Abstract
The integration of transcriptomic and proteomic data from lung tissue with chronic obstructive pulmonary disease (COPD)–associated genetic variants could provide insight into the biological mechanisms of COPD. Here, we assessed associations between lung transcriptomics and proteomics with COPD in 98 subjects from the Lung Tissue Research Consortium. Low correlations between transcriptomics and proteomics were generally observed, but higher correlations were found for COPD-associated proteins. We integrated COPD risk SNPs or SNPs near COPD-associated proteins with lung transcripts and proteins to identify regulatory cis-quantitative trait loci (QTLs). Significant expression QTLs (eQTLs) and protein QTLs (pQTLs) were found regulating multiple COPD-associated biomarkers. We investigated mediated associations from significant pQTLs through transcripts to protein levels of COPD-associated proteins. We also attempted to identify colocalized effects between COPD genome-wide association studies and eQTL and pQTL signals. Evidence was found for colocalization between COPD genome-wide association study signals and a pQTL for RHOB and an eQTL for DSP. We applied weighted gene co-expression network analysis to find consensus COPD-associated network modules. Two network modules generated by consensus weighted gene co-expression network analysis were associated with COPD with a false discovery rate lower than 0.05. One network module is related to the catenin complex, and the other module is related to plasma membrane components. In summary, multiple cis-acting determinants of transcripts and proteins associated with COPD were identified. Colocalization analysis, mediation analysis, and correlation-based network analysis of multiple omics data may identify key genes and proteins that work together to influence COPD pathogenesis.
Keywords: chronic obstructive pulmonary disease, multi-omics analyses, quantitative trait locus, weighted gene co-expression network analysis
Clinical Relevance
We observed low overall correlations between lung tissue transcriptomics and proteomics in chronic obstructive pulmonary disease (COPD) cases and control subjects, indicating that these omics levels provide different biological information. Genetic determinants of proteomic and transcriptomic COPD biomarkers were identified, and we found evidence for colocalization between COPD genetic association signals and a protein quantitative trait locus for RHOB and an expression quantitative trait locus for DSP. Correlation-based network analysis of lung transcriptomics and proteomics identified COPD-associated network modules related to the catenin complex and plasma membrane components.
Chronic obstructive pulmonary disease (COPD) is a complex lung disease defined by persistent airflow obstruction (1). Although cigarette smoking is the major environmental risk factor for COPD, genetic determinants including AAT (α-1 antitrypsin) deficiency (2) influence COPD risk. The development of COPD among smokers is highly variable, and the molecular basis of this varied susceptibility is not well understood.
The development of high-throughput omics technologies has enabled large-scale assessments of multiple biological levels, including genetics, transcriptomics, and proteomics. Progressively larger genome-wide association studies (GWASs) of COPD have been performed, and Sakornsakolpat and colleagues identified 82 loci associated with COPD at genome-wide significance (3). High-throughput transcriptomic and proteomic assays have also been performed, but, with a few notable exceptions (e.g., the association of sRAGE with COPD and emphysema [4]), replication of gene expression and protein biomarkers for COPD has been limited. Small sample sizes and studies focusing on blood rather than lung samples likely contribute to the challenges of identifying transcriptomic and proteomic biomarkers for COPD.
Multi-omics analyses may assist in overcoming the challenges in COPD biomarker identification. Analyzing multiple omics data types together can reduce technical variability related to each omics level, provide insights into biological mechanisms for genetic variation, reflect different biological time scales, and identify interactions between molecular levels (5). Multiple omics integration could also assist in understanding disease pathogenesis.
Several previous multi-omics analyses of COPD pathogenesis have been reported. For example, Li and colleagues used similarity network fusion with mRNA, microRNA, proteomics, and metabolomics from different biospecimens to attempt to discriminate patients with COPD from controls based on their molecular signatures (6). Using whole-genome sequencing, RNA sequencing, and mass spectrometry–based proteomics, we assessed the relationships between these omics data types in lung tissue samples and used genetic association, colocalization analysis, mediation analysis, and correlation-based networks. We hypothesized that integrated multi-omics analysis in lung tissue samples from COPD cases and control subjects would provide new insights into COPD pathogenesis and discover consistent and divergent COPD biomarkers across omics levels.
Methods
Study Population
Ninety-eight subjects from the Lung Tissue Research Consortium (LTRC) with matched whole-genome sequencing and lung tissue transcriptomics and proteomics data were included. COPD cases and control subjects were defined as follows: 1) COPD cases required a forced expiratory volume in 1 second (FEV1) <80% predicted and FEV1/forced vital capacity ratio lower than 0.7 based on postbronchodilator spirometry (prebronchodilator spirometry was used if postbronchodilator spirometry was not available) and 2) control subjects had an FEV1 of ⩾80% predicted and FEV1/forced vital capacity ratio of at least 0.7.
Multi-Omics Data Preprocessing
Whole-genome sequencing data from 1,542 LTRC subjects were provided by the Trans Omics for Precision Medicine (TOPMed) program. Genetic variants were extracted from the Freeze 9 version of whole-genome sequencing data. The genetic data of 98 subjects with matched transcriptomics and proteomics data were extracted and then filtered by minor allele frequency of at least 0.05 and Hardy-Weinberg equilibrium (excluding variants with P < 1 × 10−5). mRNA sequencing was performed through the TOPMed program at the University of Washington. In brief, transcriptomics data were quantified at gene level with RNA-Seq by Expectation Maximization (RSEM; v1.3.1) (7) using Gencode GTF (v29) (8). The gene-level quantified RNA-sequencing data were filtered and log2–transformed for further analyses.
Proteomics data were obtained by mass spectrometry analysis of lung tissue as previously reported (9). Proteomics data were calibrated and generalized logarithm (glog2)–transformed, and missing values were imputed using k-nearest neighbor imputation. Detailed descriptions of multi-omics sample selection and data preprocessing are included in the data supplement.
Batch effects for transcriptomic and proteomic datasets were removed using the Combat function from R package sva v3.38.0 (10). COPD-associated transcripts/proteins were tested following batch effects removal: transcript/protein = COPD + age + sex.
Correlations between Transcriptomics and Proteomics
Pearson’s correlations between transcriptomics and proteomics for each gene were calculated using residuals after removing COPD affection status, age, sex, and batch effects. Pearson’s correlations between gene pairs were calculated at transcriptomic and proteomic levels. Statistical comparisons between gene pair correlations were performed using the R package cocor v1.1-3 (11).
Integration of Genetic Variants with Lung Transcripts and Proteins
Expression and protein quantitative trait locus analyses within genomic regions of interest
Quantitative trait locus (QTL) analyses were performed using the R package MatrixEQTL v2.3 (12). SNPs for QTL analyses were selected within a 1-MB window (±500 kb): 1) around top SNPs in 82 previously reported genome-wide significant COPD GWAS loci (3) and 2) around the top proteins with FDR lower than 0.1 associated with COPD [±500 kb from the lowest start value and the highest end value of all available transcripts from Ensembl BioMart (13)].
Target genes (proteins or transcripts) for cis-QTL analyses were selected as all available proteins (for protein QTL [pQTL] analyses) or transcripts (for expression QTL [eQTL] analyses) within genomic regions of interest. Principal components of genetic ancestry were generated from all TOPMed LTRC whole-genome sequencing data using LASER (14). Age, sex, and the top two principal components of genetic ancestry were included as covariates in QTL analyses.
To find functional variants within COPD GWAS loci, a credible set of 6,509 variants in 82 reported COPD GWAS loci was previously generated with statistical fine mapping using PICS (Probabilistic Identification of Causal SNPs) (15). We used this fine-mapped set of SNPs for QTL analysis of COPD GWAS regions. FDR adjustment was performed with SNPs around top proteins or fine-mapping COPD-risk SNPs only. FDRs for significant QTL effects were controlled at 0.05.
Mediation analysis
Mediation analysis is a statistical method to decompose the total effects of the exposure into a direct effect and an indirect effect through the mediator. We observed a significant association between pQTLs and matched COPD-associated proteins (total effect). Because the protein is encoded by its respective transcript, we examined the mediator role of the transcripts of COPD-associated proteins for selected pQTLs. All mediation analyses were adjusted for age, sex, and the top two principal components of genetic ancestry as covariates using the R medflex package v0.6-7 (16).
Colocalization analysis
Colocalization with COPD GWAS results (3) and regional transcripts or proteins within 1-MB genomic regions around 85 COPD-associated proteins (FDR < 0.1) or 82 previously reported COPD GWAS loci was assessed using R package coloc v5.1.0 (17). R package coloc calculates the posterior probabilities under a single causal variant assumption and estimates the posterior probability for each SNP in the colocalization analysis region. LocusZoom plots were drawn using R package LocusCompareR (18) to present colocalized regions with high probability of colocalization, which means both traits are associated with their respective phenotype and shared a single causal variant.
Weighted Gene Co-expression Network Analysis
Consensus weighted gene co-expression network analyses (also known as weighted gene correlation network analyses [WGCNA]) were performed comparing transcriptomics and proteomics residuals with batch effect removal using the R package WGCNA v1.70-3 (19). The minimal module size was set as 10 genes, and the soft threshold (power) was set at 8 to optimize connectivity between each network node. COPD-associated network modules were identified using linear regression models with age and sex as covariates: module expression = COPD + age + sex.
Gene Ontology enrichment analyses were performed on genes from COPD-associated modules using R package topGO v2.46.0 (20).
Associations between the top SNPs in 82 previously reported genome-wide significant COPD GWAS loci and COPD-associated network modules were evaluated using linear regression models with age, sex, and the top two principal components of genetic ancestry as covariates.
Detailed descriptions of WGCNA methods are presented in the data supplement.
Cell-Type Proportion Estimation and Adjustment on Omics Associations
R package BisqueRNA estimates cell proportion using single-cell RNA sequencing data as a reference to decompose bulk RNA sequencing data. The Human Lung Cell Atlas (https://hlca.ds.czbiohub.org/) provided single-cell RNA sequencing data of approximately 75,000 human lung and blood cells clustered in 41 known cell types and 14 new clusters (21). Here, we estimated the cell proportion of four groups of cell types—epithelial cells, endothelial cells, stromal cells, and immune cells—in our 98 subjects using BisqueRNA and single-cell RNA sequencing data from the Human Lung Cell Atlas (21) as reference. We compared the cell group proportions between patients with COPD and controls. The effects of cell type proportion on shared omics biomarkers’ association with COPD were evaluated using linear regression models with cell proportion as covariates as shown below: transcript/protein = COPD + age + sex + cell type proportions (epithelial cells + endothelial cells + stromal cells).
To represent the cell proportion effects and avoid potential collinearity of cell proportion variables, we used three cell groups (epithelial cells, endothelial cells, and stromal cells) to represent the effects of cell type proportion on gene expression at the transcriptomics or proteomics level. QTL analyses with cell proportion metrics as covariates were also performed on significant QTL–gene associations.
Results
Clinical Characteristics of the Study Population
Clinical characteristics of 98 LTRC subjects with matched genetics, transcriptomics, and proteomics data are presented in Table 1. The longer smoking history (in pack-years) and lower lung function parameters (FEV1 percentage predicted and FEV1/forced vital capacity ratio) in COPD were expected. Suspected lung cancer was a frequent reason for control subjects to undergo thoracic surgery, whereas patients with COPD often underwent thoracic surgery for lung transplant or lung volume reduction surgery. The lung tissue samples obtained by the LTRC were selected to be distant from tumor (if present).
Table 1.
Clinical Information of 98 Lung Tissue Research Consortium Subjects with Matched Whole-Genome Sequencing, Lung Tissue Transcriptomics, and Lung Tissue Proteomics Data for Multi-Omics Analyses
| Controls | COPD Cases | |
|---|---|---|
| Number of subjects | 25 | 73 |
| Age* | 66.1 ± 8.6 | 63.1 ± 7.9 |
| Sex (male/all) | 44% (11/25) | 44% (32/73) |
| Pack-years of smoking* | 30.6 ± 19.5 | 48.7 ± 28.3 |
| FEV1, % predicted* | 104.9 ± 18.7 | 35.0 ± 17.0 |
| FEV1/FVC* | 0.77 ± 0.08 | 0.39 ± 0.14 |
| Race (White/all) | 100% (25/25) | 93% (68/73) |
| BMI* | 29.0 ± 6.3 | 26.1 ± 4.9 |
| Lung cancer (cancer/all)* | 68% (17/25) | 32% (23/73) |
Definition of abbreviations: BMI = body mass index; COPD = chronic obstructive pulmonary disease; FEV1 = forced expiratory volume in 1 second; FVC = forced vital capacity.
Values presented as mean ± SD where applicable.
P < 0.05 based on t test for quantitative variables and χ2 test for categorical variables comparing COPD cases and control subjects.
Comparisons of Transcriptomics and Proteomics in Lung Tissue
COPD-associated biomarker identification
COPD-associated proteins and transcripts were identified based on multiple linear regression analysis with FDR lower than 0.05. Within the 98 LTRC samples analyzed, we observed 55 COPD-associated proteins. Among them, 15 shared biomarkers were identified for transcriptomics and proteomics. The comparison between COPD-associated proteins (FDR < 0.1) at transcriptomics and proteomics levels is presented in a Venn diagram in Figure E1 in the data supplement, and the shared significant biomarkers are presented in Table 2 (FDR < 0.05) and Table E1 in the data supplement (FDR < 0.1).
Table 2.
Shared Chronic Obstructive Pulmonary Disease-Associated Biomarkers at Transcriptomics and Proteomics Levels with False Discovery Rate Lower than 0.05
| Gene Name | UniProt ID | Proteomics |
Transcriptomics |
||||
|---|---|---|---|---|---|---|---|
| P Value | FDR | β (SE) | P Value | FDR | β (SE) | ||
| FOLR1 | P15328 | 1.36 × 10−6 | 2.85 × 10−3 | −1.738 (0.337) | 6.41 × 10−5 | 1.17 × 10−2 | −0.444 (0.106) |
| AQP4 | P55087 | 1.10 × 10−5 | 1.13 × 10−2 | −1.102 (0.237) | 4.32 × 10−4 | 2.21 × 10−2 | −0.46 (0.126) |
| RDX | P35241 | 3.78 × 10−5 | 1.45 × 10−2 | −0.461 (0.107) | 1.91 × 10−5 | 8.64 × 10−3 | −0.413 (0.092) |
| AGER | Q15109 | 4.16 × 10−5 | 1.45 × 10−2 | −0.871 (0.202) | 8.78 × 10−5 | 1.31 × 10−2 | −0.736 (0.18) |
| CA3 | P07451 | 1.43 × 10−4 | 2.43 × 10−2 | 0.746 (0.188) | 7.71 × 10−4 | 2.65 × 10−2 | 0.827 (0.238) |
| CAV1 | Q03135 | 1.45 × 10−4 | 2.43 × 10−2 | −0.537 (0.136) | 2.50 × 10−4 | 1.94 × 10−2 | −0.509 (0.134) |
| COL14A1 | Q05707 | 1.58 × 10−4 | 2.43 × 10−2 | 0.818 (0.208) | 3.52 × 10−4 | 2.17 × 10−2 | 0.781 (0.211) |
| SUSD2 | Q9UGT4 | 1.53 × 10−4 | 2.43 × 10−2 | −0.643 (0.163) | 2.20 × 10−3 | 3.56 × 10−2 | −0.396 (0.126) |
| PCYOX1 | Q9UHG3 | 1.36 × 10−4 | 2.43 × 10−2 | −0.349 (0.088) | 3.13 × 10−4 | 2.15 × 10−2 | −0.47 (0.126) |
| IFIT3 | O14879 | 2.57 × 10−4 | 2.87 × 10−2 | −0.748 (0.197) | 8.06 × 10−6 | 8.64 × 10−3 | −0.554 (0.117) |
| VAMP3 | Q15836 | 2.92 × 10−4 | 3.02 × 10−2 | −0.618 (0.164) | 1.28 × 10−3 | 3.08 × 10−2 | −0.255 (0.077) |
| FARP1 | Q9Y4F1 | 3.84 × 10−4 | 3.58 × 10−2 | −0.805 (0.218) | 5.74 × 10−4 | 2.41 × 10−2 | −0.289 (0.081) |
| ALCAM | Q13740 | 4.13 × 10−4 | 3.76 × 10−2 | −0.864 (0.236) | 2.94 × 10−5 | 9.13 × 10−3 | −0.479 (0.109) |
| S100A10 | P60903 | 4.97 × 10−4 | 4.25 × 10−2 | −0.408 (0.113) | 4.87 × 10−3 | 4.86 × 10−2 | −0.238 (0.082) |
| ARRB1 | P49407 | 5.10 × 10−4 | 4.28 × 10−2 | −0.463 (0.129) | 2.21 × 10−3 | 3.56 × 10−2 | −0.385 (0.122) |
Definition of abbreviations: FDR = false discovery rate; SE = standard error.
Correlations between transcriptomics and proteomics
The correlations between transcriptomics and proteomics were evaluated using residuals that removed effects of COPD affection status, age, sex, and batch. Principal component analysis results for the transcriptomic and proteomic residuals are shown in Figure E2 in the data supplement, with comparisons versus raw datasets and residuals removing batch effects only. We tested 4,039 pairs of matched transcripts and proteins.
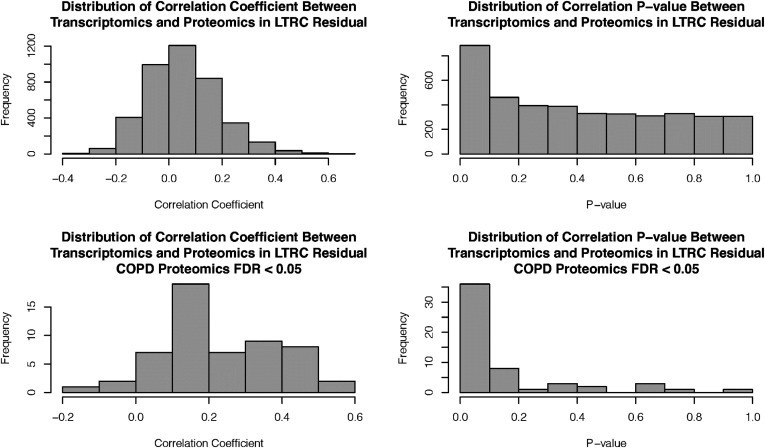
The distributions of correlation coefficients and P values between transcriptomics and proteomics of all matched genes and genes with proteomic associations with COPD are shown in Figure 1 (FDR < 0.05) and Figure E3 in the data supplement (FDR < 0.1). Comparisons between correlation coefficients of COPD-associated proteins, non–COPD-associated proteins, and all matched genes are shown in Figure E4 in the data supplement.
Figure 1.
Correlations between transcriptomics and proteomics. Correlations between all proteins and transcripts and correlations between genes with proteomics COPD–associated FDR lower than 0.05 are presented. COPD = chronic obstructive pulmonary disease; FDR = false discovery rate; LTRC = Lung Tissue Research Consortium.
The mean correlation coefficient between transcriptomics and proteomics in all genes was low (0.054), with a standard deviation of 0.134 and standard error of the mean of 0.002, but the correlation coefficients between omics pairs are significantly greater than zero (one-sample Wilcoxon signed-rank test, P < 5 × 10−113). There were 606 genes (among 4,039 matched genes) correlated with a P value lower than 0.05 and 172 genes with an FDR lower than 0.05; the median correlation value is 0.045. For genes with proteomic associations with COPD (FDR < 0.05), the mean correlation coefficient is higher (0.232; standard deviation, 0.159; standard error of the mean, 0.021; significantly greater than zero with P < 4.4 × 10−10), and the median value is 0.196. The omics correlations for COPD-associated proteins and non–COPD-associated proteins were significantly different, with Wilcoxon rank-sum test P < 1.1 × 10−15.
Because of the small sample size of our study population, we estimated the correlation test power for COPD and control populations. The estimated power for the correlation test between transcriptomics and proteomics is greater than 0.8, with a correlation coefficient threshold of 0.5, even in the control group (25 subjects), as shown in Table E2 in the data supplement.
Correlations between COPD-associated gene pairs at different omics levels
We assessed correlations between transcripts and proteins for gene pairs associated with COPD at FDR lower than 0.05 in the transcriptomics and/or proteomics datasets; 47 gene pairs were correlated at proteomic and/or transcriptomic levels with correlation coefficients greater than 0.8. Statistical comparisons between gene pair correlations in transcriptomics and proteomics datasets were performed using the cocor test with FDR controlled at 0.05 (11). For five consistent gene pairs (similar correlations in proteomics and transcriptomics), the differences between their correlations at the transcriptomics and proteomics levels are not significant, as shown in Table 3. Correlations between gene pairs associated with FDR lower than 0.1, with 8 consistent gene pairs and 51 inconsistent gene pairs, are shown in Tables E3 and E4 in the data supplement.
Table 3.
Consistent Correlations between Gene Pairs of Omics Biomarkers with False Discovery Rate Lower than 0.05 at Transcriptomics and Proteomics Levels Ranking by Correlation Coefficient at Proteomics Level
| Gene A | Gene A UniProt ID | Gene B | Gene B UniProt ID | Correlation Coefficient r |
Omics Cocor Test |
||
|---|---|---|---|---|---|---|---|
| Transcript | Protein | P Value | q-Value (FDR) | ||||
| EHD2 | Q03135 | CAV1 | Q9NZN4 | 0.718 | 0.821 | 7.22 × 10−2 | 1.54 × 10−1 |
| AGER | Q15109 | GPRC5A | Q8NFJ5 | 0.753 | 0.803 | 3.52 × 10−1 | 5.04 × 10−1 |
| AGER | Q03135 | CAV1 | Q15109 | 0.848 | 0.776 | 1.04 × 10−1 | 2.03 × 10−1 |
| SUSD2 | Q15109 | AGER | Q9UGT4 | 0.805 | 0.757 | 3.56 × 10−1 | 5.08 × 10−1 |
| RALA | P11233 | CALCRL | Q16602 | 0.803 | 0.716 | 1.43 × 10−1 | 2.59 × 10−1 |
Gene pairs with Pearson’s correlation coefficient ⩾0.8 at the transcriptomic and/or proteomic level were included.
Integration of Genetic Variants with Lung Transcripts and Proteins
eQTL and pQTL analyses on selected genomic regions
Separate eQTL and pQTL analyses were performed for local, cis-acting effects located within a 1-MB window of the 82 top COPD GWAS SNPs or 85 COPD-associated protein-coding genes. Descriptive information for all QTL analyses is shown in Tables E5 and E6 in the data supplement. Q-Q plots for QTL analyses are presented in Figure E5 in the data supplement.
In the 82 COPD GWAS regions, multiple significant cis-QTLs were associated with fine-mapped COPD risk SNPs at FDR lower than 0.1. Significant cis-QTLs with FDR lower than 0.05 are presented in Tables 4 and 5 (eQTLs and pQTLs, respectively). Significant cis-eQTL and cis-pQTL effects were identified on chromosome 17 near the MAPT gene with different top SNPs. This genomic region has a common large chromosomal inversion (22). No COPD GWAS regions with shared eQTL and pQTL determinants of genes encoding COPD-associated proteins were found.
Table 4.
Significant cis-Expression Quantitative Trait Locus Results with False Discovery Rate Lower than 0.05 for 6,509 Chronic Obstructive Pulmonary Disease Fine-Mapped Risk SNPs around 82 Chronic Obstructive Pulmonary Disease Genome-wide Association Study Loci
| SNP* | Ref | Alt | SNP Location | GWAS SNP Region (LD r2) | Top Protein Region | Gene Name (UniProt) | Gene Location | QTL |
||
|---|---|---|---|---|---|---|---|---|---|---|
| P Value† | FDR‡ | β (SE)† | ||||||||
| rs17572893 | G | A | chr17:45986842 | rs12373142 (0) | NA | MAPT (P10636) | chr17:45894527-46028334 | 2.10 × 10−11 | 2.67 × 10−9 | −0.493 (0.065) |
| rs1153594 | C | T | chr3:25518437 | rs1529672 (0.622) | NA | TOP2B (Q02880) | chr3:25597984-25664907 | 1.05 × 10−3 | 7.70 × 10−3 | −0.201 (0.059) |
| rs12362740 | A | T | chr11:86765648 | rs117261012 (0.726) | NA | HIKESHI (Q53FT3) | chr11:86302211-86345943 | 1.21 × 10−3 | 8.85 × 10−3 | 0.136 (0.041) |
| rs12522114 | C | A | chr5:52891207 | rs1551943 (0.629) | NA | ITGA1 (P56199) | chr5:52787916-52959209 | 1.22 × 10−3 | 8.86 × 10−3 | −0.35 (0.105) |
| rs2399796 | G | A | chr10:12214560 | rs7068966 (0.704) | NA | CAMK1D (Q8IU85) | chr10:12349547-12835545 | 1.89 × 10−3 | 1.37 × 10−2 | −0.176 (0.055) |
| rs798498 | T | G | chr7:2756248 | rs798565 (0.677) | NA | SNX8 (Q9Y5X2) | chr7:2251770-2354318 | 4.93 × 10−3 | 3.54 × 10−2 | 0.135 (0.047) |
| rs3131064 | T | C | chr6:30796116 | rs2284174 (0.721) | NA | NRM (Q8IXM6) | chr6:30688047-30691420 | 6.09 × 10−3 | 4.36 × 10−2 | 0.151 (0.054) |
Definition of abbreviations: eQTL = expression quantitative trait locus; GWAS = genome-wide association study; LD r2 = linkage disequilibrium squared coefficient of correlation; NA = not applicable; QTL = quantitative trait locus.
For genomic regions around each protein, only one significant top eQTL association is presented.
Positive β-coefficients mean higher expression level for the alternative allele.
FDRs were adjusted with COPD risk SNPs.
Table 5.
Significant cis-Protein Quantitative Trait Locus Results with False Discovery Rate Lower than 0.05 for 6,509 Chronic Obstructive Pulmonary Disease Fine-Mapped Risk SNPs around 82 Chronic Obstructive Pulmonary Disease Genome-wide Association Study Loci
| SNP* | Ref | Alt | SNP Location |
GWAS SNP Region (LD r2) |
Top Protein Region |
Gene Name (UniProt) | Gene Location | QTL |
||
|---|---|---|---|---|---|---|---|---|---|---|
| P Value† | FDR‡ | β (SE)† | ||||||||
| rs3785884 | G | A | chr17:45980229 | rs12373142 (0) | NA | MAPT (P10636) | chr17:45894527-46028334 | 8.84 × 10−4 | 4.37 × 10−2 | −0.705 (0.205) |
| rs10788650 | A | G | chr1:17036364 | rs9435731 (0.599) | NA | RCC2 (Q9P258) | chr1:17406760-17439677 | 1.89 × 10−3 | 4.37 × 10−2 | 0.387 (0.121) |
| rs10947233 | G | T | chr6:32156647 | rs2070600 (0.928) | AGER (Q15109) | C4A (P0C0L4) | chr6:31982057-32002681 | 3.61 × 10−3 | 4.59 × 10−2 | −0.759 (0.254) |
| rs2895504 | G | A | chr10:12202613 | rs7068966 (0.656) | NA | NUDT5 (Q9UKK9) | chr10:12165330-12196144 | 4.70 × 10−3 | 4.59 × 10−2 | −0.443 (0.153) |
| rs1182199 | C | A | chr7:2822908 | rs798565 (0.654) | NA | EIF3B (P55884) | chr7:2354086-2380745 | 5.75 × 10−3 | 4.59 × 10−2 | 0.445 (0.157) |
| rs34562262 | G | C | chr6:32053184 | rs2070600 (0.857) | AGER (Q15109) | HLA-DRA (P01903) | chr6:32439878-32445046 | 5.85 × 10−3 | 4.59 × 10−2 | −0.359 (0.127) |
| rs798560 | A | G | chr7:2718675 | rs798565 (0.696) | NA | SNX8 (Q9Y5X2) | chr7:2251770-2354318 | 6.41 × 10−3 | 4.73 × 10−2 | 0.608 (0.218) |
Definition of abbreviation: pQTL = protein quantitative trait locus.
For genomic regions around each GWAS loci and each target gene, only one significant top eQTL association is presented.
Positive β-coefficients mean higher expression level for the alternative allele.
FDRs were adjusted with COPD risk SNPs.
For genomic regions around genes encoding COPD-associated proteins (FDR < 0.05), significant QTL results influencing those COPD-associated proteins with FDR lower than 0.05 are shown in Tables 6 and 7 (eQTLs and pQTLs, respectively). Multiple cis-eQTLs were associated with COPD-associated biomarker gene expression, including PPIL3, AQP1, LGMN, ENO1, and HPCAL1. A smaller number of significant cis-pQTLs were found, which included associations with COPD-associated proteins (ARRB1 and COLGALT1). Significant shared cis-eQTLs and cis-pQTLs near COPD-associated proteins were found for C4A and HLA-DRB5, but they were not related to COPD-associated protein levels. The cis-QTL results with FDR lower than 0.1 for COPD GWAS regions and COPD-associated proteins (FDR < 0.1) are presented in Tables E7–E10 in the data supplement.
Table 6.
Significant cis-Expression Quantitative Trait Locus Results with False Discovery Rate Lower than 0.05 Regulating Chronic Obstructive Pulmonary Disease-Associated Proteins (False Discovery Rate < 0.05)
| SNP* | Ref | Alt | SNP Location | GWAS SNP Region (LD r2) | Top Protein Region | Gene Name (UniProt) | Gene Location | QTL |
||
|---|---|---|---|---|---|---|---|---|---|---|
| P Value† | FDR‡ | β (SE)† | ||||||||
| rs7559150 | T | C | chr2:200889340 | NA | PPIL3 (Q9H2H8) | PPIL3 (Q9H2H8) | chr2:200870907-200889303 | 6.76 × 10−9 | 3.30 × 10−6 | 0.452 (0.071) |
| rs10929625 | A | G | chr2:9945502 | NA | HPCAL1 (P37235) | HPCAL1 (P37235) | chr2:10302889-10427617 | 1.08 × 10−5 | 2.38 × 10−3 | 0.342 (0.073) |
| rs38498 | G | A | chr7:30587132 | NA | AQP1 (P29972) | AQP1 (P29972) | chr7:30911853-30925517 | 3.68 × 10−5 | 7.37 × 10−3 | 0.377 (0.087) |
| rs61977290 | G | A | chr14:92440176 | rs72699855 (0.019) | LGMN (Q99538) | LGMN (Q99538) | chr14:92703807-92748679 | 1.00 × 10−4 | 1.85 × 10−2 | −0.452 (0.111) |
| rs12091891 | A | G | chr1:8940161 | NA | ENO1 (P06733) | ENO1 (P06733) | chr1:8861000-8879190 | 1.22 × 10−4 | 2.23 × 10−2 | −0.314 (0.078) |
For genomic regions around each COPD-associated proteins and each target gene, only one significant top eQTL association is presented.
Positive β-coefficients mean higher expression level for the alternative allele.
FDRs were adjusted with COPD risk SNPs.
Table 7.
Significant cis-Protein Quantitative Trait Locus Results with False Discovery Rate Lower than 0.05 Regulating Chronic Obstructive Pulmonary Disease-Associated Proteins (False Discovery Rate < 0.05)
| SNP* | Ref | Alt | SNP Location | GWAS SNP Region (LD r2) | Top Protein Region | Gene Name (UniProt) | Gene Location | QTL |
||
|---|---|---|---|---|---|---|---|---|---|---|
| P Value† | FDR‡ | β (SE)† | ||||||||
| rs111516029 | C | T | chr11:75578285 | NA | ARRB1 (P49407) | ARRB1 (P49407) | chr11:75260122-75351705 | 2.63 × 10−5 | 2.31 × 10−2 | 0.785 (0.177) |
| rs4808102 | A | G | chr19:17909876 | NA | COLGALT1 (Q8NBJ5) | COLGALT1 (Q8NBJ5) | chr19:17555649-17583162 | 4.75 × 10−5 | 4.01 × 10−2 | 0.819 (0.192) |
For genomic regions around each COPD-associated proteins and each target gene, only one significant top pQTL association is presented.
Positive β-coefficients mean higher expression level for the alternative allele.
FDR were adjusted with COPD risk SNPs.
Mediation analysis
The directed acyclic graph used for the mediation analysis is presented in Figure E6 in the data supplement. Mediated associations linking cis-pQTLs (as exposure), the transcripts of COPD-associated proteins (as mediator), and the COPD-associated protein levels (as outcome) based on significant cis-pQTL results with FDR lower than 0.1 are presented in Table E11 in the data supplement. The total causal effects for all candidate associations were significant, reflecting the significant associations between cis-pQTLs and proteins. However, no evidence of mediated effects from transcripts to proteins was observed at an FDR lower than 0.1.
Colocalization of QTLs near COPD-associated proteins and COPD GWAS signals
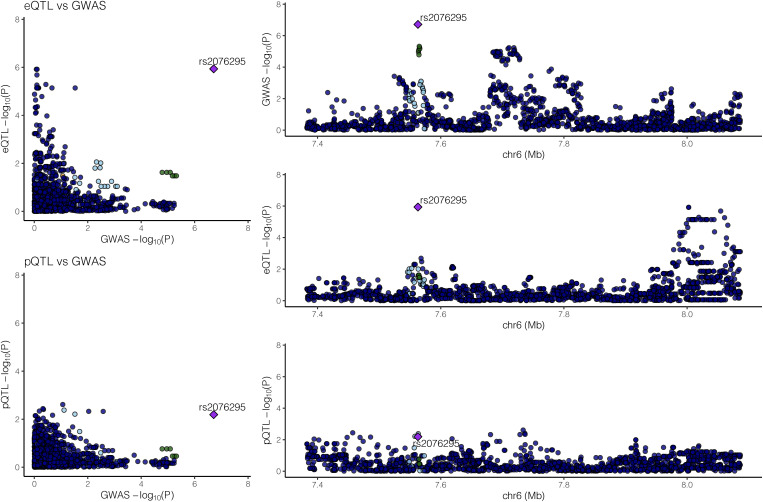
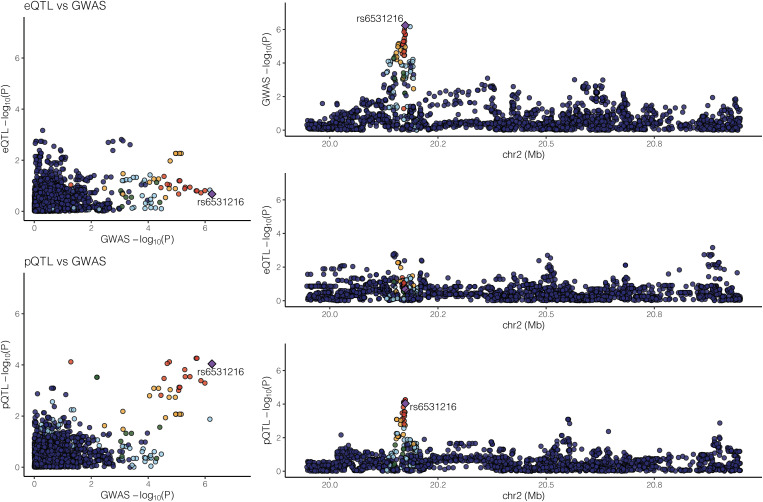
With 85 COPD-associated proteins and 82 COPD GWAS genomic regions, two genomic regions showed supportive evidence for colocalization as shown in Figures 2 and 3. First, a colocalized effect between COPD GWAS and eQTL effects (regulating DSP) around COPD-associated protein TXNDC5 was observed with a colocalization probability of 0.907. This region is also near COPD GWAS SNP rs1334576. The top COPD GWAS SNP in DSP (rs2076295) is a significant eQTL for DSP, as shown in Table E9. Second, another genomic region near RHOB, which encodes a COPD-associated protein, showed evidence for colocalized effects between COPD GWAS and pQTLs with a probability of 0.579. The top COPD GWAS SNP (rs6531216) of the RHOB region is in strong linkage disequilibrium with a significant cis-pQTL (rs11096641) in the same region (Table E10), with an r2 of 1 in Europeans in the 1000 Genomes Project (23). Interestingly, no significant eQTLs in the RHOB region or significant pQTLs in the TXNDC5/DSP region at an FDR lower than 0.05 were detected. The previously reported pQTLs for AGER in blood samples were not observed in lung tissue samples (Figure E7 in the data supplement).
Figure 2.
Significant colocalized effects between COPD genome-wide associated study (GWAS) and expression quantitative trait locus (eQTL) signal for DSP. Probability of colocalization between eQTL and COPD GWAS is 0.907; no evidence for colocalization between protein quantitative trait loci and COPD GWAS was found. pQTL = protein quantitative trait locus. DSP = desmoplakin.
Figure 3.
Significant colocalized effects between GWAS and pQTL signals for RHOB (UniProt ID: P62745, COPD proteomic FDR: 0.096). Probability of colocalization between pQTL and COPD GWAS is 0.579; no evidence for colocalization between eQTL and COPD GWAS was found.
WGCNA
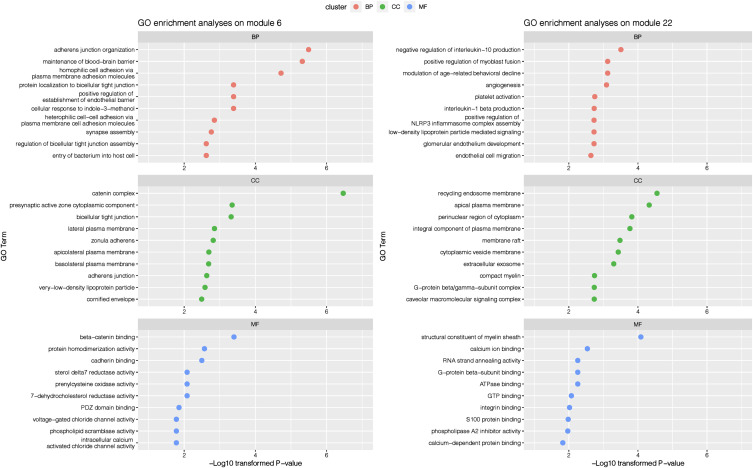
We selected optimized parameters to identify consensus correlation networks between transcriptomics and proteomics as described in Figure E8 in the data supplement. With a minimum module size of 10, we identified 53 candidate modules, and 2 modules (modules 6 and 22) were shown to be associated with COPD. Associations between COPD and genes from COPD-associated modules at the transcriptomics and proteomics levels are presented in Tables E12 and E13 in the data supplement.
Plots presenting the functional enrichment results of genes from these two consensus COPD-associated modules are shown in Figure 4. The most significantly associated biological processes were the catenin complex for one of the correlation network modules and plasma membrane components (including endosomes and exosomes) for the other correlation network module.
Figure 4.
Gene Ontology (GO) enrichment analysis results on genes from Weighted Gene Co-Expression Network Analysis consensus modules between transcriptomics and proteomics levels. Top 10 enriched terms for each GO cluster are presented.
Nominally significant associations (P < 0.05) between several of the 82 previously reported COPD GWAS loci and COPD-associated consensus network modules (modules 6 and 22) are shown in Table E14 in the data supplement. One shared significant SNP–module association between transcriptomics and proteomics levels for rs11655567 (on chromosome 17 near SOX9) and module 6 was found.
Cell Type Proportion Estimate and Adjustment on Omics Associations
We estimated the proportion of four groups of cell types (epithelial cells, endothelial cells, stromal cells, and immune cells) based on the bulk RNA sequencing data of our 98 subjects with matched transcriptomics and proteomics data. The distribution of four major cell group proportions across subjects is shown in Figure E9 in the data supplement. The proportions of cell groups were compared between COPD and control groups as shown in Figure E10 in the data supplement. Differential cell type proportions between patients with COPD and controls were observed for endothelial and stromal cells but not immune or epithelial cells.
The P values and β-coefficients of COPD associations with shared omics biomarkers after adding cell type group proportions as covariates are shown in Tables E15 and E16 in the data supplement. Many of these associations are attenuated, suggesting that they are at least partly attributed to differences in cellular composition between COPD and control lung tissue samples.
We selected two significant QTL genes: MAPT (eQTLs and pQTLs) and RHOB (pQTLs) to investigate cell type proportion effects on QTL associations. The Pearson’s correlations between the omics expression levels for these genes (after adjusting for COPD, age, and sex, with batch effects removed) and cell proportions are shown in Figure E11 in the data supplement. Nominally significant association between RHOB and endothelial cell proportion at the transcriptomics level and between RHOB and immune cell proportion at the proteomics level were observed with a P value threshold of 0.05.
The QTL analysis results with or without cell proportion adjustment (Table E17 in the data supplement) of MAPT (Microtubule-associated protein tau) are not significantly affected by cell proportion adjustments, but association between genetic determinants and proteomic expression of RHOB was attenuated by cell group proportion adjustment.
Discussion
By integrating whole-genome sequencing with lung tissue transcriptomics and proteomics data, we found a subset of 15 shared transcriptomic and proteomic analytes associated with COPD that are likely valid lung tissue COPD biomarkers. We found weak correlations overall between lung tissue transcriptomics and proteomics, but stronger omics correlations for COPD-associated genes. One COPD GWAS signal was associated with MAPT expression and could implicate the key gene (MAPT) for that GWAS locus. Multiple cis-acting QTLs for transcripts and proteins associated with COPD were detected. Colocalized effects for RHOB and DSP were observed, but evidence for colocalization was not consistent between eQTLs and pQTLs. Finally, we identified concordant correlation network modules in transcriptomics and proteomics that implicated key pathways in COPD pathogenesis, including β-catenin signaling and membrane-based transport.
Approaches for multi-omics analysis methods can be broadly divided into three scientific goals: 1) to identify COPD-associated pathogenic factors with QTL analyses integrating genetic data and quantitative omics data, 2) to perform disease subtyping, and 3) to identify networks of interacting genes and proteins. In 2015, Kim and colleagues developed an integrative phenotyping framework for COPD subphenotype identification by integrating phenotypes and other omics information (24). In 2018, Li and colleagues published multi-omics analyses integrating omics data from mRNA, microRNA, proteomics, and metabolomics using similarity network fusion (25). Quantitative relationships between multi-omics data were evaluated, and improved COPD classification was observed using multi-omics data compared with data from single omics levels (25). We focused on the first and third goals because of our limited sample size. Larger sample sizes of lung tissue multi-omics data will be needed for well-powered efforts to reclassify COPD into endotypes: subtypes based on molecular etiologies.
Previous studies have attempted to integrate COPD-associated omics biomarkers. In 2020, Mastej and colleagues applied SmCCNet analysis (Sparse Multiple Canonical Correlation Network) (26), which integrated proteomics and metabolomics data to identify novel regulatory networks connecting different omics levels (27). Although these approaches implicated different COPD biomarkers than we identified, they were similar in emphasizing the importance of network relationships in COPD pathogenesis.
We observed low correlations between transcriptomics and proteomics in lung tissue. In 2002, Chen and colleagues tested correlations between transcriptomics and proteomics levels. Correlation coefficients for 69 tested genes varied between −0.467 and 0.442, with a gene-level mean r of 0.103 (28). In 2009, Gry and colleagues observed correlations between cDNA or oligo-based microarray–detected RNA and protein across 23 human cancer cell lines, yielding low mean correlation coefficients of 0.2 (cDNA) and 0.25 (oligo), respectively (29). In 2019, Wang and colleagues performed systematic analyses of the correlations between transcriptomics and proteomics in 29 paired healthy tissues and reported a linear relationship with a correlation coefficient of 0.52 (30). In our study, approximately 15% (606 of 4,039) of all matched genes are correlated at transcriptomics and proteomics levels with correlation coefficients statistically different from zero. Various factors including translational efficiency, codon bias, ribosome density, posttranslational modification, mRNA/protein half-life, and experimental measurement bias may lead to low correlations between transcriptomics and proteomics (31).
With only 98 subjects for QTL analyses, we identified multiple cis-QTL effects for COPD-associated proteins; however, lead SNPs in COPD GWAS regions were rarely QTLs in lung tissue, which is consistent with the concept that most COPD GWAS signals are not associated with gene expression changes that can be detected in lung tissue (32). Such expression differences might be detectable in larger samples, single cell types, or specific developmental stages. One COPD GWAS signal (rs12373142) was associated with MAPT expression and could implicate the key gene for that locus. A common genomic inversion of the MAPT genomic region has been previously reported (22). MAPT itself has also been recognized as a potential high-priority eQTL-regulated gene with a genomic variant (rs2532349) associated with FEV1 (33, 34).
A cis-pQTL (but not cis-eQTL) near RHOB, which encodes a COPD protein biomarker, colocalized with a sub-genome-wide significant COPD GWAS association, suggesting that the genetic determinants of this COPD protein biomarker also influence COPD susceptibility. RHOB is a mediator for hypoxia-induced pulmonary vascular remodeling (35), which has been reported to be associated with COPD (36). The DSP eQTL for rs2076295 was previously reported to colocalize with a COPD GWAS signal (37); interestingly, no pQTL effect was found in that region. These results confirm the independent value of transcriptomics and proteomics in determining the functional impact of GWAS signals.
Mediation analyses for significant cis-pQTL results validated the associations between pQTLs and proteins but failed to recognize the mediator role of matched transcripts between SNPs and proteins in our small dataset, implying that pQTLs may not only regulate the protein expression level directly through protein-coding transcripts. Further studies with larger sample sizes may help identify more COPD multi-omics biomarkers and regulatory mechanisms between different omics.
We used concordant relationships in transcriptomics and proteomics to identify COPD-related correlation networks. By using WGCNA, we found evidence that shared network modules for transcriptomics and proteomics are associated with COPD and implicate important biological processes in COPD, including β-catenin signaling and membrane-based transport. Wnt/β-catenin signaling has been reported to be associated with COPD pathogenesis at transcriptomics (38) and proteomics (39) levels. We observed a nominal association between COPD-associated WGCNA modules and several reported COPD GWAS loci. The biological mechanisms for these associations will require further investigation.
Differential cellular composition of lung tissue could contribute to the observed transcriptomic and proteomic associations with COPD. Using cellular deconvolution, we observed significantly lower proportions of endothelial cells and higher proportions of stromal cells in COPD lung tissue samples. The statistical significance of many of the COPD-associated transcriptomic and proteomic biomarkers was partially attenuated with cell type proportion adjustment. For QTL genes MAPT and RHOB, after cell proportion adjustment, the MAPT QTL associations remained significant and the RHOB pQTL was attenuated. Thus, cellular composition may influence COPD omics biomarkers and their QTLs. Future investigations should assess cell type–specific transcriptomic and proteomic biomarkers for COPD.
Limitations of the Present Study
Although we have conducted multi- omics analyses integrating genetics, transcriptomics, and proteomics of COPD and control lung tissues, our study has several limitations. Although we have found some associations between COPD risk SNPs and protein biomarkers and colocalized effects comparing GWAS and QTL signals, our dataset (98 subjects) is underpowered to find connections between different omics levels and to identify mediated associations linking significant QTLs, genes, and COPD affection status. QTL analyses were used to find downstream transcripts or proteins regulated by COPD GWAS loci. However, not all of the previously reported COPD GWAS loci–associated proteins were detected in our mass spectrometry proteomics dataset. Associations between COPD GWAS loci and reported functional proteins like HHIP and FAM13A may be missed because those proteins were not detected in our lung tissue proteomics dataset. Use of bulk gene expression and proteomic analysis limited detection of cell type–specific effects, although cellular deconvolution methods can help to overcome this limitation.
Conclusions
In our lung tissue multi-omics analyses, a weak overall correlation between lung transcriptomics and proteomics values was observed, but some highly significant correlations were found. Biological connections between genetic variants and omics expression levels are likely to be complex network relationships, which will require larger sample sizes to study comprehensively. Mediation analyses and correlation-based network analyses of multiple omics data identified potential genes and proteins that may influence COPD pathogenesis. With limited correlations between transcripts and proteins, additional studies of COPD proteomics are warranted.
Acknowledgments
Acknowledgment
The authors thank the participants who provided biological samples and data for LTRC and TOPMed.
Footnotes
Supported by research project grants from the National Institutes of Health, including R01 HL133135, R01 HL147148, R01 HL137927, R01 HL124233, R01 GM087221, and P01HL114501. This study used biological specimens and data provided by the Lung Tissue Research Consortium supported by the National Heart, Lung, and Blood Institute (NHLBI). Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by NHLBI. Core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL117626-02S1; contract HHSN268201800002I). Core support including phenotype harmonization, data management, sample-identity quality control, and general program coordination were provided by the TOPMed Data Coordinating Center (R01HL120393; U01HL120393; contract HHSN268201800001I).
Author Contributions: Conception and design: Y.-H.Z., R.L.M., and E.K.S. Analyses and interpretation: all authors. Drafting the manuscript for important intellectual content: all authors.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2022-0302OC on February 13, 2023
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Han MK, Agusti A, Calverley PM, Celli BR, Criner G, Curtis JL, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med . 2010;182:598–604. doi: 10.1164/rccm.200912-1843CC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stockley RA. Alpha-1 antitrypsin deficiency: the learning goes on. Am J Respir Crit Care Med. 2020;202:6–7. doi: 10.1164/rccm.202004-0922ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sakornsakolpat P, Prokopenko D, Lamontagne M, Reeve NF, Guyatt AL, Jackson VE, et al. SpiroMeta Consortium; International COPD Genetics Consortium Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nat Genet . 2019;51:494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pratte KA, Curtis JL, Kechris K, Couper D, Cho MH, Silverman EK, et al. Soluble receptor for advanced glycation end products (sRAGE) as a biomarker of COPD. Respir Res . 2021;22:127. doi: 10.1186/s12931-021-01686-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Silverman E, Hobbs B. Integrating genetics and omics to understand chronic obstructive pulmonary disease. Barcelona Respir Netw Rev . 2020;6:104–117. [Google Scholar]
- 6. Li CX, Wheelock CE, Sköld CM, Wheelock ÅM. Integration of multi-omics datasets enables molecular classification of COPD. Eur Respir J . 2018;51:1701930. doi: 10.1183/13993003.01930-2017. [DOI] [PubMed] [Google Scholar]
- 7. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics . 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res . 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang YH, Hoopmann MR, Castaldi PJ, Simonsen KA, Midha MK, Cho MH, et al. Lung proteomic biomarkers associated with chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol . 2021;321:L1119–L1130. doi: 10.1152/ajplung.00198.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics . 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diedenhofen B, Musch J. cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One . 2015;10:e0121945. doi: 10.1371/journal.pone.0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics . 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) . 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am J Hum Genet . 2015;96:926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benway CJ, Liu J, Guo F, Du F, Randell SH, Cho MH, et al. International COPD Genetics Consortium Chromatin landscapes of human lung cells predict potentially functional chronic obstructive pulmonary disease genome-wide association study variants. Am J Respir Cell Mol Biol . 2021;65:92–102. doi: 10.1165/rcmb.2020-0475OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steen J, Loeys T, Moerkerke B, Vansteelandt S. medflex: an R package for flexible mediation analysis using natural effect models J Stat Softw 2017. 76 1 46 36568334 [Google Scholar]
- 17. Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet . 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu B, Gloudemans MJ, Rao AS, Ingelsson E, Montgomery SB. Abundant associations with gene expression complicate GWAS follow-up. Nat Genet . 2019;51:768–769. doi: 10.1038/s41588-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics . 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics . 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 21. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature . 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. A common inversion under selection in Europeans. Nat Genet . 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 23. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature . 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S, Herazo-Maya JD, Kang DD, Juan-Guardela BM, Tedrow J, Martinez FJ, et al. Integrative phenotyping framework (iPF): integrative clustering of multiple omics data identifies novel lung disease subphenotypes. BMC Genomics . 2015;16:924. doi: 10.1186/s12864-015-2170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li C-X, Wheelock CE, Sköld CM, Wheelock ÅM. Integration of multi-omics datasets enables molecular classification of COPD. Eur Respir J . 2018;51:1701930. doi: 10.1183/13993003.01930-2017. [DOI] [PubMed] [Google Scholar]
- 26. Shi WJ, Zhuang Y, Russell PH, Hobbs BD, Parker MM, Castaldi PJ, et al. Unsupervised discovery of phenotype-specific multi-omics networks. Bioinformatics . 2019;35:4336–4343. doi: 10.1093/bioinformatics/btz226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mastej E, Gillenwater L, Zhuang Y, Pratte KA, Bowler RP, Kechris K. Identifying protein–metabolite networks associated with copd phenotypes. Metabolites . 2020;10:124. doi: 10.3390/metabo10040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen G, Gharib TG, Huang C-C, Taylor JM, Misek DE, Kardia SL, et al. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics . 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 29. Gry M, Rimini R, Strömberg S, Asplund A, Pontén F, Uhlén M, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics . 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang D, Eraslan B, Wieland T, Hallström B, Hopf T, Zolg DP, et al. A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Syst Biol . 2019;15:e8503. doi: 10.15252/msb.20188503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haider S, Pal R. Integrated analysis of transcriptomic and proteomic data. Curr Genomics . 2013;14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrow JD, Zhou X, Lao T, Jiang Z, DeMeo DL, Cho MH, et al. Functional interactors of three genome-wide association study genes are differentially expressed in severe chronic obstructive pulmonary disease lung tissue. Sci Rep . 2017;7:44232. doi: 10.1038/srep44232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Soler Artigas M, et al. UK Brain Expression Consortium (UKBEC); OxGSK Consortium Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med . 2015;3:769–781. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wain LV, Shrine N, Artigas MS, Erzurumluoglu AM, Noyvert B, Bossini-Castillo L, et al. Understanding Society Scientific Group; Geisinger-Regeneron DiscovEHR Collaboration Genome-wide association analyses for lung function and chronic obstructive pulmonary disease identify new loci and potential druggable targets. Nat Genet . 2017;49:416–425. doi: 10.1038/ng.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wojciak-Stothard B, Zhao L, Oliver E, Dubois O, Wu Y, Kardassis D, et al. Role of RhoB in the regulation of pulmonary endothelial and smooth muscle cell responses to hypoxia. Circ Res . 2012;110:1423–1434. doi: 10.1161/CIRCRESAHA.112.264473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeffery PK. Remodeling in asthma and chronic obstructive lung disease. Am J Respir Crit Care Med . 2001;164:S28–S38. doi: 10.1164/ajrccm.164.supplement_2.2106061. [DOI] [PubMed] [Google Scholar]
- 37. Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet . 2017;49: 426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carlier FM, Dupasquier S, Ambroise J, Detry B, Lecocq M, Biétry-Claudet C, et al. Canonical WNT pathway is activated in the airway epithelium in chronic obstructive pulmonary disease. EBioMedicine . 2020;61:103034. doi: 10.1016/j.ebiom.2020.103034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baarsma HA, Menzen MH, Halayko AJ, Meurs H, Kerstjens HAM, Gosens R. β-Catenin signaling is required for TGF-β1-induced extracellular matrix production by airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol . 2011;301:L956–L965. doi: 10.1152/ajplung.00123.2011. [DOI] [PubMed] [Google Scholar]