Abstract
Fusarium ear rot (FER) is a destructive fungal disease of maize caused by Fusarium verticillioides. FER resistance is a typical complex quantitative trait controlled by micro‐effect genes, leading to difficulty in identifying the host resistance genes. SIZ1 encodes a SUMO E3 ligase regulating a wide range of plant developmental processes and stress responses. However, the function of ZmSIZ1 remains poorly understood. In this study, we demonstrate that ZmSIZ1a and ZmSIZ1b possess SUMO E3 ligase activity, and that the Zmsiz1a/1b double mutant, but not the Zmsiz1a or Zmsiz1b single mutants, exhibits severely impaired resistance to FER. Transcriptome analysis showed that differentially expressed genes were significantly enriched in plant disease resistance‐related pathways, especially in plant–pathogen interaction, MAPK signalling, and plant hormone signal transduction. Thirty‐five candidate genes were identified in these pathways. Furthermore, the integration of the transcriptome and metabolome data revealed that the flavonoid biosynthesis pathway was induced by F. verticillioides infection, and that accumulation of flavone and flavonol was significantly reduced in the Zmsiz1a/1b double mutant. Collectively, our findings demonstrate that ZmSIZ1a and ZmSIZ1b play a redundant, but indispensable role against FER, and provide potential new gene resources for molecular breeding of FER‐resistant maize cultivars.
Keywords: flavonoid, Fusarium ear rot (FER), maize (Zea mays), SUMO E3 ligase, ZmSIZ1a/1b
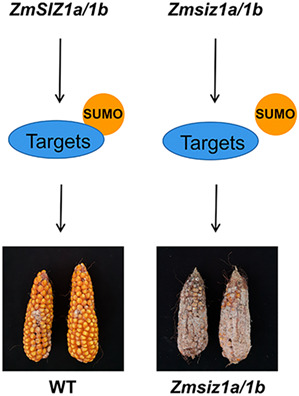
ZmSIZ1a and ZmSIZ1b are involved in the resistance to Fusarium ear rot in maize via the regulation of multiple pathways, including early defence responses and the synthesis of flavonoid metabolites.
1. INTRODUCTION
Maize ear rot, mainly caused by a variety of fungi, is one of the most destructive diseases of maize in the world. Maize ear rot not only significantly reduces yield, but the mycotoxins produced by the pathogen, such as aflatoxin (AF), fumonisin (FM), deoxynivalenol (DON), and zearalenone (ZEA), also pose a serious health risk to humans and livestock (van Egmond et al., 2007; Missmer et al., 2006). While maize ear rot can be partially reduced by improved agronomic management, such as minimizing insect damage, early harvesting, and drying immediately after harvest, so far the most effective strategy is to identify resistant genotypes and develop resistant maize varieties.
Ear rot resistance is a typical complex quantitative trait that is mainly controlled by minor‐effect genes and strongly affected by the environment. For decades, researchers have performed numerous studies on the mechanisms of resistance to Fusarium ear rot (FER) and Gibberella ear rot (GER) in maize. Extensive quantitative trait locus (QTL) analyses have been conducted using different maize populations by many research groups, and the identified resistance QTLs are widespread over all 10 chromosomes of maize (Chen et al., 2012; Ding et al., 2008; Enrico Pè et al., 1993; Galiano‐Carneiro et al., 2020; Martin et al., 2012; Maschietto et al., 2017). Although some hotspot regions of QTLs have been identified, ZmAuxRP1 is the only gene that has been map‐based cloned and functionally validated as of now due to the generally low phenotypic variance explained by these resistance QTLs (Ye et al., 2019).
With technological advances, new approaches to mine candidate resistance genes have been exploited, such as genome‐wide association study (GWAS) and RNA sequencing (RNA‐Seq) analysis. Three and seven single‐nucleotide polymorphisms (SNPs) were identified using a population of 267 inbred lines and a population of 1687 US inbred lines, respectively (Zila et al., 2013, 2014). Recently, using a combination of GWAS and RNA‐Seq analysis, researchers have identified a number of loci and candidate genes for FER resistance, including cytochrome P450 family genes, phenylpropanoid pathway genes, heat shock protein genes, and plant hormone signal transduction genes (Chen et al., 2016; Yao et al., 2020). However, there are few genetic resources that can be directly used in ear rot resistance breeding. Identification of effective and broad‐spectrum ear rot resistance genes still remains a pressing challenge in maize breeding.
SUMOylation is a reversible posttranslational modification of eukaryotic proteins. During the SUMOylation process, the small ubiquitin‐like modifier (SUMO) molecule is attached to the substrate protein with the assistance of the E1 activating enzyme complex, the E2 binding enzyme, and the E3 ligase (Augustine & Vierstra, 2018; Novatchkova et al., 2012). Of these, the main role of SUMO E3 ligase is to specifically recognize the substrate protein and transfer SUMO molecules from the subunit of the E2 binding enzyme to the target protein. Previous studies have shown that SIZ1 encodes a class of SUMO E3 ligases with the SAP and MIZ domains, and that its homologues regulate a wide range of plant developmental process and stress responses, such as flowering time (Jin et al., 2008; Wang et al., 2011), nutrient signalling (Miura et al., 2005; Park et al., 2011), hormone signalling (Kim et al., 2015; Miura et al., 2009; Zheng et al., 2012), and abiotic stress responses and disease resistance (Catala et al., 2007; Miura et al., 2007, 2013; Zhang et al., 2017).
Maize contains three ZmSIZ1 genes, ZmSIZ1a, ZmSIZ1b, and ZmSIZ1c, and each of them is able to rescue the dwarf phenotype of Atsiz1 mutant plants (Lai et al., 2022). Moreover, in vitro experiments have shown that all three ZmSIZ1s can be auto‐SUMOylated by ZmSUMO1a, indicating their potential activity as SUMO E3 ligases (Lai et al., 2022). Further analysis showed that ZmSIZ1a and ZmSIZ1b, but not ZmSIZ1c, were involved in the response to multiple stresses, such as salt, heat, drought, and abscisic acid (ABA) treatment, suggesting that there is some functional differentiation between ZmSIZ1a/1b and ZmSIZ1c (Lai et al., 2022). Nevertheless, the function of theZmSIZ1a/1b genes has not been experimentally tested.
In this study, we generated the Zmsiz1a/Zmsiz1b (Zmsiz1a/1b) double mutant and two single mutants using the CRISPR/Cas9 technology. We verified the SUMO E3 ligase function of ZmSIZ1a and ZmSIZ1b in maize and revealed that they play pivotal roles in maize development and ear rot resistance. Furthermore, we identified potential pathways and mined candidate resistance genes against FER in maize using transcriptome and metabolome analyses. Our results provide insights into the molecular mechanism of FER resistance in maize.
2. RESULTS
2.1. Sequence analysis of ZmSIZ1a and ZmSIZ1b
Sequence analysis showed that all three ZmSIZ1 proteins contain the key functional domains of SUMO E3 ligase, including the SAP (scaffold attachment factor A/B/acinus/PIAS domain), PHD (plant homeodomain), PINIT (proline‐isoleucine‐asparagine‐isoleucine‐threonine), SP‐RING (SIZ/PIASRING), and SXS (serine‐X‐serine) domains (Figure 1a). ZmSIZ1a and ZmSIZ1b are more closely related to each other and they are clustered in the same branch in the phylogenetic tree (Figure 1a; Lai et al., 2022). Previous study revealed that ZmSIZ1a and ZmSIZ1b probably play a role in abiotic stress and defence responses in maize (Lai et al., 2022), so we selected them for the following functional studies.
FIGURE 1.
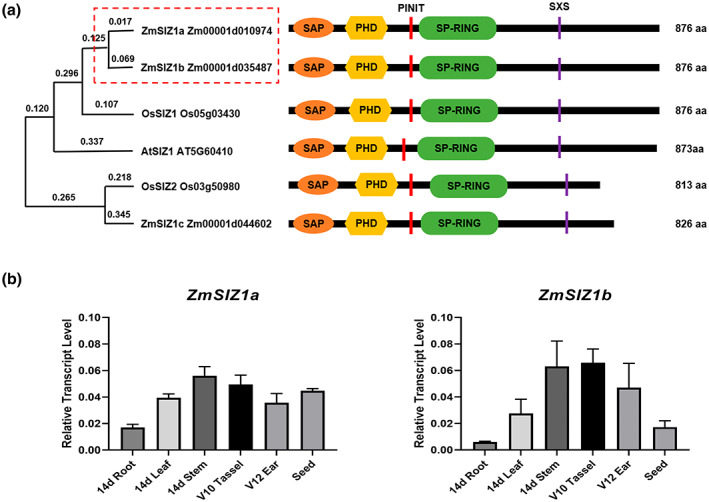
Sequence analysis and the expression patterns of ZmSIZ1a and ZmSIZ1b. (a) Phylogenetic tree of ZmSIZ1 and its Arabidopsis and rice homologues. ZmSIZ1a and ZmSIZ1b are marked with a red dashed rectangle. Five conserved domains in SIZ1 proteins are indicated as boxes and shown on the right. SAP, scaffold attachment factor A/B/acinus/PIAS domain; PHD, plant homeodomain; PINIT, proline‐isoleucine‐asparagine‐isoleucine‐threonine; SP‐RING, SIZ/PIASRING; SXS, serine‐X‐serine. (b) Relative expression levels of ZmSIZ1a and ZmSIZ1b in different tissues
2.2. ZmSIZ1a and ZmSIZ1b function as SUMO E3 ligases in maize
We first examined the expression profiles of ZmSIZ1a and ZmSIZ1b in different tissues of maize using reverse transcription‐quantitative PCR (RT‐qPCR) and found that both of them were highly expressed in maize stem, leaf, tassel, ear, and seed, while relatively low in the root (Figure 1b). To investigate the function of ZmSIZ1a and ZmSIZ1b in maize, we generated single knockout mutants of Zmsiz1a and Zmsiz1b, as well as Zmsiz1a/1b double mutants using the CRISPR/Cas9 technology in the maize inbred line ZC01 background (Figures 2a,b and S1a). We selected two target sites on two exons in the 5′ end of ZmSIZ1a and ZmSIZ1b to ensure disruption of their functional domains (Figure 2a). Mutants with large fragment deletion or frameshift mutations were selected. Previous studies showed that heat shock treatment can rapidly induce accumulation of SUMO conjugates in plants, and this accumulation is significantly reduced in the Arabidopsis siz1 mutant (Liu et al., 2015a; Park et al., 2010; Yoo et al., 2006). To further confirm the function of ZmSIZ1a and ZmSIZ1b as the SUMO E3 ligases in vivo, we examined the level of heat shock‐induced SUMO conjugates in the wild type and the Zmsiz1 mutants. The results showed that the amount of heat shock‐induced SUMO conjugates was signficantly reduced in the Zmsiz1a/1b double mutant, but not in the Zmsiz1a and Zmsiz1b single mutants (Figures 2c and S1b). These observations suggest that ZmSIZ1a and ZmSIZ1b probably play a redundant, but essential, role in mediating heat‐induced SUMOylation of substrate proteins.
FIGURE 2.
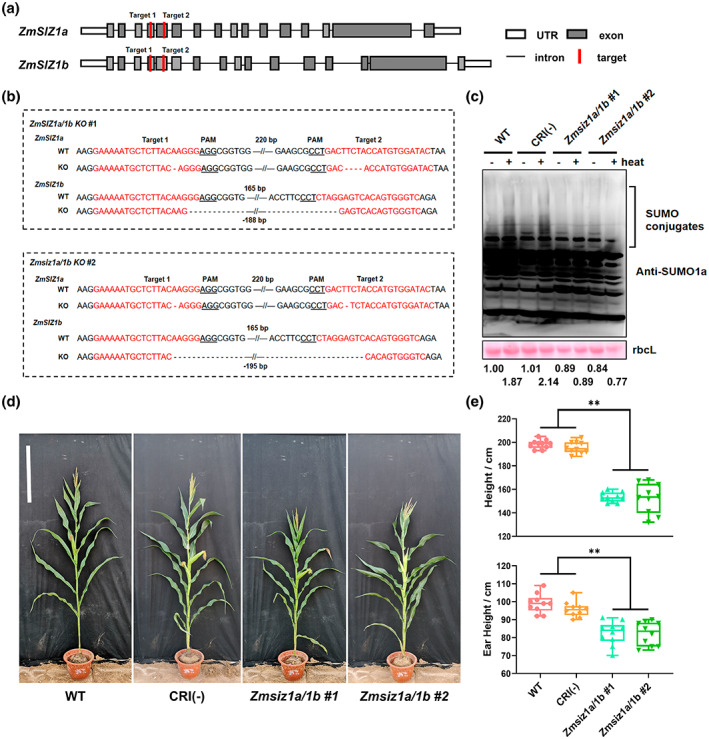
Identification of ZmSIZ1a/1b double knockout mutants. (a) Schematic diagram of two Cas9 targets on ZmSIZ1a and ZmSIZ1b. (b) Sequence analysis of the target sites (red) in two Zmsiz1a/1b double knockout lines. The wild‐type sequence (WT) is shown at the top, knockouts (KO) below. The protospacer‐adjacent motifs (PAM) are underlined. Deletions are indicated by dashes and the length of the sequence gap is shown. (c) The heat shock‐induced accumulation of SUMO conjugates was impaired in Zmsiz1a/1b double mutant seedlings. The immunoblot was probed with an anti‐AtSUMO1 antibody. Ponceau S‐stained RuBisCO large subunit (rbcL) bands are shown as a loading control. The numbers below the gel blot refer to the quantification of SUMO conjugates relative to the rbcL loading control and then normalizing to the WT without heat shock. CRI(−), the corresponding unedited control plants. (d) The morphologic phenotype of WT, CRI (−), and Zmsiz1a/1b double mutant lines. Bar = 50 cm. (e) Quantification of plant height and the ear height as in (d). Mean ± SD, n = 10, **p < 0.01, Student's t test
2.3. ZmSIZ1a and ZmSIZ1b are required for the resistance to FER
To examine the effects of ZmSIZ1a and ZmSIZ1b on plant development, we grew the Zmsiz1a/1b double mutant, and the Zmsiz1a and Zmsiz1b single mutants, together with their wild‐type plants, in Ledong (18°N, 116°E), Hainan province, in the winter of 2020. Phenotypic investigation showed that the Zmsiz1a/1b double mutants displayed significantly lower plant height and ear height in the field trial (Figure 2d,e), but the Zmsiz1a and Zmsiz1b single mutant plants had no significant difference from the wild type (Figure S1c,d). These observations suggest that ZmSIZ1a and ZmSIZ1b play a redundant role in regulating plant development in maize.
Strikingly, the ears of the Zmsiz1a/1b double mutant were heavily infested with fungal pathogens, but not the two single mutants or the wild type (Figure S2). These observations suggest that the Zmsiz1a/1b double mutant is probably prone to maize ear rot in the natural field conditions. To determine the nature of the fungal pathogen, we sequenced the internal transcribed spacer of RNA Pol1 (ITS) of the isolated fungi. Sequencing analysis revealed that the pathogens were mainly Fusarium and Penicillium (Appendix S1). Considering that Fusarium verticillioides is one of the main causal pathogens of maize ear rot, we selected F. verticillioides for the follow‐up experiments. To verify this phenotype, we analysed the transcript levels of ZmSIZ1a and ZmSIZ1b after artificial inoculation with F. verticillioides. Both ZmSIZ1a and ZmSIZ1b in kernels were rapidly induced 1.5 h after inoculation (Figure 3a). Furthermore, the kernel infection assay showed that more fungal mycelia and conidia were observed on the Zmsiz1a/1b double mutant kernels than on other variants at 72 h postinoculation (hpi) (Figure 3b,c). To further verify the role of ZmSIZ1a/1b in FER resistance, we planted the wild‐type and ZmSIZ1 mutant plants side by side in the field in Ledong and Langfang (39°N, 116°E, Hebei province, China) in the winter and summer of 2021, respectively. About 15 days after pollination, 2 ml of a F. verticillioides spore suspension containing 5 × 106 conidia/ml and 0.01% Tween 20 was injected into the middle of each ear using a continuous syringe. The results revealed that the resistance to FER was severely impaired in the Zmsiz1a/1b double mutants, but not in the Zmsiz1a and Zmsiz1b single mutant plants (Figure 3d,e). These results verified that ZmSIZ1a and ZmSIZ1b play a redundant, but indispensable, role in resistance to FER in maize.
FIGURE 3.
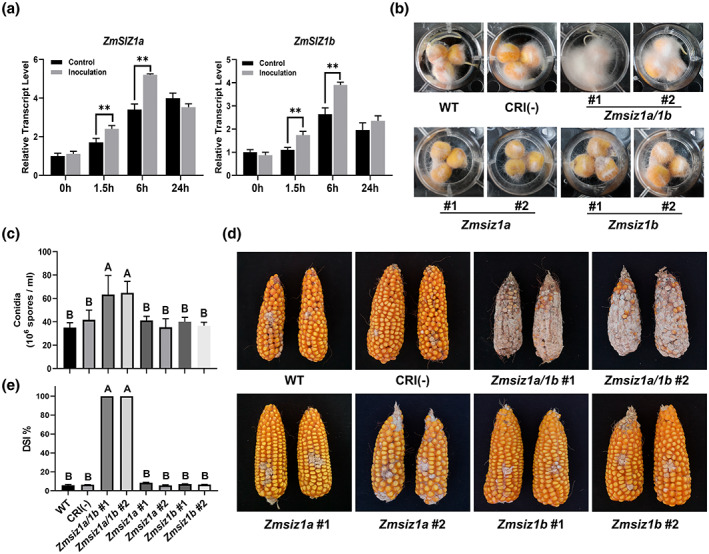
The Zmsiz1a/1b double mutant is more susceptible to Fusarium ear rot (FER). (a) Relative expression of ZmSIZ1a and ZmSIZ1b in wild‐type kernels at 0, 1.5, 6, and 24 h postinoculation. Mean ± SD, n = 3; **p < 0.01, Student's t test. (b) The visible colonization of Fusarium‐infected kernels. (c) The conidia count for the kernel infection analysis shown in (b). Mean ± SD, n = 3; significant differences are indicated by letters, p < 0.01, Tukey's multiple comparisons test. (d) The disease phenotypes of wild‐type plants (WT), unedited control plants (CRI(−)), double mutant lines, and single mutant lines after FER field inoculation. (e) The disease severity index (DSI) of plants indicated in (d). Mean ± SD, n = 20; significant differences are indicated by letters, p < 0.01, Tukey's multiple comparisons test
We also generated overexpression lines of ZmSIZ1a and ZmSIZ1b and examined their FER resistance. An inoculation assay showed that the ZmSIZ1a and ZmSIZ1b overexpression lines only displayed a marginal increase in FER resistance (fall below a significant level), compared to the wild‐type plants (Figure S3).
2.4. RNA‐Seq analysis of the Zmsiz1a/1b double mutant
The diminished resistance of the Zmsiz1a/1b double mutant to FER suggests that the ZmSIZ1a‐ and ZmSIZ1b‐mediated SUMOylation are involved in the regulation of resistance to FER. To identify the downstream genes potentially affected by the knockout of ZmSIZ1a and ZmSIZ1b, we performed RNA‐Seq analysis of the wild‐type and Zmsiz1a/1b double mutant (#1) kernels inoculated with or without F. verticillioides at 0, 1.5, 6, 24, and 72 hpi. In total, 1,164,081,642 clean reads were obtained, and an average of 76.8% of the reads could be mapped to the reference genome (B73 AGPv4) (Table S1). Principal component analysis (PCA) revealed that the PC1 explained 73.40% of the overall variances, and the samples were mainly clustered by different inoculation times (Figure 4a).
FIGURE 4.
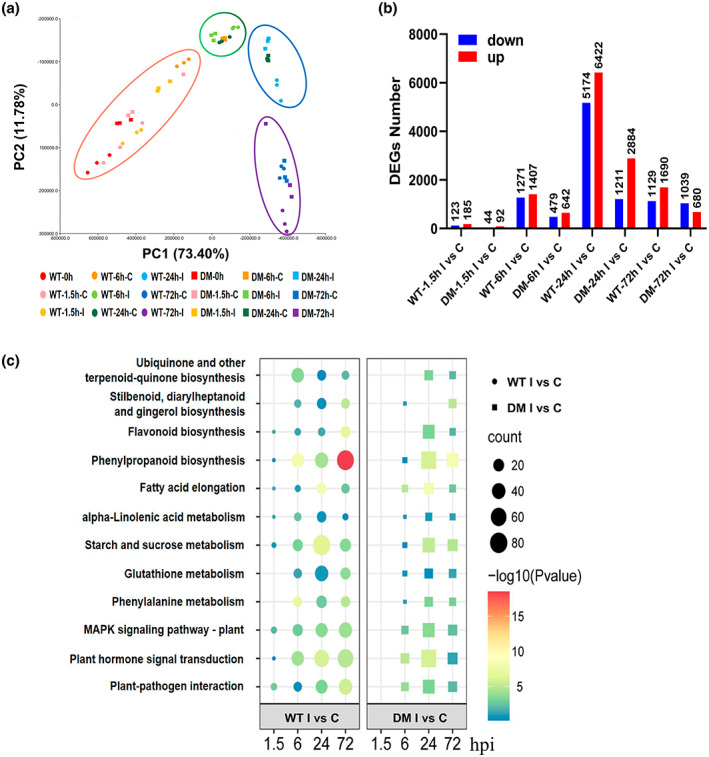
Transcriptome analysis in the wild type and Zmsiz1a/1b double mutant kernels at different time points after inoculation with Fusarium verticillioides. (a) The principal component analysis (PCA) of transcriptome data from the wild type (WT) and Zmsiz1a/1b double mutant (DM) at the indicated time after mock treatment (C) or inoculation (I). PCA was performed based on FPKM values. (b) The number of differentially expressed genes (DEGs) that were up‐ and down‐regulated between mock treatment (C) and inoculation (I) at different time points after inoculation for the WT and DM, respectively. (c) Top enriched KEGG pathways of DEGs regulated by mock treatment (C) versus inoculation (I) at different time points after inoculation for the WT, respectively. hpi, hours postinoculation
Differentially expressed genes (DEGs) between the inoculated and noninoculated conditions were identified in the wild type and the Zmsiz1a/1b double mutant (Figure 4b). Among these, DEGs of both the wild type and Zmsiz1a/1b double mutant were induced soon after inoculation (1.5 hpi) and the number of DEGs reached a peak at 24 hpi, then decreased (Figure 4b). Additionally, the wild type had more DEGs than the Zmsiz1a/1b double mutant at each time point (Figure 4b). In summary, the expression dynamics of DEGs suggest that the Zmsiz1a/1b double mutant has a dampened response to pathogen infection compared with the wild type. Further KEGG analysis revealed that DEGs were largely enriched in pathways that are closely associated with pathogen defence and stress responses, such as plant–pathogen interaction, MAPK signalling pathways (plant), plant hormone signal transduction, phenylpropanoid biosynthesis, flavonoid biosynthesis, and starch/sucrose metabolism (Figure 4c). In the wild type, three pathways, MAPK signalling pathways (plant), plant hormone signal transduction, and plant–pathogen interaction, were enriched at 1.5 hpi, and the number of enriched DEGs increased with time thereafter, whereas no DEGs were enriched in these three pathways in the Zmsiz1a/1b double mutant at 1.5 hpi, but the DEG amount matched that of the wild type at 6 and 24 hpi, followed by a decrease at 72 hpi (Figure 4c).
Based on the results of KEGG enrichment, 35 candidate genes in these three pathways were identified that were up‐regulated in the wild type at least one time point after inoculation (Table S2). These candidate genes are distributed on 10 chromosomes of maize, with eight on chromosome 5 and five on each of chromosomes 6 and 8 (Table S2). Further analysis indicated that the plant–pathogen interaction pathway mainly contains three classes of genes: WRKY family transcription factors, calcium signalling pathway, and pathogenesis‐related (PR) proteins (Figure 5a). Three WRKY family genes, containing two highly conserved domains with AtWRKY33, were triggered at 1.5 hpi in the wild type but at or after 6 hpi in the mutant (Figures 5a and S4). Calcium signalling‐related genes were mainly induced at 6 and 24 hpi in the wild type, while PR genes were largely expressed at 72 hpi (Figure 5a). Moreover, other genes related to Ser/Thr protein kinase pathway (Zm00001d022179), auxin signalling (Zm00001d17397), ABA signalling (Zm00001d47037, Zm00001d10445, Zm00001d38436), ethylene signalling (Zm00001d50861), jasmonic acid (JA) signalling (Zm00001d09714), cytokinin response (Zm00001d26594), and some transcription factors (MYC2, bHLH, bZIP, VQ‐motif) were also identified (Figure 5b). In addition, 12 of these 35 genes were further tested by RT‐qPCR to validate our RNA‐Seq data. Expression patterns of these genes from RT‐qPCR data were largely similar to those from the RNA‐Seq data, confirming the reliability of the RNA‐Seq data (Figure S5). These results suggest that ZmSIZ1a and ZmSIZ1b are extensively involved in multiple defence pathways in response to F. verticillioides invasion at different stages.
FIGURE 5.
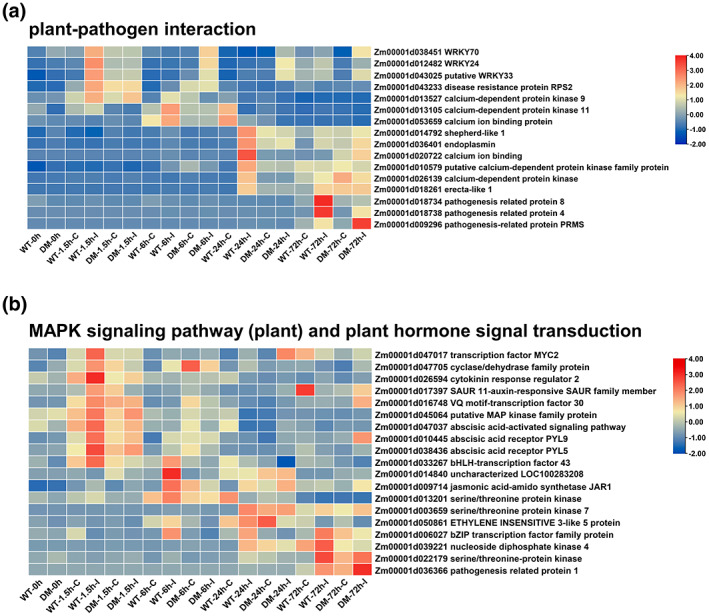
Dynamic expression patterns of differentially expressed genes (DEGs) in KEGG pathways related to disease resistance. The expression level of DEGs in (a) plant–pathogen interaction pathways and in (b) the MAPK signalling pathway (plant) and the plant hormone signal transduction pathway in the wild type (WT) and the Zmsiz1a/1b double mutant (DM) kernels after inoculation with F. verticillioides (I) and mock treatment (C) over time. Heatmaps were created using FPKM expression values. The log2 fold‐change values of the genes are shown
2.5. ZmSIZ1a and ZmSIZ1b regulate flavonoids synthesis in response to F. verticillioides
Flavonoids are well‐known defence‐related secondary metabolites that play a key role in the disease resistance of plants (Treutter, 2006). Besides the above‐mentioned pathways, DEGs in the wild type were also significantly enriched in the KEGG categories of flavonoids biosynthesis at 72 hpi (Figure 4c); therefore, the accumulation of metabolites induced by F. verticillioides inoculation was examined to validate the transcriptome data. Kernels of the wild type and Zmsiz1a/1b double mutant were treated as those for transcriptome analysis. Metabolic profiles were characterized via metabolomic analysis at 72 h after F. verticillioides inoculation (Data S1).
Correlation analysis and PCA results indicated the good reproducibility and high quality of the obtained data (Figure 6a,b). In the wild type, KEGG analysis of differentially accumulated metabolites (DAMs) between the inoculated and noninoculated samples revealed noticeable enrichment in the flavonoids biosynthesis pathways, including flavonoid biosynthesis, isoflavonoid biosynthesis, and flavone and flavonol biosynthesis (Figure 6c), whereas only the flavonoid biosynthesis pathway was identified with a relatively low enrichment factor in the Zmsiz1a/1b double mutant (Figure 6d). Furthermore, the KEGG association analysis of the transcriptomic data and metabolomic data also showed that both genes and metabolites were significantly enriched in the flavonoid biosynthesis pathway in the wild type, but not in the Zmsiz1a/1b double mutant (Figure 6e,f).
FIGURE 6.
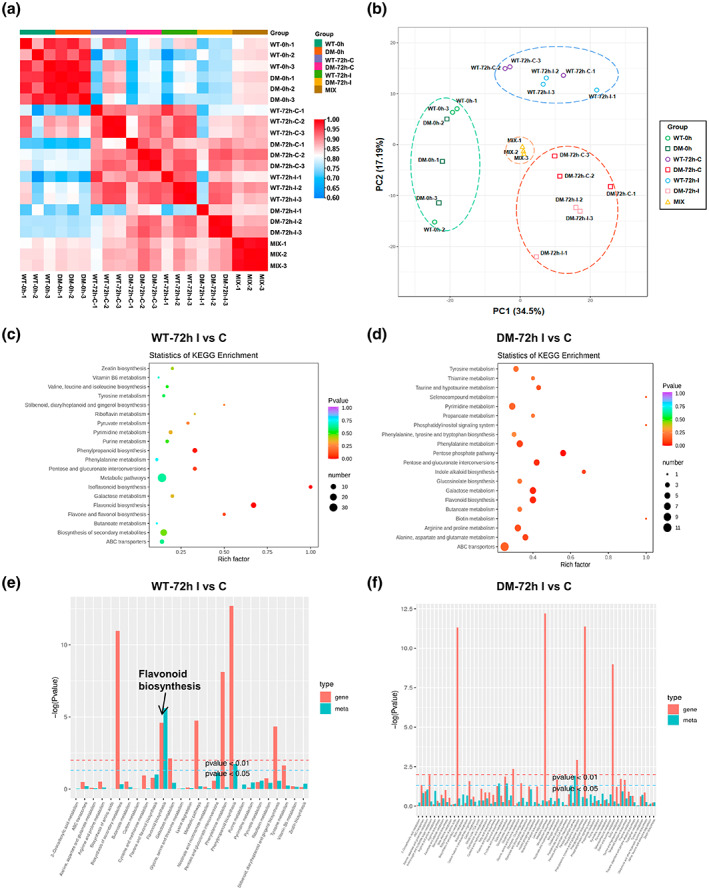
Metabolome analysis in the wild type and Zmsiz1a/1b double mutant kernels after inoculation with Fusarium verticillioides. The correlation heat map and (b) the principal component analysis (PCA) score plot of metabolome data from the wild type (WT) and the Zmsiz1a/1b double mutant (DM) at 0 and 72 h after mock treatment (C) or inoculation (I). The MIX groups were mixed samples and analysed as a control. (c, d) KEGG enrichment analyses of metabolites altered by F. verticillioides infection in the WT and DM kernels. (e, f) DEG and DAM enrichment in KEGG pathways in the WT and DM, respectively
Next, we constructed the pathway network of flavonoids biosynthesis in detail by combining the transcriptome and metabolome data to further explore the differences in response to F. verticillioides infection between the wild type and the Zmsiz1a/1b double mutant kernels. As shown in Figure 7, most dihydroflavonoids, dihydroflavonols, isoflavonoids, and their derivatives were strongly induced by inoculation and accumulated more in the mutant, except for homotrienols and hesperidin, which were only induced in the wild type. Consistently, the transcriptomic data showed similar trends in the expression levels of relevant key genes (Figure 7). However, it is interesting that some downstream compounds (flavones and flavonols), such as salcolin A, salcolin B, tricin, diosmetin, dihydroxy‐dimethoxy flovone, and their derivatives, were induced to accumulate in large amounts in the wild type after inoculation, but not in the Zmsiz1a/1b double mutant (Figure 7). These results suggest that ZmSIZ1a and ZmSIZ1b may mediate the biosynthesis pathway of these flavonoids in maize kernels, thereby having an impact on the resistance to FER.
FIGURE 7.
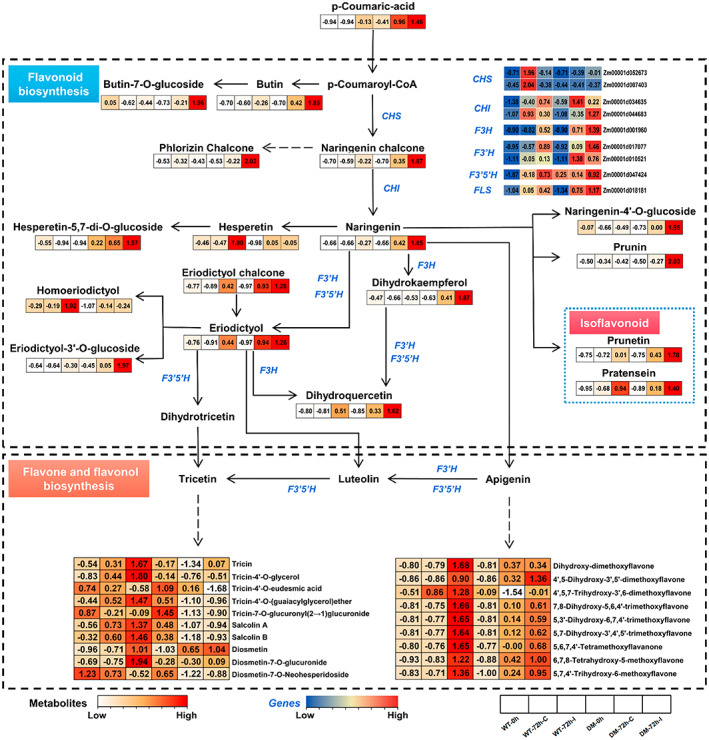
Effects of Fusarium verticillioides inoculation on the biosynthesis network for flavonoid metabolites. The heatmap represents relative expression levels of indicated genes (from blue to red) and metabolites (white to red). The heatmap of gene expression was created by the FPKM values from RNA‐Seq. Values represent log2 fold‐change values among different sample groups. Solid arrows represent a direct step, while dotted arrows represent multiple steps. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavonoid 3‐hydroxylase; F3′H, flavonoid 3′‐hydroxylase; F3′5′H, flavonoid‐3′,5′‐hydroxylase; FLS, flavonol synthase
3. DISCUSSION
3.1. ZmSIZ1a and ZmSIZ1b play an indispensable role in FER resistance in maize
SIZ1 is the SUMO E3 ligase that is highly conserved in various plants. Although ZmSIZ1a/1b/1c has been previously studied by sequence analysis and heterologous expression in Arabidopsis, the function of ZmSIZ1a/1b/1c in maize has not been reported (Augustine et al., 2016; Lai et al., 2022). In this study, we validated the function of ZmSIZ1a and ZmSIZ1b in maize mutants. The heat‐induced SUMOylation assay on ZmSIZ1a/1b mutants revealed that ZmSIZ1a and ZmSIZ1b directly regulate SUMOylation in maize, and there is functional redundancy between them in SUMO E3 ligase activity (Figures 2 and S1). Meanwhile, it should be noted that ZmSIZ1c can rescue the dwarf phenotype of the Arabidopsis siz1 mutant, but it cannot rescue the phenotype and heat shock‐induced SUMOylation defect of the Zmsiz1a/1b double mutant (Figure 2). This is consistent with the previous report and further confirms the functional differentiation between ZmSIZ1a/1b and ZmSIZ1c in maize (Lai et al., 2022).
SIZ1‐mediated SUMOylation is broadly involved in the processes of plant development and stress responses. Previous studies reported that stress tolerance was severely impaired in the Arabidopsis siz1 mutant plants (Catala et al., 2007; Miura et al., 2007; Rytz et al., 2018). In this study, both the FER resistance evaluation in the field and the kernel infection assay showed that the ZmSIZ1a/1b double mutant is extremely susceptible to FER, suggesting that ZmSIZ1a and ZmSIZ1b‐mediated SUMOylation play an indispensable role in FER resistance in maize (Figure 3).
3.2. ZmSIZ1a and ZmSIZ1b regulate the early defence response to F. verticillioides invasion
The early defence responses are very important for FER resistance in maize (Yao et al., 2020). Based on our transcriptome analysis, we identified 35 candidate DEGs in the KEGG categories of plant–pathogen interaction, MAPK signalling pathways (plant), and plant hormone signal transduction pathway that are closely related to the plant early defence response. Fifteen of the 35 candidate genes were induced at 1.5 hpi in the wild type (Figure 5), whereas these early responses were postponed in the Zmsiz1a/1b mutant, indicating that rapidly activated defence reactions are important for the response to pathogen invasion and that these responses are regulated by ZmSIZa/1b‐mediated SUMOylation.
Specifically, one MAPK (Zm00001d045064) and three WRKY (Zm00001d012482, Zm00001d038451, and Zm00001d043025) genes were identified, which are important parts of the early defence signalling pathway (Figure 5; Lanubile et al., 2017). MAPKs are involved in the regulation of both pattern‐triggered immunity and effector‐triggered immunity by mediating the phosphorylation of downstream genes (Lanubile et al., 2017). These three WRKY genes are homologues of AtWRKY33, a core transcription factor in the plant immune response regulated by MPK3/6 in Arabidopsis. Numerous studies have shown that the MPK3/6‐WRKYs pathway modulates plant immunity by regulating the synthesis of defence‐related substances, such as ABA (Liu et al., 2015b), ethylene (Li et al., 2012), camalexin (Mao et al., 2011; Zhou et al., 2020), and pipecolic acid (Wang et al., 2018). For example, ZmWRKY83 (Zm00001d038023), another maize homologue of AtWRKY33, was identified to regulate Gibberella stalk rot resistance (Bai et al., 2021). Furthermore, a recent study revealed that the SUMOylation of WRKY33 is necessary for its MPK3/6‐mediated phosphorylation (Verma et al., 2021). It will be interesting to test whether ZmSIZ1a/1b is involved in posttranslational regulation of ZmWRKY83 SUMOylation to modulate FER resistance in maize in future studies.
3.3. ZmSIZ1a and ZmSIZ1b are involved in regulating the synthesis of flavonoid metabolites
Flavonoids are well‐known plant secondary metabolites that regulate plant disease resistance through different mechanisms, such as limiting the growth and colonization of pathogens, and alleviating cellular damage caused by pathogen‐induced reactive oxygen species burst (Cho & Lee, 2015; Jia et al., 2010). Integration of our transcriptomic and metabolic data showed that DEGs and DAM in the wild type were significantly enriched in the flavonoid biosynthesis pathway after F. verticillioides inoculation, but those were not found in the mutant (Figure 6e,f), suggesting that ZmSIZ1a and ZmSIZ1b may play a major role in regulating the flavonoid biosynthesis pathway to modulate FER resistance.
The key intermediate dihydroflavonoids, such as naringenin chalcone, naringenin, eriodictyol, dihydrokaempferol, and dihydroquercetin, are often considered as markers for the synthesis of flavonoid compounds. In the present study, it is interesting that these key intermediates were heavily induced in the mutant after inoculation, but more downstream metabolites (flavone and flavonol) accumulated in the wild type (Figure 7). This implies, on the one hand, that the downstream metabolites may play a more critical role in disease resistance and, on the other hand, suggests that ZmSIZ1a/1b‐mediated SUMOylation may be involved in the conversion of dihydroflavonoids to downstream metabolites. These pathways were disrupted in the Zmsiz1a/1b mutant, leading to the accumulation of these intermediates. However, the target proteins of ZmSIZ1a and ZmSIZ1b in these processes are still unknown. Hence, it is worthwhile to look into the target proteins of ZmSIZ1a/1b‐mediated SUMOylation to regulate the synthesis of flavonoids in the response to FER in maize.
3.4. Potential applications of ZmSIZ1a/1b genes in crop breeding
Some studies have reported that overexpression of SIZ1 improves plant tolerance to abiotic stresses such as cold, heat, drought, and salt stress (Mishra et al., 2018; Miura & Nozawa, 2014; Zhang et al., 2017). We observed that the ZmSIZ1 overexpression plants only exhibited marginal increase in FER resistance compared to the wild‐type plants (Figure S3). One possible explanation is that SIZ1‐mediated SUMOylation is indispensable but not sufficient for FER resistance. Another possibility is that the high resistance of the transgenic background line ZC01 may prevent further enhancement of resistance by overexpression of ZmSIZ1a and ZmSIZ1b (Figure 3). Therefore, further evaluation of overexpressing ZmSIZ1a and ZmSIZ1b in susceptible varieties is merited. In addition, Fusarium spp. are also pathogens of various rot diseases in other crops, such as wheat, tomato, soybean, peanut, and sunflower. Hence, it is of great importance to test whether SIZ1 and its homologous genes can be used to improve disease resistance in these crops.
In summary, we demonstrate that ZmSIZ1a/ZmSIZ1b‐mediated SUMOylation plays an indispensable role in the comprehensive defence responses of maize against FER by mediating several signalling pathways, especially the early defence responses and flavonoids synthesis. We identified a number of candidate genes for FER resistance, which provide potential gene resources for FER resistance improvement breeding. Further efforts aimed at identifying the target proteins of ZmSIZ1a/ZmSIZ1b will deepen our understanding of the molecular mechanisms of FER resistance in maize and provide new targets for breeding enhanced FER‐resistant maize cultivars.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material and growth conditions
The maize inbred line ZC01 (China National Seed Group Co., Ltd) was used as the wild type. All the mutant lines were generated in the ZC01 background. ZC01 and various Zmsiz knockout mutants were planted in Langfang (39°N, 116°E, Hebei province, China) during the summer and in Ledong (18°N, 116°E, Hainan province, China) during the winter from 2019 to 2021. One row of ZC01 was planted every 20 rows. Thirteen plants were planted per row with a 15 cm plant spacing, and the distance between two rows was 30 cm. Each genotype was planted in two to four rows. The identified heterozygous plants were self‐pollinated to generate homozygous mutants. For phenotyping, 10 plants were randomly selected in each row for artificial inoculation or the measurement of plant height and ear height.
4.2. Generation and identification of CRISPR/Cas9 knockout lines of Zmsiz1
The CRISPR/Cas9 knockout construct was generated as detailed by Wu et al. (2019) with minor modifications. Briefly, two identical target sequences (target 1 and target 2) in exons of both ZmSIZ1a and ZmSIZ1b genes were selected for Cas9 cleavage according to the criteria of 5′‐G(N)19NGG‐3′. These sgRNA fragments driven by the maize ubiquitin U6‐1 and U6‐2 promoter were cloned into the CPB vector (Zhao et al., 2016). The construct was transformed into the ZC01 via Agrobacterium tumefaciens‐mediated transformation. At least two independent lines of each mutant were verified by PCR and DNA sequencing for further studies. Primers for construction and identification are listed in Table S3.
4.3. Sequences alignment and phylogenetic analysis
The amino acid sequences of ZmSIZ1a (Zm00001d010974), ZmSIZ1b (Zm00001d035487), ZmSIZ1c (Zm00001d044602), and WRKYs (Zm00001d012482, Zm00001d038451, and Zm00001d043025) were obtained from MaizeGDB (https://www.maizegdb.org). The amino acid sequences of AtSIZ1 (AT5G60410), OsSIZ1 (Os05g03430), OsSIZ2 (Os03g50980), and AtWRKY33 (AT2G38470) were obtained from the NCBI protein database. Amino acid sequence alignment was conducted using the DNAMAN software. The phylogenetic tree was performed by MEGA 10 software using the maximum‐likelihood method with default settings.
4.4. Kernel infection and spore enumeration
The kernel infection experiments were carried out as reported by Gao et al. (2009). Briefly, the kernels of the wild type and mutants were surface disinfected with 0.5% sodium hypochlorite for 10 min and then rinsed five times with distilled water. Three kernels of each genotype were wounded by a razor blade and placed into the 12‐well clear tissue culture‐treated plates, followed by inoculation with 120 μl of F. verticillioides spore suspension containing 5 × 106 conidia/ml and 0.01% Tween 20. The plates were covered with aluminium foil and incubated at 28°C for 3 days. Spores were counted with a haemocytometer and at least three independent replicate experiments were applied.
4.5. Artificial inoculation in the field
The artificial inoculation was undertaken as described by Yao et al. (2020). Briefly, we planted the wild type and various Zmsiz mutant plants side by side in the field in Ledong (18°N, 116°E, Hainan province, China) and Langfang (39°N, 116°E, Hebei province, China) in the winter and summer of 2021, respectively. F. verticillioides spore suspension was prepared as described above. At approximately 15 days after pollination, 2 ml of spore suspension containing 5 × 106 conidia/ml and 0.01% Tween 20 was inoculated in the middle of each ear using a continuous syringe. The disease level of each ear was evaluated after maturation. The disease level is classified into five grades based on the size of the infected area: 0–1% = 0, 2%–10% = 0.25, 11%–25% = 0.5, 26%–50% = 0.75, and 50%–100% = 1. Then, the disease severity index (DSI) was calculated according to this grade as follows: Σ(disease grade × number of plants with that grade) × 100/(1 × total number of plants).
4.6. In vivo heat shock‐induced SUMOylation analysis
The heat shock‐induced SUMOylation analysis in maize was performed as described by Augustine et al. (2016) with modifications. Seven‐day‐old seedlings were treated at 42°C for 1 h. Approximately 150 mg of leaves was fine ground in liquid nitrogen and transferred into a 1.5‐ml centrifuge tube containing 200 μl of 4× loading buffer. The mixture was boiled at 95°C for 5 min and then centrifuged transiently using a desktop centrifuge. The supernatants were separated by 10% SDS‐PAGE followed by western blotting and immunodetection using the anti‐SUMO1 antibodies (ab5316; Abcam). RuBisCO large subunit (rbcL) stained by Ponceau S was used as a loading control on western blots.
4.7. RNA extraction and RT‐qPCR
Total RNA was extracted from the indicated maize tissues using the Hipure plant RNA mini Kit (Megan) and then the reverse transcription was performed using the Hifair III 1st strand cDNA Synthesis SuperMix Kit (Yeasen Biotechnology) according to the manufacturer's instructions. Quantitative PCRs were run on a LightCycler 96 real‐time PCR instrument (Roche) using the Hieff UNICON qPCR SYBR Green Master Mix (Yeasen Biotechnology) following the manufacturer's instructions. Three biological repeats and three technical repeats were applied. Tubulin5 (Zm00001d006651) was used as the internal reference to normalize the expression of target genes. Primers for RT‐qPCR are listed in Table S3.
4.8. RNA sequencing and detection of DEGs
Wild‐type and Zmsiz1a/1b (#1) kernels were infected as described above. Inoculated or mock‐inoculated kernels were harvested at the indicated time (0, 1.5, 6, 24 and 72 hpi) and frozen in liquid nitrogen immediately. Three biological replicates for each sample were collected. Samples were stored at −80°C until RNA extraction. Total RNA was extracted as described above. The library preparation, RNA sequencing, and bioinformatics analyses were performed by Genewiz (www.genewiz.com.cn) using standard procedures. The sequence data were mapped to the Zea mays reference genome (B73 AGPv4) from the MaizeGDB database. The gene expression level was calculated using fragments per kilobases per million reads (FPKM). The DEGs between different groups were identified with the discriminant threshold value (|log2 (fold change)| > 1 and q‐value(FDR, p adj) ≤ 0.05) using the Bioconductor package DEseq2. The KEGG pathway enrichment of DEGs was determined by hypergeometric tests and p ≤ 0.05.
4.9. Metabolites measurement and data analysis
Samples were harvested at 72 h postinoculation and immediately frozen in liquid nitrogen. Sample preparation, extraction, and the identification and quantification of metabolome analysis were carried out by Jiaxing MetWare Biotechnology Co., Ltd (www.metware.cn) using standard procedures. The sample extracts were analysed using an ultraperformance liquid chromatography‐electrospray ionization‐tandem mass spectrometry (UPLC‐ESI‐MS/MS) system (UPLC, SHIMADZU Nexera X2, www.shimadzu.com.cn; MS, Applied Biosystems 4500 Q TRAP, www.appliedbiosystems.com.cn). The variable importance in projection (VIP) values were extracted from the orthogonal partial least squares‐discriminant analysis results using the R package MetaboAnalystR. Differential metabolites between groups were determined by VIP ≥1 and |log2 (fold change)| ≥ 1. Identified metabolites were annotated using the KEGG compound database and mapped to the KEGG pathway database (http://www.kegg.jp). The KEGG pathway enrichment of metabolites was determined by hypergeometric tests and p ≤ 0.05.
4.10. Statistical analysis and data visualization
Statistical analysis of the data was performed on GraphPad Prism software. The FPKM, relative gene expression levels, and relative metabolite levels were visualized in heatmaps by TBtools (Chen et al., 2020).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Appendix S1 rDNA internal transcribed spacer sequencing results of the isolated fungi
Data S1 List of identified metabolites in metabolomic analysis
Figure S1 Identification of ZmSIZ1a and Zmsiz1b knockout mutants
Figure S2 The Zmsiz1a/1b double mutants are susceptible to Fusarium ear rot under natural field conditions
Figure S3 The ZmSIZ1a and ZmSIZ1b overexpression lines show a marginal increase in Fusarium ear rot resistance
Figure S4 Sequence alignment analysis of identified ZmWRKY proteins
Figure S5 Validation of differentially exporessed genes identified by RNA‐Seq
Table S1 Clean reads and mapping ratios of RNA‐Seq data
Table S2 Candidate genes in the KEGG pathways of plant–pathogen interaction, MAPK signalling pathways, and plant hormone signal transduction
Table S3 List of the primers used in this study
ACKNOWLEDGEMENTS
We thank Professor Jianyu Wu (Henan Agricultural University) for providing F. verticillioides. This work was supported by the National Natural Science Foundation of China (32001852, 31901550) and the China Postdoctoral Science Foundation (2020M672648).
Liao, X. , Sun, J. , Li, Q. , Ding, W. , Zhao, B. , Wang, B. et al. (2023) ZmSIZ1a and ZmSIZ1b play an indispensable role in resistance against Fusarium ear rot in maize. Molecular Plant Pathology, 24, 711–724. Available from: 10.1111/mpp.13297
DATA AVAILABILITY STATEMENT
Additional supporting information may be found online in the Supporting Information section at the end of the article. The raw data of RNA‐Seq has been submitted to National Genomics Data Center of China (https://ngdc.cncb.ac.cn/) under the Bioproject ID: PRJCA011369.
REFERENCES
- Augustine, R.C. & Vierstra, R.D. (2018) SUMOylation: re‐wiring the plant nucleus during stress and development. Current Opinion in Plant Biology, 45, 143–154. [DOI] [PubMed] [Google Scholar]
- Augustine, R.C. , York, S.L. , Rytz, T.C. & Vierstra, R.D. (2016) Defining the SUMO system in maize: SUMOylation is up‐regulated during endosperm development and rapidly induced by stress. Plant Physiology, 171, 2191–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, H. , Si, H. , Zang, J. , Pang, X. , Yu, L. , Cao, H. et al. (2021) Comparative proteomic analysis of the defense response to Gibberella stalk rot in maize and reveals that ZmWRKY83 is involved in plant disease resistance. Frontiers in Plant Science, 12, 694973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala, R. , Ouyang, J. , Abreu, I.A. , Hu, Y. , Seo, H. , Zhang, X. et al. (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. The Plant Cell, 19, 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Ding, J. , Li, H. , Li, Z. , Sun, X. , Li, J. et al. (2012) Detection and verification of quantitative trait loci for resistance to Fusarium ear rot in maize. Molecular Breeding, 30, 1649–1656. [Google Scholar]
- Chen, J. , Shrestha, R. , Ding, J. , Zheng, H. , Mu, C. , Wu, J. et al. (2016) Genome‐wide association study and QTL mapping reveal genomic loci associated with Fusarium ear rot resistance in tropical maize germplasm. G3: Genes, Genomes, Genetics, 6, 3803–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y. et al. (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Cho, M.H. & Lee, S.W. (2015) Phenolic phytoalexins in rice: biological functions and biosynthesis. International Journal of Molecular Sciences, 16, 29120–29133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J.‐Q. , Wang, X.‐M. , Chander, S. , Yan, J.‐B. & Li, J.‐S. (2008) QTL mapping of resistance to Fusarium ear rot using a RIL population in maize. Molecular Breeding, 22, 395–403. [Google Scholar]
- van Egmond, H.P. , Schothorst, R.C. & Jonker, M.A. (2007) Regulations relating to mycotoxins in food: perspectives in a global and European context. Analytical and Bioanalytical Chemistry, 389, 147–157. [DOI] [PubMed] [Google Scholar]
- Enrico Pè, M. , Gianfranceschi, L. , Taramino, G. , Tarchini, R. , Angelini, P. , Dani, M. et al. (1993) Mapping quantitative trait loci (QTLs) for resistance to Gibberella zeae infection in maize. Molecular and General Genetics, 241, 11–16. [DOI] [PubMed] [Google Scholar]
- Galiano‐Carneiro, A.L. , Kessel, B. , Presterl, T. , Gaikpa, D.S. , Kistner, M.B. & Miedaner, T. (2020) Multi‐parent QTL mapping reveals stable QTL conferring resistance to Gibberella ear rot in maize. Euphytica, 217, 2. [Google Scholar]
- Gao, X. , Brodhagen, M. , Isakeit, T. , Brown, S.H. , Gobel, C. , Betran, J. et al. (2009) Inactivation of the lipoxygenase ZmLOX3 increases susceptibility of maize to Aspergillus spp. Molecular Plant‐Microbe Interactions, 22, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Z. , Zou, B. , Wang, X. , Qiu, J. , Ma, H. , Gou, Z. et al. (2010) Quercetin‐induced H(2)O(2) mediates the pathogen resistance against Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis thaliana . Biochemical and Biophysical Research Communications, 396, 522–527. [DOI] [PubMed] [Google Scholar]
- Jin, J.B. , Jin, Y.H. , Lee, J. , Miura, K. , Yoo, C.Y. , Kim, W.Y. et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid‐mediated floral promotion pathway and through affects on FLC chromatin structure. The Plant Journal, 53, 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.I. , Park, B.S. , Kim, D.Y. , Yeu, S.Y. , Song, S.I. , Song, J.T. et al. (2015) E3 SUMO ligase AtSIZ1 positively regulates SLY1‐mediated GA signalling and plant development. The Biochemical Journal, 469, 299–314. [DOI] [PubMed] [Google Scholar]
- Lai, R. , Jiang, J. , Wang, J. , Du, J. , Lai, J. & Yang, C. (2022) Functional characterization of three maize SIZ/PIAS‐type SUMO E3 ligases. Journal of Plant Physiology, 268, 153588. [DOI] [PubMed] [Google Scholar]
- Lanubile, A. , Maschietto, V. , Borrelli, V.M. , Stagnati, L. , Logrieco, A.F. & Marocco, A. (2017) Molecular basis of resistance to Fusarium ear rot in maize. Frontiers in Plant Science, 8, 1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Meng, X. , Wang, R. , Mao, G. , Han, L. , Liu, Y. et al. (2012) Dual‐level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genetics, 8, e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Wang, X. , Su, M. , Yu, M. , Zhang, S. , Lai, J. et al. (2015a) Functional characterization of DnSIZ1, a SIZ/PIAS‐type SUMO E3 ligase from dendrobium. BMC Plant Biology, 15, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kracher, B. , Ziegler, J. , Birkenbihl, R.P. & Somssich, I.E. (2015b) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife, 4, e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, G. , Meng, X. , Liu, Y. , Zheng, Z. , Chen, Z. & Zhang, S. (2011) Phosphorylation of a WRKY transcription factor by two pathogen‐responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis . The Plant Cell, 23, 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M. , Miedaner, T. , Schwegler, D.D. , Kessel, B. , Ouzunova, M. , Dhillon, B.S. et al. (2012) Comparative quantitative trait loci mapping for Gibberella ear rot resistance and reduced deoxynivalenol contamination across connected maize populations. Crop Science, 52, 32–43. [Google Scholar]
- Maschietto, V. , Colombi, C. , Pirona, R. , Pea, G. , Strozzi, F. , Marocco, A. et al. (2017) QTL mapping and candidate genes for resistance to Fusarium ear rot and fumonisin contamination in maize. BMC Plant Biology, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N. , Srivastava, A.P. , Esmaeili, N. , Hu, W. & Shen, G. (2018) Overexpression of the rice gene OsSIZ1 in Arabidopsis improves drought‐, heat‐, and salt‐tolerance simultaneously. PLoS One, 13, e0201716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missmer, S.A. , Suarez, L. , Felkner, M. , Wang, E. , Merrill, A.H., Jr. , Rothman, K.J. et al. (2006) Exposure to fumonisins and the occurrence of neural tube defects along the Texas‐Mexico border. Environmental Health Perspectives, 114, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. & Nozawa, R. (2014) Overexpression of SIZ1 enhances tolerance to cold and salt stresses and attenuates response to abscisic acid in Arabidopsis thaliana . Plant Biotechnology, 31, 167–172. [Google Scholar]
- Miura, K. , Rus, A. , Sharkhuu, A. , Yokoi, S. , Karthikeyan, A.S. , Raghothama, K.G. et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proceedings of the National Academy of Sciences of the United States of America, 102, 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Jin, J.B. , Lee, J. , Yoo, C.Y. , Stirm, V. , Miura, T. et al. (2007) SIZ1‐mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis . The Plant Cell, 19, 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Lee, J. , Jin, J.B. , Yoo, C.Y. , Miura, T. & Hasegawa, P.M. (2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences of the United States of America, 106, 5418–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K. , Okamoto, H. , Okuma, E. , Shiba, H. , Kamada, H. , Hasegawa, P.M. et al. (2013) SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid‐induced accumulation of reactive oxygen species in Arabidopsis . The Plant Journal, 73, 91–104. [DOI] [PubMed] [Google Scholar]
- Novatchkova, M. , Tomanov, K. , Hofmann, K. , Stuible, H.P. & Bachmair, A. (2012) Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. The New Phytologist, 195, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.C. , Kim, H. , Koo, S.C. , Park, H.J. , Cheong, M.S. , Hong, H. et al. (2010) Functional characterization of the SIZ/PIAS‐type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant, Cell & Environment, 33, 1923–1934. [DOI] [PubMed] [Google Scholar]
- Park, B.S. , Song, J.T. & Seo, H.S. (2011) Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nature Communications, 2, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz, T.C. , Miller, M.J. , Mcloughlin, F. , Augustine, R.C. , Marshall, R.S. , Juan, Y.T. et al. (2018) SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. The Plant Cell, 30, 1077–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutter, D. (2006) Significance of flavonoids in plant resistance: a review. Environmental Chemistry Letters, 4, 147–157. [Google Scholar]
- Verma, V. , Srivastava, A.K. , Gough, C. , Campanaro, A. , Srivastava, M. , Morrell, R. et al. (2021) SUMO enables substrate selectivity by mitogen‐activated protein kinases to regulate immunity in plants. Proceedings of the National Academy of Sciences of the United States of America, 118, e2021351118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Makeen, K. , Yan, Y. , Cao, Y. , Sun, S. & Xu, G. (2011) OsSIZ1 regulates the vegetative growth and reproductive development in rice. Plant Molecular Biology Reporter, 29, 411–417. [Google Scholar]
- Wang, Y. , Schuck, S. , Wu, J. , Yang, P. , Doring, A.C. , Zeier, J. et al. (2018) A MPK3/6‐WRKY33‐ALD1‐pipecolic acid regulatory loop contributes to systemic acquired resistance. The Plant Cell, 30, 2480–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G. , Zhao, Y. , Shen, R. , Wang, B. , Xie, Y. , Ma, X. et al. (2019) Characterization of maize phytochrome‐interacting factors in light signaling and photomorphogenesis. Plant Physiology, 181, 789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, L. , Li, Y. , Ma, C. , Tong, L. , Du, F. & Xu, M. (2020) Combined genome‐wide association study and transcriptome analysis reveal candidate genes for resistance to Fusarium ear rot in maize. Journal of Integrative Plant Biology, 62, 1535–1551. [DOI] [PubMed] [Google Scholar]
- Ye, J. , Zhong, T. , Zhang, D. , Ma, C. , Wang, L. , Yao, L. et al. (2019) The auxin‐regulated protein ZmAuxRP1 coordinates the balance between root growth and stalk rot disease resistance in maize. Molecular Plant, 12, 360–373. [DOI] [PubMed] [Google Scholar]
- Yoo, C.Y. , Miura, K. , Jin, J.B. , Lee, J. , Park, H.C. , Salt, D.E. et al. (2006) SIZ1 small ubiquitin‐like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiology, 142, 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S. , Zhuang, K. , Wang, S. , Lv, J. , Ma, N. & Meng, Q. (2017) A novel tomato SUMO E3 ligase, SlSIZ1, confers drought tolerance in transgenic tobacco. Journal of Integrative Plant Biology, 59, 102–117. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Zhang, C. , Liu, W. , Gao, W. , Liu, C. , Song, G. et al. (2016) An alternative strategy for targeted gene replacement in plants using a dual‐sgRNA/Cas9 design. Scientific Reports, 6, 23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Schumaker, K.S. & Guo, Y. (2012) SUMOylation of transcription factor MYB30 by the small ubiquitin‐like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America, 109, 12822–12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Wang, X. , He, Y. , Sang, T. , Wang, P. , Dai, S. et al. (2020) Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis . The Plant Cell, 32, 2621–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zila, C.T. , Samayoa, L.F. , Santiago, R. , Butron, A. & Holland, J.B. (2013) A genome‐wide association study reveals genes associated with Fusarium ear rot resistance in a maize core diversity panel. G3: Genes, Genomes, Genetics, 3, 2095–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zila, C.T. , Ogut, F. , Romay, M.C. , Gardner, C.A. , Buckler, E.S. & Holland, J.B. (2014) Genome‐wide association study of Fusarium ear rot disease in the USA maize inbred line collection. BMC Plant Biology, 14, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 rDNA internal transcribed spacer sequencing results of the isolated fungi
Data S1 List of identified metabolites in metabolomic analysis
Figure S1 Identification of ZmSIZ1a and Zmsiz1b knockout mutants
Figure S2 The Zmsiz1a/1b double mutants are susceptible to Fusarium ear rot under natural field conditions
Figure S3 The ZmSIZ1a and ZmSIZ1b overexpression lines show a marginal increase in Fusarium ear rot resistance
Figure S4 Sequence alignment analysis of identified ZmWRKY proteins
Figure S5 Validation of differentially exporessed genes identified by RNA‐Seq
Table S1 Clean reads and mapping ratios of RNA‐Seq data
Table S2 Candidate genes in the KEGG pathways of plant–pathogen interaction, MAPK signalling pathways, and plant hormone signal transduction
Table S3 List of the primers used in this study
Data Availability Statement
Additional supporting information may be found online in the Supporting Information section at the end of the article. The raw data of RNA‐Seq has been submitted to National Genomics Data Center of China (https://ngdc.cncb.ac.cn/) under the Bioproject ID: PRJCA011369.


