Abstract
Peroxisome‐localized oxo‐phytodienoic acid (OPDA) reductases (OPR) are enzymes converting 12‐OPDA into jasmonic acid (JA). However, the biochemical and physiological functions of the cytoplasmic non‐JA producing OPRs remain largely unknown. Here, we generated Mutator‐insertional mutants of the maize OPR2 gene and tested its role in resistance to pathogens with distinct lifestyles. Functional analyses showed that the opr2 mutants were more susceptible to the (hemi)biotrophic pathogens Colletotrichum graminicola and Ustilago maydis, but were more resistant to the necrotrophic fungus Cochliobolus heterostrophus. Hormone profiling revealed that increased susceptibility to C. graminicola was associated with decreased salicylic acid (SA) but increased JA levels. Mutation of the JA‐producing lipoxygenase 10 (LOX10) reversed this phenotype in the opr2 mutant background, corroborating the notion that JA promotes susceptibility to this pathogen. Exogenous SA did not rescue normal resistance levels in opr2 mutants, suggesting that this SA‐inducible gene is the key downstream component of the SA‐mediated defences against C. graminicola. Disease assays of the single and double opr2 and lox10 mutants and the JA‐deficient opr7opr8 mutants showed that OPR2 negatively regulates JA biosynthesis, and that JA is required for resistance against C. heterostrophus. Overall, this study uncovers a novel function of a non‐JA producing OPR as a major negative regulator of JA biosynthesis during pathogen infection, a function that leads to its contrasting contribution to either resistance or susceptibility depending on pathogen lifestyle.
Keywords: anthracnose leaf blight, corn smut, OPDA reductase, oxylipins, SA‐JA antagonism, southern corn leaf blight
ZmOPR2 functions as a key component of salicylic acid‐mediated defences via suppressing jasmonic acid biosynthesis during pathogen infection, leading to its contrasting contribution to either resistance or susceptibility depending on pathogen lifestyle.
1. INTRODUCTION
Anthracnose leaf blight (ALB) and stalk rot (ASR) diseases caused by the hemibiotrophic fungal pathogen Colletotrichum graminicola are maize diseases that have economic importance worldwide (Belisário et al., 2022; Bergstrom & Nicholson, 1999; Dean et al., 2012). Combined, both diseases are major threats to maize production, accounting for yield losses from 1% to 5% in the United States annually and as high as 10% to 20% worldwide (Crop Protection Network, 2021; Deleon et al., 2021; Frey et al., 2011; White, 1999). Infection by the hemibriotrophic C. graminicola has been reported to initiate with spore germination and appressoria formation within 24 h. The pathogen invades the epidermal cell and grows biotrophically, spreading to adjacent host cells within 48 h. A switch to necrotrophic growth occurs approximately after 48–72 h, causing the death of host cells (Belisário et al., 2022; Bergstrom & Nicholson, 1999; Mims & Vaillancourt, 2002; O'Connell et al., 1985; Vargas et al., 2012; Wharton et al., 2001). The biotrophic fungal pathogen Ustilago maydis causes corn smut disease and is able to induce tumours on all the aerial parts of its host plants maize and teosinte (Basse & Steinberg, 2004). Common smut occurs in most places where maize is grown but does not usually cause significant economic losses, ranging from a trace up to 10% in localized areas (White, 1999). Under favourable conditions, the diploid spores germinate to form the infectious hyphae and appressorium for penetration, which is probably facilitated by cell wall‐degrading enzymes. During the early infection stage, invading hyphae remain intracellular and are surrounded by the host plasma membrane while the hyphae grow both intra‐ and intercellularly at later stages and plant tumours are formed that are associated with plant cell enlargement and increased cell divisions (Banuett & Herskowitz, 1996; Callow & Ling, 1973; Doehlemann et al., 2008b; Lanver et al., 2017). Other than ALB, ASR, and corn smut, southern corn leaf blight (SCLB), caused by the necrotrophic fungal pathogen Cochliobolus heterostrophus (amorph Bipolaris maydis), is one of the most devastating diseases on maize, reducing crop yield and grain quality (Bruns, 2017). SCLB can be found everywhere maize is grown throughout the world and it is one of the most prevalent and severe diseases in tropical and subtropical maize‐producing areas, causing annual losses ranging from 10% to 68% under favourable conditions. An epidemic in 1970 destroyed an estimated 15% of the maize crop, resulting in approximately $1 billion in losses in the United States and southern Canada (Bruns, 2017; Dai et al., 2016; Ngoko et al., 2002). In a humid and warm environment, the spores can germinate in 6 h and penetrate the plants directly or through natural openings such as the stomata (Mendgen et al., 1996; Singh & Sricastava, 2012). C. heterostrophus can produce a group of chemically diverse and low molecular weight host‐selective toxins that serve as virulence or pathogenicity factors, leading to leaf chlorosis and lesion formation (Turgeon & Baker, 2007; Wu et al., 2012).
Plants produce an array of small signalling molecules to mediate defence against pathogens. Among the best studied are defence phytohormones, such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET). In response to pathogen infection, phytohormones interact with each other synergistically and antagonistically. SA has been extensively reported to antagonize JA and govern resistance (Kumar, 2014) against biotrophic and hemibiotrophic pathogens (Boatwright & Pajerowska‐Mukhtar, 2013). JA and ET play important roles in resistance to necrotrophic pathogens (Glazebrook, 2005). Both JA and SA accumulate in response to C. graminicola infection and are implicated in plant immunity against this pathogen (Balmer et al., 2013; Miranda et al., 2017). Recently, however, a report showed that green leaf volatiles and JA enhance susceptibility to C. graminicola in maize, whereas increased SA is associated with higher resistance levels (Gorman et al., 2020). SA plays an essential role in plant defence against biotrophic pathogens (Glazebrook, 2005). U. maydis is known to down‐regulate SA biosynthesis genes within 12 h of plant infection (Doehlemann et al., 2008a) and secrete the effector protein Jsi1, which interacts with several members of the plant corepressor family Topless/Topless related (TPL/TPR) and hijacks the plant JA/ET signalling pathway that leads to biotrophic susceptibility (Darino et al., 2021) during compatible interactions. JA and ET play important roles in resistance to necrotrophic pathogens (Glazebrook, 2005). However, whether JA is required for resistance against necrotrophic C. heterostrophus has not been well reported thus far.
JA and its derivatives, including methyl jasmonate (MeJA) and the biologically active jasmonoyl‐isoleucine (JA‐Ile), are fatty acid‐derived compounds ubiquitously found in higher plant species (Farmer et al., 2003). Jasmonates play essential roles in modulating a variety of biological processes, such as root growth (Gasperini et al., 2015; Sirhindi et al., 2020; Yan et al., 2014), seed germination (Linkies & Leubner‐Metzger, 2012; Pan et al., 2020; Singh et al., 2017), senescence (Hu et al., 2017; Qi et al., 2015; Schommer et al., 2008), trichome formation (Boughton et al., 2005; Traw & Bergelson, 2003), and anther development (Ishiguro et al., 2001; Saito et al., 2015; Sanders et al., 2000). In addition, JA also regulates defence against insects and necrotrophic pathogens (Browse, 2009; Wasternack, 2007; Wasternack & Strnad, 2016; Yan et al., 2014). JA biosynthesis starts with the oxygenation of α‐linolenic acid (C18:3) by 13‐lipoxygenase (13‐LOX), followed by the actions of allene oxide synthase (AOS) and allene oxide cyclase (AOC), leading to production of 12‐oxo‐phytodienoic acid (12‐OPDA) (Feussner & Wasternack, 2002; Wasternack & Hause, 2013). 12‐OPDA is transported into the peroxisome and the subsequent reduction of the cyclopentenone ring catalysed by peroxisome‐localized OPDA reductase (OPR) generates 3‐oxo‐2‐(2′‐pentenyl)‐cyclopentane‐1‐octanoic acid (OPC‐8:0) (Schaller, 2001; Vick & Zimmerman, 1984). OPC‐8:0 is converted to JA after three cycles of β‐oxidation that occur in the peroxisome (Turner et al., 2002).
To date, multiple members of OPR gene families have been identified in several plant species: three OPR genes in Arabidopsis (Biesgen & Weiler, 1999; Schaller et al., 2000; Stintzi & Browse, 2000), eight genes in maize (Zhang et al., 2005), 13 genes in rice (Agrawal et al., 2003), three isoforms in tomato (Strassner et al., 2002), 48 genes in wheat (Mou et al., 2019), and five genes in watermelon (Guang et al., 2021). Plant OPRs are classified into two groups (I and II) depending on their substrate specificity (Schaller et al., 1998). Members of the OPRI group preferentially catalyse the reduction of cis‐(−) OPDA rather than cis‐(+) OPDA, and therefore are not involved in JA biosynthesis (Schaller et al., 2000; Strassner et al., 1999). Members of the OPRII group catalyse the conversion of cis‐(+) OPDA, the natural precursor of JA, and therefore are involved with JA biosynthesis (Schaller et al., 1998). To date, only a few members of the plant OPRII group have been well characterized. Arabidopsis OPR3 is required for JA biosynthesis and lack of OPR3 function confers male fertility (Stintzi & Browse, 2000). Overexpression of rice OsOPR7 was able to complement the phenotypes of male sterility and JA production in the Arabidopsis opr3 mutant (Tani et al., 2008). Maize ZmOPR7 and ZmOPR8 provide redundant function in JA production and the double mutant opr7opr8 displays complete JA‐deficiency, resulting in the tasselseed phenotype, and is extremely susceptible to insect herbivory and root‐rotting necrotrophic oomycete Pythium spp. (Yan et al., 2012). The biochemical and physiological functions of most plant OPRI enzymes remain largely unknown.
To explore the function of the non‐JA producing OPRI subfamily in maize, we generated Mutator‐insertional mutants in one of the six members of the OPRI subfamily, ZmOPR2 (Borrego & Kolomiets, 2016; Zhang et al., 2005). ZmOPR2 was chosen based on its strong induction by the fungal elicitor chitooligosaccharide, SA, and infections with Cochliobolus carbonum, C. heterostrophus and Fusarium verticillioides (Zhang et al., 2005), suggesting a role in SA‐dependent defence responses against pathogens. Functional analyses of three mutant alleles, opr2‐1, opr2‐2, and opr2‐3, revealed that ZmOPR2 contributes to resistance to the hemibiotrophic pathogen C. graminicola and the biotrophic maize smut pathogen U. maydis by influencing ZmLOX10‐mediated SA and JA antagonism. Conversely, ZmOPR2 facilitates disease by the necrotroph C. heterostrophus via suppression of JA synthesis.
2. RESULTS
2.1. Characterization of opr2 mutants
Full‐length ZmOPR2 cDNA clones have been identified by sequence analyses of the DuPont/Pioneer and publicly available EST collection from Zea mays inbred line B73 as described in Zhang et al. (2005). Full‐length cDNA of ZmOPR2 (Zm00001d044906) was aligned with the corresponding genomic DNA sequences from NCBI and Maize Genetics and Genomics Database (MaizeGDB: https://www.maizegdb.org/) and the results show ZmOPR2 contains two exons and one intron (Figure 1a). The maize genome encodes another OPRI family member, ZmOPR1, that shares over 96% identity with ZmOPR2 (Zhang et al., 2005). A transposon insertional reverse genetics approach was used to generate opr1 and opr2 mutants (McCarty et al., 2005; Meeley & Briggs, 1995). Approximately 42,000 Mutator (Mu)‐insertional individual plants (Meeley & Briggs, 1995), available at Corteva (Johnston, IA, USA), were screened using gene‐specific primers and Mu terminal inverted repeat (TIR)‐specific primers. One allele with the Mu element inserted in the ZmOPR2 first exon (PV 03 80 A‐05) and one in the intron (PV 03145 D‐08) were identified by PCR‐based screening and were named opr2‐1 and opr2‐2, respectively (Figure 1a). Recently, we found a new exonic transposon insertional mutant (mu1079063::Mu), named opr2‐3, from the UniformMu Transposon Resource (https://www.maizegdb.org/documentation/uniformmu/index.php). Unfortunately, our multiple attempts to identify Mu insertions in the ZmOPR1 gene failed. To test whether Mu insertions impaired gene expression and to determine their suitability for subsequent functional analyses, semiquantitative reverse transcription (RT)‐PCR was conducted using ZmOPR2 gene‐specific primers to detect ZmOPR2 transcripts in the opr2‐1, opr2‐2, and opr2‐3 mutants and their wild types (WT) using ubiquitin carrier protein (UBCP) as an internal control. The results showed that Mu insertions in exon I and II of ZmOPR2 resulted in a complete lack of detectable transcripts and thus opr2‐1 and opr2‐3 represent knockout mutant alleles, while opr2‐2 is a knockdown allele exhibiting a reduced ZmOPR2 transcript level compared to WT (Figure 1b). To eliminate unrelated mutations, opr2‐1 was backcrossed seven times into a B73 genetic background (BC7) and opr2‐2 and opr2‐3 were at BC4 and BC1 stages in the B73 background, respectively. Next, transcript accumulation of ZmOPR2 in various tissues was assessed from publicly available datasets (Walley et al., 2016). This analysis showed that silk had the highest ZmOPR2 transcript level followed by pericarp/aleurone, root cortex, and secondary root (Figure S1).
FIGURE 1.
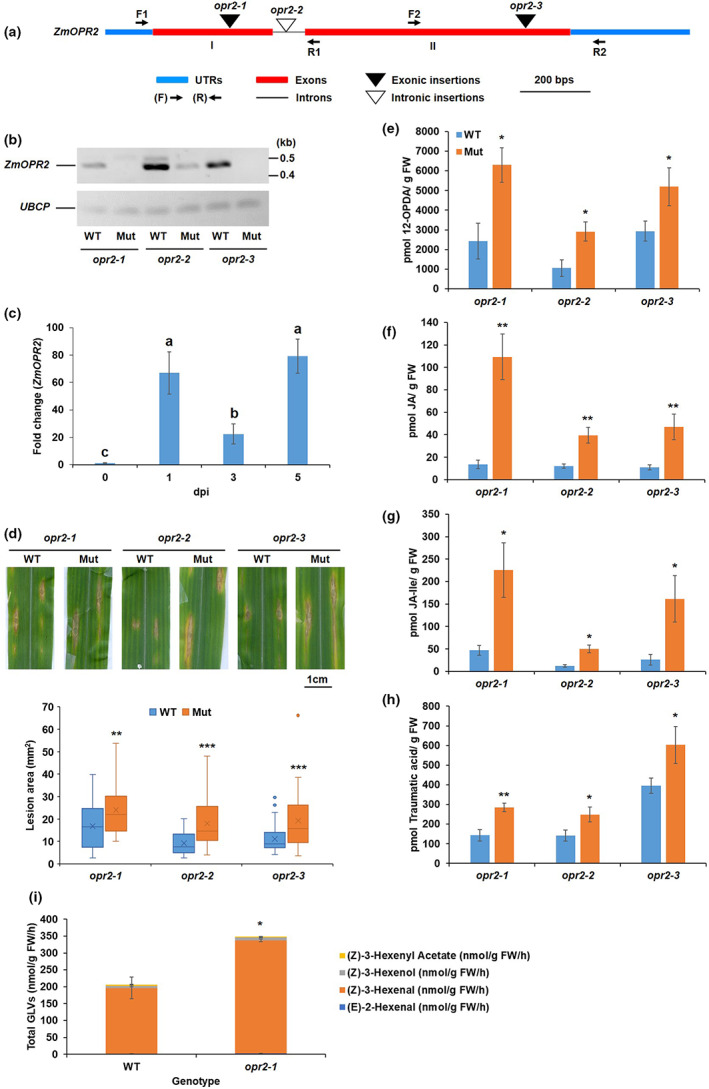
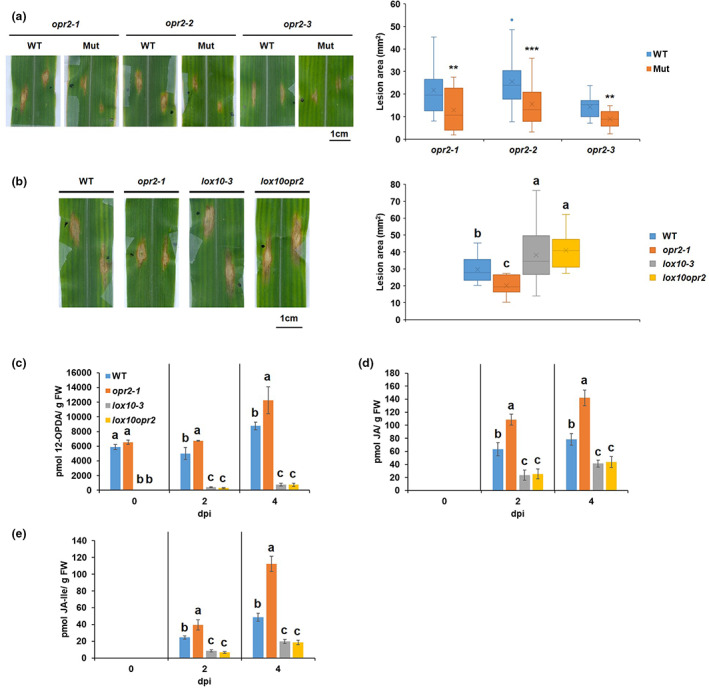
Disruption of ZmOPR2 reduced resistance to Colletotrichum graminicola. (a) Schematic representation of the genomic structure of ZmOPR2 showing the Mutator (Mu)‐element insertion sites. (b) Reverse transcription‐PCR analysis of ZmOPR2 gene expression in opr2‐1, opr2‐2, and opr2‐3 mutants and their corresponding wild type (WT). ZmOPR2 gene expression in opr2‐1 and opr2‐2 was checked using primer pair F1 and R1 while primer pair F2 and R2 was used in opr2‐3. UBCP (Ubiquitin carrier protein) represents a reference gene. (c) Expression of ZmOPR2 at 1, 3, and 5 days postinoculation (dpi) in response to C. graminicola infection relative to uninfected control at day 0. Bars are mean ± SEM (n = 5 maize plants of each genotype as biological replicates, no technical replicate). Different letters indicate statistically significant differences among the samples (Tukey's HSD test, p < 0.05). (d) Disease symptoms of opr2‐1, opr2‐2, and opr2‐3 mutants and their WT 7 dpi with C. graminicola. Disease symptoms were scanned and lesion areas were measured using ImageJ software. The data are shown in the box and whisker plot and × indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Outliers are represented by dots. The experiments were repeated at least two times with similar results. Contents of (e) 12‐OPDA, (f) jasmonic acid (JA), (g) JA‐Ile, and (h) traumatic acid were measured at 7 dpi. Bars are mean ± SEM (n = 6 maize plants of each genotype as biological replicates). (i) Quantification of total green leaf volatiles (GLVs) emissions in opr2‐1 mutant and its WT leaves 7 dpi with C. graminicola. Measurement of selected volatile emissions (z)‐3‐henxenyl acetate, (Z)‐3‐hexenol, (z)‐3‐hexenal, and (E)‐2‐hexenal. Bars are sum of mean ± SEM of each volatile (n = 4 maize plants of each genotype as biological replicates). Asterisks represent statistically significant differences between WT and mutant (Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001).
2.2. Disruption of ZmOPR2 reduced resistance to C. graminicola
Previously, ZmOPR2 was reported to be the most strongly induced maize OPR gene in response to diverse pathogens and SA treatment (Zhang et al., 2005). This prompted us to test whether ZmOPR2 is involved in resistance to C. graminicola. Because the response to C. graminicola was not tested before, we first examined the expression level of ZmOPR2 transcripts in maize inbred line B73 leaves in response to this pathogen. C. graminicola infection strongly induced ZmOPR2 transcript accumulation to 66‐, 22‐, and 79‐fold at 1, 3, and 5 days postinoculation (dpi), respectively, compared to uninfected control at day 0 (Figure 1c), therefore we tested the hypothesis that ZmOPR2 plays a role in maize defence against C. graminicola. Comparison of lesion areas between mutants and their respective WT revealed that all three opr2 mutant alleles are more susceptible to C. graminicola, with lesion areas approximately 1.5–2 times larger in the mutants compared to WT (Figure 1d), suggesting that ZmOPR2 contributes to defence against this pathogen.
2.3. Reduced resistance to C. graminicola is associated with increased JA and reduced levels of defensive ketols
Green leaf volatiles (GLVs) and JA have been shown to enhance susceptibility while SA promotes resistance to C. graminicola in maize (Gorman et al., 2020). To test whether reduced resistance of opr2 mutants to C. graminicola is associated with the changes in the defence hormones, we initially quantified the accumulation of diverse defence‐related metabolites in leaf tissues at 7 dpi. The results showed that opr2 mutants accumulated significantly higher concentrations of 12‐OPDA (Figure 1e), JA (Figure 1f), and JA‐Ile (Figure 1g), which is in line with the reported role of JA as a susceptibility factor (Gorman et al., 2020). Also, opr2 mutants accumulated significantly higher levels of traumatic acid (Figure 1h), which indicates higher LOX10 activity in opr2 mutants after pathogen infection because LOX10 is the sole LOX isoform responsible for producing traumatic acid (Christensen et al., 2013; He et al., 2020). To strengthen the evidence that opr2 mutants increased LOX10 activity after pathogen infection, we measured GLV production in C. graminicola‐infected leaves of opr2‐1 mutant and its WT at 7 dpi because LOX10 is the only LOX isoform providing substrates for GLV production (Christensen et al., 2013) and the results showed that indeed C. graminicola‐infected opr2‐1 leaves produced significantly greater amounts of GLV compared to WT (Figure 1i). In addition, significantly lower levels of multiple α‐ and γ‐ketol molecular species, including the recently identified mobile signal 9,10‐KODA, required for induced systemic resistance (Wang et al., 2020a), were found in opr2 mutants compared to WT (Figure S2a–h). Previous reports showed that exogenous treatment with several of these α‐ and γ‐ketols increased resistance against C. graminicola in maize (Wang et al., 2020a, 2020b). Moreover, increased resistance to ALB by treatment with pentyl leaf volatiles was associated with increased levels of these defensive ketols (Gorman et al., 2021). Therefore, both increased JA and reduced levels of defensive ketols in the opr2 mutant may explain at least in part its increased susceptibility to this pathogen.
2.4. Disruption of the JA‐producing lipoxygenase 10 in the opr2 mutant background increases resistance to C. graminicola
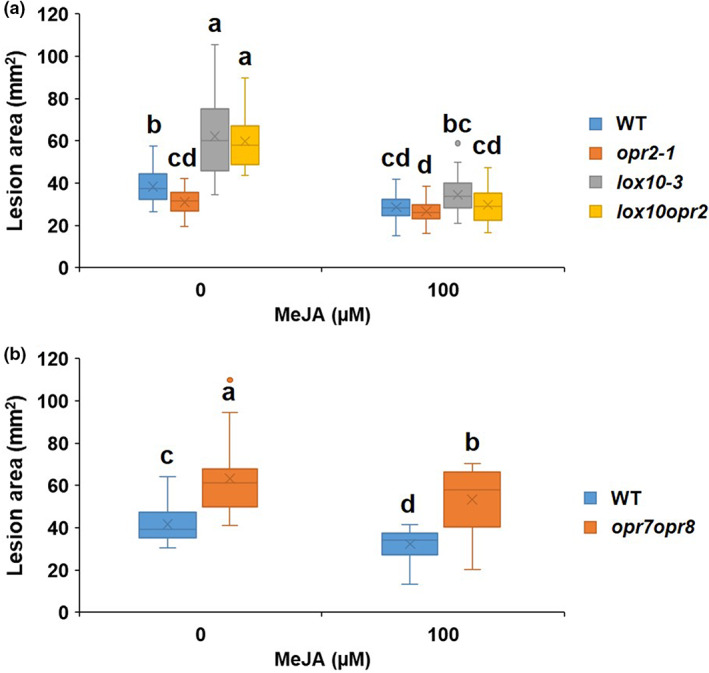
Previously, we showed that the LOX10 isoform is involved in GLV, traumatin, and JA biosynthesis (Christensen et al., 2013; He et al., 2020) and that lox10 mutants are remarkably more resistant to C. graminicola (Gorman et al., 2020). To test the hypothesis that opr2 mutants are more susceptible due to higher LOX10 activity (evident by increased content of traumatic acid and GLV production in opr2 mutants in Figure 1h,i), which in turn results in increased JA levels, we generated lox10‐3opr2‐1 double mutants (referred to as lox10opr2). ALB disease assays showed that the lox10opr2 mutant was as resistant as the single lox10‐3 mutant and displayed increased resistance, as evidenced by smaller lesion areas as compared to either WT or single opr2‐1 mutant (Figure 2a). These results indicate that ZmLOX10‐mediated JA production is associated with reduced resistance to C. graminicola in opr2 mutants. Hormone and oxylipin profiling of infected leaves at 0, 1, 3, and 5 dpi demonstrated that the susceptible opr2‐1 mutant accumulated between 2‐ to 6‐fold greater amounts of all three jasmonates, 12‐OPDA, JA, and JA‐Ile, compared to WT, lox10‐3 or lox10opr2 double mutants (Figure 2c–e). The transcript levels of two JA‐responsive genes, ZmMyc7 and ZmJAZ1, were in line with the higher amount of JA accumulation in opr2‐1 mutant in response to C. graminicola infection (Figure S3a,b). Moreover, the levels of jasmonates in the lox10opr2 mutants were similar to those in the single lox10‐3 mutant, which correlated with similarly increased levels of resistance to ALB for these two genotypes. These results support our hypothesis that increased JA content contributes to increased susceptibility in the opr2 mutants.
FIGURE 2.
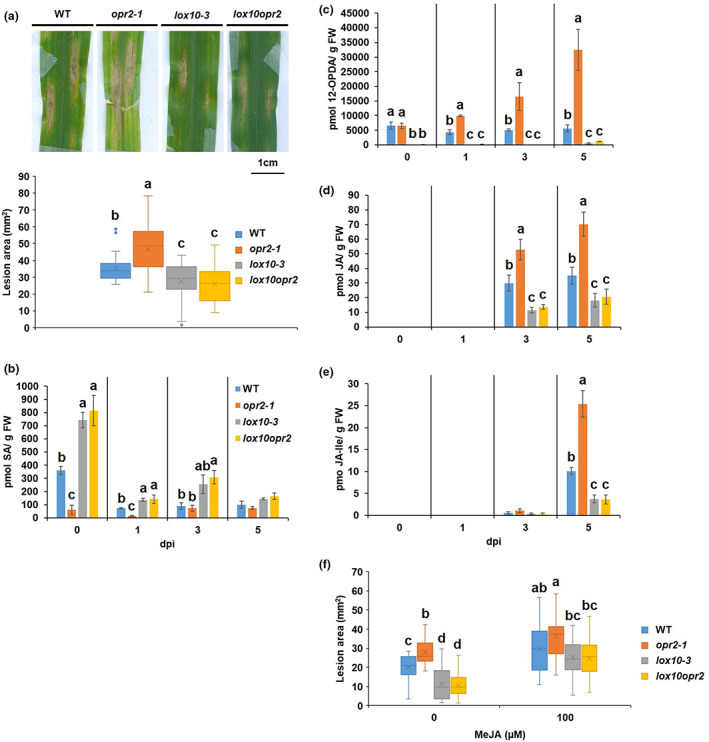
ZmLOX10 functions to suppress salicyclic acid (SA) and enhance jasmonic acid (JA) accumulation resulting in reduced resistance of opr2‐1 to Colletotrichum graminicola. (a) Disease symptoms of wild type (WT), opr2‐1, lox10‐3, and lox10opr2 7 days postinoculation (dpi) with C. graminicola. Disease symptoms were scanned and lesion areas were measured using ImageJ software. The data are shown in the box and whisker plot and X indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Outliers are represented by dots. This experiment was repeated at least three times with similar results. Contents of (b) SA, (c) 12‐OPDA, (d) JA, and (e) JA‐Ile were measured at 0, 1, 3, and 5 dpi of the WT, opr2‐1, lox10‐3, and lox10opr2 mutants. JA and JA‐Ile were at undetectable levels at 0 and 1 dpi. Bars are mean ± SEM (n = 5 maize plants of each genotype as biological replicates). (f) opr2‐1, lox10‐3, and lox10opr2 mutants and their WT were sprayed with 2 ml of mock or 100 μM MeJA per plant 1 h prior to inoculation with C. graminicola. The data are shown in the box and whisker plot and X indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Different letters indicate statistically significant differences among the genotypes (Tukey's HSD test, p < 0.05). Metabolite measurement was repeated at least two times with similar results.
Interestingly, the results clearly showed that maize plants respond to initial biotrophic infection and the later necrotrophic phase via differential production of SA and JA, with the transition approximately at 3 dpi. In addition to higher JA at necrotrophic stages, the susceptible opr2‐1 mutant accumulated significantly lower SA at the biotrophic stage, with only approximately one‐sixth and one‐fifth at 0 and 1 dpi, respectively, compared to WT (Figure 2b). This concurs with previous studies showing that SA plays an important role in defence against hemibiotrophic pathogens (Boatwright & Pajerowska‐Mukhtar, 2013; Gorman et al., 2020). The analysis showed that increased resistance of the lox10opr2 mutant was associated with increased SA (Figure 2b) at 0, 1, and 3 dpi and reduced JA (Figure 2d) and JA‐Ile (Figure 2e) at 3 and 5 dpi, mirroring the levels of SA and JAs in the lox10‐3 single mutant. To confirm that reduced JA in lox10‐3 and lox10opr2 confers increased resistance to C. graminicola, the plants were exogenously treated with 100 μM MeJA 1 h prior to C. graminicola infection. The results clearly showed that MeJA treatment restored the normal susceptibility of both lox10‐3 and lox10opr2 mutants to the WT level (Figure 2f). Together, these data indicate that increased LOX10 activity in opr2 mutants functions to suppress SA and enhance JA accumulation in response to C. graminicola infection, resulting in reduced resistance in the opr2‐1 mutant.
2.5. ZmOPR2 functions downstream of the SA‐dependent pathway for resistance to C. graminicola
To test whether reduced resistance of opr2 mutants is due to lower SA content, opr2‐1 and opr2‐3 mutants and WT were exogenously treated with 100 μM SA solution 1 h prior to C. graminicola infection. The results showed that although SA treatment moderately increased resistance to C. graminicola in both opr2‐1 and opr2‐3 mutants and WT, SA‐treated opr2‐1 and opr2‐3 mutants were still more susceptible, displaying significantly larger lesions than both mock‐ and SA‐treated WT (Figure 3a). Exogenous treatment of SA at a higher concentration (500 μM) 24 h prior to C. graminicola infection showed similar results (Figure 3b). Hormone and oxylipin profiling revealed that exogenous SA treatment significantly suppressed 12‐OPDA, JA, and JA‐Ile accumulation, causing 45%, 64%, and 48% reductions, respectively, in WT leaves after C. graminicola infection at 5 dpi compared to mock‐treated WT leaves while there was lack of a statistically significant reduction of 12‐ODPA (Figure 3c) and JA‐Ile (Figure 3e) contents and only moderate reduction of JA (Figure 3d) in SA‐treated opr2‐1 compared to mock‐treated opr2‐1 leaves. We suspect that the moderate reduction of JA is due to the presence of intact ZmOPR1. Together, these data suggest that exogenous SA treatment could not fully restore resistance of opr2 mutants to the WT level, indicating that this SA‐induced gene (Zhang et al., 2005) functions downstream of SA and is one of the targets of SA‐mediated signalling in defence against this pathogen via modulating SA–JA antagonism.
FIGURE 3.
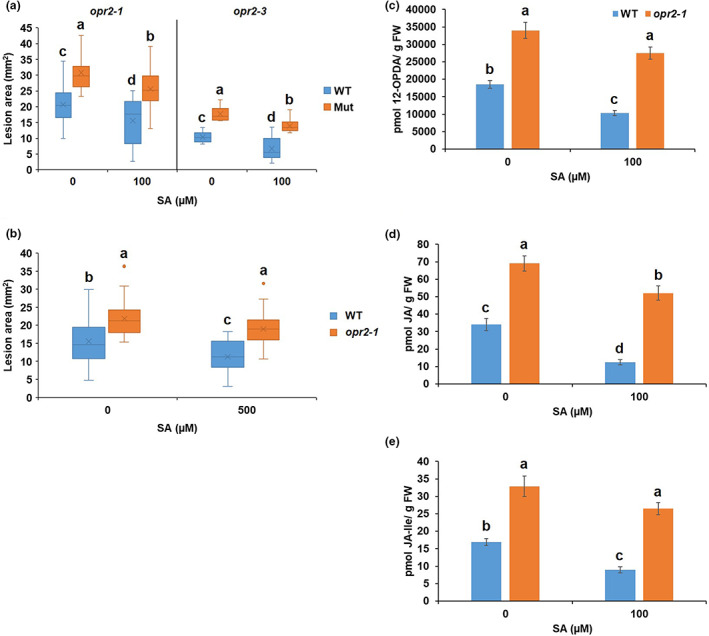
Salicylic acid (SA) treatment failed to fully restore the resistance of opr2 mutants to wild‐type (WT) level against Colletotrichum graminicola. (a) opr2‐1 and opr2‐3 mutants and their WT were sprayed with 2 ml of mock or 100 μM SA 1 h prior to inoculation with C. graminicola. (b) opr2‐1 and WT were sprayed with mock or 500 μM SA 24 h prior to inoculation with C. graminicola. Disease symptoms were scanned and lesion areas were measured using ImageJ software at 7 days postinoculation (dpi). The data are shown in the box and whisker plot and X indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Outliers are represented by dots. Different letters indicate statistically significant differences within the same allele on log‐transformed data (Tukey's HSD test, p < 0.05). These experiments were repeated at least two times with similar results. Contents of (c) 12‐OPDA, (d) JA, and (e) JA‐Ile were measured of the mock or 100 μM SA‐treated WT, opr2‐1, lox10‐3, and lox10opr2 mutants 5 dpi with C. graminicola. Bars are mean ± SEM (n = 5 maize plants of each genotype as biological replicates). Different letters indicate statistically significant differences (Tukey's HSD, p < 0.05).
2.6. Mutation of ZmOPR2 leads to reduced resistance to corn smut
SA plays a pivotal role in defence against biotrophic pathogens, including U. maydis (Djamei et al., 2011; Rabe et al., 2013). To test whether ZmOPR2 and ZmLOX10 are also involved in defence against U. maydis, opr2‐1 and lox10‐2 mutants and their respective NIL WT were infected with U. maydis and disease symptoms were analysed at 13 dpi. The results demonstrated that the opr2‐1 mutant has increased susceptibility to U. maydis resulting in overall heavier tumour formation and the appearance of dead plants (>30%) in response to infection compared to no death among WT plants (Figure 4). This result reveals that ZmOPR2 contributes to defence against U. maydis. No clear difference in disease progression was observed between lox10‐2 mutant and WT (Figure 4).
FIGURE 4.
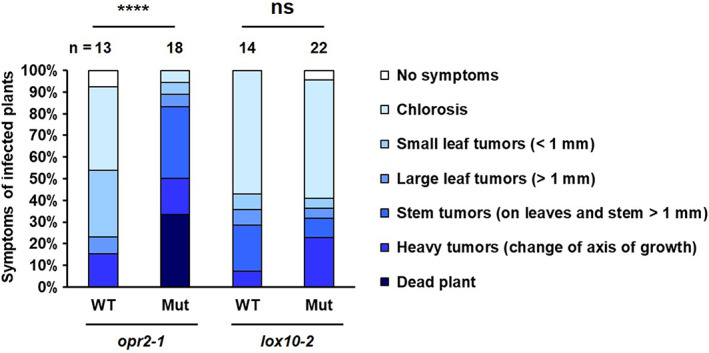
The opr2‐1 mutant is more susceptible to Ustilago maydis while the lox10‐2 mutant displays no difference to the wild type (WT). opr2‐1 and lox10‐2 mutants and their WT were inoculated with biotrophic pathogen U. maydis strain SG200. Disease symptoms were analysed at 13 days postinoculation (dpi). The severity of the disease symptoms on each inoculated plant was scored using a 0 to 6 rating scale. The statistical analysis was performed by comparing the disease scores between mutants and WT (Student's t test, ****p < 0.0001; ns, not significant). n = the total number of plants scored.
2.7. JA plays an essential role in defence against C. heterostrophus
JA plays a central role in defence against necrotrophic pathogens (Glazebrook, 2005; Thomma et al., 1998). Very little is known about the significance of JA for resistance to SCLB caused by the necrotrophic fungal pathogen C. heterostrophus. Higher JA accumulation after pathogen infection in opr2 mutants prompted us to test the role of ZmOPR2 in the maize–C. heterostrophus interaction, therefore we inoculated opr2 mutants with C. heterostrophus spores, and disease symptoms and lesion areas were measured 7 dpi. As predicted, the opr2 mutants were more resistant to C. heterostrophus, displaying significantly smaller lesions than WT (Figure 5a). To test whether ZmLOX10‐mediated JA production is associated with enhanced resistance in the opr2 mutant, we measured resistance levels in lox10‐3 and lox10opr2 with C. heterostrophus. The results showed that both the single and double mutants were more susceptible (Figure 5b). Hormone analysis of infected leaves at 0, 2, and 4 dpi showed that opr2‐1 mutant accumulated higher levels of 12‐ODPA (Figure 5c), JA (Figure 5d), and JA‐Ile (Figure 5e) compared to WT, lox10‐3, or lox10opr2 double mutants. In addition, the amounts of 12‐ODPA and JA‐Ile in the lox10opr2 mutants was similar to those in the single lox10‐3 mutant, which correlated with decreased levels of resistance to SCLB of these two genotypes. These results support our hypothesis that increased JA content in the opr2 mutant contributes to increased resistance to SCLB.
FIGURE 5.
Mutation of ZmLOX10 increased susceptibility of opr2 mutant to Cochliobolus heterostrophus. (a) Disease symptoms of opr2‐1, opr2‐2, and opr2‐3 mutants and their wild type (WT) 7 days postinoculation (dpi) with C. heterostrophus. The data are shown in the box and whisker plot and X indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Outliers are represented by dots. Asterisks represent statistically significant differences between WT and mutant (Student's t test, **p < 0.01, ***p < 0.001). (b) Disease symptoms of opr2‐1, lox10‐3, and lox10opr2 and their WT 7 dpi with C. heterostrophus. Disease symptoms were scanned and lesion areas were measured using ImageJ software. The data are shown in the box and whisker plot and X indicates means (n = 30 lesions from six different plants of each genotype as biological replicates). Contents of (c) 12‐OPDA, (d) jasmonic acid (JA), and (e) JA‐Ile were measured at 0, 2, and 4 dpi of the WT, opr2‐1, lox10‐3, and lox10opr2 mutants. JA and JA‐Ile were at undetectable levels at 0 dpi. Bars are mean ± SEM (n = 5 maize plants of each genotype as biological replicates). Different letters indicate statistically significant differences among the genotypes (Tukey's HSD test, p < 0.05). These experiments were repeated at least two times with similar results.
To further confirm that increased susceptibility of lox10‐3 and lox10opr2 mutants resulted from reduced JA contents, we exogenously treated opr2‐1, lox10‐3, and lox10opr2 mutants and WT with 100 μM MeJA 1 h prior to inoculation with C. heterostrophus. This treatment successfully rescued resistance of lox10‐3 and lox10opr2 mutants to WT levels (Figure 6a). To obtain further genetic evidence that JA is required for resistance to C. heterostrophus, JA‐deficient opr7opr8 mutant and WT plants were inoculated with C. heterostrophus after mock or MeJA treatment. The results showed that the JA‐deficient opr7opr8 mutant is more susceptible to C. heterostrophus, displaying significantly larger lesions (1.5‐fold larger) than WT (Figure 6b). Although exogenous MeJA treatment enhanced resistance to C. heterostrophus in both opr7opr8 mutant and WT, MeJA treatment failed to fully restore resistance of opr7opr8 to the WT level, presumably due to the hormone treatment applied only once before infection while the infection process lasted 7 days. Collectively, these data provided strong pharmacological and genetic evidence that JA is indeed required for defence against necrotrophic C. heterostrophus and that ZmOPR2 negatively regulates JA production during infection.
FIGURE 6.
Jasmonic acid (JA) promotes resistance to necrotrophic Cochliobolus heterostrophus. Lesion areas of (a) opr2‐1, lox10‐3, and lox10opr2 mutants and wild type (WT) and (b) JA‐deficient opr7opr8 mutant and WT. Plants were sprayed with 2 ml of mock or 100 μM MeJA per plant 1 h prior to inoculation with C. heterostrophus. Disease symptoms were scanned and lesion areas were measured using ImageJ software. The data are shown in the box and whisker plot and X indicates means (n = 36 lesions from six different plants of each genotype as biological replicates). Outliers are represented by dots. Different letters indicate statistically significant differences (Tukey's HSD test, p < 0.05). These experiments were repeated at least two times with similar results.
3. DISCUSSION
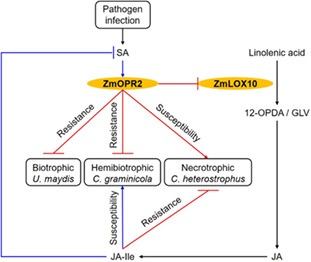
In stark contrast to the well‐known functions of the JA‐producing peroxisome‐localized OPRs belonging to the OPRII subfamily, the physiological functions of non‐JA‐producing OPRI subfamily enzymes are largely unknown (Borrego & Kolomiets, 2016; Zhang et al., 2005). While the exact in vivo biochemical substrates of OPRI group enzymes are not known, they are capable of reducing double bonds in α, β‐unsaturated aldehydes or ketones that are potent Michael acceptor systems and often toxic to cells (Kohli & Massey, 1998). ZmOPR2 belongs to the OPRI group that preferentially catalyses the reduction of cis‐(−) OPDA over cis‐(+) OPDA, the precursor of JA (Schaller et al., 1998), and localizes in the cytoplasm (Tolley et al., 2018). ZmOPR2 is therefore unlikely to be involved in JA biosynthesis and disruption of both ZmOPR7 and ZmOPR8 as in opr7opr8 double mutants results in JA deficiency, reduced resistance to insects, and complete abolition of immunity against soilborne necrotrophic pathogens such as Pythium spp. and Fusarium spp. (Christensen et al., 2014; Yan et al., 2012). Here, we present strong evidence showing that the ZmOPR2 isoform is involved in maize defence against both biotrophic (U. maydis) and hemibiotrophic (C. graminicola) fungal pathogens, but promotes susceptibility to the necrotrophic C. heterostrophus. Importantly, regardless of the pathogen lifestyle, both functions are due to the ZmOPR2 activity in suppressing JA biosynthesis, which in turn results in increased SA production (Figure 7).
FIGURE 7.
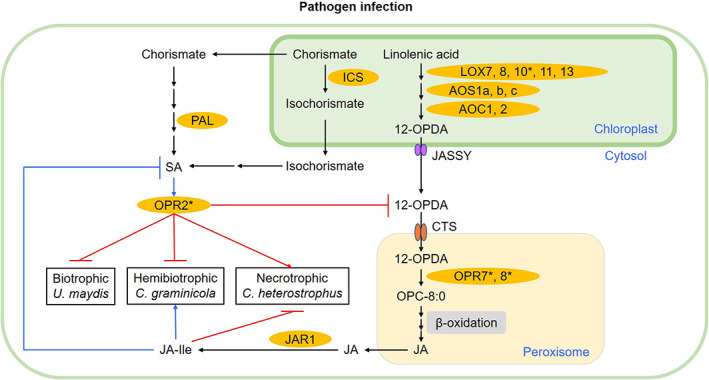
Working model of ZmOPR2‐mediated defence responses against fungal pathogens. During pathogen infection, salicylic acid (SA)‐inducible ZmOPR2 plays a pivotal role in SA‐mediated defence responses against (hemi)biotrophic pathogens and suppresses jasmonic acid (JA)‐Ile accumulation, resulting in increased resistance to biotrophic Ustilago maydis and hemibiotrophic Colletotrichum graminicola, but facilitates disease progression of necrotrophic Cochliobolus heterostrophus. Major enzyme names are indicated in orange and are abbreviated as follows: LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR, OPDA reductase; JAR1, jasmonoyl amino acid conjugate synthase; ICS, isochorismate synthase; PAL, phenylalanine ammonia‐lyase. Abbreviations for compounds: 12‐OPDA, cis‐(+)‐12‐oxo‐phytodienoic acid; OPC:8–0, 3‐oxo‐2‐(2′‐pentenyl)‐cyclopentane‐1‐octanoic acid; JA, jasmonic acid; JA‐Ile, jasmonic acid isoleucine conjugate. Abbreviations for proteins: JASSY, a chloroplast outer membrane protein responsible for transport of 12‐OPDA from chloroplast; CTS, an ABC transporter of the peroxisomal membrane responsible for transporting 12‐OPDA into peroxisome. Black arrows indicate the biosynthesis pathways. Blue arrows and blocks represent positive (promotion) or negative (suppression) interactions, respectively. Red arrows and blocks represent the OPR2‐mediated positive (promotion) or negative (suppression) interactions demonstrated in this study, respectively. Enzymes with asterisks were tested in this study.
Specifically, we showed that opr2 mutants are susceptible to C. graminicola and displayed abnormally low levels of SA at the biotrophic stage of infection and increased JA at the necrotrophic phase. This hemibiotrophic pathogen infects plants with a brief biotrophic phase before switching into a more destructive necrotrophic lifestyle that occurs for approximately 48–72 h after inoculation (Mims & Vaillancourt, 2002). The molecular details and factors regulating the transition have yet to be identified (Dean et al., 2012; Vargas et al., 2012). Our analysis showed that JA and JA‐Ile contents increased around the same time as C. graminicola switched to necrotrophic growth followed by extensive lesion progression. We previously reported that ZmLOX10 is involved in the synthesis of GLVs and JA (Christensen et al., 2013; He et al., 2020), the two oxylipins that facilitate maize susceptibility to C. graminicola by suppressing the levels of SA, the major defence hormone against this hemibiotrophic pathogen (Gorman et al., 2020). Higher levels of traumatic acid and GLV production in opr2‐1 leaves after C. graminicola infection indicate increased ZmLOX10 activity resulting in enhanced JA accumulation. We therefore tested the hypothesis that opr2 mutants are more susceptible to this pathogen because of increased levels of JA and found that, indeed, disruption of ZmLOX10 in the opr2 mutant background abolished pathogen‐induced JA accumulation, which in turn resulted in increased resistance. These results, once again, provided solid genetic evidence for JA as a susceptibility factor in maize–C. graminicola interactions. Similarly, recent studies of the interactions between maize and F. graminearum provided strong evidence based on the transcriptome, metabolome, and functional analyses of lox5, opr7opr8, and JAZ mutants that JA contributes to disease progression against this hemibiotrophic pathogen (Ma et al., 2021; Sugimoto et al., 2022; Wang et al., 2021).
JA and SA are widely reported to interact antagonistically during infection processes, with plants prioritizing induction of either SA or JA signalling depending on whether the virulent pathogen has a (hemi)biotrophic or necrotrophic mode of infection, respectively (Aerts et al., 2021; Kazan & Manners, 2008; Koornneef & Pieterse, 2008; Pieterse et al., 2009; Spoel & Dong, 2008). The strong inhibitory effect of the SA pathway on JA synthesis and signalling is mediated by a number of proteins with regulatory roles in SA–JA cross‐talk. These include the lipase‐like proteins EDS1 and PAD4 (Brodersen et al., 2006), SA receptor complex protein NPR1 (Spoel et al., 2003), the fatty acid desaturase SSI2 (Kachroo et al., 2001), the glutaredoxin GRX480 (Ndamukong et al., 2007), and WRKY transcription WRKY70 (Li et al., 2004). In addition to these reported key regulators, this study uncovered a novel mechanism in which SA antagonizes JA during plant–pathogen interactions by employing a member of the OPRI subfamily to suppress JA biosynthesis (Figure 7). The following reasoning lays the foundation for this notion. First, ZmOPR2 expression is strongly induced by exogenous SA treatment (Zhang et al., 2005), suggesting that ZmOPR2 is a component of SA‐dependent resistance mechanisms. Second, as a result of higher JA levels, lower SA contents were found in opr2 mutants and disruption of ZmLOX10 in opr2 mutants greatly increased SA accumulation, leading to elevated resistance to C. graminicola. Third, we showed that exogenous SA application failed to restore the WT‐level resistance, which confirms that ZmOPR2 functions downstream in SA‐mediated defence responses. The exact mechanism by which SA‐regulated ZmOPR2 negatively regulates pathogen‐induced JA is unclear as the exact in vivo substrates and final products of the enzymatic action of OPRI enzymes are currently unknown.
Multiple reports have shown that JA plays a major role in defence against necrotrophic pathogens in several dicot species, but no clear genetic evidence has been reported regarding the role of JA in maize defence against necrotrophic C. heterostrophus. Here, we demonstrate that JA confers resistance to necrotrophic C. heterostrophus using JA‐deficient opr7opr8 mutant and exogenous MeJA treatment. In line with our finding, overexpression of ZmAPX1 and biocontrol agent Trichoderma harzianum treatment enhanced resistance to SCLB via activating the JA‐mediated defence signalling pathways (Wang et al., 2019; Zhang et al., 2022). Zhang et al. (2005) reported that expression of ZmOPR2 is strongly induced by an HC‐toxin‐producing strain of Cochliobolus carbonum in the susceptible maize Pr inbred line, whereas the avirulent, toxin‐deficient strain is unable to induce appreciable levels of its transcripts. This suggests that ZmOPR2 may contribute to susceptibility towards this necrotrophic pathogen. Also, C. heterostrophus infection induces ZmOPR2 expression (Zhang et al., 2005), suggesting that this necrotrophic pathogen may potentially modulate expression of the JA‐suppressing OPRI enzymes to facilitate disease progression. In agreement with this hypothesis, here we showed that opr2 mutants overproduced JA and displayed enhanced resistance to C. heterostrophus, and that reduced JA due to disruption of ZmLOX10 greatly reduced resistance of opr2‐1. Exogenous application of MeJA successfully restored WT levels of resistance of both lox10‐3 and lox10opr2 mutants, indicating that JA produced by ZmLOX10 is required for normal defence against C. heterostrophus. Overall, these data demonstrate that JA contributes to defence against C. heterostrophus and that ZmLOX10 enhances resistance while ZmOPR2 promotes SCLB disease progression by influencing JA biosynthesis.
It is noteworthy that the maize genome contains one other SA‐inducible OPRI gene, ZmOPR1, sharing 96.5% amino acid sequence identity with ZmOPR2 (Zhang et al., 2005), and both ZmOPR1 (Zm00001d044908) and ZmOPR2 (Zm00001d044906) genes are located on chromosome 9 with only one other gene found between them. Thus, ZmOPR1 and ZmOPR2 are linked and essentially tandem duplicate genes, and public transcriptome datasets reveal that these two genes are often co‐expressed (Figure S1) (Forestan et al., 2016; Johnston et al., 2014; Kakumanu et al., 2012; Stelpflug et al., 2016; Walley et al., 2016; Waters et al., 2017). Despite the fact that the expression patterns of these two genes are highly similar, suggesting gene redundancy, our results clearly show that disruption of the ZmOPR2 gene results in a strong phenotypic difference from WT. One potential reason behind these discernible phenotypes of opr2 mutants is that ZmOPR2 has a higher expression level than ZmOPR1 in response to SA treatment (Zhang et al., 2005), suggesting a greater impact on SA‐mediated pathogen defence. However, the intact ZmOPR1 gene may be a reason behind the partial rescue of opr2‐1 resistance levels. A similar scenario can be found in the other pair of duplicated OPR genes, ZmOPR7 and ZmOPR8, that is, that the opr8‐2 single mutant accumulated lower 12‐OPDA and JA, especially in roots, than WT while the single opr7‐5 mutant had similar 12‐ODPA and JA contents (Yan et al., 2012), suggesting the ZmOPR8 gene plays a preferential role in JA. Unfortunately, we were unable to identify any Mu insertions in ZmOPR1 for its functional analysis. We hypothesize that if both genes were mutated, the disease and biochemical phenotypes would have been even stronger compared to single opr2 mutants. A potential caveat associated with the ZmOPR2 locus is that a full‐length cDNA that runs in the opposite direction covering the entire length of the ZmOPR2 gene has been reported (Soderlund et al., 2009), yet it is still unclear if the protein made by the antiparallel transcript does or does not accumulate.
To date, several plant OPR gene families have been reported, including Arabidopsis (Biesgen & Weiler, 1999; Schaller et al., 2000; Stintzi & Browse, 2000), cotton (Liu et al., 2020), maize (Zhang et al., 2005), rice (Li et al., 2011), tomato (Strassner et al., 2002), watermelon (Guang et al., 2021), and wheat (Mou et al., 2019) and most of the reported studies propose the functions of OPR genes in different plant species relying on the analyses of expression profiles in different tissues or under phytohormone and stress treatments. This study demonstrated that ZmOPR2, one of the OPRI group members, modulates defence responses to hemibiotrophic C. graminicola but enhances susceptibility to necrotrophic C. heterostrophus via differential regulation of ZmLOX10‐mediated JA and SA antagonism (Figure 7).
4. EXPERIMENTAL PROCEDURES
4.1. Plant and fungal materials
Mutator‐insertional opr2‐1 (PV 03 80 A‐05) and opr2‐2 (PV 03145 D‐08) alleles were obtained from Dupont‐Pioneer, Inc. (currently Corteva) and identified by PCR‐based screening for insertions in ZmOPR2; opr2‐3 (mu1079063::Mu; stock ID UFMu‐08953) was obtained from the maize genetics and genomics database (http://www.maizegdb.org). All three alleles had been backcrossed to the B73 inbred line and advanced to the BC7, BC4, and BC1 stages, respectively. Mutants and their WT were identified by PCR‐based genotyping using the Mu‐terminal inverted repeat‐specific and gene‐specific primers listed in Table S1. Identification of the lox10‐2 and lox10‐3 alleles was previously described by Christensen et al. (2013). The double mutant lox10opr2 was generated by crossing lox10‐3 and opr2‐1 single mutants in B73 background at the BC7 stage. The B73 line is used as a WT for opr2‐1, lox10‐3, and lox10opr2 mutants at the BC7 stage. The data for opr2‐2 and opr2‐3 presented in this study were analysed by comparing the mutant to its WT identified from the BC4F2 and BC1F2 segregating populations, respectively. Maize seedlings were grown in conical pots (20.5 × 4 cm) filled with commercial potting mix (Jolly Gardener Pro Line C/20) on light shelves at room temperature (22–24°C) with a 16‐h light period. C. graminicola strain 1.001 and C. heterostrophus local Texas isolate (provided by Dr Thomas Isakeit, Texas A&M University) were cultured from glycerol stocks kept in a freezer at −80°C and grown on potato dextrose agar (PDA) plates for at least 2–3 weeks to allow sporulation. U. maydis strain SG200 was prepared and maize plants were inoculated as described in Schirawski et al. (2010).
4.2. RNA extraction, semiquantitative RT‐PCR, and RT‐quantitative PCR analyses
Maize third fully expanded leaves at V3 stage were used for pathogen infection. Control or infected leaf tissues were frozen in liquid nitrogen and ground into powder by mortar and pestle. About 100 mg of plant tissue was used for RNA extraction using TRI reagent (Molecular Research Center Inc.) according to the manufacturer's protocol. DNase I (Thermo Fisher Scientific) treatment and cDNA synthesis (RevertAid H Minus First Strand cDNA Synthesis Kit; Thermo Fisher Scientific) were performed following the manufacturer's instructions. Gene‐specific forward and reverse primers of ZmOPR2 and ubiquitin carrier protein (UBCP, GRMZM2G102471) (Manoli et al., 2012) were used to perform semiquantitative PCR. For RT‐quantitative PCR analysis, a Verso 1‐step RT‐qPCR kit, SYBR Green, ROX (Thermo Fisher Scientific) was used following the manufacturer's procedure with 10‐μl volume reactions in a StopOne Plus Real‐Time PCR system (Applied Biosystems) using the following conditions: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 54°C for 30 s, and 72°C for 30 s. Melt curve conditions were 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. Relative expression was determined as fold change compared to control sample using the 2−ΔΔCt method after normalizing to reference gene α‐TUBULIN (Rao et al., 2013). All primers are listed in Table S1. PCR genotyping and semiquantitative PCR were performed with following conditions: 94°C for 5 min, followed by 20–35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30–60 s.
4.3. ALB and SCLB assays and phytohormone treatment
ALB and SCLB assays were performed following the protocol described in Gao et al. (2007). To harvest C. graminicola and C. heterostrophus spores, 10 ml of sterile water was added to the PDA plate and scraped with a sterile cell scraper to free the conidia then filtered through several layers of sterile cheesecloth to remove mycelia. The concentration of spores was determined using a haemocytometer and then diluted to a concentration of 106 conidia/ml. The plants were laid down in trays (78.5 × 63 × 7 cm) lined with paper towels and the third leaves were taped down flat and drop‐inoculated with 10 μl of conidial suspension (106 conidia/ml) with six droplets per leaf. The tray was sealed with Press'n Seal plastic wrap (Glad Products Company) after adding 200 ml of sterile water to form a humid chamber and placed in darkness overnight. The droplets were allowed to dry on the leaf surface after the plastic wrap was removed the next day before the plants were returned to an upright position on light shelves at approximately 24 h after inoculation and grown under the same conditions as before inoculation. The disease symptoms were scanned and lesion areas were measured using ImageJ software (v. 1.50i; NIH) at 7 dpi. The data shown in this study are presented using box and whisker plots with all lesions measured in the experiment and × indicating means. Outliers are represented by dots. For SA treatment, mutant plants and their WTs were sprayed with 2 ml of mock (0.01% ethanol) or 100 μM SA per plant 1 h prior to pathogen infection or mutant plants and their WT were sprayed with 2 ml of mock or 500 μM SA per plant 24 h prior to C. graminicola infection. For MeJA treatment, plants were exogenously applied with 2 ml of mock (0.01% ethanol) or 100 μM MeJA per plant 1 h prior to pathogen infection. For measuring the expression of genes in response to C. graminicola infection, the leaf‐inoculated area, approximately a 2‐cm region with six infection sites, was harvested in liquid nitrogen at different time points for RNA extraction and RT‐qPCR analysis.
4.4. Corn smut assays
U. maydis strain SG200 was prepared and maize plants were inoculated as described in Schirawski et al. (2010). In brief, the U. maydis SG200 is a solopathogenic haploid strain that can induce tumours on infected plants without prior fusion with a mating partner (Bölker et al., 1995; Kämper et al., 2006). The strain was grown from frozen glycercol stocks on potato dextrose agar and cultivated in YEPS‐light (0.4% yeast extract, 0.4% peptone, 2% sucrose) on a rotary shaker at 220 rpm at 28°C. Plant inoculations were performed as described previously (Gillissen et al., 1992) by syringe‐assisted leaf‐whorl inoculation of fungal cell suspensions in water at a calculated optical density at 600 nm of 2.0. Disease rating was done at 13 dpi as described (Kämper et al., 2006) by scoring only the strongest symptom displayed by the inoculated plant. The severity of the disease symptoms on each inoculated plant was scored using a 0 to 6 rating scale, where 0 = no visible chlorosis or symptoms, 1 = chlorosis, 2 = small leaf tumours (<1 mm), 3 = larger leaf tumours (>1 mm), 4 = stem tumours (on leaf and stem >1 mm), 5 = heavy tumours (change of axis of growth), and 6 = dead plant. The statistical analysis was performed by comparing the disease scores between mutant and its WT via Student's t test. The data shown in this study are presented using 100% stacked bar charts.
4.5. Phytohormone and oxylipin profiling
For hormone and oxylipin profiling in maize leaves in response to pathogen infection, a 2‐cm region with six infection sites on the third leaf were harvested in 2‐ml screw‐cap Fast‐Prep tubes (Qbiogene) in liquid nitrogen. Zirconia beads and 500 μl of phytohormone extraction buffer (1‐propanol/water/HCl [2:1:0.002 vol/vol/vol]) containing 5 μM internal standards of d‐JA (2,4,4‐d3; acetyl‐2,2‐d2 JA; CDN Isotopes) and d6‐SA (Sigma‐Aldrich) were added into each tube and homogenized and agitated for 30 min at 4°C under darkness. Then, 500 μl of dichloromethane was added and agitated for another 30 min at 4°C under darkness then centrifuged for 10 min at 15,871 × g. The bottom organic phase was transferred into 1.8‐ml glass vials (VWR International), evaporated by continuous N2 gas, and then dissolved in 150 μl of methanol. The dissolved samples were transferred into a 1.5‐ml microcentrifuge tube and centrifuged at 15,871 × g for 5 min then 100 μl of the sample was transferred into autosampler vials with glass inserts for liquid chromatography‐tandem mass spectrometry (LC–MS/MS). For each sample, a 10 μl aliquot was injected into an API 3200 LC–MS/MS system (Sciex) connected to an Ascentis Express C‐8 column (3 cm × 2.1 mm, 2.7 μm) (Sigma‐Aldrich) using electrospray ionization in negative mode with multiple reaction monitoring. This is a highly specific and sensitive mass spectrometry technique that selectively quantifies compounds within complex mixtures. The mobile phase was set at 600 μl/min consisting of solution A (0.2% acetic acid in water) and solution B (0.2% acetic acid in acetonitrile) with a gradient consisting of 0.5 min 10% B, 1 min 20% B, 21 min 70% B, 24.6 min 100% B, and 29 min end. Integration was done with Analyst v. 1.6.3 (Sciex). All hormones and oxylipins were quantified by comparing levels of endogenous metabolites to isotopically labelled standards with appropriate response factors (Müller & Munn‐Bosch, 2011). The amount of 12‐OPDA presented in this study represents the levels of cis‐(+)‐12‐OPDA because cis‐(−)‐12‐OPDA is not found in plants and the endogenous cis‐12‐OPDA is exclusively the (+)‐enentiomer (Laudert et al., 1997; Stelmach et al., 1998). The data shown in this study are presented using bar charts in which the bars represent mean ± SEM.
4.6. GLV measurement
GLV measurement was performed following the methods described in He et al. (2020) and Gorman et al. (2021). In brief, leaves of the opr2‐1 mutant and its WT were infected with C. graminicola for 7 days and excised for imaging, with a delay of approximately 40 min between scanning and volatile collection. Then, leaves were cut into 1‐cm pieces and immediately placed into 800‐ml jars for volatile collection. HaySepQ filter traps containing 80–100 mesh (Supelco) were used to collect volatiles via a dynamic airflow that was approximately 1.5 L/min filtered by activated carbon while entering the system. Volatiles were collected for 30 min and eluted off the HaySepQ trap with 250 μl of dichloromethane containing 100 μM of the (Z)‐4‐hexenol as internal standard. Samples were then analysed via gas chromatography‐mass spectrometry. An Agilent 7890B gas chromatograph connected to a 5977B quadrupole mass spectrometer (Agilent) was used to detect volatiles. Two microlitres of sample was injected splitless into a HP‐5 ms Ultra Inert column (Agilent). The inlet temperature was set to 240°C for the duration of the run. The oven temperature was as follows: 40°C hold for 2 min, 3°C/min ramp to 160°C, 15°C/min ramp to 280°C, 280°C/min hold for 2 min. The solvent delay was 2.5 min. Analytes were fragmented by positive EI (230°C source, 150°C quadrupole, ionization energy 70 eV, scan range 25–500 amu). Most compounds were identified and quantified based on retention times and the spectra of pure external standards purchased from Sigma‐Aldrich. The compound tentatively referred to as (Z)‐2‐pentenal was identified by almost identical spectral matching to E‐2‐pentenal, and a close retention proximity characteristic of (E/Z)‐isomers of other lipoxygenase‐produced volatiles. All volatiles were quantified based on the use of internal and external standards. The data shown in this study are presented using stacked bar charts.
4.7. Statistical analysis
Statistically significant differences were determined by Student's t test for comparisons between two sample groups or one‐way analysis of variance (ANOVA) followed by Tukey's honestly significant differences (HSD) test (p < 0.05) using software JMP Pro v. 16 (SAS Institute Inc.).
AUTHOR CONTRIBUTIONS
P.‐C.H. and M.V.K. designed the research and analysed data. P.‐C.H. performed most of the experiments and drafted the article. M.T. helped with plant genotyping and ALB and SCLB disease assays. K.M.B.‐F. helped with oxylipin profiling and volatile measurement. S.A.C. and J.Z. contributed to backcrossing of opr2 mutants. J.S. performed the corn smut disease resistance assay. R.M. identified the Mu insertional alleles of OPR2. P.‐C.H. and M.V.K. wrote the article with input from and revisions by S.A.C. and J.S. There is no conflict of interest among authors of this study.
CONFLICT OF INTEREST STATEMENT
There is no conflict of interest among the authors of this study.
Supporting information
Figure S1 RNA‐Seq expression analysis of ZmOPR2 in different tissues of the maize inbred line B73 (Walley et al., 2016)
Figure S2 Accumulation of α‐ and γ‐ketols after Colletotrichum graminicola infection. Contents of (a) 9,10‐KOMA, (b) 9,10‐KODA, (c) 13,12‐KOMA, (d) 13,12‐KODA, (e) 13,10‐KOMA, (f) 13,10‐KODA, (g) 9,12‐KOMA, and (h) 9,12‐KODA were measured at 7 days postinoculation of the opr2‐1, opr2‐2, and opr2‐3 mutants and their wild type (WT). Bars are means ± SEM (n = 6 maize plants of each genotype as biological replicates). Asterisks represent statistically significant differences between WT and mutant. (Student’s t test, *p < 0.05, **p < 0.01)
Figure S3 Reverse transcription‐quantitative PCR analyses of the expression levels of jasmonic acid (JA)‐responsive ZmMyc7 and ZmJAZ1 in response to Colletotrichum graminicola infection. Expression of (a) ZmMyc7 and (b) ZmJAZ1 at 1, 3, and 5 days postinoculation (dpi) after C. graminicola infection compared to uninfected control B73 plants at day 0 in opr2‐1, lox10‐3, lox10opr2 mutants and their wild type (WT). Bars are means ± SEM (n = 5 maize plants of each genotype as biological replicates with two technical replicates per sample). Different letters indicate statistically significant differences among the samples (Tukey’s HSD test, p < 0.05)
Table S1 Primers used in this study
ACKNOWLEDGEMENTS
We thank Katja Zuther, University of Göttingen, Germany, for help with the U. maydis infection experiments, and Ivo Feussner, University of Göttingen, for greenhouse support. Thomas Isakeit, Texas A&M University, is acknowledged for providing us with the cultures of Cochliobolus heterostrophus local Texas isolate used in this study. We thank Katherine Berg for help with phytohormone and oxylipin profiling via LC–MS/MS. This study was supported by USDA‐NIFA 2017‐67013‐26524 and 2021‐67013‐33568 grants awarded to M.V.K. and the German Research Foundation (DFG) through the German Initiative of Excellence, grant number DFG ZUK45/1 to J.S.
Huang, P.‐C. , Tate, M. , Berg‐Falloure, K.M. , Christensen, S.A. , Zhang, J. , Schirawski, J. et al. (2023) A non‐JA producing oxophytodienoate reductase functions in salicylic acid‐mediated antagonism with jasmonic acid during pathogen attack. Molecular Plant Pathology, 24, 725–741. Available from: 10.1111/mpp.13299
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- Aerts, N. , Pereira Mendes, M. & Van Wees, S.C.M. (2021) Multiple levels of crosstalk in hormone networks regulating plant defense. The Plant Journal, 105, 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, G.K. , Jwa, N.S. , Shibato, J. , Han, O. , Iwahashi, H. & Rakwal, R. (2003) Diverse environmental cues transiently regulate OsOPR1 of the “octadecanoid pathway” revealing its importance in rice defense/stress and development. Biochemical and Biophysical Research Communications, 310, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Balmer, D. , de Papajewski, D.V. , Planchamp, C. , Glauser, G. & Mauch‐Mani, B. (2013) Induced resistance in maize is based on organ‐specific defence responses. The Plant Journal, 74, 213–225. [DOI] [PubMed] [Google Scholar]
- Banuett, F. & Herskowitz, I. (1996) Discrete developmental stages during teliospore formation in the corn smut fungus, Ustilago maydis . Development, 122, 2965–2976. [DOI] [PubMed] [Google Scholar]
- Basse, C.W. & Steinberg, G. (2004) Ustilago maydis, model system for analysis of the molecular basis of fungal pathogenicity. Molecular Plant Pathology, 5, 83–92. [DOI] [PubMed] [Google Scholar]
- Belisário, R. , Robertson, A.E. & Vaillancourt, L.J. (2022) Maize anthracnose stalk rot in the genomic era. Plant Disease, 106, 2281–2298. [DOI] [PubMed] [Google Scholar]
- Bergstrom, G.C. & Nicholson, R.L. (1999) The biology of corn anthracnose: knowledge to exploit for improved management. Plant Disease, 83, 596–608. [DOI] [PubMed] [Google Scholar]
- Biesgen, C. & Weiler, E.W. (1999) Structure and regulation of OPR1 and OPR2, two closely related genes encoding 12‐oxophytodienoic acid‐10, 11‐reductases from Arabidopsis thaliana . Planta, 208, 155–165. [DOI] [PubMed] [Google Scholar]
- Boatwright, J.L. & Pajerowska‐Mukhtar, K. (2013) Salicylic acid: an old hormone up to new tricks. Molecular Plant Pathology, 14, 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bölker, M. , Böhnert, H.U. , Braun, K.H. , Görl, J. & Kahmann, R. (1995) Tagging pathogenicity genes in Ustilago maydis by restriction enzyme‐mediated integration (REMI). Molecular and General Genetics, 248, 547–552. [DOI] [PubMed] [Google Scholar]
- Borrego, E.J. & Kolomiets, M.V. (2016) Synthesis and functions of jasmonates in maize. Plants, 5, 41–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughton, A.J. , Hoover, K. & Felton, G.W. (2005) Methyl jasmonate application induces increased densities of glandular trichomes on tomato, Lycopersicon esculentum . Journal of Chemical Ecology, 31, 2211–2216. [DOI] [PubMed] [Google Scholar]
- Brodersen, P. , Petersen, M. , Bjørn Nielsen, H. , Zhu, S. , Newman, M.A. , Shokat, K.M. et al. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid‐ and jasmonic acid/ethylene‐dependent responses via EDS1 and PAD4. The Plant Journal, 47, 532–546. [DOI] [PubMed] [Google Scholar]
- Browse, J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annual Review of Plant Biology, 60, 183–205. [DOI] [PubMed] [Google Scholar]
- Bruns, H.A. (2017) Southern corn leaf blight: a story worth retelling. Agronomy Journal, 109, 1218–1224. [Google Scholar]
- Callow, J.A. & Ling, I.T. (1973) Histology of neoplasms and chlorotic lesions in maize seedlings following the injection of sporidia of Ustilago maydis (DC) corda. Physiological Plant Pathology, 3, 489–494. [Google Scholar]
- Christensen, S.A. , Nemchenko, A. , Borrego, E. , Murray, I. , Sobhy, I.S. , Bosak, L. et al. (2013) The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore‐induced plant volatile production for defense against insect attack. The Plant Journal, 74, 59–73. [DOI] [PubMed] [Google Scholar]
- Christensen, S.A. , Nemchenko, A. , Park, Y.S. , Borrego, E. , Huang, P.C. , Schmelz, E.A. et al. (2014) The novel monocot‐specific 9‐lipoxygenase ZmLOX12 is required to mount an effective jasmonate‐mediated defense against Fusarium verticillioides in maize. Molecular Plant‐Microbe Interactions, 27, 1263–1276. [DOI] [PubMed] [Google Scholar]
- Crop Protection Network . (2021) Tools: yield loss calculator .
- Dai, Y.L. , Yang, X.J. , Gan, L. , Chen, F.R. , Ruan, H.C. , Du, Y.X. et al. (2016) First report of southern leaf blight caused by Cochliobolus heterostrophus on corn (Zea mays) in Fujian Province, China. Plant Disease, 100, 1781. [Google Scholar]
- Darino, M. , Chia, K.‐S. , Marques, J. , Aleksza, D. , Soto‐Jiménez, L.M. , Saado, I. et al. (2021) Ustilago maydis effector Jsi1 interacts with topless corepressor, hijacking plant jasmonate/ethylene signaling. New Phytologist, 229, 3393–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R. , Van Kan, J.A.L. , Pretorius, Z.A. , Hammond‐Kosack, K.E. , Di Pietro, A. , Spanu, P.D. et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleon, A.M. , Fengler, K.A. , Thatcher, S. & Wolters, P.J. (2021) Methods of identifying, selecting, and producing anthracnose stalk rot resistant crops. World Intellectual Property Organization, patent no. WO 2021/041077. [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. , Ghosh, A. , Vincon, V. , Kahnt, J. et al. (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Wahl, R. , Horst, R.J. , Voll, L.M. , Usadel, B. , Poree, F. et al. (2008a) Reprogramming a maize plant: transcriptional and metabolic changes induced by the fungal biotroph Ustilago maydis . The Plant Journal, 56, 181–195. [DOI] [PubMed] [Google Scholar]
- Doehlemann, G. , Wahl, R. , Vranes, M. , de Vries, R.P. , Kämper, J. & Kahmann, R. (2008b) Establishment of compatibility in the Ustilago maydis/maize pathosystem. Journal of Plant Physiology, 165, 29–40. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E. , Alméras, E. & Krishnamurthy, V. (2003) Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Current Opinion in Plant Biology, 6, 372–378. [DOI] [PubMed] [Google Scholar]
- Feussner, I. & Wasternack, C. (2002) The lipoxygenase pathway. Annual Review of Plant Biology, 53, 275–297. [DOI] [PubMed] [Google Scholar]
- Forestan, C. , Aiese Cigliano, R. , Farinati, S. , Lunardon, A. , Sanseverino, W. & Varotto, S. (2016) Stress‐induced and epigenetic‐mediated maize transcriptome regulation study by means of transcriptome reannotation and differential expression analysis. Scientific Reports, 6, 30446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, T.J. , Weldekidan, T. , Colbert, T. , Wolters, P.J.C.C. & Hawk, J.A. (2011) Fitness evaluation of Rcg1, a locus that confers resistance to Colletotrichum graminicola (Ces.) G.W. Wils. using near‐isogenic maize hybrids. Crop Science, 51, 1551–1563. [Google Scholar]
- Gao, X. , Shim, W.B. , Göbel, C. , Kunze, S. , Feussner, I. , Meeley, R. et al. (2007) Disruption of a maize 9‐lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Molecular Plant‐Microbe Interactions, 20, 922–933. [DOI] [PubMed] [Google Scholar]
- Gasperini, D. , Chételat, A. , Acosta, I.F. , Goossens, J. , Pauwels, L. , Goossens, A. et al. (2015) Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genetics, 11, e1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillissen, B. , Bergemann, J. , Sandmann, C. , Schroeer, B. , Bölker, M. & Kahmann, R. (1992) A two‐component regulatory system for self/non‐self recognition in Ustilago maydis . Cell, 68, 647–657. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology, 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gorman, Z. , Christensen, S.A. , Yan, Y. , He, Y. , Borrego, E. & Kolomiets, M.V. (2020) Green leaf volatiles and jasmonic acid enhance susceptibility to anthracnose diseases caused by Colletotrichum graminicola in maize. Molecular Plant Pathology, 21, 702–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman, Z. , Tolley, J.P. , Koiwa, H. & Kolomiets, M.V. (2021) The synthesis of pentyl leaf volatiles and their role in resistance to anthracnose leaf blight. Frontiers in Plant Science, 12, 719587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang, Y. , Luo, S. , Ahammed, G.J. , Xiao, X. , Li, J. , Zhou, Y. et al. (2021) The OPR gene family in watermelon: genome‐wide identification and expression profiling under hormone treatments and root‐knot nematode infection. Plant Biology, 23(Suppl 1), 80–88. [DOI] [PubMed] [Google Scholar]
- He, Y. , Borrego, E.J. , Gorman, Z. , Huang, P.C. & Kolomiets, M.V. (2020) Relative contribution of LOX10, green leaf volatiles and JA to wound‐induced local and systemic oxylipin and hormone signature in Zea mays (maize). Phytochemistry, 174, 112334. [DOI] [PubMed] [Google Scholar]
- Hu, Y. , Jiang, Y. , Han, X. , Wang, H. , Pan, J. & Yu, D. (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. Journal of Experimental Botany, 68, 1361–1369. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S. , Kawai‐Oda, A. , Ueda, J. , Nishida, I. & Okada, K. (2001) The defective in anther dehiscience gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis . The Plant Cell, 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, R. , Wang, M. , Sun, Q. , Sylvester, A.W. , Hake, S. & Scanlon, M.J. (2014) Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. The Plant Cell, 26, 4718–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, P. , Shanklin, J. , Shah, J. , Whittle, E.J. & Klessig, D.F. (2001) A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proceedings of the National Academy of Sciences of the United States of America, 98, 9448–9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu, A. , Ambavaram, M.M.R. , Klumas, C. , Krishnan, A. , Batlang, U. , Myers, E. et al. (2012) Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA‐seq. Plant Physiology, 160, 846–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämper, J. , Kahmann, R. , Bölker, M. , Ma, L.‐J. , Brefort, T. , Saville, B.J. et al. (2006) Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature, 444, 97–101. [DOI] [PubMed] [Google Scholar]
- Kazan, K. & Manners, J.M. (2008) Jasmonate signaling: toward an integrated view. Plant Physiology, 146, 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli, R.M. & Massey, V. (1998) The oxidative half‐reaction of old yellow enzyme. The role of tyrosine 196. The Journal of Biological Chemistry, 273, 32763–32770. [DOI] [PubMed] [Google Scholar]
- Koornneef, A. & Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiology, 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, D. (2014) Salicylic acid signaling in disease resistance. Plant Science, 228, 127–134. [DOI] [PubMed] [Google Scholar]
- Lanver, D. , Tollot, M. , Schweizer, G. , Lo Presti, L. , Reissmann, S. , Ma, L.‐S. et al. (2017) Ustilago maydis effectors and their impact on virulence. Nature Reviews Microbiology, 15, 409–421. [DOI] [PubMed] [Google Scholar]
- Laudert, D. , Hennig, P. , Stelmach, B.A. , Müller, A. , Andert, L. & Weiler, E.W. (1997) Analysis of 12‐oxo‐phytodienoic acid enantiomers in biological samples by capillary gas chromatography‐mass spectrometry using cyclodextrin stationary phases. Analytical Biochemistry, 246, 211–217. [DOI] [PubMed] [Google Scholar]
- Li, J. , Brader, G. & Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. The Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Zhou, F. , Liu, B. , Feng, D. , He, Y. , Qi, K. et al. (2011) Comparative characterization, expression pattern and function analysis of the 12‐oxo‐phytodienoic acid reductase gene family in rice. Plant Cell Reports, 30, 981–995. [DOI] [PubMed] [Google Scholar]
- Linkies, A. & Leubner‐Metzger, G. (2012) Beyond gibberellins and abscisic acid: how ethylene and jasmonates control seed germination. Plant Cell Reports, 31, 253–270. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Sun, R. , Zhang, X. , Feng, Z. , Wei, F. , Zhao, L. et al. (2020) Genome‐wide analysis of OPR family genes in cotton identified a role for GhOPR9 in Verticillium dahliae resistance. Genes, 11, 1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L. , Sun, Y. , Ruan, X. , Huang, P.C. , Wang, S. , Li, S. et al. (2021) Genome‐wide characterization of jasmonates signaling components reveals the essential role of ZmCOI1a‐ZmJAZ15 action module in regulating maize immunity to Gibberella stalk rot. International Journal of Molecular Sciences, 22, 870–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoli, A. , Sturaro, A. , Trevisan, S. , Quaggiotti, S. & Nonis, A. (2012) Evaluation of candidate reference genes for qPCR in maize. Journal of Plant Physiology, 169, 807–815. [DOI] [PubMed] [Google Scholar]
- McCarty, D.R. , Settles, A.M. , Suzuki, M. , Tan, B.C. , Latshaw, S. , Porch, T. et al. (2005) Steady‐state transposon mutagenesis in inbred maize. The Plant Journal, 44, 52–61. [DOI] [PubMed] [Google Scholar]
- Meeley, R. & Briggs, S. (1995) Reverse genetics for maize. Maize Newsletter, 69, 67–82. [Google Scholar]
- Mendgen, K. , Hahn, M. & Deising, H. (1996) Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annual Review of Phytopathology, 34, 367–386. [DOI] [PubMed] [Google Scholar]
- Mims, C.W. & Vaillancourt, L.J. (2002) Ultrastructural characterization of infection and colonization of maize leaves by Colletotrichum graminicola, and by a C. graminicola pathogenicity mutant. Phytopathology, 92, 803–812. [DOI] [PubMed] [Google Scholar]
- Miranda, V.D.J. , Porto, W.F. , Fernandes, G.D.R. , Pogue, R. , Nolasco, D.O. , Araujo, A.C.G. et al. (2017) Comparative transcriptomic analysis indicates genes associated with local and systemic resistance to Colletotrichum graminicola in maize. Scientific Reports, 7, 2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou, Y. , Liu, Y. , Tian, S. , Guo, Q. , Wang, C. & Wen, S. (2019) Genome‐wide identification and characterization of the OPR gene family in wheat (Triticum aestivum L.). International Journal of Molecular Sciences, 20, 1914–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M. & Munné‐Bosch, S. (2011) Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods, 7, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong, I. , Abdallat, A.A. , Thurow, C. , Fode, B. , Zander, M. , Weigel, R. et al. (2007) SA‐inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA‐responsive PDF1.2 transcription. The Plant Journal, 50, 128–139. [DOI] [PubMed] [Google Scholar]
- Ngoko, Z. , Cardwell, K.F. , Marasas, W.F.O. , Wingfield, M.J. , Ndemah, R. & Schulthess, F. (2002) Biological and physical constraints on maize production in the humid forest and Western highlands of Cameroon. European Journal of Plant Pathology, 108, 893–902. [Google Scholar]
- O'Connell, R.J. , Bailey, J.A. & Richmond, D.V. (1985) Cytology and physiology of infection of Phaseolus vulgaris by Colletotrichum lindemuthianum . Physiological Plant Pathology, 27, 75–98. [Google Scholar]
- Pan, J. , Hu, Y. , Wang, H. , Guo, Q. , Chen, Y. , Howe, G.A. et al. (2020) Molecular mechanism underlying the synergetic effect of Jasmonate on abscisic acid signaling during seed germination in Arabidopsis . The Plant Cell, 32, 3846–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. & Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nature Chemical Biology, 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Qi, T. , Wang, J. , Huang, H. , Liu, B. , Gao, H. , Liu, Y. et al. (2015) Regulation of jasmonate‐induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis . The Plant Cell, 27, 1634–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe, F. , Ajami‐Rashidi, Z. , Doehlemann, G. , Kahmann, R. & Djamei, A. (2013) Degradation of the plant defence hormone salicylic acid by the biotrophic fungus Ustilago maydis . Molecular Microbiology, 89, 179–188. [DOI] [PubMed] [Google Scholar]
- Rao, X. , Huang, X. , Zhou, Z. & Lin, X. (2013) An improvement of the 2^(−delta delta CT) method for quantitative real‐time polymerase chain reaction data analysis. Biostatistics, Bioinformatics and Biomathematics, 3, 71–85. [PMC free article] [PubMed] [Google Scholar]
- Saito, H. , Oikawa, T. , Hamamoto, S. , Ishimaru, Y. , Kanamori‐Sato, M. , Sasaki‐Sekimoto, Y. et al. (2015) The jasmonate‐responsive GTR1 transporter is required for gibberellin‐mediated stamen development in Arabidopsis . Nature Communications, 6, 6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, P.M. , Lee, P.Y. , Biesgen, C. , Boone, J.D. , Beals, T.P. , Weiler, E.W. et al. (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. The Plant Cell, 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, F. (2001) Enzymes of the biosynthesis of octadecanoid‐derived signalling molecules. Journal of Experimental Botany, 52, 11–23. [PubMed] [Google Scholar]
- Schaller, F. , Hennig, P. & Weiler, E.W. (1998) 12‐Oxophytodienoate‐10,11‐reductase: occurrence of two isoenzymes of different specificity against stereoisomers of 12‐oxophytodienoic acid. Plant Physiology, 118, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, F. , Biesgen, C. , Müssig, C. , Altmann, T. & Weiler, E.W. (2000) 12‐oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta, 210, 979–984. [DOI] [PubMed] [Google Scholar]
- Schirawski, J. , Mannhaupt, G. , Münch, K. , Brefort, T. , Schipper, K. , Doehlemann, G. et al. (2010) Pathogenicity determinants in smut fungi revealed by genome comparison. Science, 330, 1546–1548. [DOI] [PubMed] [Google Scholar]
- Schommer, C. , Palatnik, J.F. , Aggarwal, P. , Chételat, A. , Cubas, P. , Farmer, E.E. et al. (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology, 6, e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. & Sricastava, R.P. (2012) Southern corn leaf blight ‐ an important disease of maize: an extension fact sheet. Indian Research Journal of Extension Education Special Issue, I, 334–337. [Google Scholar]
- Singh, P. , Dave, A. , Vaistij, F.E. , Worrall, D. , Holroyd, G.H. , Wells, J.G. et al. (2017) Jasmonic acid‐dependent regulation of seed dormancy following maternal herbivory in Arabidopsis . New Phytologist, 214, 1702–1711. [DOI] [PubMed] [Google Scholar]
- Sirhindi, G. , Mushtaq, R. , Gill, S.S. , Sharma, P. , Abd_Allah, E.F. & Ahmad, P. (2020) Jasmonic acid and methyl jasmonate modulate growth, photosynthetic activity and expression of photosystem II subunit genes in Brassica oleracea L. Scientific Reports, 10, 9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C. , Descour, A. , Kudrna, D. , Bomhoff, M. , Boyd, L. , Currie, J. et al. (2009) Sequencing, mapping, and analysis of 27,455 maize full‐length cDNAs. PLoS Genetics, 5, e1000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. & Dong, X. (2008) Making sense of hormone crosstalk during plant immune responses. Cell Host & Microbe, 3, 348–351. [DOI] [PubMed] [Google Scholar]
- Spoel, S.H. , Koornneef, A. , Claessens, S.M. , Korzelius, J.P. , Van Pelt, J.A. , Mueller, M.J. et al. (2003) NPR1 modulates cross‐talk between salicylate‐ and jasmonate‐dependent defense pathways through a novel function in the cytosol. The Plant Cell, 15, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelmach, B.A. , Müller, A. , Hennig, P. , Laudert, D. , Andert, L. & Weiler, E.W. (1998) Quantitation of the octadecanoid 12‐oxo‐phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry, 47, 539–546. [DOI] [PubMed] [Google Scholar]
- Stelpflug, S.C. , Sekhon, R.S. , Vaillancourt, B. , Hirsch, C.N. , Buell, C.R. , de Leon, N. et al. (2016) An expanded maize gene expression atlas bbased on RNA sequencing and its use to explore root development. The Plant Genome, 9. Available from: 10.3835/plantgenome2015.04.0025 [DOI] [PubMed] [Google Scholar]
- Stintzi, A. & Browse, J. (2000) The Arabidopsis male‐sterile mutant, opr3, lacks the 12‐oxophytodienoic acid reductase required for jasmonate synthesis. Proceedings of the National Academy of Sciences of the United States of America, 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner, J. , Fürholz, A. , Macheroux, P. , Amrhein, N. & Schaller, A. (1999) A homolog of old yellow enzyme in tomato. Spectral properties and substrate specificity of the recombinant protein. The Journal of Biological Chemistry, 274, 35067–35073. [DOI] [PubMed] [Google Scholar]
- Strassner, J. , Schaller, F. , Frick, U.B. , Howe, G.A. , Weiler, E.W. , Amrhein, N. et al. (2002) Characterization and cDNA‐microarray expression analysis of 12‐oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. The Plant Journal, 32, 585–601. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K. , Allmann, S. & Kolomiets, M.V. (2022) Editorial: oxylipins: the front line of plant interactions. Frontiers in Plant Science, 13, 878765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani, T. , Sobajima, H. , Okada, K. , Chujo, T. , Arimura, S. , Tsutsumi, N. et al. (2008) Identification of the OsOPR7 gene encoding 12‐oxophytodienoate reductase involved in the biosynthesis of jasmonic acid in rice. Planta, 227, 517–526. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Penninckx, I.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P. et al. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences of the United States of America, 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley, J.P. , Nagashima, Y. , Gorman, Z. , Kolomiets, M.V. & Koiwa, H. (2018) Isoform‐specific subcellular localization of Zea mays lipoxygenases and oxo‐phytodienoate reductase 2. Plant Gene, 13, 36–41. [Google Scholar]
- Traw, M.B. & Bergelson, J. (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis . Plant Physiology, 133, 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon, B.G. & Baker, S.E. (2007) Genetic and genomic dissection of the Cochliobolus heterostrophus Tox1 locus controlling biosynthesis of the polyketide virulence factor T‐toxin. Advances in Genetics, 57, 219–261. [DOI] [PubMed] [Google Scholar]
- Turner, J.G. , Ellis, C. & Devoto, A. (2002) The jasmonate signal pathway. The Plant Cell, 14(Suppl), S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas, W.A. , Martín, J.M.S. , Rech, G.E. , Rivera, L.P. , Benito, E.P. , Díaz‐Mínguez, J.M. et al. (2012) Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiology, 158, 1342–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick, B.A. & Zimmerman, D.C. (1984) Biosynthesis of jasmonic acid by several plant species. Plant Physiology, 75, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley, J.W. , Sartor, R.C. , Shen, Z. , Schmitz, R.J. , Wu, K.J. , Urich, M.A. et al. (2016) Integration of omic networks in a developmental atlas of maize. Science, 353, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.‐Q. , Ma, J. , Wang, M. , Wang, X.‐H. , Li, Y.‐Q. & Chen, J. (2019) Combined application of Trichoderma harzianum SH2303 and difenoconazole‐propiconazolein controlling southern corn leaf blight disease caused by Cochliobolus heterostrophus in maize. Journal of Integrative Agriculture, 18, 2063–2071. [Google Scholar]
- Wang, K.D. , Borrego, E.J. , Kenerley, C.M. & Kolomiets, M.V. (2020a) Oxylipins other than Jasmonic acid are xylem‐resident signals regulating systemic resistance induced by Trichoderma virens in maize. The Plant Cell, 32, 166–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K.D. , Gorman, Z. , Huang, P.C. , Kenerley, C.M. & Kolomiets, M.V. (2020b) Trichoderma virens colonization of maize roots triggers rapid accumulation of 12‐oxophytodienoate and two ᵧ‐ketols in leaves as priming agents of induced systemic resistance. Plant Signaling & Behavior, 15, 1792187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Sun, Y. , Wang, F. , Huang, P.C. , Wang, Y. , Ruan, X. et al. (2021) Transcriptome and oxylipin profiling joint analysis reveals opposite roles of 9‐oxylipins and jasmonic acid in maize resistance to Gibberella stalk rot. Frontiers in Plant Science, 12, 699146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany, 100, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. & Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany, 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. & Strnad, M. (2016) Jasmonate signaling in plant stress responses and development ‐ active and inactive compounds. New Biotechnology, 33, 604–613. [DOI] [PubMed] [Google Scholar]
- Waters, A.J. , Makarevitch, I. , Noshay, J. , Burghardt, L.T. , Hirsch, C.N. , Hirsch, C.D. et al. (2017) Natural variation for gene expression responses to abiotic stress in maize. The Plant Journal, 89, 706–717. [DOI] [PubMed] [Google Scholar]
- Wharton, P.S. , Julian, A.M. & O'Connell, R.J. (2001) Ultrastructure of the infection of Sorghum bicolor by Colletotrichum sublineolum . Phytopathology, 91, 149–158. [DOI] [PubMed] [Google Scholar]
- White, D.G. (1999) Compendium of corn diseases. St Paul, MN: American Phytopathological Society. [Google Scholar]
- Wu, D. , Oide, S. , Zhang, N. , Choi, M.Y. & Turgeon, B.G. (2012) ChLae1 and ChVel1 regulate T‐toxin production, virulence, oxidative stress response, and development of the maize pathogen Cochliobolus heterostrophus . PLoS Pathogens, 8, e1002542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y. , Christensen, S. , Isakeit, T. , Engelberth, J. , Meeley, R. , Hayward, A. et al. (2012) Disruption of OPR7 and OPR8 reveals the versatile functions of jasmonic acid in maize development and defense. The Plant Cell, 24, 1420–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y. , Huang, P.‐C. , Borrego, E. & Kolomiets, M. (2014) New perspectives into jasmonate roles in maize. Plant Signaling & Behavior, 9, e970442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Simmons, C. , Yalpani, N. , Crane, V. , Wilkinson, H. & Kolomiets, M. (2005) Genomic analysis of the 12‐oxo‐phytodienoic acid reductase gene family of Zea mays . Plant Molecular Biology, 59, 323–343. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Jia, X. , Wang, G.F. , Ma, S. , Wang, S. , Yang, Q. et al. (2022) Ascorbate peroxidase 1 confers resistance to southern corn leaf blight in maize. Journal of Integrative Plant Biology, 64, 1196–1211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 RNA‐Seq expression analysis of ZmOPR2 in different tissues of the maize inbred line B73 (Walley et al., 2016)
Figure S2 Accumulation of α‐ and γ‐ketols after Colletotrichum graminicola infection. Contents of (a) 9,10‐KOMA, (b) 9,10‐KODA, (c) 13,12‐KOMA, (d) 13,12‐KODA, (e) 13,10‐KOMA, (f) 13,10‐KODA, (g) 9,12‐KOMA, and (h) 9,12‐KODA were measured at 7 days postinoculation of the opr2‐1, opr2‐2, and opr2‐3 mutants and their wild type (WT). Bars are means ± SEM (n = 6 maize plants of each genotype as biological replicates). Asterisks represent statistically significant differences between WT and mutant. (Student’s t test, *p < 0.05, **p < 0.01)
Figure S3 Reverse transcription‐quantitative PCR analyses of the expression levels of jasmonic acid (JA)‐responsive ZmMyc7 and ZmJAZ1 in response to Colletotrichum graminicola infection. Expression of (a) ZmMyc7 and (b) ZmJAZ1 at 1, 3, and 5 days postinoculation (dpi) after C. graminicola infection compared to uninfected control B73 plants at day 0 in opr2‐1, lox10‐3, lox10opr2 mutants and their wild type (WT). Bars are means ± SEM (n = 5 maize plants of each genotype as biological replicates with two technical replicates per sample). Different letters indicate statistically significant differences among the samples (Tukey’s HSD test, p < 0.05)
Table S1 Primers used in this study
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.


