Abstract
AlphaFold2 (AF2) has revolutionized the field of protein structural prediction. Here, we test its ability to predict the tertiary and quaternary structure of a previously undescribed scaffold with new folds and unusual architecture, the monotopic membrane protein caveolin-1 (CAV1). CAV1 assembles into a disc-shaped oligomer composed of 11 symmetrically arranged protomers, each assuming an identical new fold, and contains the largest parallel β-barrel known to exist in nature. Remarkably, AF2 predicts both the fold of the protomers and the interfaces between them. It also assembles between seven and 15 copies of CAV1 into disc-shaped complexes. However, the predicted multimers are energetically strained, especially the parallel β-barrel. These findings highlight the ability of AF2 to correctly predict new protein folds and oligomeric assemblies at a granular level while missing some elements of higher-order complexes, thus positing a new direction for the continued development of deep-learning protein structure prediction approaches.
Significance
Recent advances in computational biology like AlphaFold2 make it possible to accurately predict the structure of proteins from their primary sequence based on deep-learning algorithms extensively trained on known protein structures. New scaffolds with low homology to the training set sequences may challenge AlphaFold2. Here, we test the ability of AlphaFold2 to predict the structure of an unusual, previously undescribed oligomeric assembly of a monotopic membrane protein lacking close homologs: caveolin-1. High similarity between the overall architecture and contacts of the predicted and experimental caveolin-1 structures indicates that AlphaFold2 can perform well even in the absence of close homologs. Further, AlphaFold2 predicts multiple oligomeric states exist in addition to the experimentally determined 11-mer, providing new insights into its oligomerization mechanisms.
Introduction
A long-standing goal of computational biology is to accurately predict the three-dimensional structure of proteins from their primary sequence. This goal appears now within reach using deep-learning algorithms trained on a database of known proteins, most prominently AlphaFold2 (AF2) (1). AF2 can predict protein structure even in the absence of high-homology templates (1,2). Another advantage of AF2 is its ability to predict multimeric assemblies without prior knowledge of the interaction sites of partner proteins or subunits (3,4). This feature is useful when the quaternary structure of a multimeric protein is unknown.
Despite the many successes of AF2 (5,6,7,8), its performance with structures for which there are no homologs present in the PDB remains an open question. Although AF2 does not rely on templates for structure prediction, the AF2 algorithm was trained based on available structural information from most structures in the PDB (1). This may potentially limit its ability to model structures that fall outside the scaffolds of these proteins. One such example is structures obtained through NMR, which were not included in the training set of AF2. While AF2 was mostly successful in predicting these protein structures, it had lower accuracy in some cases, especially with flexible proteins (9). In other cases, lower-accuracy predictions by AF2 were argued to be related to a less deep multiple sequence alignment, high proportion of heterotypic contacts, and high oligomerization states (10). Therefore, studies focusing on the prediction of novel protein scaffolds with AF2 can shed light on both its strengths and areas for future improvement.
We recently determined a cryoelectron microscopy (cryo-EM) structure for the protein caveolin-1 (CAV1), an integral membrane protein that plays a role in formation of membrane invaginations called caveolae (11). CAV1 has several unusual and unexpected features in its structure that make it exceptionally well suited to benchmark the ability of AF2 to predict the structure of proteins with unique scaffolds. First, CAV1 is a member of a protein family that shares little sequence or structural homology with other proteins (11), and no high-resolution structures of any CAV family members were available in the PDB at the time AF2 was trained. Second, CAV1 is a monotopic membrane protein, a class of proteins that is underrepresented in the PDB compared with other types of membrane proteins (12). Third, the CAV1 complex has a previously undescribed quaternary structure consisting of a tightly packed disc composed of 11 primarily α-helical protomers that tightly pack into a spiral arrangement. Finally, the complex contains an 11-fold symmetric parallel β-barrel, the largest parallel β-barrel to be reported to date. This distinguishes it from known examples of parallel β-barrel structures, which belong primarily to the TIM barrel family, whose members consist of eight β-strands that fold into a barrel with fourfold symmetry (13).
In this work, we used CAV1 as a test case to address two questions. The first one is whether AF2 can model the tertiary and quaternary structure of a scaffold for which it has no homologs in its training set. The second question is whether the CAV1 models generated by AF2 can be used to understand the mechanism of CAV1 oligomerization.
Methods
AF2 calculations
Prediction runs were executed either using AlphaFold v.2.2.0 with default settings via a Colab notebook named “alphafold21_predict_colab” provided by ChimeraX daily builds version (ChimeraX 1.4.0) or using AlphaFoldv2.1 via another Colab notebook named “AlphaFold2_advanced (AlphaFold2_advanced.ipynb - Colaboratory (google.com)” with default settings. Full-length human CAV1 was used for AlphaFold v.2.2.0 predictions for structures containing 2–13 protomers. Due to memory limitations, the sequence of human CAV1-β isoform (residues 32–178) was used for the predictions of 14-mer and 15-meric CAV1. For AlphaFold v.2.1, predictions for 8-mer complexes were based on the sequence of human CAV1-β isoform.
The figures were analyzed, rendered, and exported with either ChimeraX 1.4.0 (daily builds version) or ChimeraX1.3. Root-mean-square deviation values were calculated using ChimeraX1.3. Chain A from the model was used as a reference for all structure matching.
Rosetta cryo-EM relax calculations
The experimental CAV1 structure was relaxed with the Rosetta cryo-EM relax protocol (14). A single protomer was relaxed with 11-fold symmetry based on symmetry files generated using the 11-meric experimental CAV1 structure. The ref2015_cart score function with residue weights adjusted for cryo-EM calculations was used to score the structures. An elec_dens_fast value of 50 was selected as the restraint weight for the relax calculations based on the resolution of the CAV1 structure. A total of 500 structures were generated, and the lowest-scoring structure was used for the analyses based on lowest total score.
Per-residue energy calculations
The per-residue calculations were run with Rosetta per_residue_energies application (15). For each oligomeric assembly, membrane coordinates were calculated using the positioning proteins in membranes server (16). The per-residue energies of the aligned protein structures were calculated with the mpframework_smooth_fa_2012 score function (17,18). For the comparison of the three β-barrels from PDB: 5BVL, 2AO9, and 7SC0, the soluble membrane score function ref2015 was used to calculate the per-residue energies since these structures are not embedded in membrane (19). The resulting energies from the per-residue energy calculations were used to color each oligomeric assembly using the define attribute module of UCSF Chimera. A three-color coloring scheme from blue to red was used to represent the negative and positive per-residue scores, respectively.
Residue contact map predictions
The residue-residue contact map predictions were made and plotted with the Bio.PDB module of the BioPython software (20). Two neighboring protomers of each oligomeric structure were used for the contact map analyses. The α-carbon coordinates were calculated for every residue in the system, and the pairwise distances were calculated for each residue. Of the calculated distances, each pair within 12 Å of each other was considered a contact, and only the pairs of residues in contact were plotted.
Results
The unique experimental CAV1 structure presents a formidable test case for AF2
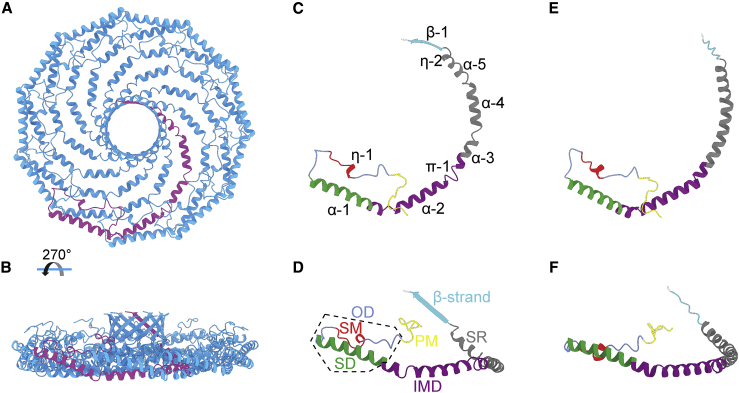
We previously determined the only experimental structure of the human CAV1 complex (PDB: 7SC0). It consists of 11 CAV1 protomers, each of which assumes an identical new fold, that are symmetrically arranged into a tightly packed disc-shaped complex with a central pore formed by an 11-stranded parallel β-barrel (11) (Fig. 1, A and B). Residues 1–48 of the 178-residue-long protomers are absent from the cryo-EM structure and are predicted to be unstructured (21,22). This region of CAV1 contains several phosphorylation, ubiquitination, and protein interaction sites (23,24,25,26,27,28,29,30). It also differs in length between the α- and β-isoforms of CAV1 (31,32). Structurally well-defined regions of the protein include the pin motif (PM; residues 49–60), a loop that envelopes and stabilizes the interactions between neighboring protomers; the CAV signature motif (residues 68–75), a highly conserved region across all CAVs composed of short α-helical region; and the scaffolding domain (residues 82–101), comprising a portion of α-helix 1 at the periphery of the CAV1 8S disc that assists with oligomerization as part of a larger oligomerization domain (residues 61–101). Other notable domains include a region traditionally referred to as the “intramembrane” domain (IMD; residues 102–134) consisting of a helical region that defines one boundary of the membrane-facing surface of the protein, a long C-terminal amphipathic α-helix termed the spoke region (SR; residues 135–169), and a C-terminal β-strand (residues 170–176) that contributes to the assembly of the central β-barrel in the complex (11).
Figure 1.
The structure of a CAV1 monomer predicted by AF2.1 closely resembles that of a CAV1 protomer experimentally determined by cryo-EM. (A and B) Top and side view of human CAV1 8S complex cryo-EM structure (PDB: 7SC0) with a single protomer colored purple. (C and D) Top and side views of a single CAV1 protomer extracted from the experimental structure. The secondary structure assignments for the cryo-EM structure are labeled. Specific regions of CAV1 are labeled and colored as follows: PM, pin motif (yellow); SM, signature motif (red); SD, scaffolding domain (green); IMD, intermembrane domain (purple); SR, spoke region (gray); and β-strand (cyan). The OD, which contains the SM and SD, is indicated by the dashed box. (E and F) Top and side views of the AF2- predicted CAV1 monomer (AF-Q03135-F1).
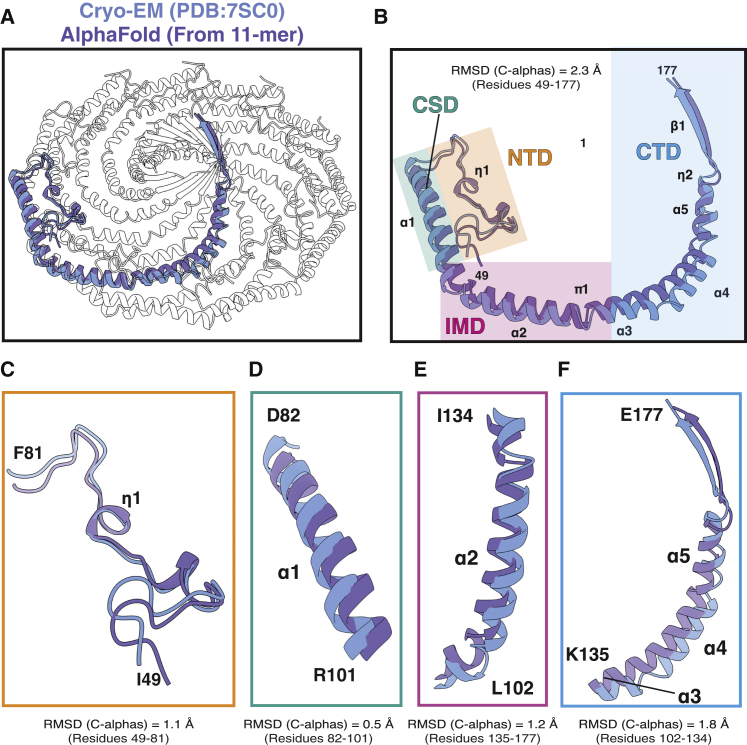
AF2 predicts the tertiary structure of the CAV1 protomer at a root-mean-square deviation of 2.3 Å
To evaluate the ability of AF2 to predict the tertiary structure of CAV1 protomers, we compared the structure of a single CAV1 protomer extracted from the experimentally determined structure (PDB: 7SC0) with a model of the CAV1 monomer structure predicted by AF2 (AF-Q03135-F1; Fig. 1). Several regions of CAV1 (residues 1–48 and 178) were predicted with low confidence. These regions overlap with the regions of CAV1 that were not resolved in the experimental structure (11). We thus focused our comparison on the well-resolved/high-confidence regions of the experimental and predicted structures. Overall, there was marked similarity between the predicted and experimental structures, with a few notable exceptions (Fig. 1). First, the experimental protomer contains two α-helices (α3 and α4) separated by a short loop close to the boundary between the IMD and SR, whereas the predicted CAV1 AF2 monomer forms a continuous α-helix with a slight kink near residue P132 in the same region. The predicted structure also is missing a loop between α4 and α5. The absence of these loops gives rise to a difference in curvature between the two structures (Fig. S1). Furthermore, the last 10 residues are predicted to form a coiled C-terminus in the AF2 monomer structure, in contrast to the experimentally observed β-strand geometry of the protomer (Fig. 1). Nevertheless, considering that the AF2 model for CAV1 was predicted in the absence of an actual structure for CAV1 in the PDB, combined with the fact that the CAV1 monomer exists in nature within a higher-order complex, the degree of similarity between the two structures is remarkable.
AF2 predicts the overall quaternary structure of CAV1 oligomers
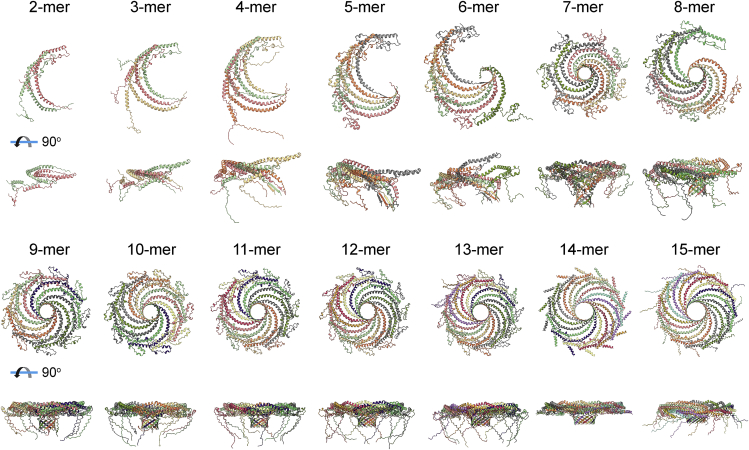
We next asked whether AF2 is capable of assembling CAV1 protomers into closed discs, and, if so, if it correctly predicts that CAV1 exists as an 11-mer. To test this, we generated n-mers containing between two and 15 copies of CAV1 using AF2.2 (Figs. 2 and S2). The probability of forming a closed structure varied across n-mers (Figs. S2 and S3). Oligomers containing six or fewer CAV1s were unable to generate closed assemblies where the PM was properly positioned (Figs. 2 and S2). A closed assembly was observed for the 7-mer, but its structure was highly curved compared with the experimental Cav-1 structure, and the best prediction for the 8-mer failed to form a closed disc. However, the best-fitting models of oligomers containing between nine and 15 protomers were predicted to form closed discs with an architecture similar to the experimental CAV1 11-mer structure (Figs. 2 and S2). The top model predictions were highly reproducible for the case of the 11-mer (Fig. S4). The complexes formed by the 9- to 15-mers share several structural features. Similar to the experimental structure, the PM of each protomer locks neighboring CAV1 protomers in place along the outer rim of the complex. The predicted structures also exhibit extensive interactions between the hydrophobic residues of α-helices α2, α3, and α4. The last nine amino acids of the C-termini of each protomer form β-strands that assemble into a parallel β-barrel. This β-strand was not observed in the CAV1 monomer structure predicted by AF2 as measured from the Φ and Ψ angles at this region (Figs. 1 F and S5), suggesting that it forms as a consequence of the oligomerization process.
Figure 2.
Best fit predictions by AF2.2 for CAV1 n-mers ranging in size from two to 15. En face (top) and side views (bottom) are shown for each case. Closed discs are predicted to form for n = 7 and 9–15. For the 2- to 13-mers, the predictions were based on full-length alpha isoform (1–178) of human CAV1. Predictions for the 14- and 15-mers were based on the sequence of the β-isoform (32–178) of human CAV1. Each protomer is depicted in a different color. All structures are shown to scale.
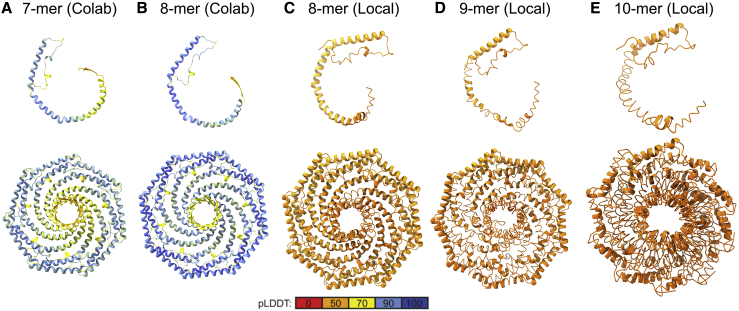
We also compared the predictions of AF2.2 and AF2.1. Like AF2.2, AF2.1 predicted that CAV1 can assemble into closed discs, but in this case, only seven or eight CAV1 protomers were required. Furthermore, oligomers composed of nine or more protomers exhibited significant clashes and unrealistic structures (Fig. 3). Thus, AF2.2 was used for all subsequent analysis.
Figure 3.
CAV1 oligomers predicted by AF2.1 become increasingly distorted as a function of increasing number of protomers in the complex. Predicted structures for CAV1 oligomers generated with the Colab version of AF2.1 (modeled as 32–178) and a local installation of AF2.1 (modeled as 1–178). (A and B) 7- and 8-mer CAV1 generated with the Colab version. (C–E) 8-, 9-, and 10-mers generated with a local installation of AF2.1. A protomer belonging to each oligomer is shown in the top row, and the whole complex viewed en face is shown in the bottom row. Only residues 49–178 are shown for clarity. All structures are shown to scale.
Conserved critical interactions at protomer-protomer interfaces stabilize CAV1 oligomers with 10–12 copies
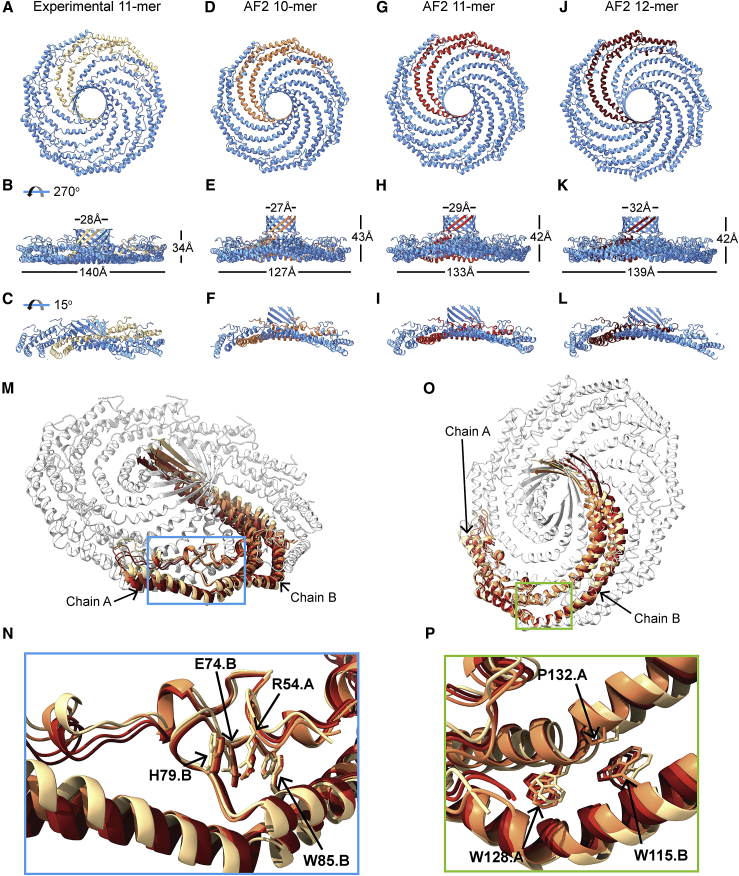
We next investigated the protomer-protomer interactions that underlie the formation of the CAV1 disc structure repeatedly observed among the AF2-predicted structures in more detail (Fig. 4). For these studies, we chose to analyze 10-, 11-, and 12-meric models generated by AF2 due to their similarity in size to the 11-meric experimental CAV1 structures (Fig. 4, A–L). Alignment of the AF2-predicted protomers showed a similar tertiary structure that mainly differed in terms of the curvature of the α-helical regions, as well as shifts in the positioning of the PM region that locks the neighboring protomer in place at their turning point (Fig. 4, M and O). The curvature differences result from the presence of several turns in the SR in the experimental CAV1 protomer that are not observed in the predicted protomer structure (Figs. 5 and S1), similar to the case of the AF2 CAV1 monomer. The differences between the curvature of the SR explains how the predicted and experimental CAV1 structures can assemble differently at the quaternary level. Several additional differences between the experimental and predicted structures were noted. In contrast to the flat membrane-facing surface of the cryo-EM structure, the membrane-facing surface of the predicted complexes bows in toward the center of the complex (Fig. 4, C, F, I, and L). The β-barrels of the AF2 models are positioned above the outer rim of the complex (Fig. 4, B, E, H, and K). Finally, the overall diameter of the predicted complexes is also slightly smaller than that of the experimental complex (133 Å for the predicted 11-mer versus 140 Å for the cryo-EM structure).
Figure 4.
Packing and interactions of neighboring protomers in predicted CAV1 complexes are similar to their counterparts in the cryo-EM-based structure. (A–L) Comparison of en face and side views of secondary structure model of cryo-EM-based CAV1 11-mer complex with structures of CAV1 10-, 11- and 12-mer predicted by AF2.2. Dimensions of complexes are labeled in (B), (E), (H), and (K). Cut-through views of each model are shown in (C), (F), (I), and (L). Residues 1–48 and 178 were hidden from the AF2.2-predicted structures to make them more comparable to the cryo-EM structure. Two neighboring protomers from each model are highlighted in gradually warming colors. All structures are shown to scale. (M and O) Overlay of two neighboring dimers extracted from each of the models shown in (A)–(L). Protomers from the cryo-EM structure are shown in beige or white. (N and P) Close up of boxed regions from (M) and (O) showing zoomed views of the key residues in the pin motif (N) and in surrounding P132 (P).
Figure 5.
Comparison of the CAV1 protomers from the cryo-EM structure and 11-meric complex of CAV1 predicted by AF2.2. (A) The CAV1 8S complex (PDB: 7SC0) is shown with a single protomer colored blue and overlayed with an aligned AF2.2 protomer taken from the AF2.2-predicted CAV1 11-mer in purple. (B) Overlay of a single protomer from the cryo-EM structure and the AlphaFold2 CAV1 protomer. The secondary structure assignments for the cryo-EM structure are labeled. The coordinates were aligned by their alpha carbons, and their root-mean-square deviation values are shown. The first 48 residues were not included in the alignments. Each region is highlighted by a different color. Orange, residues 49–81 of the N-terminal domain; teal, SD; pink, intramembrane domain; and blue, residues 135–177 of the C-terminal domain. (C–F) Each of the indicated regions was aligned individually and root-mean-square deviation values calculated. Experimental structures are shown in blue, and predicted structures are shown in purple.
We next focused on the neighborhood of two residues important for CAV1 structural integrity and function. The first one is R54 on the N-terminal α-helix forming the PM domain (Fig. 4 N). In the experimental structure, this arginine is sandwiched between the residues W85 and H79 of the neighboring protomer through π-π stacking interactions and forms additional electrostatic interactions with E74. Mutations of R54 were shown to abolish the ability of CAV1 to form disc-shaped assemblies in vivo, suggesting a key role for the stabilization of protomer-protomer interactions (11). The other residue is P132, which is located close to the transition between the IMD and SR (Fig. 4 P). A P132L mutation at this site results in disruption of CAV1 structure in both monomeric and oligomeric environments (33,34).
Comparisons of the three AF2 models and experimental structure showed that the vicinities of R54 and P132 are almost identical to the wild-type structure with all key interactions conserved. All three binding partners of R54 had nearly identical configurations among the three oligomeric states investigated (Fig. 4 N). In the case of P132, the relative coordinates of α-helix α3 shifted due to the changes in the curvature at the turning point of P110 connecting the IMD α-helices α2 and α3. However, hydrophobic interactions of P132 with W115 of the neighboring protomer were conserved in all three cases (Fig. 4 P). Further, the residue-residue interactions between neighboring protomers were nearly identical for all predicted CAV1 oligomers formed by 9–15 protomers, which was also consistent with the contacts calculated for the experimental 11-meric CAV1 structure with minor differences at the PM region (Fig. S6).
CAV1 oligomers of different sizes show similar energetics at α-helical regions but vary in their ability to form stable β-barrels
The finding that AF2 predicts that between nine and 15 CAV1 protomers can assemble into oligomeric complexes with a conserved architecture raises the question of whether these n-mers differ in stability, and, if so, what regions of the protein contribute most importantly to these differences. To this end, we calculated the per-residue energies of each complex with Rosetta (15). To account for interactions of CAV1 with membranes, we incorporated an implicit membrane model (17,18) based on the distribution of residues in contact with detergent in the experimental structure (11). The membrane-embedded residues consist of the helical domains of CAV1, whereas the β-barrel is predicted to be completely solvent exposed. Because the first 48 residues of CAV1 are not observed in the experimental structure and are also predicted with low confidence by AF2, we confined our analyses to the CAV1 residues 49–178.
Both the experimental 11-meric CAV1 structure and the AF2 models had favorable energies overall, as indicated by blue color throughout the per-residue energy distributions (Fig. S7). A closer inspection of the individual domains showed that the PM had the largest number of residues with high energy, followed by the β-barrel and α4 as indicated by red color. Increasing oligomer size had a slight but noticeable effect on the stabilities of the helices α1 and α2 whereby these domains had lower energies in larger assemblies such as the 14- and 15-meric structures. The most striking difference was observed at the β-barrel region. Here, larger assemblies with 13–15 protomers showed higher calculated scores, suggesting issues with the stability of their β-barrels (Fig. S8). Further, the experimental CAV1 structure had lower energies in the β-barrel region compared with the AF2-generated models including the 11-meric AF2 model (Fig. 6).
Figure 6.
Comparison of per-residue energies for the experimental and AF2-generated 11-meric CAV1 structures. (A and B) Per-residue energies mapped onto the (A) experimental 11-meric CAV1 structure versus the (B) AF2-predicted structures the 11-meric complex. (C and D) Close-up view of energetics of CAV1 β-barrel residues for experimental and predicted structures. Red indicates unfavorable energies, and blue indicates favorable energies. All scores are in Rosetta energy units. Structures in (A) and (B) are shown to scale.
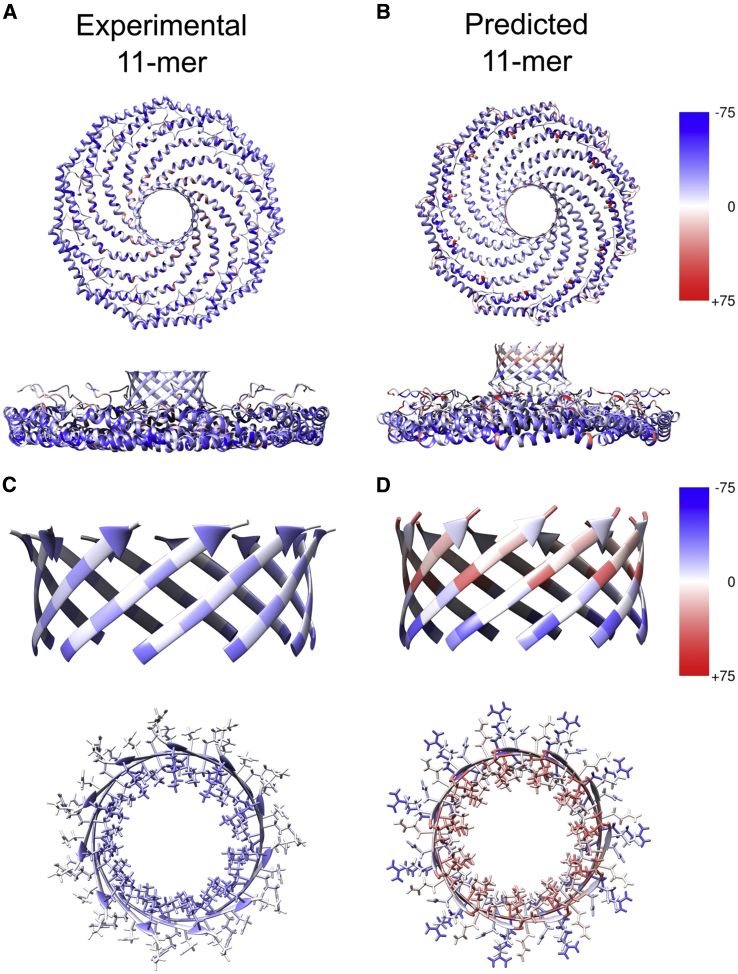
Unusual design principles underlie CAV1’s 11-stranded parallel β-barrel
The central parallel β-barrel of CAV1 is formed by alternating hydrophobic residues lining the cavity and hydrophilic residues facing outward. One exception to this pattern is an inwardly facing lysine residue at position 176, a highly conserved residue among CAV1s. An evolutionary analysis of 315 CAV1 proteins suggests that the propensity to form β-barrels with a similar pattern of residues is conserved across different species, highlighting the importance of the alternating hydrophobicity motif for the structure and function of the protein (Fig. S9). In all AF2 models, the residues facing the barrel pore had higher energies compared with the outer-facing residues (Fig. S8, bottom views). This reflects the tight packing of the inner barrel residues, which are positioned at the same level for each strand as a consequence of their shear number of 22. Another interesting finding was the visible distortions observed for the barrels larger than 12 strands, with accompanying high calculated energies (Fig. S8). The experimental CAV1 β-barrel had a more uniform distribution of scores for the inside- and outside-facing residues with a lower overall energy compared with the predicted models. The experimental β-barrel also had a lower overall energy compared with the predicted structures, which may partly be attributed to its larger barrel diameter (∼26.5 Å in predicted 11-mer vs. ∼30 Å in experimental) and more favorable alignment of the pore-facing residue rotamers compared with the predicted structures.
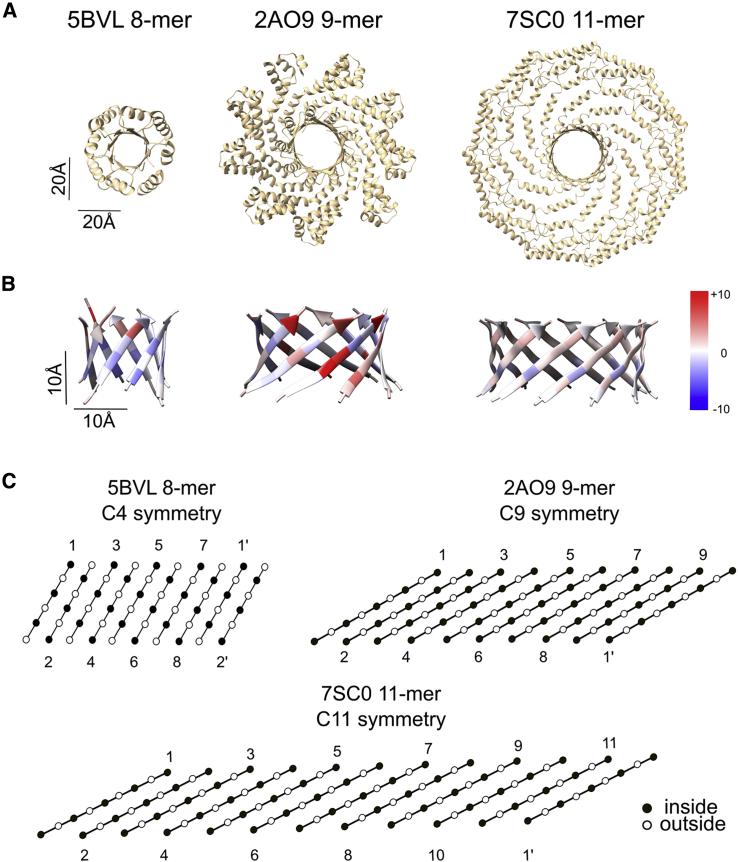
Lastly, we compared the energetics of the CAV1 barrel with two other parallel β-barrels to assess the effects of barrel size on their stability (Fig. 7; Table 1). Most parallel β-barrels in nature belong to the TIM barrel family, whose members consist of eight β-strands that fold typically into a barrel with C4 symmetry (35) (Fig. 7). Another example was recruited from a soluble phage protein (Bacillus cereus) whose C-termini folds into a central 9-meric parallel β-barrel (2AO9) (Fig. 7). Compared with the 9-meric barrel, a representative 8-meric TIM barrel (PDB: 5BVL) shows a more stable energy profile. The terminal residues of both the selected 8- and 9-mer barrels are asymmetric, likely to help prevent clashes caused by the proximity of pore-facing residues in these tight assemblies. In contrast, the CAV1 barrel is perfectly symmetric, suggesting that its larger radius allows less strain between the residues pointing into the barrel (Fig. 7). Although the inner lining of the TIM barrel example and CAV1 had mostly polar residues deeper in the pore, the 9-meric β-barrel had no clear preference. The 8-meric structure had a tight pore caused by the tightly packed hydrophobic residues at the center of the pore and arginine residues at its C-terminus. The pore size was larger for the 9- and 11-meric β-barrels, as expected. Comparison of the 9-meric β-barrel with the AF2-predicted 9-meric CAV1 structure showed marked similarities at the barrel region, potentially due to the constrained nature of a parallel β-barrel, but the geometry of other domains had marked differences (Fig. S10).
Figure 7.
Comparison of structural features of CAV1 β-barrel with other parallel β-barrels. (A) The full structures of the assemblies from which the analyzed parallel β-barrels were taken viewed in the direction of the barrel pore. All structures are shown to scale. (B) Structures and the per-residue energy breakdowns of three parallel β-barrels calculated with Rosetta. Left panel, a representative TIM barrel with C4 symmetry (PDB: 5BVL); middle panel, a phage protein from Bacillus cereus with C9 symmetry (PDB: 2AO9); and right panel, experimentally determined CAV1 β-barrel with C11 symmetry (PDB: 7SC0). Red indicates positive scores (destabilizing), and blue indicates negative scores (stabilizing). (C) Amino acid alignments of the 8-, 9-, and 11-meric β-barrel structures. Black circles indicate pore-facing residues, and white circles indicate residues facing away from the pore. Individual strands are labeled. Primes denote the same strand as the starting strand and are included to show the relative levels of the first and last strands in the barrel.
Table 1.
Characteristics of three different parallel β-barrels analyzed
Number of strands (n), shear number (S), register shift (t), barrel radius (R) measured between the farthest α-carbons, and the angle between an individual β-strand and the major helix axis (θ).
Discussion
Even for an isolated monomer, AF2 was able to predict the tertiary structure of CAV1. This is surprising as the biophysical context was absent: it folds in the absence of other protomers required to form the assembly and also lacks an explicit membrane environment, which is likely to play a significant role in CAV1 folding considering its role as a membrane-embedded curvature-inducing protein. It is particularly astounding that AF2 predicts this fold unambiguously, also reflected by the uniformity of the monomeric predictions in our calculations, as one would intuitively expect a large ensemble of possible conformations with rather similar energy. Apparently, local conformational preference is sufficient to determine the structure of the protomer even in isolation. Interestingly, and consistent with the absence of hydrogen-bonding partners to form the β-barrel, AF2 folded the C-terminal residues into a coil. It is also notable that AF2 predicts the structure of residues 1–48 of CAV1 with low confidence, suggesting that they are disordered. This is in agreement with previous predictions and the absence of these residues in the cryo-EM structure (11,21,22). It is also consistent with the presence of phosphorylation sites, often located in intrinsically disordered regions of proteins (36,37), within this domain of CAV1 (24,25,26,27,28).
The 11-meric CAV1 structure predicted by AF2 was consistent with the general scaffold of the experimental CAV1 structure. This is remarkable given the unusual topology of CAV1 revealed by cryo-EM combined with the fact that AF2 does not incorporate any explicit membrane system or otherwise account for the lipid environment other than the information contained within the sequence of the protein itself. Interestingly, while some detailed aspects of the structure were predicted consistently at atomic-detail accuracy, such as helix-helix interactions, other overarching features were predicted in an inconsistent manner, such as the disc-shaped symmetric arrangement and the curvature and length of the α-helical regions. We argue that this hints at an important metric of AF2 multimer: it is achieving a high local accuracy driven by the hundreds of thousands of protein structures it has been trained with while sometimes struggling with global aspects of novel arrangements of these local features (10).
Although the experimental structure of CAV1 suggests that it prefers to exist as an 11-mer, AF2 predicts that the protein has the capacity to assemble into oligomers containing between seven and 15 protomers. This range of values overlaps remarkably well with early biochemical estimates of the oligomeric state of CAV1 as well as more recent superresolution analyses of CAV1 scaffolds in cells lacking caveolae (38,39,40,41,42). Interestingly, AF2 predicts that the varied n-mers may not only differ in size but also in curvature. Comparison of the predicted models reveals how the complex could potentially accommodate different copy numbers of CAV1. Many interactions between the domains of neighboring protomers are independent of the number of protomers forming the oligomers in the range of 9–15 protomers. All oligomers successfully formed the PM that locks neighboring CAV1 protomers. Hydrophobic interactions between the neighboring IMDs were also conserved, although the angles at the turning points varied among the different protomer numbers, which caused curvature differences at the SR. These findings suggest that other oligomeric forms of CAV1 could exist in nature and may be selected under different physiological conditions. For example, complexes of variable size and curvature could potentially enable their more efficient packing in caveolae, contribute to the formation of caveolae of differing shapes or orientations, or facilitate the formation of larger more complex structures such as the diverse CAV1 scaffolds found in cells lacking caveolae. Alternatively, caveolae might only incorporate CAV1 oligomers within a certain size range, leading to the degradation of other n-mers.
Formation of a closed and well-defined β-barrel likely helps to define the oligomeric state of the CAV1 complex. Of the predicted 7- to 15-meric complexes, all but 8-meric CAV1 are capable of doing so. Oligomers containing 10–12 protomers had the highest probability of forming discs, and the β-barrel was most stable with 9–12 copies. Further, the experimental 11-meric CAV1 structure had a more stable β-barrel compared with the AF2-generated models. These results may indicate that while interactions at the PM may be important for the formation of CAV1 oligomers, the overall size and stability of the complex is heavily dependent on the stability of the β-barrel. Consistent with this possibility, disease-associated mutations of CAV1 that disrupt the C-terminus form discs that are less regular and biochemically stable than those generated by wild-type CAV1 (43,44).
Our findings also uncover design features that enable CAV1 to assemble into such large parallel β-barrels. By their nature, β-barrels are constrained by the hydrogen-bonding requirements for the formation of the barrel shape (45). The majority of the parallel β-barrels found in the PDB are TIM barrels, i.e., β8α8-barrels, formed by eight β-strands and eight α-helices. Interestingly, β8α8-barrels have (only) a fourfold symmetry, as side chains from uneven β-strands point inward, while side chains from even β-strands point outward, and vice versa. The highest symmetric β-barrel in the PDB prior to CAV1 is a β-barrel consisting of nine β-strands that is formed by the C-terminal regions of a protein from B. cereus (PDB: 2AO9; Fig. S10). Comparison of these β-barrels with the experimental CAV1 structure showed that the side chains of the former structures break the symmetry of the system by a shifting of the pore-facing side chains to adapt to the tight packing environment inside the pore, whereas the CAV1 β-barrel showed full symmetry likely due to its larger pore size (∼19, ∼24, and ∼28 Å). These findings, when taken together with the AF2 predictions for CAV1 oligomers with different numbers of protomers, suggest that parallel β-barrel sizes formed by 10–12 β-strands are best tolerated when the same residue of each β-strand is pointing inside simultaneously.
Overall, these results show the effectiveness of AF2 as a tool to predict the structures of novel overall oligomeric protein architectures even if the protomers have no close homologs in the PDB, despite some limitations regarding the prediction of secondary, tertiary, and quaternary structures. Extension of analyses such as ours to all the putative members of the CAV family may not only enable us to better understand the conserved motifs among this family but also provide a useful screening tool to distinguish sequences that are annotated as CAVs based on their tendencies to form disc-shaped structures similar to those observed for human CAV1. Importantly, while the experimental structures of CAV1 display an 11-mer, other arrangements of open and closed discs are likely to be biologically relevant and might be similar to the AF2-proposed models. Such structures could, for example, potentially correspond to intermediate complexes formed as the newly synthesized protein first assembles into oligomers. Conversely, it will also be important to apply other approaches to determine how CAV1 sits in membranes and whether the complexes impose curvature, as suggested by the predicted structures.
Finally, improvements to the oligomeric prediction capabilities of AF2 will drastically improve our ability to predict protein-protein complex structures in the absence of experimental data regarding the interaction sites of such multimeric assemblies.
Author contributions
Conceptualization, A.G., B.H., H.S.M., J.M., and A.K.K.; methodology, A.G., B.H., and J.C.P.; investigation, A.G., B.H., and J.C.P.; data curation, A.G., B.H., and J.C.P.; formal analysis, A.G., B.H., and J.C.P.; writing – original draft, A.G., B.H., and A.K.K.; writing – review & editing, A.G., B.H., J.C.P., H.S.M., J.M., and A.K.K.; visualization, A.G., B.H., and J.C.P.; supervision, A.K.K., H.S.M., and J.M.; project administration, A.K.K. and J.M.; funding acquisition, A.K.K. and J.M.
Acknowledgments
We thank Erkan Karakas for helpful discussions. This work was supported by National Institutes of Health grants R01 HL144131 (A.K.K.), R01GM080403 (J.M.), R01HL122010 (J.M.) and R01GM129261 (J.M.) and the Humboldt Professorship of the Alexander von Humboldt Foundation (J.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
The authors declare that they have no competing interests.
Editor: Alexandr Kornev.
Footnotes
Alican Gulsevin and Bing Han contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.11.011.
Contributor Information
Jens Meiler, Email: jens@meilerlab.org.
Anne K. Kenworthy, Email: akk7hp@virginia.edu.
Supporting material
References
- 1.Jumper J., Evans R., et al. Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kryshtafovych A., Schwede T., et al. Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins. 2021;89:1607–1617. doi: 10.1002/prot.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tunyasuvunakool K., Adler J., et al. Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao M., Nakajima An D., et al. Skolnick J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022;13:1744. doi: 10.1038/s41467-022-29394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Alamo D., Govaerts C., McHaourab H.S. AlphaFold2 predicts the inward-facing conformation of the multidrug transporter LmrP. Proteins. 2021;89:1226–1228. doi: 10.1002/prot.26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Alamo D., Sala D., et al. Meiler J. Sampling alternative conformational states of transporters and receptors with AlphaFold2. Elife. 2022;11 doi: 10.7554/eLife.75751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegedűs T., Geisler M., et al. Farkas B. Ins and outs of AlphaFold2 transmembrane protein structure predictions. Cell. Mol. Life Sci. 2022;79:73. doi: 10.1007/s00018-021-04112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant P., Pozzati G., Elofsson A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 2022;13:1265. doi: 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler N.J., Williamson M.P. The accuracy of protein structures in solution determined by AlphaFold and NMR. Structure. 2022;30:925–933.e2. doi: 10.1016/j.str.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jumper J., Evans R., et al. Hassabis D. Applying and improving AlphaFold at CASP14. Proteins. 2021;89:1711–1721. doi: 10.1002/prot.26257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porta J.C., Han B., et al. Ohi M.D. Molecular architecture of the human caveolin-1 complex. Sci. Adv. 2022;8:eabn7232. doi: 10.1126/sciadv.abn7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen K.N., Entova S., et al. Imperiali B. Monotopic membrane proteins join the fold. Trends Biochem. Sci. 2019;44:7–20. doi: 10.1016/j.tibs.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wierenga R.K. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 2001;492:193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 14.Wang R.Y.R., Song Y., et al. DiMaio F. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife. 2016;5 doi: 10.7554/eLife.17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nivón L.G., Moretti R., Baker D. A Pareto-optimal refinement method for protein design scaffolds. PLoS One. 2013;8 doi: 10.1371/journal.pone.0059004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomize M.A., Pogozheva I.D., et al. Lomize A.L. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barth P., Schonbrun J., Baker D. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. USA. 2007;104:15682–15687. doi: 10.1073/pnas.0702515104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alford R.F., Koehler Leman J., et al. Gray J.J. An integrated framework advancing membrane protein modeling and design. PLoS Comput. Biol. 2015;11 doi: 10.1371/journal.pcbi.1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alford R.F., Leaver-Fay A., et al. Gray J.J. The Rosetta all-atom energy function for macromolecular modeling and design. J. Chem. Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamelryck T., Manderick B. PDB file parser and structure class implemented in Python. Bioinformatics. 2003;19:2308–2310. doi: 10.1093/bioinformatics/btg299. [DOI] [PubMed] [Google Scholar]
- 21.Ariotti N., Rae J., et al. Parton R.G. Molecular characterization of caveolin-induced membrane curvature. J. Biol. Chem. 2015;290:24875–24890. doi: 10.1074/jbc.M115.644336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Root K.T., Julien J.A., Glover K.J. Secondary structure of caveolins: a mini review. Biochem. Soc. Trans. 2019;47:1489–1498. doi: 10.1042/BST20190375. [DOI] [PubMed] [Google Scholar]
- 23.Kirchner P., Bug M., Meyer H. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. J. Biol. Chem. 2013;288:7363–7372. doi: 10.1074/jbc.M112.429076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glenney J.R., Jr., Zokas L. Novel tyrosine kinase substrates from Rous sarcoma virus-transformed cells are present in the membrane skeleton. J. Cell Biol. 1989;108:2401–2408. doi: 10.1083/jcb.108.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S., Seitz R., Lisanti M.P. Phosphorylation of caveolin by src tyrosine kinases. The alpha-isoform of caveolin is selectively phosphorylated by v-Src in vivo. J. Biol. Chem. 1996;271:3863–3868. [PubMed] [Google Scholar]
- 26.Vainonen J.P., Aboulaich N., et al. Vener A.V. N-terminal processing and modifications of caveolin-1 in caveolae from human adipocytes. Biochem. Biophys. Res. Commun. 2004;320:480–486. doi: 10.1016/j.bbrc.2004.05.196. [DOI] [PubMed] [Google Scholar]
- 27.Corley Mastick C., Sanguinetti A.R., et al. Newcomb L.F. Caveolin-1 and a 29-kDa caveolin-associated protein are phosphorylated on tyrosine in cells expressing a temperature-sensitive v-Abl kinase. Exp. Cell Res. 2001;266:142–154. doi: 10.1006/excr.2001.5205. [DOI] [PubMed] [Google Scholar]
- 28.Mastick C.C., Saltiel A.R. Insulin-stimulated tyrosine phosphorylation of caveolin is specific for the differentiated adipocyte phenotype in 3T3-L1 cells. J. Biol. Chem. 1997;272:20706–20714. doi: 10.1074/jbc.272.33.20706. [DOI] [PubMed] [Google Scholar]
- 29.Parr R.D., Martin G.G., et al. Schroeder F. A new N-terminal recognition domain in caveolin-1 interacts with sterol carrier protein-2 (SCP-2) Biochemistry. 2007;46:8301–8314. doi: 10.1021/bi7002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doucey M.A., Bender F.C., et al. Bron C. Caveolin-1 interacts with the chaperone complex TCP-1 and modulates its protein folding activity. Cell. Mol. Life Sci. 2006;63:939–948. doi: 10.1007/s00018-005-5551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherer P.E., Tang Z., et al. Lisanti M.P. Caveolin isoforms differ in their N-terminal protein sequence and subcellular distribution. Identification and epitope mapping of an isoform-specific monoclonal antibody probe. J. Biol. Chem. 1995;270:16395–16401. doi: 10.1074/jbc.270.27.16395. [DOI] [PubMed] [Google Scholar]
- 32.Kogo H., Fujimoto T. Caveolin-1 isoforms are encoded by distinct mRNAs. Identification of mouse caveolin-1 mRNA variants caused by alternative transcription initiation and splicing. FEBS Lett. 2000;465:119–123. doi: 10.1016/s0014-5793(99)01730-5. [DOI] [PubMed] [Google Scholar]
- 33.Rieth M.D., Lee J., Glover K.J. Probing the caveolin-1 P132L mutant: critical insights into its oligomeric behavior and structure. Biochemistry. 2012;51:3911–3918. doi: 10.1021/bi3001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han B., Gulsevin A., et al. Kenworthy A.K. Structural characterization of a breast cancer-associated mutation in caveolin-1. bioRxiv. 2022 doi: 10.1101/2022.05.23.493104. Preprint at. [DOI] [Google Scholar]
- 35.Romero-Romero S., Costas M., et al. Fernández-Velasco D.A. The stability landscape of de novo TIM barrels explored by a modular design approach. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2021.167153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bah A., Forman-Kay J.D. Modulation of intrinsically disordered protein function by post-translational modifications. J. Biol. Chem. 2016;291:6696–6705. doi: 10.1074/jbc.R115.695056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iakoucheva L.M., Radivojac P., et al. Dunker A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez I., Ying Y., et al. Anderson R.G.W. Mechanism of caveolin filament assembly. Proc. Natl. Acad. Sci. USA. 2002;99:11193–11198. doi: 10.1073/pnas.172196599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monier S., Parton R.G., et al. Kurzchalia T.V. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sargiacomo M., Scherer P.E., et al. Lisanti M.P. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khater I.M., Meng F., et al. Hamarneh G. Super resolution network analysis defines the molecular architecture of caveolae and caveolin-1 scaffolds. Sci. Rep. 2018;8:9009. doi: 10.1038/s41598-018-27216-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khater I.M., Liu Q., et al. Nabi I.R. Super-resolution modularity analysis shows polyhedral caveolin-1 oligomers combine to form scaffolds and caveolae. Sci. Rep. 2019;9:9888. doi: 10.1038/s41598-019-46174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han B., Porta J.C., et al. Kenworthy A.K. Structure and assembly of CAV1 8S complexes revealed by single particle electron microscopy. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han B., Copeland C.A., et al. Kenworthy A.K. Characterization of a caveolin-1 mutation associated with both pulmonary arterial hypertension and congenital generalized lipodystrophy. Traffic. 2016;17:1297–1312. doi: 10.1111/tra.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lasters I., Wodak S.J., et al. van Cutsem E. Structural principles of parallel beta-barrels in proteins. Proc. Natl. Acad. Sci. USA. 1988;85:3338–3342. doi: 10.1073/pnas.85.10.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.