 recommendation to include the X chromosome in GWASs had an impact on the practice of our field, we examined temporal changes. Figure 2 shows the average number of studies per Mb with at least one genome-wide-significant finding, separately for the autosomes and the X chromosome, from prior to 2008 to November 29, 2022. Unfortunately, the gap between the autosomes and the X chromosome appears to be widening in recent years. Between 2009 and 2021, the average number of studies with genome-wide-significant findings on the X chromosome grew at approximately 0.012 studies/Mb/year, remaining below 0.3/Mb every year (Figures 2 and S2). In contrast, the numbers increased consistently for the autosomes by approximately 0.086 studies/Mb/year. For comparison, Figure S1 shows the total number of studies reporting one or more signals per chromosome over time.
Figure 2.
Average number of studies per Mb reporting at least one genome-wide significant finding (p value < 5 × 10−8) over time, separately for the autosomes and X chromosome, from the NHGRI-EBI GWAS Catalog
Genetic associations were indexed by unique PubMed IDs up to November 29, 2022. Studies reporting associations with multiple traits were only counted once.
Examining GWAS array and sequencing studies (GWAS-by-WES [whole-exome sequencing], GWAS-by-WGS [whole-genome sequencing]) separately, using the “genotyping technology” variable, revealed that 4.6% of GWAS loci came from studies that included sequencing and only 0.76% of those loci (12 out of 1,576) were on the X chromosome; six of those 12 loci came from a single study.44 Additionally, many of the studies that employed sequencing also used data from genotyping arrays.
Ironically, one of the most comprehensive X chromosome-wide studies (XWASs45) is not included in the NHGRI-EBI GWAS Catalog, presumably because it only reported the X chromosome association results, which does not meet the catalog inclusion criteria requiring genome-wide results. Although they showed that the contribution of X chromosome loci to trait variability may be smaller than similar-sized autosomes, for height in males, the X chromosome h2 estimate is similar to that for many shorter autosomes, including chromosomes 13 and 18.45 These authors also observed interesting sex differences in X chromosome heritability across 20 quantitative traits in the UK Biobank on the basis of the central imputation from the Affymetrix arrays. Specifically, NPR X chromosome h2 estimates were on average twice as high for males as for females (0.63% vs. 0.30%), with the noticeable exception of educational attainment. When the XWASs were performed, hundreds of X chromosomal loci were identified across these 20 quantitative traits, with twice as many signals detected in males than females, and some loci had remarkable male-specific effects across numerous traits.
Genetic associations on the Y chromosome were even more rarely documented. Out of all 447,939 associations (p value < 1 × 10−5), only nine, arising from two studies, were on the Y chromosome; among the 293,170 genome-wide-significant findings, only one was from the Y chromosome.
Lack of X and Y chromosome results in genome-wide summary statistics in the NHGRI-EBI GWAS Catalog
To address whether lack of sex chromosomal GWAS results were due to lack of appropriate (or any) analysis of the sex chromosomes, we calculated the proportion of genome-wide summary statistics that included sex chromosome results, regardless of if there were significant findings.
There were 19,935 genome-wide summary statistics published in 2021 and posted at the NHGRI-EBI GWAS Catalog5,6 (web resources). These GWAS submissions came from 136 publications, of which most provided one to two sets of summary statistics, but four provided >1,000 sets (Data S1). To avoid analyzing multiple submissions from the same publication, we randomly selected one submission from each of the 136 publications (Data S1).
Out the 136 GWAS summary statistics, only 34 (25%) contained X chromosome results (of the 34, only four also included Y chromosome results), which is less than the 33% based on the survey of GWASs conducted in 2010 and 2011.1 Thus, exclusion has become more rather than less prevalent, contrary to the intent of the initial commentary! Further, exclusion appears to be an exclusionary problem, where both sex chromosomes have been routinely neglected in whole-genome studies.
If we assume that the 136 studies with summary statistics in the NHGRI-EBI GWAS Catalog are a random sample of all GWASs in 2021 and recall from the previous section that there is a 6-fold difference in the average findings between chromosome 1 and the X chromosome (Figure 1B), it is then reasonable to hypothesize that much of the paucity would be resolved if the X chromosome were actually analyzed across all GWASs.
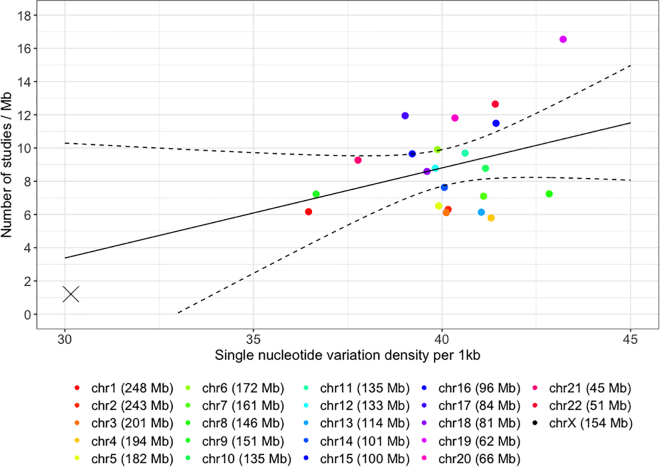
Some well-known contributing factors include the smaller effective population size (Ne) and X chromosome inactivation in females, which reduce power to detect associations compared to autosomes.45,46 Variation in single-nucleotide diversity47,48 can be another contributing factor. For example, chromosome 19 has the highest density of single-nucleotide variations of 43.21/kb (based on the 1000 Genomes Project) among all chromosomes, while chromosome 1 has the lowest of all autosomes at 36.46/kb. However, cumulatively as of December 2022, there is no statistically significant linear relationship (slope = 0.54; p value = 0.13; Figure 3) between nucleotide diversity and the average number of genome-wide-significant findings among the autosomes. Even if we were willing to extrapolate the linearly fitted line to 30.16/kb, the nucleotide diversity of the X chromosome, the expected research yield on the X chromosome is 3.47/Mb, almost thrice the actual output of 1.21/Mb.
Figure 3.
Average number of studies per Mb reporting at least one genome-wide significant finding (p value < 5 × 10−8) per chromosome, from the NHGRI-EBI GWAS Catalog cumulatively, compared to chromosome-specific nucleotide diversity48
Genetic associations were indexed by unique PubMed IDs up to November 29, 2022. Studies reporting associations with multiple traits were only counted once. The solid slope was fitted using the autosomal data only, and the dashed curves are the 95% confidence bands.
Based on high-coverage whole-genome sequencing of TOPMed15 cohorts, the X chromosome has lower density of variants in coding sequences compared to the autosomes.49 This can be an additional contributing factor to the paucity of signals on the X chromosome.
Lack of X and Y chromosome results in the PolyGenic Score (PGS) Catalog
Based on the above results from the GWAS Catalog, there is also a lack of sex chromosome results in the PGS Catalog, as expected. We downloaded PGS scoring files from the PGS Catalog50 (web resources), focusing on the 430 files (PGS001802 to PGS002231) all uploaded on January 10, 2022. Unsurprisingly, none of the 430 files contained any results from the sex chromosomes, confirming the current exclusionary practice in PGS research as well.
Other emerging exclusionary issues: Quality control, association analysis and reporting, results interpretation, the Y chromosome, and clinical implications
Quality control
In addition to the QC discussed by Wise et al.,1 many data quality pipelines and imputation tools15,16,17,18,23,24,25,26,27,28,29,30 have been developed for GWASs. However, most are autosome-centric, ignoring the sex chromosomes either explicitly or implicitly. In 2014, König et al.11 highlighted “the steps in which the X chromosome requires specific attention, and [gave] tentative advice for each of these,” including sex-stratified minor allele frequencies (MAFs) and missing rates, as well as testing for differential missingness. However, these recommendations have not been followed in practice. For example, sex-specific variant call rates are rarely reported.
There has been little work on chromosome-specific imputation quality. However, a recent study that compared the X chromosome with the autosomes51 examined imputation from the Affymetrix 500k array in an admixed population with the Illumina MEGA array as the gold standard. They showed that, using the Michigan Imputation Server with the 1000 Genomes Project phase 3 data, the X chromosome had 70% imputation accuracy compared to 84% on the autosomes. Further, they showed that imputation quality scores were also lower on the X chromosome across all imputation approaches. It would be interesting to study sex-specific imputation quality on the X chromosome.
Sex difference in minor allele frequency as QC revisited
Checking for sex difference in minor allele frequency (sdMAF) is rarely formed as part of GWAS QC. However, it was already noted a decade ago that “MAF checks might need to be conducted separately for the X chromosome because the expected frequencies are sex dependent,” based on an informal poll of leading statistical geneticists working in GWASs in 2013.1 Others also suggested to include an sdMAF test as part of the QC for the X chromosome.11 However, a recent work has shown that there are possible causes of sdMAF: genotyping errors and biology.52 Delineating the two causes for each X chromosomal SNP is not straightforward, creating challenges in QC pipelines.
The recent study analyzed the high-coverage whole-genome sequencing data of the 1000 Genomes Project48 and gnomAD v3.1.253 and identified many SNPs with genome-wide-significant sdMAF across the X chromosome, particularly at the boundaries between PAR and NPR.52 Further, the study concluded that region-specific sdMAF at the PAR-NPR boundaries is most likely a biological phenomenon, possibly due to sex-specific linkage.54,55,56 This illustrates the challenges of including sdMAF as a QC measure.
As sdMAF is statistically equivalent to GWAS of sex, both evaluating whether there is allele frequency difference between sexes, there is also a connection between the sdMAF study52 and a recent GWAS of sex.57 This GWAS of sex used data from 2.46 million customers of 23andMe but did not examine the X chromosome.57 Although their main conclusion was that sdMAF is a result of participation bias, they also noted that 55% of their significant findings on the autosomes are most likely results of genotyping errors, further illustrating the importance and challenges of separating genotyping errors from biology (and other causes) that could lead to sdMAF.
Hardy-Weinberg equilibrium (HWE) test as QC revisited
Departure from HWE is routinely used as part of GWAS QC for autosomes,17 as SNPs with severe Hardy-Weinberg disequilibrium (HWD) are typically believed to have genotyping errors.58 However, how to evaluate HWE for the X chromosome is unclear and it remains debatable whether testing for HWD should be used at all as part of data QC, both of which we discuss next.
The standard HWE test is Pearson’s test, testing for the difference between the observed and expected genotype counts based on HWE.59,60 This test is typically applied to sex-combined genotype counts, which is reasonable for an autosomal SNP. But applying such an HWE test to an X chromosomal PAR or NPR SNP requires additional considerations.61 For example, König et al.11 recommended performing the HWE test with only females.
Alternatively, Graffelman and Weir21,62 suggested using both females and males, and they proposed a new HWE test for an NPR SNP that includes the deviation of male genotype counts from the expected, based on sex-pooled allele frequency estimate. However, this alternative test has been shown to be simultaneously testing for HWD in females and sdMAF between males and females.61 Therefore, if sdMAF were present, this sex-combined HWE test can be misleading. Instead of the Pearson’s test, testing for model fit has also been proposed.63
Regardless of the specific HWE test used, screening out variants with HWD is questionable for the X chromosome for two other reasons. First, it has been long (but not well) known that it takes several generations to achieve HWE on the X chromosome in contrast to a single generation for the autosomes under the same set of assumptions such as random mating.22 Second, for the autosomes, recent works64,65,66 have shown that association power can be improved by leveraging the difference in HWD between cases and controls while remaining robust to HWD caused by genotyping errors, but this has yet to be explored for the X chromosome.
X-inactivation uncertainty and association results interpretation
Until very recently, the statistical genetics community believed that X inactivation was the main analytical challenge to achieving X chromosome-inclusive GWASs.1,11 Therefore, most of the association methods developed so far have focused on X inactivation.67,68,69,70,71,72 As the true model can be escaping, random, or skewed X inactivation, existing analytic methods include using minimum p value,67 model selection,69 or Bayesian model averaging.72
We note, however, that these statistical approaches rarely address the practical limitation that X inactivation can vary by cell and tissue, and until an association is identified, the relevant cells and tissues cannot even be guessed. Additionally, skewed X inactivation is confounded with non-additive genetic effect, statistically, based on GWAS data alone.9 While these observations helped to develop a new association test that is robust to X-inactivation uncertainty,9 both SNP and sex-effect estimates are biased if the model assumptions were incorrect.73 As SNP-effect estimates are the bases for constructing PGS or PRSs, future research should consider how to correct for the biases when X chromosomal variants are included in PRS.
Heterogeneous reporting of summary statistics and the X chromosome results
A workshop has resulted in recommendations for improving the standardization of genome-wide summary statistics,74,75 having acknowledged that there has been large variation in reporting practices.76 In one specific analysis, 127 unique formats were present among 327 summary statistics files analyzed. The authors then developed MungeSumstats, a Bioconductor package to standardize and perform QC of GWAS summary statistics. Interestingly, #27 among the total of 31 checks “for SNPs on chromosome X, Y and mitochondrial SNPs, [and] if any are found these are removed,” even though an option of retaining them was provided.
We further examined the reporting standard in the original publications of the X chromosomal signals documented in the NHGRI-EBI GWAS Catalog5,6 (downloaded on 2020-03-08) with the genome-wide significance level of p value < 5 × 10−8. Out of the 3,869 studies available at that time (male-only studies excluded), 195 reported a total of 253 genome-wide-significant loci on the X chromosome. To streamline the analysis, we selected only one SNP from each associated region by retaining the SNP with the smallest association p value (Data S2).
We then extracted information on the analyses performed from the original publications; in total, there are 36 columns in Data S2. These details are crucial to the analysis and reporting of the X chromosome but are largely irrelevant to the autosomes. They include, for example, whether (1) the analysis was sex stratified (70% did sex-combined analysis); (2) for sex-combined analysis, sex was included as a covariate (57% did not); and (3) the genotype coding was documented (75% did not, presumably used the default X-inactivation assumption), because if X inactivation was assumed, males are typically coded 0 and 2 for the two hemizygous genotypes. These considerations were not included in the guidelines recommended by Little et al.77 Not surprisingly, there was much heterogeneity in both the analysis and reporting among the 195 studies we examined. Such heterogeneity creates challenges for meta-analyses since the lack of necessary details may impact power if assumptions about how the analysis was performed are incorrect. We suggest sex-chromosome-aware research guidelines to be developed by the community.
The Y chromosome
Non-recombining Y chromosome haplotypes (haplogroups) have a long history in population genetics and genealogy,78 since these haplotypes can be determined without ambiguity, making them the patrilineal equivalent to mitochondrial haplogroups.79,80 However, the Y chromosome has long been a thorn in the side of human geneticists81: more than half of the Y chromosome is absent from GRCh38.82 Two recent papers used combinations of multiple long-read next-generation-sequencing technologies to generate much more complete sequence of the Y chromosome, and they also described a high degree of heterogeneity in chromosome length and content between individuals.82,83
The Telomere-to-Telomere Consortium has reported the sequence of an approximately 62 Mb long human Y chromosome,82 which includes >30 Mb that were missing from the reference sequence. Human geneticists can often be criticized for exaggeration, claiming that their phenotype or gene of interest has extensive complexity, but the recent analysis of 43 diverse Y chromosomes takes the crown.83 For example, some Y chromosomes are only 45 Mb, while others are as long as 85 Mb, in part as a result of large duplications and inversions.
It has been shown that standard sequencing alignment methods may be problematic for females, without masking the Y chromosome from the reference genome.84 For example, more variants were called after masking the Y chromosome in females, particularly in PARs. Similarly, for variant calling in PARs in males, it was recommended to provide only one PAR reference sequence from the two sex chromosomes (i.e., either the X or Y chromosome). Prior to variant calling, the authors recommended using read depths for the X and Y chromosomes, relative to the autosomes, to determine the sex chromosome composition of a sample, similar to that proposed for GWAS arrays.16
Additionally, in the past few years, age-dependent clonal loss of the Y chromosome has been reported in leukocytes.85,86 This phenomenon may further affect data quality and analysis of PAR and Y chromosomal variants.
Clinical implications
The exclusion of sex chromosomes from analysis and reporting also has significant clinical implications. Chief among these is failure to identify disease-associated SNPs or regions important in pathophysiology, prevention, diagnosis, or treatment. While common GWAS-identified SNPs tend to have small estimated effect sizes, this is not necessarily true for SNPs affecting drug responses, which have not generally been subjected to strong selective pressures.
Current pharmacogenetic guidelines such as those of the Clinical Pharmacogenetics Implementation Consortium87 do not include genes on the sex chromosomes (quite possibly because of exclusion of these genes from analyses); were such variants to be identified, guidelines for screening or drug dosing might need to be modified on the basis of a patient’s biologic sex. Similarly, adequate identification and inclusion of sex chromosome variants in PRSs might mandate stratification of these predictions by sex. It will be difficult, if not impossible, to assess these sex-stratified risks accurately until the dearth of analyses of sex chromosomes in clinically important traits is rectified.
Recommendations and future directions
After 15 years, several authors have observed that GWASs are “realizing the promise”88 with “no signs of slowing down.”89 Interestingly, sex chromosomes were not discussed in the 5-year,90 10-year,4 and 15-year88,89 reviews of GWASs. Here, we revealed that, for example, sex chromosomes are omitted from ∼75% of the GWASs in 2021, which is likely the major cause of the paucity of signals on the sex chromosomes. Given these observations, to achieve sex-chromosome-inclusive research, we make several recommendations and discuss related future research directions.
First, the existing bioinformatic and sequencing pipelines need to be revised for the sex chromosomes, from variant calling84 to imputation, so that the downstream analyses improve the integrity and robustness of sex chromosome analyses and provide greater confidence in conclusions drawn from them.
Second, QC procedures need to follow previously recommended sex-stratified approaches.1,11 Additionally, sex difference in MAF52,91,92 needs to be examined, but whether attributing significant sdMAF solely to genotyping errors (then screening out such variants) is appropriate warrants future research. This is because sdMAF could also be a result of sex-specific linkage, particularly at the PAR-NPR boundaries.55
Third, the distinction between association testing and effect size estimation is particularly important for the X chromosome9,73 Because of X-inactivation uncertainty, genetic effects may be more reliably estimated in a sex-stratified fashion to construct sex-specific PRSs,93 conceptually analogous to population-specific PRSs.94
Fourth, obtaining and then incorporating SNP/gene/tissue/individual-specific X inactivation could improve association methods. To this end, recent advances in long-read next-generation-sequencing technology, enabling phased allele-specific methylation, could be useful.95 Additionally, gene expression data such as the GTEx resource can be utilized.10,96
Fifth, many other existing statistical genetics analyses require sex-chromosome-aware development and implementation. These include, for example, rare variants,97,98 meta-analysis,99 LD score regression,100 pleiotropy,101 and causal inference via Mendelian randomization.102 More work is also need to better understand trait heritability103 attributed to the sex chromosomes,45,46 including the effect of imputation quality.
In summary, 10 years after the seminal work by Wise et al.,1 the exclusion of the X and Y chromosomes from whole-genome analysis persists. Until the sex chromosomes are indeed included in a whole-genome study, instead of GWASs, we propose they be more properly referred to as “AWASs” for “autosome-wide scans.”
Acknowledgments
The authors would like to thank Karl Broman, Sara Good, Anthony Herzig, Inke König, Michael Schatz, Bhooma Thiruvahindrapuram, Melissa Wilson, Stacey Winham, and Andreas Ziegler for valuable discussions. This research was funded by the Canadian Institutes of Health Research (CIHR, PJT-180460) and a University of Toronto Data Sciences Institute (DSI) Catalyst Grant.
Author contributions
L.S., A.D.P., and T.A.M. conceptualized the study. L.S. and A.D.P. supervised the study and drafted the manuscript. Z.W. and T.L. performed the analyses and summarized the results. Z.W., T.L., and T.A.M. reviewed and edited the manuscript.
Declaration of interests
T.L. is an employee and shareholder of 5 Prime Sciences Inc.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.04.009.
Contributor Information
Lei Sun, Email: lei.sun@utoronto.ca.
Andrew D. Paterson, Email: andrew.paterson@sickkids.ca.
Web resources
Genome-wide summary statistics reported in the NHGRI-EBI GWAS Catalog, https://www.ebi.ac.uk/gwas/downloads/summary-statistics
Significant SNPs reported in the NHGRI-EBI GWAS Catalog, https://www.ebi.ac.uk/gwas/docs/file-downloads
The PolyGenic Score (PGS) Catalog, https://www.pgscatalog.org/downloads/
Supplemental information
The columns contain the following information:
Column A; PubMed ID
Column B: Ftp Link to NHGRI-EBI GWAS Catalog summary statistics for one trait selected from each PubMed ID for examination of summary statistics.
Column C: Number of GWAS summary statistics present within each PubMed ID.
Column D: Indicator whether results were present for X chromosome SNPs: 0 = No, 1 = Yes Column E: Indicator whether results were present for Y chromosome SNPs: 0 = No, 1 = Yes
Detailed information about the column descriptions are provided in the Header Description tab
Data and code availability
All data used are publicly available. The specific downloads of the time-stamped datasets and codes used for the different analyses are all available at https://github.com/Paterson-Sun-Lab/eXclusionarY/.
References
- 1.Wise A.L., Gyi L., Manolio T.A. eXclusion: toward integrating the X chromosome in genome-wide association analyses. Am. J. Hum. Genet. 2013;92:643–647. doi: 10.1016/j.ajhg.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uffelmann E., Huang Q.Q., Munung N.S., De Vries J., Okada Y., Martin A.R., Martin H.C., Lappalainen T., Posthuma D. Genome-wide association studies. Nat. Rev. Methods Primers. 2021;1 59-21. [Google Scholar]
- 3.Agler C.S., Shungin D., Ferreira Zandoná A.G., Schmadeke P., Basta P.V., Luo J., Cantrell J., Pahel T.D., Meyer B.D., Shaffer J.R. Odontogenesis. Springer; 2019. Protocols, methods, and tools for genome-wide association studies (GWAS) of dental traits; pp. 493–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sollis E., Mosaku A., Abid A., Buniello A., Cerezo M., Gil L., Groza T., Güneş O., Hall P., Hayhurst J., et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker K., Erzurumluoglu A.M., Rodriguez S. The Y Chromosome: A Complex Locus for Genetic Analyses of Complex Human Traits. Genes. 2020;11:1273. doi: 10.3390/genes11111273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Editorial Accounting for sex in the genome. Nat Med. 2017;23:1243. doi: 10.1038/nm.4445. [DOI] [PubMed] [Google Scholar]
- 9.Chen B., Craiu R.V., Strug L.J., Sun L. The X factor: A robust and powerful approach to X-chromosome-inclusive whole-genome association studies. Genet. Epidemiol. 2021;45:694–709. doi: 10.1002/gepi.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tukiainen T., Villani A.C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A., et al. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550:244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.König I.R., Loley C., Erdmann J., Ziegler A. How to include chromosome X in your genome-wide association study. Genet. Epidemiol. 2014;38:97–103. doi: 10.1002/gepi.21782. [DOI] [PubMed] [Google Scholar]
- 12.Gendrel A.V., Heard E. Fifty years of X-inactivation research. Development. 2011;138:5049–5055. doi: 10.1242/dev.068320. [DOI] [PubMed] [Google Scholar]
- 13.Clayton D.G. Sex chromosomes and genetic association studies. Genome Med. 2009;1:110. doi: 10.1186/gm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J., et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marees A.T., de Kluiver H., Stringer S., Vorspan F., Curis E., Marie-Claire C., Derks E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018;27:e1608. doi: 10.1002/mpr.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson C.A., Pettersson F.H., Clarke G.M., Cardon L.R., Morris A.P., Zondervan K.T. Data quality control in genetic case-control association studies. Nat. Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun L. In: Statistical Human Genetics: Methods and Protocols. 2nd Edition. Elston R., editor. Springer; 2017. Detecting pedigree relationship errors; pp. 25–44. [Google Scholar]
- 20.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graffelman J., Weir B.S. On the testing of Hardy-Weinberg proportions and equality of allele frequencies in males and females at biallelic genetic markers. Genet. Epidemiol. 2018;42:34–48. doi: 10.1002/gepi.22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crow J.F., Kimura M. Harper and Row; 1970. An Introduction in Population Genetics Theory. [Google Scholar]
- 23.Browning B.L., Tian X., Zhou Y., Browning S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021;108:1880–1890. doi: 10.1016/j.ajhg.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., Karlsson R., Frei O., Fan C.C., De Witte W., et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M., et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaneau O., Marchini J., 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934–3939. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Willer C.J., Ding J., Scheet P., Abecasis G.R. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet. Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 32.Mbatchou J., Barnard L., Backman J., Marcketta A., Kosmicki J.A., Ziyatdinov A., Benner C., O’Dushlaine C., Barber M., Boutkov B., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 33.Loh P.-R., Kichaev G., Gazal S., Schoech A.P., Price A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.6.4, S.A.G.E. 2016. Statistical Analysis for Genetic Epidemiology. [Google Scholar]
- 35.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis A.C.F., Green R.C. Polygenic risk scores in the clinic: new perspectives needed on familiar ethical issues. Genome Med. 2021;13:14. doi: 10.1186/s13073-021-00829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chatterjee N., Shi J., García-Closas M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.International Schizophrenia Consortium. Purcell S.M., Wray N.R., Stone J.L., Visscher P.M., O'Donovan M.C., Sullivan P.F., Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S., Abecasis G.R., Browning B.L. Genotype Imputation from Large Reference Panels. Annu. Rev. Genomics Hum. Genet. 2018;19:73–96. doi: 10.1146/annurev-genom-083117-021602. [DOI] [PubMed] [Google Scholar]
- 43.Dudbridge F., Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y., Stilp A.M., McHugh C.P., Rao S., Jain D., Zheng X., Lane J., Méric de Bellefon S., Raffield L.M., Chen M.H., et al. Whole-genome sequencing association analysis of quantitative red blood cell phenotypes: The NHLBI TOPMed program. Am. J. Hum. Genet. 2021;108:1165. doi: 10.1016/j.ajhg.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidorenko J., Kassam I., Kemper K.E., Zeng J., Lloyd-Jones L.R., Montgomery G.W., Gibson G., Metspalu A., Esko T., Yang J., et al. The effect of X-linked dosage compensation on complex trait variation. Nat. Commun. 2019;10:3009. doi: 10.1038/s41467-019-10598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., Nguyen-Viet T.A., Bowers P., Sidorenko J., Karlsson Linnér R., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 48.Byrska-Bishop M., Evani U.S., Zhao X., Basile A.O., Abel H.J., Regier A.A., Corvelo A., Clarke W.E., Musunuri R., Nagulapalli K., et al. High-coverage whole-genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Cell. 2022;185:3426–3440.e19. doi: 10.1016/j.cell.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorlov I.P., Amos C.I. Why does the X chromosome lag behind autosomes in GWAS findings? PLoS Genet. 2023;19:e1010472. doi: 10.1371/journal.pgen.1010472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambert S.A., Gil L., Jupp S., Ritchie S.C., Xu Y., Buniello A., McMahon A., Abraham G., Chapman M., Parkinson H., et al. The Polygenic Score Catalog as an open database for reproducibility and systematic evaluation. Nat. Genet. 2021;53:420–425. doi: 10.1038/s41588-021-00783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schurz H., Müller S.J., van Helden P.D., Tromp G., Hoal E.G., Kinnear C.J., Möller M. Evaluating the Accuracy of Imputation Methods in a Five-Way Admixed Population. Front. Genet. 2019;10:34. doi: 10.3389/fgene.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Z., Sun L., Paterson A.D. Major sex differences in allele frequencies for X chromosomal variants in both the 1000 Genomes Project and gnomAD. PLoS Genet. 2022;18:e1010231. doi: 10.1371/journal.pgen.1010231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Flaquer A., Fischer C., Wienker T.F. A new sex-specific genetic map of the human pseudoautosomal regions (PAR1 and PAR2) Hum. Hered. 2009;68:192–200. doi: 10.1159/000224639. [DOI] [PubMed] [Google Scholar]
- 55.Dupuis J., Van Eerdewegh P. Multipoint linkage analysis of the pseudoautosomal regions, using affected sibling pairs. Am. J. Hum. Genet. 2000;67:462–475. doi: 10.1086/303008. S0002-9297(07)62655-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rouyer F., Simmler M.C., Johnsson C., Vergnaud G., Cooke H.J., Weissenbach J. A gradient of sex linkage in the pseudoautosomal region of the human sex chromosomes. Nature. 1986;319:291–295. doi: 10.1038/319291a0. [DOI] [PubMed] [Google Scholar]
- 57.Pirastu N., Cordioli M., Nandakumar P., Mignogna G., Abdellaoui A., Hollis B., Kanai M., Rajagopal V.M., Parolo P.D.B., Baya N., et al. Genetic analyses identify widespread sex-differential participation bias. Nat. Genet. 2021;53:663–671. doi: 10.1038/s41588-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P.A., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L., Sun L. A generalized robust allele-based genetic association test. Biometrics. 2022;78:487–498. doi: 10.1111/biom.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong A.M., Blackwell T.W., LeFaive J., de Andrade M., Barnard J., Barnes K.C., Blangero J., Boerwinkle E., Burchard E.G., Cade B.E., et al. Robust, flexible, and scalable tests for Hardy–Weinberg equilibrium across diverse ancestries. Genetics. 2021;218:iyab044. doi: 10.1093/genetics/iyab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Wang Z., Paterson A.D., Sun L. A novel regression-based method for X-chromosome-inclusive Hardy-Weinberg equilibrium test. Genet. Epidemiol. 2021;45:792. [Google Scholar]
- 62.Graffelman J., Weir B.S. Testing for Hardy-Weinberg equilibrium at biallelic genetic markers on the X chromosome. Heredity. 2016;116:558–568. doi: 10.1038/hdy.2016.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wellek S., Ziegler A. Testing for goodness rather than lack of fit of an X–chromosomal SNP to the Hardy-Weinberg model. PLoS One. 2019;14:e0212344. doi: 10.1371/journal.pone.0212344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song K., Elston R.C. A powerful method of combining measures of association and Hardy–Weinberg disequilibrium for fine-mapping in case-control studies. Stat. Med. 2006;25:105–126. doi: 10.1002/sim.2350. [DOI] [PubMed] [Google Scholar]
- 65.Wang J., Shete S. A test for genetic association that incorporates information about deviation from Hardy-Weinberg proportions in cases. Am. J. Hum. Genet. 2008;83:53–63. doi: 10.1016/j.ajhg.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang L., Strug L., Sun L. Leveraging Hardy–Weinberg disequilibrium for association testing in case-control studies. Ann. Appl. Stat. 2023;17:1764–1781. doi: 10.1214/22-AOAS1695. [DOI] [Google Scholar]
- 67.Wang J., Yu R., Shete S. X-chromosome genetic association test accounting for X-inactivation, skewed X-inactivation, and escape from X-inactivation. Genet. Epidemiol. 2014;38:483–493. doi: 10.1002/gepi.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao F., Chang D., Biddanda A., Ma L., Guo Y., Zhou Z., Keinan A. XWAS: A Software Toolset for Genetic Data Analysis and Association Studies of the X Chromosome. J. Hered. 2015;106:666–671. doi: 10.1093/jhered/esv059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J., Talluri R., Shete S. Selection of X-chromosome Inactivation Model. Cancer Inform. 2017;16 doi: 10.1177/1176935117747272. 1176935117747272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z., Ng H.K.T., Li J., Liu Q., Huang H. Detecting associated single-nucleotide polymorphisms on the X chromosome in case control genome-wide association studies. Stat. Methods Med. Res. 2017;26:567–582. doi: 10.1177/0962280214551815. [DOI] [PubMed] [Google Scholar]
- 71.Özbek U., Lin H.M., Lin Y., Weeks D.E., Chen W., Shaffer J.R., Purcell S.M., Feingold E. Statistics for X-chromosome associations. Genet. Epidemiol. 2018;42:539–550. doi: 10.1002/gepi.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen B., Craiu R.V., Sun L. Bayesian model averaging for the X-chromosome inactivation dilemma in genetic association study. Biostatistics. 2020;21:319–335. doi: 10.1093/biostatistics/kxy049. [DOI] [PubMed] [Google Scholar]
- 73.Song Y., Biernacka J.M., Winham S.J. Testing and estimation of X-chromosome SNP effects: Impact of model assumptions. Genet. Epidemiol. 2021;45:577–592. doi: 10.1002/gepi.22393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.MacArthur J.A.L., Buniello A., Harris L.W., Hayhurst J., McMahon A., Sollis E., Cerezo M., Hall P., Lewis E., Whetzel P.L., et al. Workshop proceedings: GWAS summary statistics standards and sharing. Cell Genom. 2021;1:100004. doi: 10.1016/j.xgen.2021.100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hayhurst J., Buniello A., Harris L., Mosaku A., Chang C., Gignoux C.R., Hatzikotoulas K., Karim M.A., Lambert S.A., Lyon M., et al. A community driven GWAS summary statistics standard. bioRxiv. 2023 doi: 10.1101/2022.07.15.500230. Preprint at. [DOI] [Google Scholar]
- 76.Murphy A.E., Schilder B.M., Skene N.G. MungeSumstats: A Bioconductor package for the standardisation and quality control of many GWAS summary statistics. Bioinformatics. 2021;37:4593–4596. doi: 10.1093/bioinformatics/btab665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Little J., Higgins J.P.T., Ioannidis J.P.A., Moher D., Gagnon F., von Elm E., Khoury M.J., Cohen B., Davey-Smith G., Grimshaw J., et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the strengthening the reporting of observational studies in epidemiology (STROBE) statement. J. Clin. Epidemiol. 2009;62:597–608.e4. doi: 10.1016/j.jclinepi.2008.12.004. S0895-4356(08)00355-7 [pii] [DOI] [PubMed] [Google Scholar]
- 78.Hughes J.F., Page D.C. The history of the Y chromosome in man. Nat. Genet. 2016;48:588–589. doi: 10.1038/ng.3580. [DOI] [PubMed] [Google Scholar]
- 79.Wallace D.C. Mitochondrial genetic medicine. Nat. Genet. 2018;50:1642–1649. doi: 10.1038/s41588-018-0264-z. [DOI] [PubMed] [Google Scholar]
- 80.Timmers P.R.H.J., Wilson J.F. Limited Effect of Y Chromosome Variation on Coronary Artery Disease and Mortality in UK Biobank-Brief Report. Arterioscler. Thromb. Vasc. Biol. 2022;42:1198–1206. doi: 10.1161/ATVBAHA.122.317664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Anderson K., Cañadas-Garre M., Chambers R., Maxwell A.P., McKnight A.J. The Challenges of Chromosome Y Analysis and the Implications for Chronic Kidney Disease. Front. Genet. 2019;10:781. doi: 10.3389/fgene.2019.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rhie A., Nurk S., Cechova M., Hoyt S.J., Taylor D.J., Altemose N., Hook P.W., Koren S., Rautiainen M., Alexandrov I.A., et al. The complete sequence of a human Y chromosome. bioRxiv. 2022 doi: 10.1101/2022.12.01.518724. Preprint at. [DOI] [Google Scholar]
- 83.Hallast P., Ebert P., Loftus M., Yilmaz F., Audano P.A., Logsdon G.A., Bonder M.J., Zhou W., Höps W., Kim K., et al. Assembly of 43 diverse human Y chromosomes reveals extensive complexity and variation. bioRxiv. 2022 doi: 10.1101/2022.12.01.518658. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webster T.H., Couse M., Grande B.M., Karlins E., Phung T.N., Richmond P.A., Whitford W., Wilson M.A. Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. GigaScience. 2019;8:giz074. doi: 10.1093/gigascience/giz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Terao C., Momozawa Y., Ishigaki K., Kawakami E., Akiyama M., Loh P.-R., Genovese G., Sugishita H., Ohta T., Hirata M., et al. GWAS of mosaic loss of chromosome Y highlights genetic effects on blood cell differentiation. Nat. Commun. 2019;10:4719–4810. doi: 10.1038/s41467-019-12705-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thompson D.J., Genovese G., Halvardson J., Ulirsch J.C., Wright D.J., Terao C., Davidsson O.B., Day F.R., Sulem P., Jiang Y., et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature. 2019;575:652–657. doi: 10.1038/s41586-019-1765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Relling M.V., Klein T.E., Gammal R.S., Whirl-Carrillo M., Hoffman J.M., Caudle K.E. The clinical pharmacogenetics implementation consortium: 10 years later. Clin. Pharmacol. Ther. 2020;107:171–175. doi: 10.1002/cpt.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdellaoui A., Yengo L., Verweij K.J., Visscher P.M. The American Journal of Human Genetics; 2023. 15 Years of GWAS Discovery: Realizing the Promise. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loos R.J.F. 15 years of genome-wide association studies and no signs of slowing down. Nat. Commun. 2020;11:5900. doi: 10.1038/s41467-020-19653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Visscher P.M., Brown M.A., McCarthy M.I., Yang J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z., Sun L., Paterson A.D. Features of X Chromosomal SNPs Associated with Significant Sex-Difference in Allele Frequency in High Coverage Whole Genome Sequence Data. Genetic Epidemiology. 2022;46:522–523. https://onlinelibrary.wiley.com/doi/epdf/10.1002/gepi.22503 [Google Scholar]
- 92.Wang Z., Paterson A.D., Sun L. A Population-Aware Retrospective Regression to Detect Genome-Wide Variants with Sex Difference in Allele Frequency. arXiv. 2022 doi: 10.48550/arXiv.2212.12228. Preprint at. [DOI] [Google Scholar]
- 93.Zhang C., Ye Y., Zhao H. Comparison of Methods Utilizing Sex-Specific PRSs Derived From GWAS Summary Statistics. Front. Genet. 2022;13:892950. doi: 10.3389/fgene.2022.892950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Akbari V., Garant J.-M., O’Neill K., Pandoh P., Moore R., Marra M.A., Hirst M., Jones S.J.M. Megabase-scale methylation phasing using nanopore long reads and NanoMethPhase. Genome Biol. 2021;22 doi: 10.1186/s13059-021-02283-5. 68-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Derkach A., Lawless J.F., Sun L. Pooled association tests for rare genetic variants: a review and some new results. Stat. Sci. 2014;29:302–321. doi: 10.1214/13-STS456. [DOI] [Google Scholar]
- 98.Ma C., Boehnke M., Lee S., GoT2D Investigators Evaluating the Calibration and Power of Three Gene-Based Association Tests of Rare Variants for the X Chromosome. Genet. Epidemiol. 2015;39:499–508. doi: 10.1002/gepi.21935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. btq340 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stearns F.W. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–773. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burgess S., Thompson S.G. CRC Press; 2021. Mendelian Randomization: Methods for Causal Inference Using Genetic Variants. [Google Scholar]
- 103.Yang J., Zeng J., Goddard M.E., Wray N.R., Visscher P.M. Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 2017;49:1304–1310. doi: 10.1038/ng.3941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The columns contain the following information:
Column A; PubMed ID
Column B: Ftp Link to NHGRI-EBI GWAS Catalog summary statistics for one trait selected from each PubMed ID for examination of summary statistics.
Column C: Number of GWAS summary statistics present within each PubMed ID.
Column D: Indicator whether results were present for X chromosome SNPs: 0 = No, 1 = Yes Column E: Indicator whether results were present for Y chromosome SNPs: 0 = No, 1 = Yes
Detailed information about the column descriptions are provided in the Header Description tab
Data Availability Statement
All data used are publicly available. The specific downloads of the time-stamped datasets and codes used for the different analyses are all available at https://github.com/Paterson-Sun-Lab/eXclusionarY/.