Abstract
Programmed chromosome breakage occurs at 50–200 specific sites in the genome of Tetrahymena thermophila during somatic nuclear (macronuclear) differentiation. Previous studies have identified a 15 bp sequence, the Cbs (for chromosome breakage sequence), that is necessary and sufficient to specify these sites. In this study we determined the effects of mutations in the Cbs on its ability to specify the chromosome breakage site and promote new telomere formation in conjugating cells. Twenty-one constructs with single nucleotide substitutions covering all 15 positions of the Cbs were made and tested. Fourteen of them (covering 11 positions) abolished breakage entirely, six (covering six positions, including the remaining four) caused partial loss of breakage function and one showed no detectable effect. This result indicates that the Cbs has an exceptionally long and stringent sequence requirement. It offers no evidence that the Cbs contains a separate domain for promoting new telomere formation. In addition, we found that a partially functional Cbs retained in the macronucleus does not induce chromosome breakage during vegetative growth and that excess copies of this germline-specific sequence in the somatic nucleus have little deleterious effect on cell growth.
INTRODUCTION
Programmed chromosome breakage is known to occur during development of the somatic progenitor cells or nuclei of several eukaryotes, including ciliated protozoa, ascarid nematodes (1) and, possibly, hagfish (2). This developmental process converts intact chromosomes into specific smaller fragments, which are maintained throughout the organism’s somatic life. This drastic alteration is likely to have significant impact on the expression and maintenance of the genome, although little is known about its exact roles or its regulatory mechanism. Recent studies on the model ciliate Tetrahymena thermophila have begun to shed light on this issue, especially on the nature of its cis-regulatory sequences (3).
Like most ciliates, T.thermophila contains two types of nuclei in each cell: a diploid micronucleus in which transcription is not detectable during vegetative growth and a polyploid, transcriptionally active macronucleus. The micronucleus divides by mitosis whereas the macronucleus divides amitotically without detectable chromosome condensation. When cells conjugate, their micronuclei undergo a series of events including meiosis, nuclear exchange, nuclear fusion and mitosis to give rise to progenitors of the macro- and micronuclei for the succeeding asexual generation. Their old macronuclei are resorbed during this process (for a review see 4). Development of the new macronucleus involves a complex series of DNA altering events including chromosome breakage, DNA deletion and gene amplification (3,5). Chromosome breakage is known to occur at 50–200 specific sites distributed among the five germline chromosomes. The resulting chromosome fragments, which average ~800 kb in size, are maintained in the mature macronucleus throughout the following vegetative life (for a review see 6).
Analysis of DNA sequences around several breakage sites has revealed a common 15 bp sequence (5′-TAAACCAACCTCTTT), referred to as the Cbs (for chromosome breakage sequence), which is necessary and sufficient to specify chromosome breakage sites in T.thermophila (7). Plasmid constructs bearing or lacking the Cbs at specific sites have been made and introduced into developing macronuclei by microinjection. DNA breakage occurs at, and only at, sites containing the Cbs (8,9). The Cbs appears to be the only breakage signal used by T.thermophila and is highly conserved among most species of Tetrahymena (10).
Chromosome breakage in T.thermophila and other species is coupled with the formation of new telomeres. In T.thermophila, new telomeric DNA is added to the broken ends at sites 5–25 bp away from either side of the Cbs, resulting in the loss of ~25–65 bp of sequence at the breakage junction, including the Cbs (9,11). The points of telomere addition are variable for a given site. The Cbs appears to be the only signal needed for chromosome breakage and end healing to occur. Alteration of sequences surrounding the Cbs has little or no effect on breakage or telomere addition (11), suggesting a mechanistic link between these two events.
Identification of the Cbs was based mainly on its conservation among different genomic sites and different Tetrahymena species. Whether the entire 15 bp sequence is needed is not known. It is also unclear if separate regions of the Cbs control breakage and end healing. To better understand this process, we have created and tested specific mutations within the Cbs in this study and found that all 15 nt positions are important for normal function. We also found no evidence for a separate domain for telomere formation.
MATERIALS AND METHODS
Cell culture, Tetrahymena transformation and DNA analysis
Tetrahymena thermophila inbreeding line B, strains CU 427 and CU 428, were obtained from Peter Bruns and used throughout this study. Cells were grown in axenic medium as described earlier (12). Transformation of T.thermophila was carried out using the microinjection method described earlier (8,13). Synchronous conjugating cells were injected with the cloned DNA and afterwards individually cultivated. They were replicated to medium containing paromomycin to check for drug resistance as a result of transformation. The transformation rates ranged from 1 to 5%. These cells were grown in 5–10 ml of selective medium until reaching the stationary phase of growth before harvesting for DNA analysis. Isolation of DNA was carried out using a previously published method (14). Restriction enzyme digestion, ligation, gel electrophoresis, DNA labeling and Southern hybridization were performed using standard methods (15).
PCR analysis
PCR was carried out using Taq DNA polymerase (Perkin Elmer) under standard reaction conditions specified by the supplier and the following reaction temperature cycles were used: 93°C for 1 min, 53°C for 1 min, 72°C for 50 s for up to 50 cycles. Template DNAs were used at ~200 ng/ml. PCR products were separated by electrophoresis in a 3% agarose gel and transferred to a nylon filter (Gene Screen; NEN) for hybridization by standard blotting methods. The sequences of the two oligonucleotides used as primers were 5′-ACAA-AAAACCCTTCTAAAAG (rdn10026) and 5′-CCCCAACCCCAACCCCAA (Tel), and the sequence of the hybridization probe rdn10069 was 5′-TAAAAATTTATGAAAACTAC.
DNA clone construction
To construct clones with Cbs insertions, the vector FANA3 (11) was either digested with SmaI and ligated with a double-stranded oligonucleotide with the sequence 5′-GGGAA-TTAAACCAACCTCTTTGTTTAG (the Cbs is underlined) in one strand or digested with SmaI and NotI and ligated with an oligonucleotide with the same sequence but containing an additional 4 nt (5′-GGCC) at the 5′-end of the complementary strand to generate a protruding 5′-end. To introduce mutations, one of the two oligonucleotides was synthesized using mixtures of nucleotides in all positions occupied by the Cbs. In each position 92.5% of the nucleotides corresponded to the wild-type nucleotide and the remaining were equal mixtures of the other three nucleotides. Clones with inserts were identified by colony hybridization using the insert as probe and the regions surrounding the inserts were sequenced. Some clones contained additional sequences adjacent to the expected inserts, probably resulting from co-insertion of aberrant oligonucleotides during cloning. More than 100 clones were sequenced to produce most of the 21 clones described in this paper. Mutations in some locations were not obtained by this method after repeated trials and were produced directly by synthesizing the desired sequences for insertion (for clones FA11A and FA14C).
RESULTS
Construction of clones with a mutated Cbs
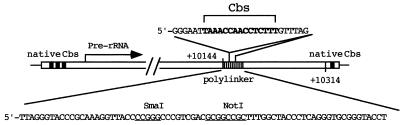
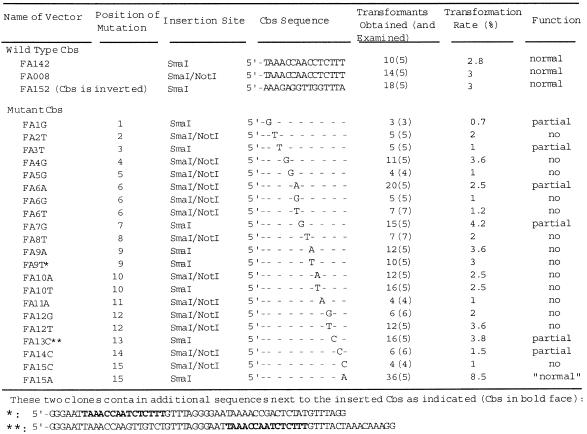
We have constructed a panel of clones with mutations in the Cbs using the modified micronuclear rDNA vector FANA3 (11) (Fig. 1) as a starting point. FANA3 contains the entire rDNA sequence plus its immediate flanking regions, including native copies of the Cbs and other sequence signals necessary for rDNA processing during formation of the macronucleus. In addition, it contains a 72 bp synthetic linker with unique SmaI and NotI sites in the 3′-spacer region of the rDNA just upstream of the native Cbs (at position 10144 of the rDNA sequence). We inserted into this SmaI site, or between the SmaI and NotI sites, a 27 bp synthetic oligonucleotide that included a copy of the Cbs with various mutations. Altogether 21 clones with single nucleotide substitutions spanning the 15 positions of the Cbs were generated and tested (Table 1).
Figure 1.
rDNA vector with mutated Cbs inserts. The micronuclear rDNA clone FANA3 contains a 72 bp synthetic polylinker in the 3′-spacer region (at position 10144) of the rDNA which serves as a site for insertion. It also contains the Cbs at its native locations at both ends of the rDNA and all sequences needed for rDNA processing and expression. A series of clones containing a mutated version of the Cbs were constructed by inserting a 27 bp oligonucleotide containing the mutated sequence into the polylinker at the SmaI site or between the SmaI and NotI sites.
Table 1. Cbs mutant clones and their transformation properties.
The insertion site is ~190 bp upstream of the native Cbs at the 3′-boundary of the rDNA. Thus breakage at the insertion site would produce an rDNA molecule with a truncated terminus, while failure of breakage at this site would produce one with a normal terminus (as a result of breakage at the native Cbs site). These two types of termini are known to be capped with new telomeric DNA and can be distinguished from each other using a PCR assay (see below). Breakage at the inserted Cbs without telomere addition, however, would produce an rDNA molecule with a broken end, which is likely to be lost. Thus no transformant should be generated (11). Alternatively, the broken end could be repaired through recombination, thus producing ends indistinguishable from those of the endogenous rDNA. To serve as positive controls, three clones (FA142, FA152 and FA008) containing a normal Cbs in the insert were also tested (Table 1). FA142 and FA152 were analyzed in a previous study and shown to be able to transform Tetrahymena to produce rDNA with the expected truncated termini (11).
Analysis of chromosome breakage
These DNA clones were injected into developing macronuclei of conjugating cells using a method developed previously (8,13). Transformants were selected and grown and their DNA were isolated for analysis. To determine the structure of the macronuclear rDNA termini, the sequences were amplified by PCR using two oligonucleotide primers, the 5′ primer matching a sequence near the rDNA 3′-end (rdn10026) and the 3′ primer matching the telomeric sequence (Tel) (Fig. 2A). We expected a 0.13 kb PCR product if breakage and telomere formation occurred at the insertion site or a 0.41 kb product if breakage failed to occur at this site. These two products were easily distinguished from the 0.31 kb product of the endogenous rDNA, which is present in varying amounts in these transformants. To verify their identities and increase the sensitivity of detection, after gel electrophoresis and blotting these PCR products were hybridized with an oligonucleotide probe (rdn10069) from the amplified region (Fig. 2A). The sensitivity of this PCR assay was tested by mixing DNA from FA142 transformants with a small quantity (a 1:100 or 1:1000 weight ratio) of DNA from FANA3 transformants. FANA3 is known to transform Tetrahymena and generate rDNA with normal termini (11). The results (Fig. 2B) show that the 0.41 kb fragment from FANA3 transformants and a small amount of the 0.31 kb fragment from endogenous rDNA are present in both cases, as expected. The 0.13 kb fragment from the FA142 transformant was also present in the 1:100 mixture (lane 1) but not in the 1:1000 mixture (lane 2). Thus the method allows the detection of a very low level of breakage activity at the insertion site.
Figure 2.

A PCR assay for detecting rDNA termini. (A) The strategy of the assay. PCR was carried out using rdn10026 and Tel as primers and the transformant DNA as template. If breakage and telomere addition occur at the inserted Cbs, the rDNA should produce a short (0.13 kb) fragment after PCR. Otherwise a 0.41 kb DNA fragment should be produced due to breakage at the native 3′ Cbs of the rDNA. PCR products from the endogenous rDNA is shorter (0.31 kb) due to absence of the linker and the insert. (B) The results of an experiment to determine the sensitivity of the PCR assay. In this experiment total DNAs extracted from FA142 and FANA3 transformants were mixed in different ratios (1:100 and 1:1000) and used as templates for PCR. The results show that the 0.13 kb fragment is detectable when its template is only 1/100 of the total (lane 1), but not when it is 1/1000 (lane 2).
Most mutations abolish Cbs function
All 21 clones with mutated Cbs inserts transformed Tetrahymena at normal rates (1–5%) (Table 1). None of them produced rDNA with only the endogenous-type termini (see below). To obtain a better representation, several (3–7) transformants derived from each clone were pooled for analysis. Examples of these results are shown in Figures 3 and 4. These clones fall into three groups. The first group includes 14 clones (Fig. 3, lanes 5, 6, 8–16, 19 and 20). Amplification of DNA of transformants derived from these clones produced only the 0.41 kb fragment and not the 0.13 kb fragment. Since each DNA sample was derived from a mixture of transformants and since the PCR assay is quite sensitive, the results indicate that little if any breakage occurs at the insertion site in these clones. We conclude that this group of mutated Cbs has no detectable function. The second group includes six clones. Their transformants produced both the 0.13 and 0.41 kb fragments (Fig. 3, lanes 7, 17 and 21, and data not shown). Thus this group of mutated Cbs are able to function, but only partially, as a chromosome breakage site. Because the assay is rather sensitive, some or all of these constructs could have only weak activities. The third group includes only one clone, whose transformants produced the 0.13 and not the 0.41 kb fragment (Fig. 3, lane 18), similar to the control construct FA142. Thus, this Cbs mutant appears to have a strong, and perhaps normal, level of function. In addition to the 0.13 and 0.41 kb fragments, the 0.31 kb fragment derived from the endogenous rDNA was also detected in most transformants, as expected.
Figure 3.
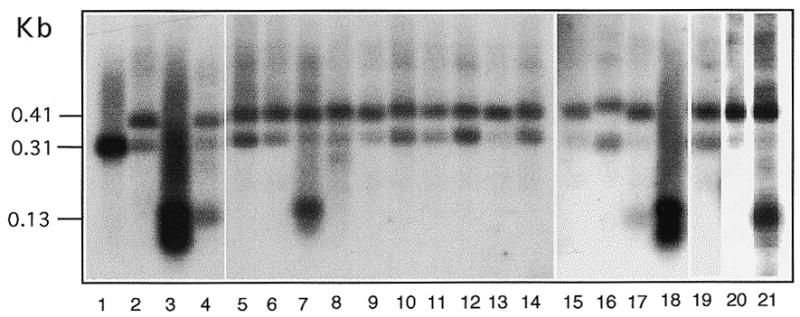
Effects of base substitutions on Cbs function. To analyze each construct listed in Table 1, total DNA from several independent transformants generated (numbers shown in parentheses in Table 1) were mixed and analyzed by the PCR assay described in Figure 2. These constructs (or controls) are: lane 1, control endogenous DNA from strain CU427; lane 2, FANA3 (without Cbs insertion); lane 3, FA142 (wild-type Cbs); lane 4, 1:100 mixture of transformants from FA142 and FANA3; lanes 5–21, FA4G, FA8T, FA6A, FA2T, FA12G, FA6G, FA6T, FA5G, FA12T, FA15C, FA9A, FA9T, FA7G, FA15A, FA10T, FA10A and FA3T, respectively. The 0.41 kb fragment is detected in FANA3 and Cbs mutant transformants. The 0.13 kb fragment is detected in transformants of FA142, FA6A, FA7G, FA15A and FA3T. The 0.31 kb fragment is generated from endogenous rDNA and found in varying amounts in some transformants.
Figure 4.
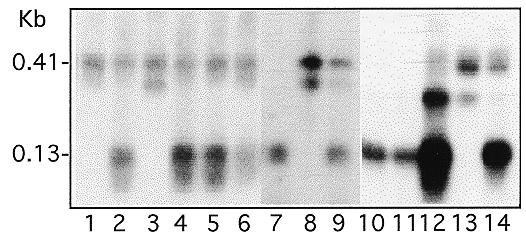
Analysis of individual transformants from clones with breakage activities. DNA from individual transformed lines derived from clones displaying some breakage activity was analyzed by the same method as described in Figure 2. Transformants from three clones are shown here as examples. Each lane contains the PCR product from an individual line. Clones used to generate these transformants were: lanes 1–6, FA6A; lanes 7–9, FA1G; lanes 10–14, FA3T.
In addition to analyzing DNA pooled from several transformants, we have also analyzed DNA from individual transformants in the second group. Three examples are shown in Figure 4. We found transformants containing either the 0.13 or 0.41 kb fragment, or both. This distribution pattern again indicates that these mutated Cbs are partially defective and that different copies of the same sequence present in the same cell or nucleus could have different fates. Analysis of subclones of these transformants further supports this point (see below).
Cbs in vegetative cells
The Cbs is normally absent from the macronucleus of Tetrahymena (7), but many transformants described above contain thousands of copies of the mutated Cbs in their macronucleus, including those with moderate function for specifying breakage sites. This situation offers a unique opportunity to study the fate of the Cbs in the macronucleus during vegetative growth. Two FA1G transformants were chosen for this analysis. One of them contained transforming rDNA that had not been broken at the insertion site and the other contained a mixture of transforming rDNA with broken and unbroken sites. Three or four single cells were isolated from each transformed line to establish subclones, which were grown in selective medium through successive transfers. After ~20 and ~120 cell doublings, DNA was extracted from these subclones and analyzed. The results show that the composition of the rDNA, as indicated by the structure of the termini, does not change significantly during this growth period (Fig. 5). Those clones that contained only the 0.41 kb band initially did not produce any of the 0.13 kb rDNA band (compare Fig. 5, lanes 1–4 with lanes 8–11). This result indicates that breakage and end healing do not occur at these sites during vegetative growth. In addition, clones that contained both types of the rDNA in the beginning were able to maintain them both after extended growth, although the rDNA with truncated termini might have decreased slightly in proportion (Fig. 5, lanes 5–7 and 12–14). It should be noted that both types of transforming rDNA confer drug resistance, thus drug selection is not expected to affect their relative abundance. This result suggests that a partially functional Cbs, even at thousands of copies in the cell, does not have significant effects on cell growth.
Figure 5.
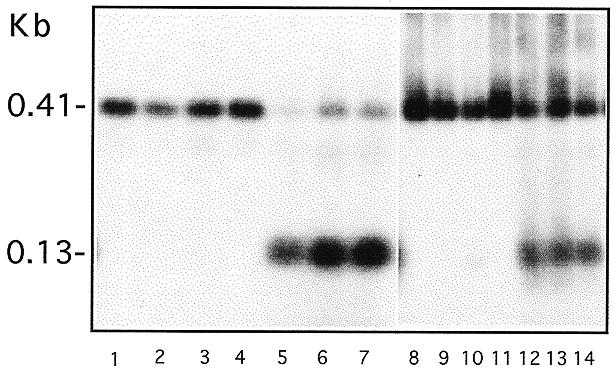
Fate of the Cbs in macronuclei of vegetative cells. Two of the FA1G transformants (see Fig. 4, lanes 8 and 9) were used to analyze the fate of rDNA with a mutated, partially functional Cbs in vegetatively growing cells. Four subclones were isolated from one and three from the other transformant and propagated for 20 and 120 generations before harvesting for the PCR analysis described in Figure 2. Lanes 1–7 are PCR products of these subclones after 20 generations of growth and lanes 8–14 are products of the same subclones (in the same order) after 120 generations of growth. Subclones analyzed in lanes 1–4 and 8–11 are from one transformed line and those in lanes 5–7 and 12–14 from the other.
DISCUSSION
The 15 bp sequence Cbs (5′-TAATTCCAACCTCTTT) is known to specify the sites of developmentally regulated chromosome breakage in Tetrahymena (9). In this study we analyzed the effects of single base substitutions within this sequence by testing at least one mutation in each of the 15 positions, and found that most of them either abolished or significantly reduced Cbs function. These results indicate that all 15 positions within this sequence are important for its function. In several positions multiple mutations were tested. For example, all three possible mutations at position 6 were analyzed; two abolished Cbs function whereas the third only reduced it. Thus different substitutions had different effects at some positions.
These results indicate that the Cbs is an unusually long and stringent DNA sequence signal. It is considerably longer than the recognition sequences of most restriction endonucleases, which are 3–8 bp in length. Although homing endonucleases have long recognition sequences (between 14 and 40 bp), they are generally more tolerant of sequence variations than the Cbs (16). This feature of the Cbs could be related to its biological role. Chromosome breakage is a tightly regulated process that fragments chromosomes. Although its exact function is not yet clear, such a drastic process is likely to have major effects on gene activities and/or maintenance. A long and stringent sequence signal would help establish a high degree of site specificity, thus reducing the chance of incurring potentially detrimental breaks at the wrong sites. In fact, no case of partial utilization or position variation of natural breakage sites has been reported in T.thermophila (9,11).
The Cbs is normally lacking from the mature macronucleus of Tetrahymena. For this reason it is not known whether Cbs-induced chromosome breakage could occur in macronuclei of vegetatively growing cells. The creation of transformed lines with a partially functional Cbs in their macronuclei offered us an opportunity to examine this issue. Our results clearly show that chromosome breakage does not occur during the extended growth period and offer support for a tight temporal regulation of this process in Tetrahymena.
Although its major role is in inducing chromosome breakage, it is conceivable that the Cbs may have an important role in micronuclear chromosome organization during vegetative growth. If this is true, multiple copies of the Cbs in the macronucleus may compete for essential proteins that recognize this sequence and thereby interfere with its normal micronuclear function. This possibility could provide one explanation for the lack of a Cbs in normal macronuclei, since unprocessed copies of the Cbs left in the macronucleus will be quickly lost during growth. We examined this issue by using the transformed cell lines with a partially functional Cbs. We found that rDNA containing this type of mutated Cbs was maintained at least as well as those without a Cbs during the 100 cell doubling period tested. Since random assortment of the macronuclear rDNA occurs normally during division, cells with varying copies of Cbs should be produced during this growth period. If cells with higher copies of the Cbs grow more slowly, rDNA with a Cbs should decrease or be lost completely. Thus, our results show that excess copies of the Cbs in the macronucleus do not affect cell growth.
A special feature of programmed chromosome breakage in ciliates and nematodes is tight coupling with chromosome end healing. Essentially all broken ends produced acquire new telomeres during this process (6). In Tetrahymena, both breakage and end healing depend on the Cbs alone as a cis-acting sequence (11). The length of the Cbs, however, raises the possibility that the sequence may contain separate domains for directing these two processes. The mutational analysis presented here offers some insights into this issue. Mutations affecting breakage but not end healing will produce the same effect in our assay as those affecting both, and cannot be distinguished from each other. However, mutations affecting end healing alone will lead to the production of broken molecules without new telomeres, which are likely to be lost (11) or repaired by recombination with the endogenous rDNA. Constructs with this type of mutation will either produce no transformant or produce transformants with only normal (endogenous-type) rDNA termini. None of the mutations tested here produce either type of result. Thus, it does not appear that the Cbs contains a separate domain to control end healing. It should be pointed out that mutations that retain some end healing activity will not be detected here. It is likely that both breakage and end healing are controlled through the same sequence signal, possibly as part of a multi-molecule complex.
The Cbs at different genomic locations is highly conserved in sequence. Of the nine copies isolated from T.thermophila, eight are identical and the remaining one differs from it by a single nucleotide (T instead of A in position 13) (7,17). This variant sequence is not among those tested here. This high level of sequence conservation is in good agreement with our mutational analysis described here. A mutant strain of T.thermophila has previously been isolated and found to contain a single nucleotide substitution in the Cbs adjacent to the rDNA (18). This mutation is thought to reduce breakage at this site, but a direct assessment of this reduction has been difficult to make. It is the same mutation present in clone FA04G tested here, which abolishes breakage in our study. The Cbs also appears to be highly conserved during the evolution of Tetrahymena spp. Studies of the Cbs site near the rDNA 3′-end in four other Tetrahymena and a related Gluacoma species show that two of them have exactly the same sequence as the major sequence in T.thermophila, while the other three differ from it by a single nucleotide (10). They are identical to those present in clones FA14C, FA15C and FA15A, of which only FA15C is completely non-functional in T.thermophila. This evolutionary conservation suggests that the Cbs is under stringent selection. Our mutational analysis indicates that the integrity of the sequence is crucial for its function, and provides a good explanation for this conservation.
Our study establishes that the Cbs is an exceptionally long and stringent sequence signal. It is comparable to the recognition sequences of Group I homing intron endonucleases (16). No other nucleases are known to have such lengthy recognition sequences. This feature raises the interesting possibility that the nucleases involved in programmed chromosome breakage in Tetrahymena and those in intron homing are evolutionarily related. It is conceivable that a homing endonuclease has evolved in this group of organisms to carry out this special function, in much the same way that the HO endonuclease of the yeast Saccharomyces cerevisiae may have evolved (19). Isolation and characterization of the endonuclease that cleaves at the Cbs may answer this interesting question.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs Caterina Randolph and Emily Wiley for their helpful comments on this manuscript. This work is supported by NIH grant GM26210 to M.C.Y.
REFERENCES
- 1.Prescott D.M. (1994) Microbiol. Rev., 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubota S., Kuro-o,M., Mizuno,S. and Kohno,S.-i. (1993) Chromosoma, 102, 163–173. [DOI] [PubMed] [Google Scholar]
- 3.Coyne R.S., Chalker,D.L. and Yao,M.C. (1996) Annu. Rev. Genet., 30, 557–578. [DOI] [PubMed] [Google Scholar]
- 4.Bruns P.J. (1986) In Gall,J.G. (ed.), The Molecular Biology of Ciliated Protozoa. Academic Press, Orlando, FL, pp. 27–44.
- 5.Yao M.-C. (1986) In Gall,J.G. (ed.), The Molecular Biology of Ciliated Protozoa. Academic Press, Orlando, FL, pp. 179–201.
- 6.Yao M.-C. (1989) In Berg,D.E. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 715–734.
- 7.Yao M.C., Zheng,K. and Yao,C.H. (1987) Cell, 48, 779–788. [DOI] [PubMed] [Google Scholar]
- 8.Yao M.C. and Yao,C.H. (1989) Mol. Cell. Biol., 9, 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao M.-C., Yao,C.-H. and Monks,B. (1990) Cell, 63, 763–772. [DOI] [PubMed] [Google Scholar]
- 10.Coyne R.S. and Yao,M.C. (1996) Genetics, 144, 1479–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan Q. and Yao,M. (1996) Mol. Cell. Biol., 16, 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorovsky M.A., Yao,M.C., Keevert,J.B. and Pleger,G.L. (1975) Methods Cell Biol., 9, 311–327. [DOI] [PubMed] [Google Scholar]
- 13.Tondravi M.M. and Yao,M.C. (1986) Proc. Natl Acad. Sci. USA, 83, 4369–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Austerberry C.F. and Yao,M.C. (1987) Mol. Cell. Biol., 7, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J., Fritsch,E.T. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Belfort M. and Roberts,R.J. (1997) Nucleic Acids Res., 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G.L. and Blackburn,E.H. (1991) Cell, 67, 823–832. [DOI] [PubMed] [Google Scholar]
- 18.Kapler G.M. and Blackburn,E.H. (1994) Genes Dev., 8, 84–95. [DOI] [PubMed] [Google Scholar]
- 19.Kostriken R., Strathern,J.N., Klar,A.J., Hicks,J.B. and Heffron,F. (1983) Cell, 35, 167–174. [DOI] [PubMed] [Google Scholar]