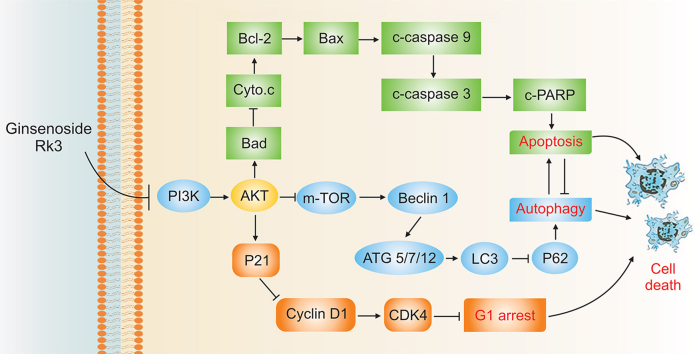
Abstract
Hepatocellular carcinoma (HCC) is the third leading cause of cancer death worldwide. Ginsenoside Rk3, an important and rare saponin in heat-treated ginseng, is generated from Rg1 and has a smaller molecular weight. However, the anti-HCC efficacy and mechanisms of ginsenoside Rk3 have not yet been characterized. Here, we investigated the mechanism by which ginsenoside Rk3, a tetracyclic triterpenoid rare ginsenoside, inhibits the growth of HCC. We first explored the possible potential targets of Rk3 through network pharmacology. Both in vitro (HepG2 and HCC-LM3 cells) and in vivo (primary liver cancer mice and HCC-LM3 subcutaneous tumor-bearing mice) studies revealed that Rk3 significantly inhibits the proliferation of HCC. Meanwhile, Rk3 blocked the cell cycle in HCC at the G1 phase and induced autophagy and apoptosis in HCC. Further proteomics and siRNA experiments showed that Rk3 regulates the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway to inhibit HCC growth, which was validated by molecular docking and surface plasmon resonance. In conclusion, we report the discovery that ginsenoside Rk3 binds to PI3K/AKT and promotes autophagy and apoptosis in HCC. Our data strongly support the translation of ginsenoside Rk3 into novel PI3K/AKT-targeting therapeutics for HCC treatment with low toxic side effects.
Keywords: Hepatocellular carcinoma, Ginsenoside Rk3, Apoptosis, Autophagy, PI3K/AKT pathway
Graphical abstract
Highlights
-
•
The possible potential targets of Rk3 were explored by network pharmacology.
-
•
Rk3 exhibits a low toxicity effect compared to the first-line drug sorafenib.
-
•
Rk3 induces HCC apoptosis and autophagy in vivo and in vitro.
-
•
Rk3 is a novel PI3K/AKT-targeting agent that regulates autophagy and apoptosis.
1. Introduction
Liver cancer is one of the most common types of cancer and has a high mortality rate, accounting for 8.3% of all cancer deaths worldwide and ranking third in the global cancer mortality rate [1]. In China, liver cancer is the second most common cancer in men and the sixth most common cancer in women, and the number of new cases reached 388,800 in 2016 [2]. Currently, treatment options for hepatocellular carcinoma (HCC) include surgical resection, chemotherapy, and liver transplantation. Yet half of all HCC patients are diagnosed at a late stage when liver resection and transplantation are not feasible [3]. Sorafenib and its derivative regorafenib are the first- and second-line targeted drugs for advanced HCC, respectively. Due to the complex etiology and heterogeneity of HCC, targeted drugs for advanced HCC remain severely limited [4]. Thus, it is essential to identify biomarkers and effective interventions for early diagnosis and treatment.
Traditional Chinese medicine (TCM) has a long history in the prevention and treatment of liver diseases. Recently, studies have shown that TCM and extracts from vegetables and fruits such as polyphenols and flavonoids have a strong inhibitory effect on HCC and other tumors, providing an important alternative treatment plan [[5], [6], [7], [8], [9]]. Ginseng, a slow-growing perennial plant, has long been used to treat various human diseases including cancer, diabetes, and traumatic brain injury [[10], [11], [12], [13]]. Ginsenoside Rk3 is a major rare saponin in heat-treated ginseng that induces a variety of biological changes. Our previous studies showed a role of Rk3 in anti-tumor, anti-anemia, anti-type 2 diabetes, and alleviation of alcohol-induced hepatotoxicity [[14], [15], [16], [17]]. However, the effect of Rk3 in HCC, as well as the underlying mechanisms, has not been fully investigated.
The phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) signaling pathway plays important roles in multiple cellular processes including cancer cell growth and survival and is highly activated in many tumors including HCC [18,19]. Indeed, many proteins in the pathway are highly susceptible to dysregulation via amplification, mutation, or translocation in cancer patients [20]. PI3K is a phosphatidylinositol kinase consisting of a regulatory subunit (p85) and a catalytic subunit (p110). AKT is a serine/threonine tyrosine kinase that can be activated by PI3K with the help of phosphatidylinositol-dependent kinase 1 (PDK1), a downstream effector of PI3K. The PI3K/AKT pathway is central for the sustained activation of AKT, which further regulates cell cycle, apoptosis, metastasis, and angiogenesis and usually also leads to aberrant signaling and uncontrolled proliferation-associated diseases [21], an important feature of many human cancers (e.g., breast [22], melanoma [23], lung [24], colorectal [25], prostate [26], liver [27], and gastric cancers [28]), making PI3K/AKT a promising important target for cancer treatment. The role of PI3K/AKT in mediating the effect of Rk3 in HCC has not been reported before.
Network pharmacology is a powerful tool to understand the pharmacological action, mechanism, and toxicity of TCM and had been used to predict drug targets and to improve drug discovery efficiency [29]. In this study, we first used such a bioinformatics approach to identify potential targets of Rk3. We then determined the anti-tumor effect of Rk3 on human HCC cell lines and investigated the possible molecular mechanisms underlying these actions. Initial in vitro findings were validated in two animal models: the HCC xenograft nude mice model and the primary liver cancer mouse model. Proteinological analysis was performed to reveal the protein causes and signaling pathways regulated by Rk3 from a systematic perspective. In addition, siRNA, molecular docking and surface plasmon resonance (SPR) were used to reveal the target mechanisms of Rk3-induced changes.
2. Materials and methods
2.1. Cell culture and reagents
HepG2 and HCC-LM3 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HL-02 cell was obtained from Fuheng Biology Co., Ltd (Shanghai, China). Both cell lines were used for in vitro assays if not specified. Cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% (V/V) fatal bovine serum (FBS) (Biological Industries, Beit Haemek, Israel) at 37 °C in a 5% CO2 humidified air incubator (Thermo Scientific, Waltham, MA, USA).
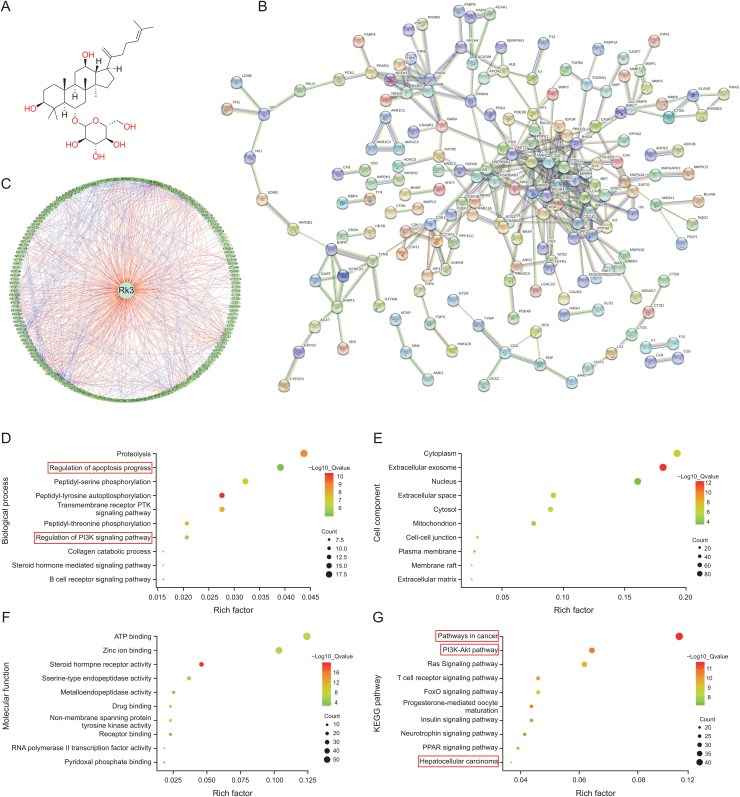
Rk3 (Fig. 1A) (purity ≥99%) and sorafenib were purchased from Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China). Rk3 was dissolved in 0.2% CMC-Na for subsequent animal study. Diethylnitrosamine (DEN), CCl4, N-hydroxysuccinimide (NHS), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC), and ethanolamine HCl were bought from Sigma-Aldrich (St. Louis, MO, USA). The inhibitor or activator 3-methyladenine (3-MA) (Cat. No. HY-19312), Z-VAD-fmk (Cat. No. HY-16658B), and recilisib (Cat. No. HY-101625) were bought from MedChemExpress (Princeton, NJ, USA).
Fig. 1.
Target prediction of ginsenoside Rk3 based on PharmMapper database and bioinformatics analysis. (A) Molecular structural formula of ginsenoside Rk3. (B) Protein-protein interactions (PPI) network diagram of target proteins. (C) Network pharmacology. (D) Biological process. (E) Cell component. (F) Molecular function. (G) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway.
2.2. 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay
The viability was determined by MTT assay as mentioned in previous literature [30]. Cells were seeded in 96-well plates at 8 × 103 cells/well for 24 h. Then, cells were treated with 0.1% dimethyl sulfoxide (DMSO) as control or Rk3 for another 24 h, followed by incubation with MTT for 4 h at 37 °C. The supernatant was removed and 150 μL of DMSO was added. The absorbance was measured at 490 nm.
2.3. Clony formation assay
Cells were seeded in 6-well plates at 500 cells/well and treated with Rk3 for 24 h. The medium was changed every 2 days for 14 days, and colonies were fixed with methyl alcohol and stained using the Giemsa stain as described [31].
2.4. Flow cytometry
Cells were seeded in 6-well plates at 5 × 105 cells/well and treated with Rk3 for 24 h. For cell cycle analysis, cells were collected, washed, and fixed with 75% ice-cold ethanol overnight at 4 °C, and then stained with propidium iodide (PI)/RNase staining buffer. The fluorescence per nucleus was measured. For apoptosis analysis, cells were collected and stained with Annexin V-FITC and PI for 15 min at room temperature.
2.5. Transmission electronic microscopy (TEM)
Cells were seeded in 6-well plates at 5 × 105 cells/well and treated with Rk3 for 24 h. Cells were collected and fixed with 2.5% glutaraldehyde. Ultrathin sections (50–60 nm) were obtained for ultrastructure observation under a TEM (JEM-1230, Japan Electron Optics Laboratory, Tokyo, Japan).
2.6. Animal models
For the HCC-LM3 xenograft nude mouse model, four-week-old female BALB/c athymic nude mice (n = 48, weighting 14 ± 2 g) were obtained from SJA Lab Animal Co., Ltd (Wuhan, China). After acclimation for one week, HCC-LM3 cells (1 × 107 cells/each) were subcutaneously inoculated into the left flanks. When the tumors reached approximately 150 mm3, mice were randomly divided into six groups (n = 8): Normal group, Normal + 100 mg/kg Rk3 (i.p.), Control group, L-Rk3 group (50 mg/kg Rk3, i.p.), H-Rk3 group (100 mg/kg Rk3, i.p.), and 100 mg/kg sorafenib (i.g.) [32]. Body weight and tumor size were measured every three days. Mice were euthanized after 28 days. Tumor tissues and primary organs were obtained, weighed, and stored at −80 °C for future analysis.
For the primary liver cancer mouse model, the DEN and CCl4 method was used as described [33]. Male C57BL/6 mice (2 weeks old, weighing 12–15 g) were bought from the Air Force Military Medical University (Xi'an, China). After acclimation for one week, all mice were randomly divided into five groups (n = 10 per group): Normal group, HCC group, L-Rk3 group (50 mg/kg Rk3, i.p.), H-Rk3 group (100 mg/kg Rk3, i.p.), and sorafenib group (100 mg/kg sorafenib, i.g.) [32]. Mice were injected with DEN (1 mg/kg in phosphate buffered saline (PBS) and then with CCl4 (0.2 mL dissolved in olive oil, two times per week beginning at 8 weeks of age, i.p.) up to 22 weeks of age. The normal group was given PBS and olive oil. At 22 weeks, mice were given Rk3 or sorafenib for another 8 weeks. At the end of the experiment, mice were euthanized, and liver samples were obtained, weighed, and stored at −80 °C for future analysis.
All animal experiments were approved by the Animal Ethics Committee of Northwest University (Approval number: NWU-AWC-20200813M) and met the requirements of the Laboratory Animal Act of the People's Republic of China.
2.7. Survival analysis
HCC-LM3 subcutaneous tumor-bearing mice were randomly divided into three groups (n = 20 for each): control group, 100 mg/kg Rk3 group, and 100 mg/kg sorafenib group. Survival was assessed daily for 45 days.
2.8. Hemogram assay and serum analysis
Blood samples from each group of the HCC-LM3 xenograft nude mouse model were collected on the 28th day. Neutrophil granulocyte (GRAN), lymphocytes (LYM), platelets (PLT), and white blood cell (WBC) were analyzed by an automatic blood cell analyzer (HC2200, Meiyilinm Electronic Instrument Co., Ltd, Jinan, China).
The serum content of alanine aminotransferase (ALT), aspartate aminotransferase (AST), urine creatinine (UCR), and urea in HCC-LM3 xenograft nude mice was determined using commercial kits according to the manufacturer's instructions (Shanghai Enzyme-linked Biotechnology Co., Ltd, Shanghai, China).
2.9. Histopathology, immunohistochemistry, and immunofluorescence staining
Pathological staining was performed according to previous studies with slight modifications [[34], [35], [36], [37]]. Liver, tumor, and other tissues were fixed with 10% formalin and then embedded in paraffin. Samples were cut into 5 μm sections and fixed on a glass slide, followed by staining with hematoxylin-eosin (H&E) or sirius red. For immunohistochemical analysis, paraffin sections were immune-stained with antibodies.
For immunofluorescence staining, tissues were treated with paraffin. Tissues or cell slides were fixed in 4% paraformaldehyde, followed by incubation with primary antibodies overnight at 4 °C in a humidified box. Horseradish peroxidase (HRP)-binding secondary antibodies were then applied, followed by detection with a Nikon TE 2000 fluorescence microscope (Nikon, Tokyo, Japan).
2.10. Western blot
Cell or tissue samples were lysed in the radio immunoprecipitation assay (RIPA) buffer (Beyotime, Shanghai, China). Proteins were quantified and then subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After transferring proteins to a polyvinylidene difluoride (PVDF) membrane, samples were incubated first with primary antibodies at 4 °C for overnight and then with secondary antibodies at room temperature for 1 h. An enhanced chemiluminescence (ECL) kit was used for signal detection (PerkinElmer, Waltham, MA, USA).
2.11. Bioinformatics
The 3D structure of Rk3 (obtained from PubChem, CID: 75412555) was uploaded to PharmMapper (covering 1,627 drug target information, of which 459 are human protein targets) for potential target identification [38]. The “human protein targets only” option was enabled, and the default was used for all other parameters. Potential targets were selected based on the fit score. Protein-protein interactions (PPI) were constructed using STRING database [39]. The herb-compound-target network was created based on the PPI data. Cytoscape v3.6.1 was used for the analysis of all networks [40]. Gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were performed using DAVID (https://david.ncifcrf.gov/) [41]. GO terms and KEGG pathways with P values < 0.05 were considered statistically significant.
2.12. siRNA transfection assay
Primers for siRNAs (5′-GCUGGUUAAAUGGCUAUAATT-3′ and 5′-UUAUAGCCAUUUAACCAGCTT-3′ for PI3K; and 5′-UCAUGCAGCAUGGCUUCUUTT-3′, and 5′-AAGAAGCGAUGCUGCAUGATT-3′ for AKT) were synthesized by GenePharma (Shanghai, China). Cells were seeded in six-well plates at a density of 5 × 105 cells per well and transfected with 150 nM siRNA using Lipofectamine™ 2000 reagents (Invitrogen Carlsbad, Carlsbad, CA, USA). After 24 h, cells were harvested for cell viability and Western blot analysis.
2.13. Proteomics
Proteomics was performed as described previously [42]. HCC-LM3 cells were treated with Rk3 for 24 h and proteins were extracted. A tandem mass tag (TMT)-based quantitative proteomics experiment was performed by Majorbio company (Shanghai, China). Significant proteins were defined using a fold change over 1.2 and P < 0.05 (Student's t-test). The UniProt GOA database (https://www.ebi.ac.uk/GOA/) was used for GO annotation. Pathway enrichment was performed using the KEGG (https://www.kegg.jp/) database.
2.14. Clinical data
Clinical data and gene expression profiles for HCC patients were downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/), covering 50 normal tissues and 374 tumor tissues. The RNA sequencing data was converted to that similar to microarray data. The results of PI3K and AKT survival curves were downloaded from the UALCAN database [43].
2.15. Molecular docking
The 3D structures (crystal structure) of PI3K (PDB ID: 1H9O) and AKT (PDB ID: 2W1C) were downloaded from the Protein Data Bank and processed with Maestro 11.9. The 3D molecular structure of Rk3 was downloaded from PubChem database (PubChem CID: 75412555). The docking study was processed with AutoDock Tools. The docking results were detected using the PyMOL Molecular Graphics System.
2.16. Binding of PI3K or AKT with Rk3
Recombinant PI3K or AKT were prepared as glutathione S-transferase (GST)-tagged fusion proteins using a kit (Beyotime Biotechnology, Shanghai, China). Briefly, the coding sequence was cloned into a pgex-6p-1 vector and transformed into Escherichia coli BL21 star (DE3) cells. After the bacterial culture grew to a 600 nm optical density (OD600) of about 0.6, isopropyl β- d-thiogalactopyranoside (IPTG) was used to induce PI3K-GST or AKT-GST expression at 37 °C for 2 h. The GST was cleaved by a PreScission Protease (Beyotime Biotechnology) to obtain PI3K and AKT proteins.
PI3K or AKT was coupled to a CM5 chip in 10 mm NaAc at pH 4.5 and 5.5, respectively. Activation was performed with NHS and EDC, and blocking was performed with ethanolamine HCl. Rk3 was dissolved in DMSO, and samples were injected into a Biacore T200 for SPR experiment (GE Healthcare Life Sciences, Uppsala, Sweden).
2.17. Statistical analysis
All data were shown as mean ± standard deviation (SD). One-way analysis of variance (ANOVA) followed by Student's t-test was conducted using SPSS version 19.0 (SPSS Inc., Chicago, IL, USA). Statistical significance was defined as P < 0.05. Graphs were constructed using GraphPad Prism (Version 7.00, GraphPad Software Inc., San Diego, CA, USA).
3. Results
3.1. Targeting prediction of Rk3 based on network pharmacology
To explore the basis of the pharmacological effect of Rk3, its potential protein targets were predicted and the associated PPI network was built (Fig. 1B). In parallel, the pharmacology diagram in TCM was also shown (Fig. 1C). Bioinformatics analysis of the potential target proteins revealed that they are enriched in cancer-related pathways and the PI3K/AKT pathway and are involved in HCC (Figs. 1D−G). Thus, Rk3 may play an important role in regulating cancer signaling, especially in HCC.
3.2. Rk3 inhibits the growth of HCC
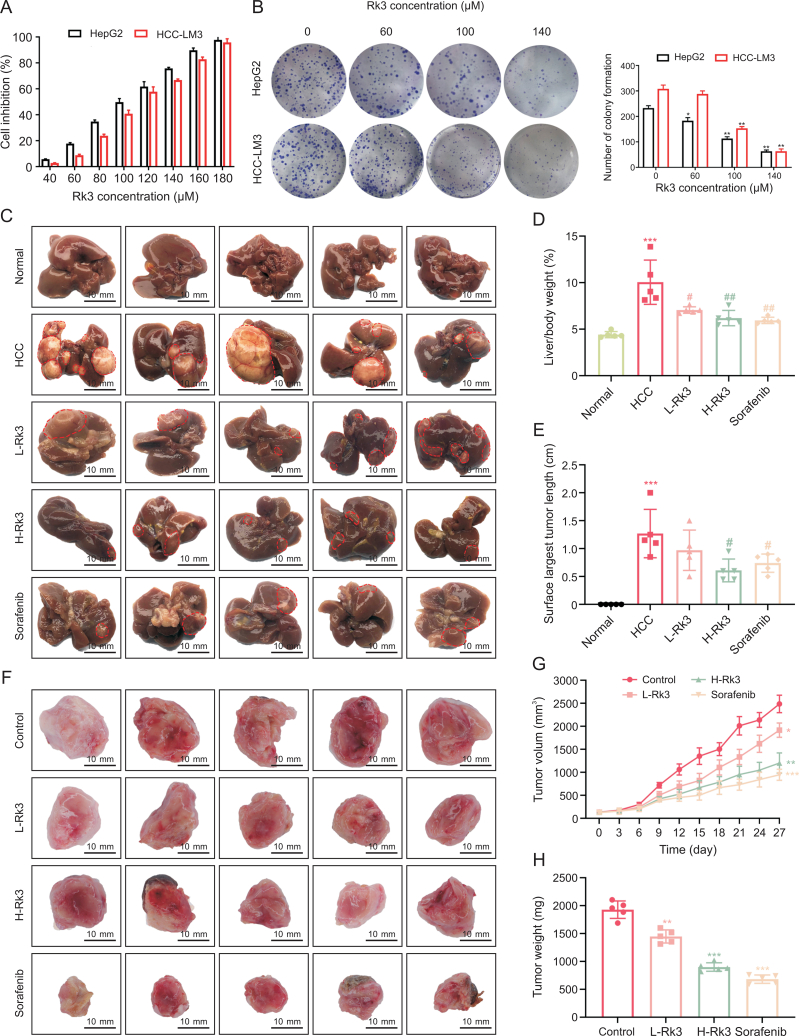
To determine whether Rk3 can inhibit the growth of HCC, the effect of Rk3 in two HCC cell lines HepG2 and HCC-LM3 was assessed by the MTT assay. As expected, Rk3 could effectively inhibit the growth of HCC cells in a dose and time-dependent manner (P < 0.05, Fig. 2A). Similarly, Rk3 significantly reduced the number of HCC cell clones (Fig. 2B).
Fig. 2.
Ginsenoside Rk3 inhibited the growth of hepatocellular carcinoma (HCC) both in vitro and in vivo. (A) Cell viability of HepG2 and HCC-LM3 treatment with ginsenoside Rk3. (B) Clone formation of HepG2 and HCC-LM3. (C) Anatomy of the liver of a mouse with primary liver cancer. (D) Liver to body weight of a mouse with primary liver cancer. (E) The surface largest tumor length of a mouse with primary liver cancer. (F) Tumor anatomy of HCC-LM3 subcutaneous tumor-bearing mice. (G) Tumor volume of HCC-LM3 subcutaneous tumor-bearing mice. (H) Tumor weight of HCC-LM3 subcutaneous tumor-bearing mice. L-Rk3: 50 mg/kg Rk3; H-Rk3: 100 mg/kg Rk3; sorafenib: 100 mg/kg. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the normal or control group, ∗∗P < 0.01 compared with the normal or control group, ∗∗∗P < 0.001 compared with the normal or control group, #P < 0.05 compared with the HCC group, ##P < 0.01 compared with the HCC group.
For in vivo test, a primary liver cancer model (induced by DEN and CCl4) was created with abundant large tumor nodules in the liver (Fig. 2C). Remarkably, the intervention of Rk3 and sorafenib resulted in a significant decrease in the number and volume of tumor nodules (Fig. S1A). Both the liver/body weight ratio and maximum tumor length on the liver surface were much reduced in the treatment groups compared to the HCC group (Figs. 2D and E). In addition, these results were also validated in an HCC-LM3 subcutaneous tumor-bearing mouse model, which showed that both Rk3 and sorafenib reduced tumor volume and tumor weight (Figs. 2F–H). Compared to a 58% tumor inhibition rate of Sorafenib, Rk3 inhibited the growth of HCC-LM3 subcutaneous tumors in a dose-dependent manner (23% and 45% in the low- and high-dose groups, respectively). Similar results had been found for tumor weight.
Moreover, survival analysis showed that both Rk3 and sorafenib significantly prolonged the survival of HCC-LM3 subcutaneous tumor-bearing mice (P < 0.05, Fig. S1B). In line with this, staining sections of liver or tumor in primary liver cancer mouse model or HCC-LM3 subcutaneous tumor-bearing mouse model showed that Rk3 reduces the proliferation of liver HCC cells (Fig. S2). In addition, Rk3 also reduced the level of liver fibrosis compared to the HCC group as revealed by sirius red staining (Fig. S3). Together, Rk3 inhibits the growth of HCC both in vivo and in vitro.
To investigate the low toxicity of Rk3, HL-02 cell viability of normal hepatocytes treated with Rk3 was examined, and the results are shown in Fig. S4. It shows that Rk3 treatment produced low toxicity to normal hepatocytes. The HCC-LM3 subcutaneous tumor-bearing mice were used to evaluate potential toxic and side effects of Rk3. First, the body weight of mice with the Rk3 treatment for 4 weeks was comparable to that of the control group (Fig. S5A). By contrast, the body weight of mice in the sorafenib group dropped sharply, indicating possible side effects of sorafenib. Next, analysis of GRAN, LYM, PLT, and WBC in the blood revealed similar levels between the Rk3 treatment and the control groups, while significant lower levels were found in the sorafenib group (Figs. S5B–E). Such effects of sorafenib are consistent with a clinical study demonstrating that sorafenib causes myelosuppression with abnormal decreases in these blood indicators (GRAN, LYM, PLT, and WBC) in patients during chemotherapy [44]. Further analysis with mouse liver injury indicators (ALT and AST) and kidney injury indicators (UCR and urea) showed similar results, where no damage and slight damage to liver/kidney were found for Rk3 and sorafenib, respectively (Figs. S5F–I). Lastly, while Rk3 did not result in any pathological changes in major organs, such features were observed in the sorafenib group including inflammatory cell infiltrations in the liver, necrosis in part of the spleen, and renal glomerular atrophy (Fig. S6). Thus, Rk3 exhibits a low toxicity effect compared to the first-line drug sorafenib.
3.3. Rk3 blocks HCC cell cycle at the G1 phase
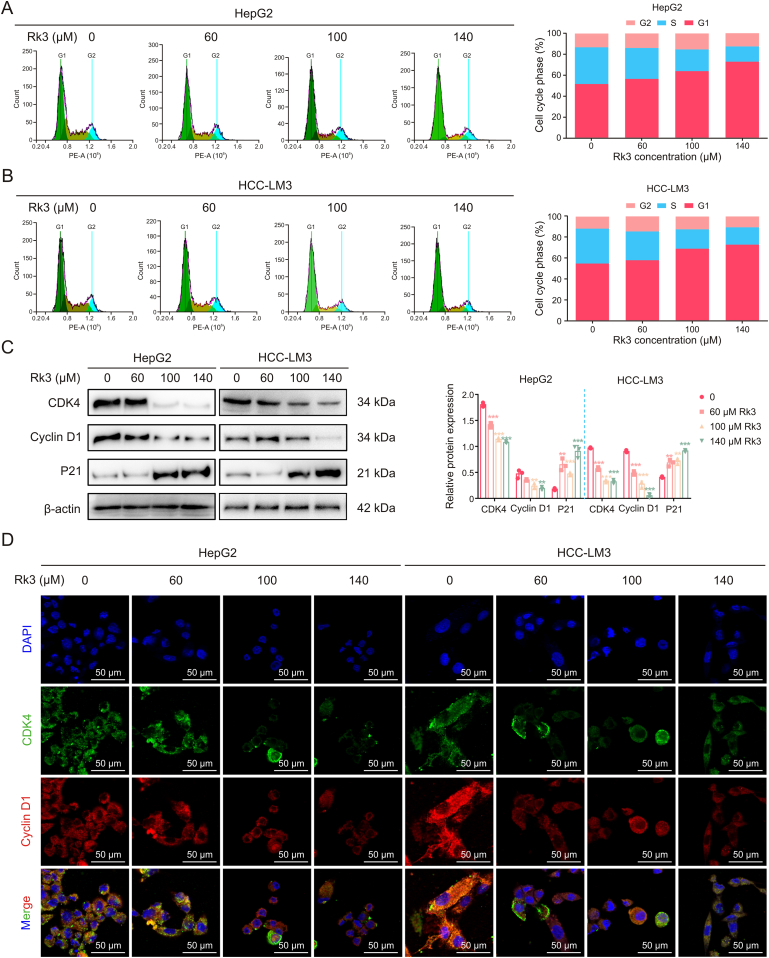
Next, we assessed the impact of Rk3 on the cell cycle in HCC cells by flow cytometry. The Rk3 treatment group showed a gradual increase of cells at the G1 phase compared to the control group (Figs. 3A and B). The possible cell cycle block at the G1 phase was further evaluated by checking phase-related proteins. As expected, Rk3 resulted in increased (P21) or decreased (cyclin-dependent kinase (CDK) 4 and cyclin D1) protein abundances for selected markers (Fig. 3C). These results by Western blot were further validated for CDK4 and cyclin D1 using immunofluorescent (IF) staining (Fig. 3D). Thus, Rk3 inhibits the cell cycle of HCC at the G1 phase in vitro.
Fig. 3.
Ginsenoside Rk3 blocked the growth of hepatocellular carcinoma (HCC) cells at G1 phase in vitro. (A) Flow cytometry cell cycle detection of HepG2 cell. (B) Flow cytometry cell cycle detection of HCC-LM3 cell. (C) Western blotting detection of G1 phase related proteins of HepG2 and HCC-LM3 cells. (D) Immunofluorescence (IF) staining of CDK4 and cyclin D1 protein of HepG2 and HCC-LM3 cells. Data are shown as mean ± standard deviation (SD). ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group. DAPI: 4’,6-diamidino-2-phenylindole.
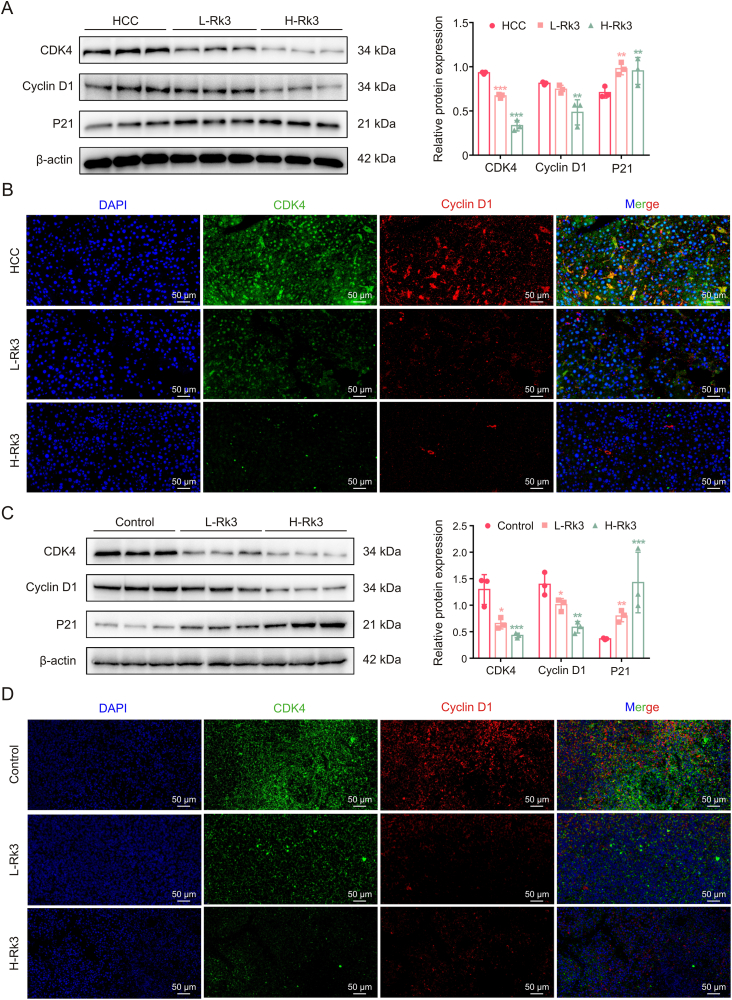
The in vivo effect was first tested in the primary liver cancer mouse model using Western blot, immunohistochemistry (IHC), and IF staining. As with the in vitro assay, Rk3 inhibited the expression of G1 phase-related proteins in a dose-dependent manner (Figs. 4A, 4B and S7A). Similar results were found in the HCC-LM3 subcutaneous tumor-bearing mouse model (Figs. 4C, 4D and S7B).
Fig. 4.
Ginsenoside Rk3 blocked the growth of hepatocellular carcinoma (HCC) at G1 phase in vivo. (A) Western blotting detection of G1 phase related proteins of primary liver cancer mice. (B) Immunofluorescence (IF) staining of CDK4 and cyclin D1 protein of primary liver cancer mice. (C) Western blotting detection of G1 phase related proteins of HCC-LM3 subcutaneous tumor-bearing mice. (D) IF staining of CDK4 and cyclin D1 protein of HCC-LM3 subcutaneous tumor-bearing mice. L-Rk3: 50 mg/kg Rk3; H-Rk3: 100 mg/kg Rk3. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the HCC or control group, ∗∗P < 0.01 compared with the HCC or control group, ∗∗∗P < 0.001 compared with the HCC or control group. DAPI: 4',6-diamidino-2-phenylindole.
3.4. Rk3 induces HCC apoptosis
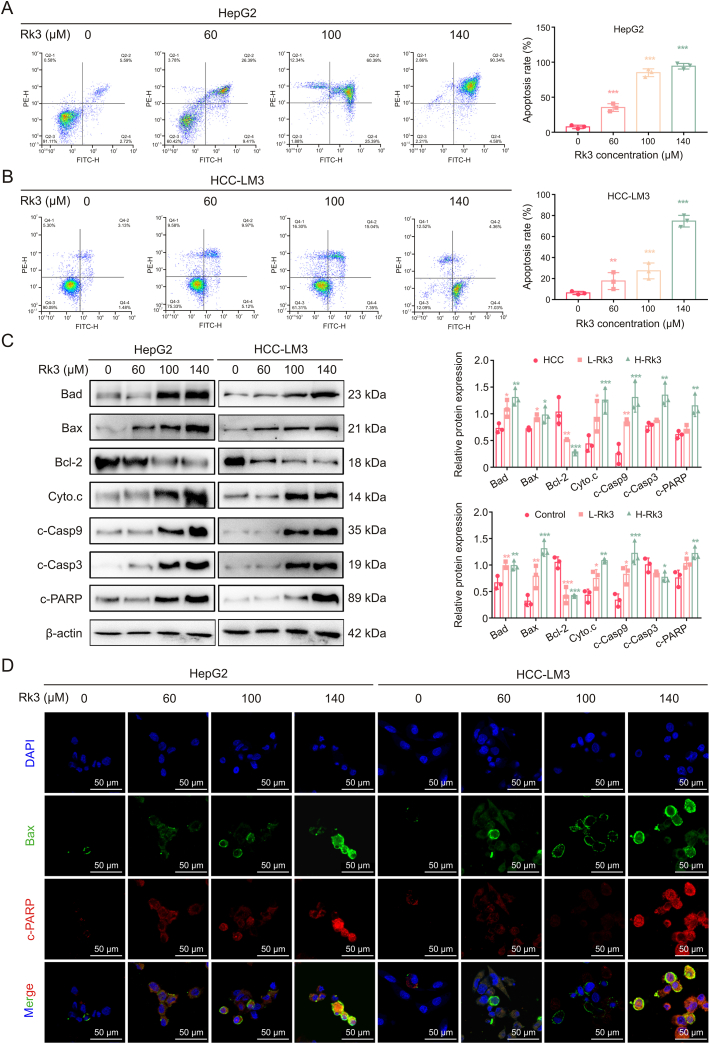
Network pharmacology (network pharmacology and pathway analysis) indicated that Rk3 may regulate cell apoptosis. This hypothesis was first tested by TEM analysis of Rk3-treated HCC cells, which revealed extensive morphological changes including cell shrinkage, more concentrated cytoplasm and chromatin, presence of pyknotic and crescent-shaped nucleus, and disappearance of the nucleoli (Fig. S8). Next, flow cytometry analysis showed that Rk3 led to a significantly increased portion of apoptotic cells (Figs. 5A and B). The impact of Rk3 on apoptosis was further evaluated by measuring a panel of apoptosis-related proteins including Bad, Bax, Cyto.c, c-Casp9, c-Casp3, and c-poly(ADP-ribose) polymerase (PARP). Indeed, the abundance of these pro-apoptotic proteins showed an increasing trend as the Rk3 dosage increased (Figs. 5C and D). By contrast, a decreasing trend was found for anti-apoptotic proteins such as Bcl-2. Thus, Rk3 could induce HCC cell apoptosis in vitro.
Fig. 5.
Ginsenoside Rk3 induces apoptosis in hepatocellular carcinoma (HCC) cells in vitro. (A) Flow cytometry apoptosis detection of HepG2 cell. (B) Flow cytometry apoptosis detection of HCC-LM3 cell. (C) Western blotting detection of apoptosis related proteins of HepG2 and HCC-LM3 cells. (D) Immunofluorescence (IF) staining of Bax and c-PARP protein of HepG2 and HCC-LM3 cells. Data are shown as mean ±standard deviation (SD). ∗P < 0.05 compared with the control group, ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group. DAPI: 4',6-diamidino-2-phenylindole.
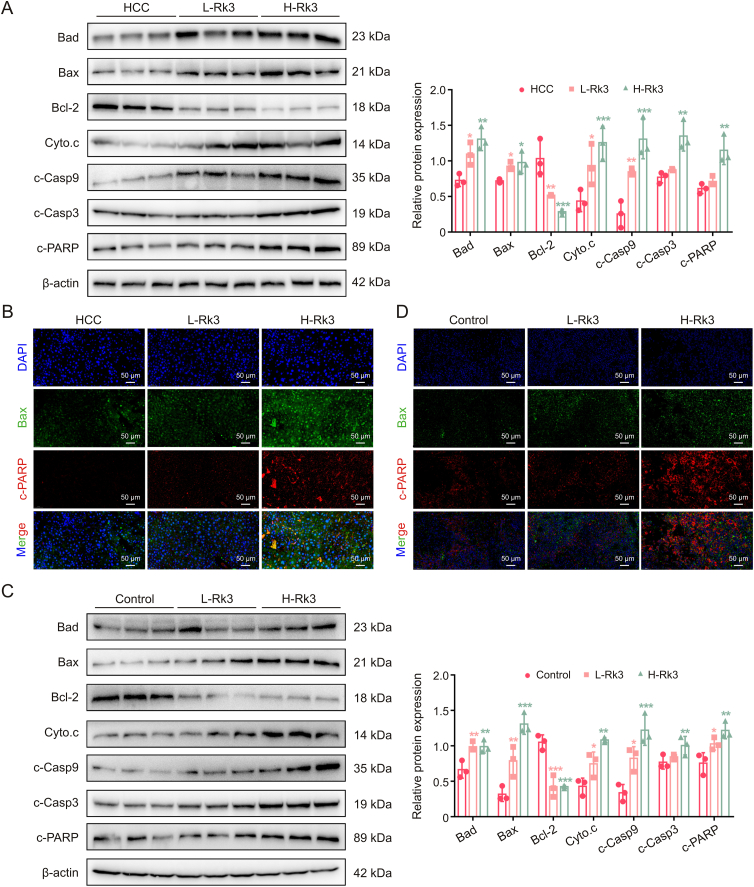
The above biochemical experiments (Western blot, IF, and IHC staining) on apoptosis-related proteins were also performed in two models (the primary liver cancer mice and the HCC-LM3 subcutaneous tumor-bearing mouse models) with similar results (Figs. 6, S9 and S10). In addition, the primary liver cancer mice model was also subjected to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, which confirmed that Rk3 induces HCC apoptosis (Figs. S11 and S12).
Fig. 6.
Ginsenoside Rk3 induced hepatocellular carcinoma (HCC) apoptosis in vivo. (A) Western blotting detection of apoptosis related proteins of primary liver cancer mice. (B) Immunofluorescence (IF) staining of Bax and c-PARP protein of primary liver cancer mice. (C) Western blotting detection of apoptosis related proteins of HCC-LM3 subcutaneous tumor-bearing mice. (D) IF staining of Bax and c-PARP protein of HCC-LM3 subcutaneous tumor-bearing mice. L-Rk3: 50 mg/kg Rk3; H-Rk3: 100 mg/kg Rk3. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the HCC or control group, ∗∗P < 0.01 compared with the HCC or control group, ∗∗∗P < 0.001 compared with the HCC or control group. DAPI: 4',6-diamidino-2-phenylindole.
3.5. Rk3 induces HCC autophagy
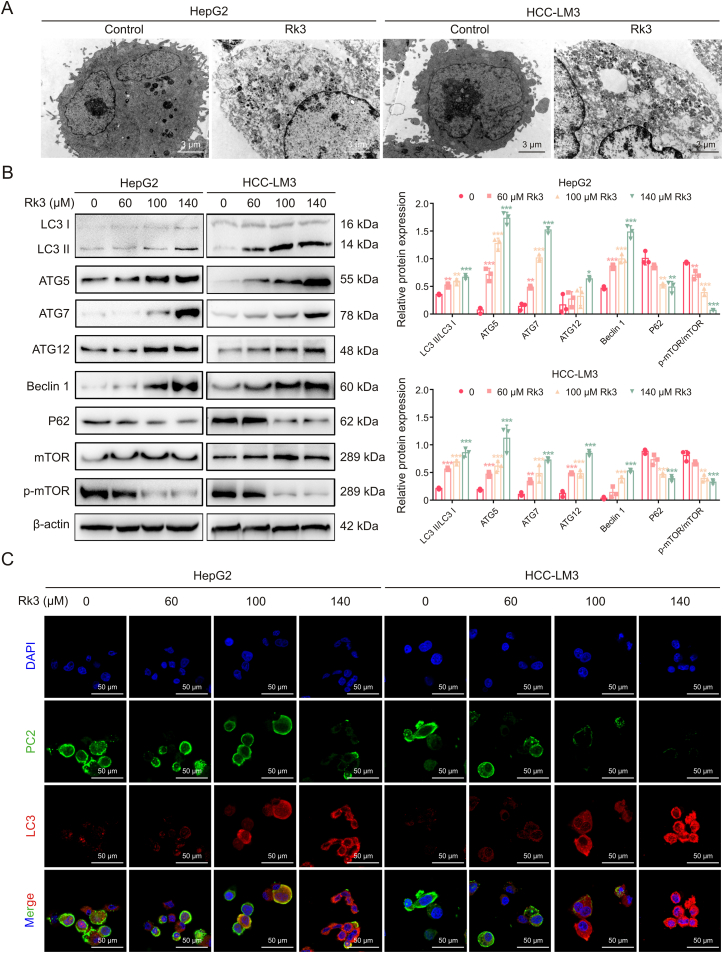
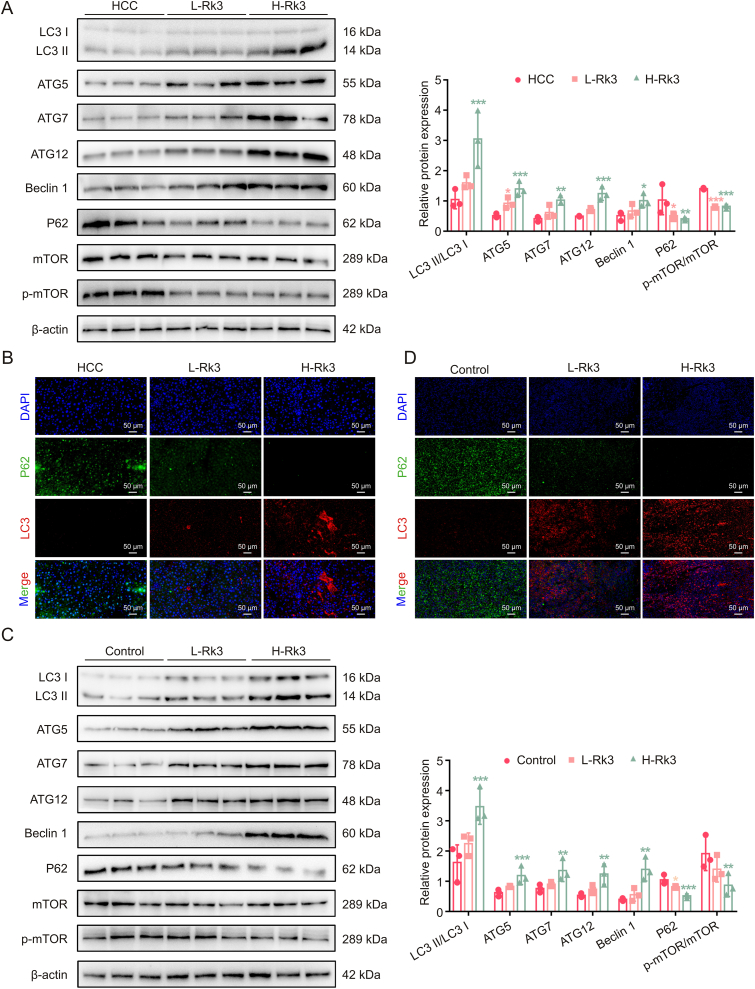
As Rk3 inhibits tumor growth in HCC, we next determined its effect on autophagy. TEM analysis showed that Rk3 promotes autophagosome formation in HCC cells (Fig. 7A). In line with this, significant up- (LC3II/LC3I, ATG5, ATG7, ATG12, Beclin 1) or down- (P62 and p-mTOR/mTOR protein) regulation of cellular autophagy marker proteins following Rk3 treatment in HCC cells was observed by Western blot (Fig. 7B). Results for LC3 and P62 proteins, key players in autophagy, were further confirmed by IF staining (Fig. 7C). More importantly, the same protein regulation patterns were observed in two animal models following either low- and high-dose of Rk3 treatment (Figs. 8, S13 and S14). Thus, Rk3 induces HCC autophagy both in vitro and in vivo.
Fig. 7.
Ginsenoside Rk3 induces autophagy in hepatocellular carcinoma (HCC) cells in vitro. (A) Transmission electron microscopy (TEM) of HCC cells for autophagy. (B) Western blotting detection of autophagy related proteins of HepG2 and HCC-LM3 cells. (C) Immunofluorescence (IF) staining of LC3 and P62 proteins of HepG2 and HCC-LM3 cells. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the control group, ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group. DAPI: 4',6-diamidino-2-phenylindole.
Fig. 8.
Ginsenoside Rk3 induced hepatocellular carcinoma (HCC) autophagy in vivo. (A) Western blotting detection of autophagy related proteins of primary liver cancer mice. (B) Immunofluorescence (IF) staining of P62 and LC3 protein of primary liver cancer mice. (C) Western blotting detection of autophagy related proteins of HCC-LM3 subcutaneous tumor-bearing mice. (D) IF staining of P62 and LC3 protein of HCC-LM3 subcutaneous tumor-bearing mice. L-Rk3: 50 mg/kg Rk3; H-Rk3: 100 mg/kg Rk3. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the HCC or control group, ∗∗P < 0.01 compared with the HCC or control group, ∗∗∗P < 0.001 compared with the HCC or control group. DAPI: 4',6-diamidino-2-phenylindole.
3.6. The interaction between apoptosis and autophagy
As both apoptosis and autophagy can lead to cell death, we next explored the potential interaction between the two for the Rk3-induced inhibition of HCC. While co-treatment with an autophagy inhibitor (3-MA) resulted in a significant decrease in cell viability, co-treatment with an apoptosis inhibitor (Z-VAD-fmk) led to a significant increase in cell viability compared to the Rk3-only group (Fig. S15A). As shown in Figs. S15B and S16A, LC3II/LC3I protein expression was decreased in the Rk3+3-MA group compared to the Rk3 group, while Bax, and c-PARP protein expression was increased in the Rk3+3-MA group. By contrast, co-treatment with Z-VAD-fmk led to a significant decrease in Bax, c-PARP, and LC3II/LC3I and an increase in p62 (P < 0.05) (Figs. S15C and S16B). Thus, it seems that Rk3-induced autophagy promotes apoptosis, while Rk3-induced apoptosis inhibits autophagy.
3.7. Proteomics and clinical data analysis
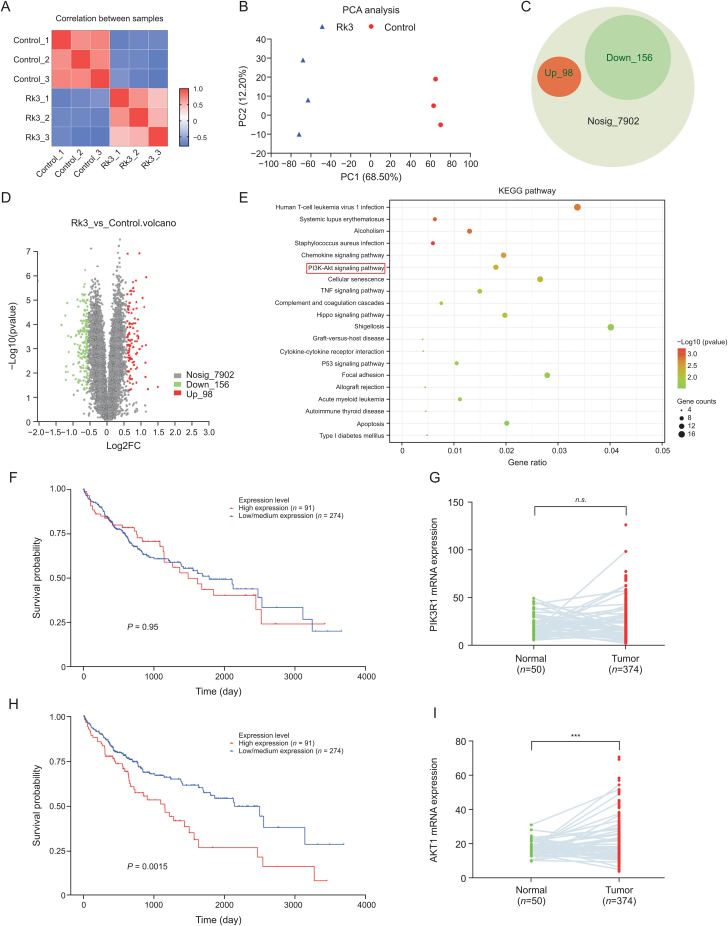
To further investigate the molecular mechanism underlying the inhibitory effect of HCC by Rk3, a proteomic analysis was performed in HCC-LM3 cells with or without Rk3 treatment (Fig. S17). Significant differences were observed at the proteome level between the two groups as revealed by correlation and principal component analysis (PCA) analysis (Figs. 9A and B). A total of 8,156 proteins were detected, of which 98 and 156 proteins were significantly up- and down-regulated, respectively (Figs. 9C and D). These differential proteins were enriched in KEGG pathways such as human T-cell leukemia virus 1 infection, systemic lupus erythematosus, alcoholism, staphylococcus aureus, and the PI3K-AKT pathway (Fig. 9E).
Fig. 9.
Proteomics and clinical data analysis. (A) Correlation analysis. (B) Principal component analysis (PCA) analysis. (C) Differential protein pie chart. (D) Differential protein volcano plots; (E) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. (F) Phosphatidylinositol-3-kinase (PI3K) clinical survival curve. (G) PI3K clinical mRNA expression. (H) Protein kinase B (AKT) clinical survival curve. (I) AKT clinical mRNA expression. ∗∗∗P < 0.001 compared with the normal group, n.s.: no significant difference between normal group and tumor group.
Of note, the PI3K-AKT pathway has been suggested to play an important role in the inhibition of HCC growth by Rk3, in line with previous network pharmacological predictions. Thus, we checked the expression levels of PI3K and AKT in clinical samples. The mRNA abundance of PI3K showed a significant difference in the survival curves of HCC patients (Fig. 9F), while the mRNA expression of PI3K was higher in HCC patients compared to normal (P < 0.05) (Fig. 9G). For AKT, the mRNA levels showed a significant difference in the survival curves of HCC patients (P < 0.05, with low AKT expression associated with longer survival, Fig. 9H). Similar to PI3K, the mRNA of AKT was higher in HCC patients compared to normal (Fig. 9I). These correlations support that PI3K/AKT could be used as a therapeutic target for HCC treatment.
3.8. Rk3 regulates PI3K/AKT to inhibit HCC growth
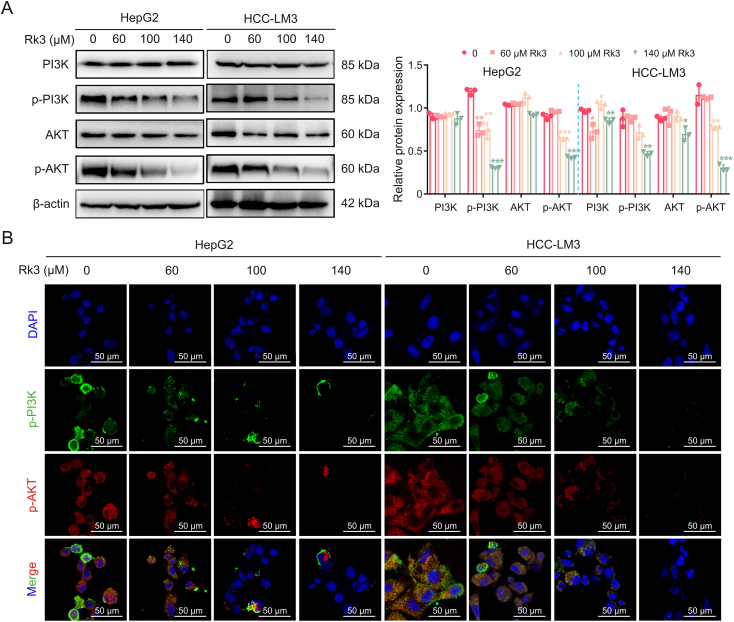
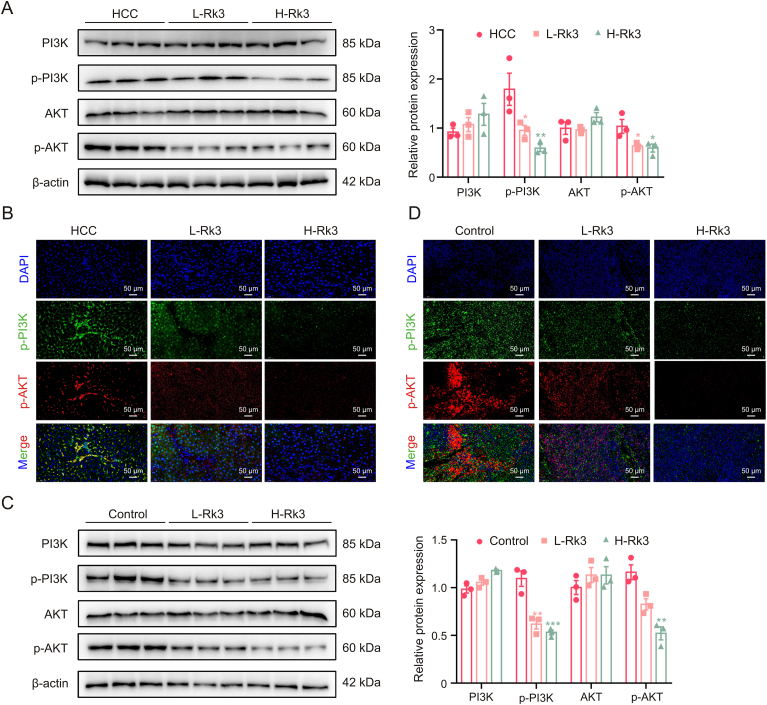
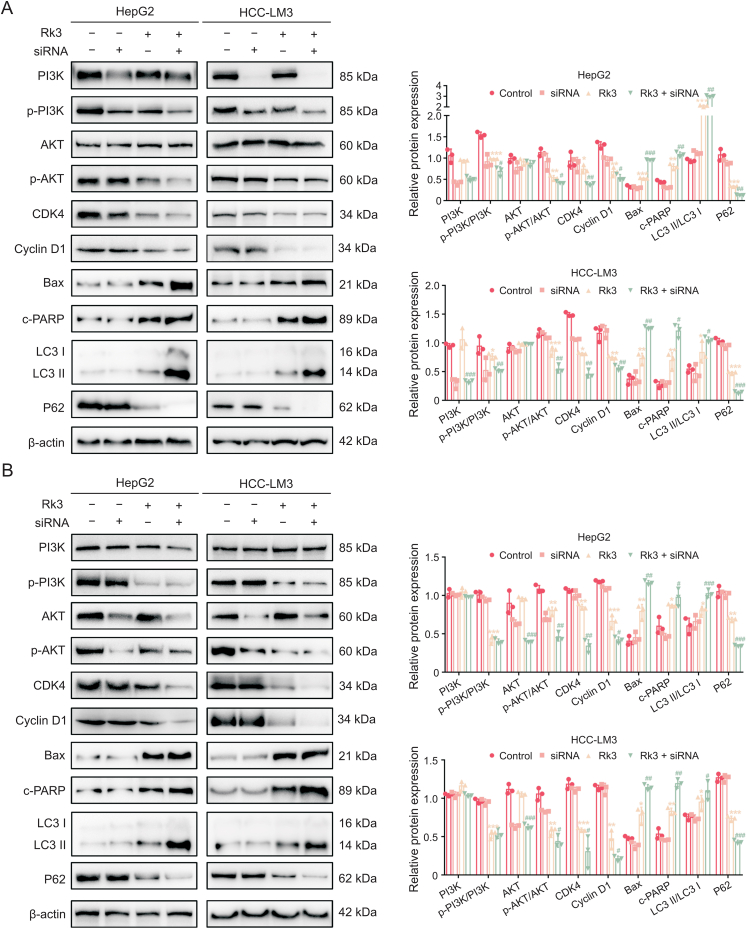
We verified the regulation of PI3K/AKT by Rk3 in both HCC cells (Fig. 10) and animal models (Figs. 11 and S18). For all systems tested, Rk3 treatment led to a significant decrease in p-PI3K and p-AKT. Thus, Rk3 inhibits the PI3K/AKT pathway both in vitro and in vivo. The involvement of PI3K/AKT was further tested by cotreatment with siRNAs in HCC cells. Remarkably, both PI3K-siRNA cotreatment and AKT-siRNA groups showed a significant decrease in cell viability compared to the Rk3 only group (P < 0.05, Fig. S19). In addition, such cotreatment with either siRNA also led to decreased levels of G1 phase-related protein (CDK4 and cyclin D1) and anti-autophagy proteins (P62) and increased levels of pro-apoptosis proteins (Bax and c-PARP) and autophagy markers (LC3II/LC3I) (Fig. 12). Thus, it is possible that inhibition of the PI3K/AKT pathway could further enhance the HCC growth inhibitory effect of Rk3.
Fig. 10.
Ginsenoside Rk3 inhibits phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) signaling pathway in vitro. (A) Western blotting detection of PI3K/AKT proteins of HepG2 and HCC-LM3 cells. (B) Immunofluorescence (IF) staining of p-PI3K and p-AKT protein of HepG2 and HCC-LM3 cells. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the control group, ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group.
Fig. 11.
Ginsenoside Rk3 inhibits phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) signaling pathway in vivo. (A) Westerrn blotting detection of PI3K/AKT proteins of primary liver cancer mice. (B) Immunofluorescence (IF) staining of p-PI3K and p-AKT protein of primary liver cancer mice. (C) Westerrn blotting detection of PI3K/AKT proteins of HCC-LM3 subcutaneous tumor-bearing mice. (D) IF staining of p-PI3K and p-AKT protein of HCC-LM3 subcutaneous tumor-bearing mice. L-Rk3; 50 mg/kg Rk3; H-Rk3: 100 mg/kg Rk3. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the HCC or control group, ∗∗P < 0.01 compared with the HCC or control group, ∗∗∗P < 0.001 compared with the hepatocellular carcinoma (HCC) or control group.
Fig. 12.
Phosphatidylinositol-3-kinase (PI3K)-siRNA and protein kinase B (AKT)-siRNA enhance the effect of ginsenoside Rk3 in inhibiting hepatocellular carcinoma (HCC) growth. (A) Westerrn blotting detection of PI3K/AKT pathway, G1 phase, autophagy, and apoptosis related proteins of HepG2 and HCC-LM3 cells treated with ginsenoside and PI3K-siRNA. (B) Westerrn blotting detection of PI3K/AKT pathway, G1 phase, autophagy, and apoptosis related proteins of HepG2 and HCC-LM3 cells treated with ginsenoside and AKT-siRNA. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the control group, ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group, #P < 0.05 compared with the ginsenoside Rk3 group, ##P < 0.01 compared with the ginsenoside Rk3 group, ###P < 0.001 compared with the ginsenoside Rk3 group.
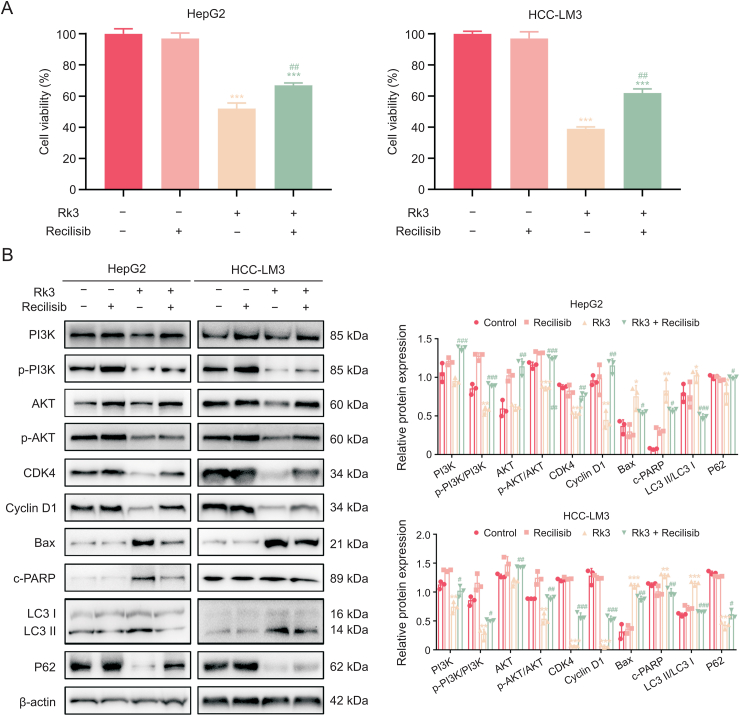
On the other hand, activating the PI3K/AKT pathway (via recilisib) led to an increased HCC cell viability in the cotreatment group compared to the Rk3 only group (Fig. 13A). The activation effect was confirmed by higher levels of PI3K, p-PI3K/PI3K, AKT, and p-AKT/AKT proteins in the cotreatment group (Fig. 13B). More importantly, analysis of marker proteins for multiple biological processes (cell cycle, apoptosis, and autophagy) in the cotreatment group showed that recilisib exerts opposite regulations of downstream proteins compared to siRNAs, resulting in up-regulation of CDK4, cyclin D1, and P62, as well as down-regulation of Bax, c-PARP and LC3II/LC3I. Thus, activation of PI3K/AKT attenuates the growth inhibitory effect of Rk3.
Fig. 13.
Phosphatidylinositol-3-kinase (PI3K)/protein kinase B (AKT) activator recilisib weaken the effect of ginsenoside Rk3 in inhibiting hepatocellular carcinoma (HCC) growth. (A) Cell viability of HepG2 and HCC-LM3 treated with or without 100 μM Rk3 and 10 μM recilisib. (B) Westerrn blotting detection of PI3K/AKT pathway, G1 phase, autophagy, and apoptosis related proteins of HepG2 and HCC-LM3 cells treated with 100 μM Rk3 and 10 μM recilisib. Data are shown as mean ± standard deviation (SD). ∗P < 0.05 compared with the control group, ∗∗P < 0.01 compared with the control group, ∗∗∗P < 0.001 compared with the control group, #P < 0.05 compared with the ginsenoside Rk3 group, ##P < 0.01 compared with the ginsenoside Rk3 group, ###P < 0.001 compared with the ginsenoside Rk3 group.
3.9. Rk3 targets PI3K/AKT
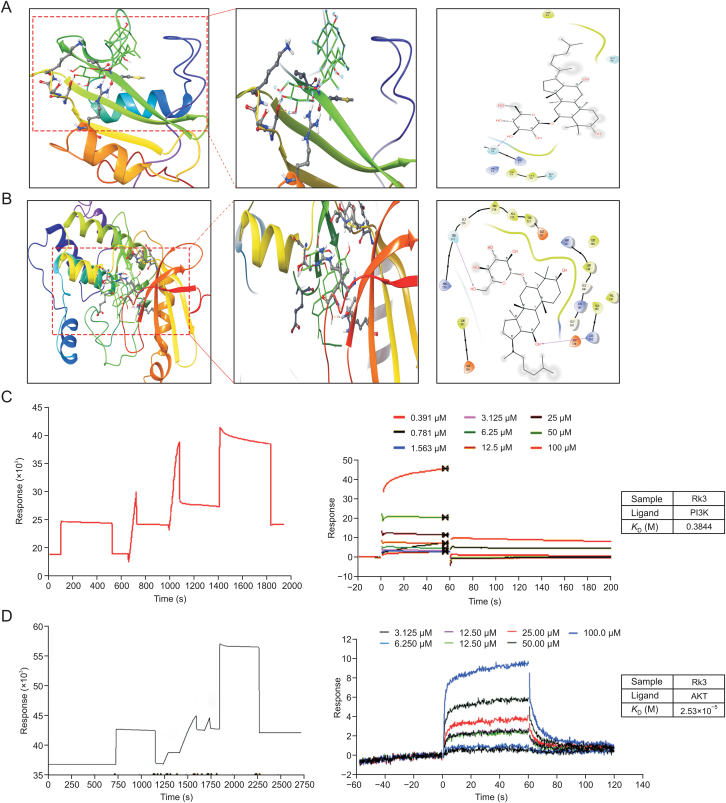
We next took a computational simulation approach to investigate the binding between Rk3 and PI3K/AKT. Docking data showed that Rk3 interacts with both PI3K (SER39) and AKT (THR217 and LYS162) via hydrogen bonds with the free energy of binding of −7.4 and −10.2 kcal/mol, respectively (Figs. 14A and B). In addition, the β-D-glucoside of Rk3 is located deep in the structural domain pocket and acts as an anchor for the interaction. The triterpene ring scaffold is surrounded by the amino acids of PI3K/AKT. Furthermore, the interactions were tested using a Biacore 3000 biosensor system, in which PI3K/AKT proteins were immobilized on a CM5 chip with a coupling capacity of approximately 25,000 RU. Rk3 bound both PI3K and AKT with KD values of 0.3844 mol/L and 2.53 × 10−5 mol/L, respectively, indicating that Rk3 could target PI3K/AKT (Figs. 14C and D).
Fig. 14.
Ginsenoside Rk3 can bind to phosphatidylinositol-3-kinase (PI3K) and protein kinase B (AKT). (A, B) Molecular docking of ginsenoside Rk3 and PI3K protein (A), and ginsenoside Rk3 and AKT protein (B). (C, D) Surface plasmon resonance results of ginsenoside Rk3 and PI3K proteins (C), and ginsenoside Rk3 and AKT proteins (D).
4. Discussion
HCC is the second leading cause of cancer-related deaths worldwide. Patients with advanced HCC have a mortality rate of up to 80% (<1 year for median survival; 18% for 5-year survival rate) [45]. The prognosis remains poor due to a high recurrence rate, aggressiveness and metastasis despite advancements in targeted therapies [46], calling for new strategies for treatment. In this study, we investigated the mechanism of Rk3 in inhibiting HCC growth, aiming to open opportunities for more effective treatment.
Cell cycle D proteins (D1, D2, and D3 in human) control the activity of CDK6 and CDK4 to regulate the cell cycle [47]. In human cancer progression, cyclin D1 is more frequently dysregulated compared to D2 and D3 and has therefore been more extensively studied [48]. CDKs are required for a complete cell division. Cyclin D and CDK4/6 play a key role in cell cycle progression by phosphorylating and inactivating Rb proteins, blocking the cell proliferation cycle at the S phase [49]. The turnover of these cyclins is regulated by various endogenous and exogenous stimuli. Here, we found that Rk3 inhibits HCC proliferation by suppressing G1 phase-related protein (CDK4, Cyclin D1, and P21) expression to block HCC cell growth at the G1 phase.
Sorafenib is the first approved drug for the treatment of patients with advanced HCC. However, it is associated with common serious side effects including leukopenia, neutropenia, thrombocytopenia, gastrointestinal and skin toxicity [50,51]. Here we showed superior anti-HCC effects of Rk3 compared to sorafenib both in vitro and in vivo. Significantly, Rk3 exhibited low toxicity in animal studies (blood analysis and liver and kidney function indexes). In line with this, recent studies have shown that compounds or drugs containing different forms of ginseng protect against chemotherapy-induced side effects [52,53].
Apoptosis is essential for the development and maintenance of tissue homeostasis, failure of which leads to the development and progression of cancer [54]. Cellular stress and apoptotic signals can cause the release of mitochondrial cytochrome c, which forms apoptosome to activate caspase-3 and a cascade of caspases to promote apoptosis [55,56]. The Bcl-2 family consists of both anti-apoptotic and pro-apoptotic proteins, inhibiting and promoting, respectively, the release of other pro-apoptotic factors such as apoptosis-inducing factors [57]. Here, we found that Rk3 significantly regulates the expression of cytochrome c and Bcl-2 family proteins, which in turn induces the caspase cascade and ultimately leads to apoptosis in HCC.
Autophagy is a dynamic cyclic process that plays a key role in degrading and recycling cellular components to maintain cellular homeostasis. In tumor cells, autophagy removes targeted components to maintain intracellular homeostasis and to promote stress tolerance. On the other hand, excessive autophagy induces autophagic cell death, leading to inhibition of tumor cell proliferation [58]. Multiple autophagy makers had been discovered including the autophagosomal membrane type of LC3 (LC3-II) and P62 that links LC3 and polyubiquitinated proteins, the abundance of which shows a positive and negative, respectively, correlation with the autophagic activity [59]. In addition, ATG family proteins are key players involved in autophagic vesicle extension and mTOR is a major negative regulator of autophagy in cancer cells. Here, we found that Rk3 significantly induces cellular autophagy in HCC by quantifying these autophagy markers. It is likely that activation of cellular autophagy is one of the mechanisms underlying the Rk3-induced inhibition of HCC.
Autophagy and apoptosis are closely related to each other. While autophagy may prevent apoptosis in some cases, it may promote apoptosis or independently induce cell death (autophagic cell death) [60]. Previous studies showed that natural phytochemicals can trigger cell death by either classical apoptosis or autophagy. In this study, our data showed that co-treatment with the autophagy inhibitor 3-MA promotes apoptosis. In addition, co-treatment with the apoptosis inhibitor Z-VAD-fmk significantly decreased the expression of autophagy marker protein LC3II/LC3I, indicating that apoptosis inhibits the autophagic process.
PI3K/AKT has been reported to be widely expressed in many types of cancer cells and is a promising target for tumor therapy [55]. An increasing number of studies have shown that the PI3K/AKT pathway is involved in the regulation of tumor cell growth, proliferation, tumor migration and invasion, and metabolism [[61], [62], [63]]. Meanwhile, PI3K/AKT plays an important role in apoptotic cell death and the control of autophagy-apoptosis crosstalk following phytochemical exposure [64,65]. Rk3 significantly inhibited the PI3K/AKT signaling pathway in HCC cells. Furthermore, pretreatment with PI3K-siRNA or AKT-siRNA enhanced Rk3-induced apoptosis and autophagy. In line with this, pretreatment with PI3K/AKT activator had the opposite effects. Moreover, molecular docking and SPR results showed that Rk3 binds to PI3K/AKT proteins. Thus, PI3K/AKT is an important target of Rk3 in inhibiting HCC growth.
5. Conclusion
In conclusion, Rk3 effectively inhibits HCC proliferation and exhibits low toxic effects and is thus a promising candidate for the treatment of HCC. Network pharmacology, proteomics, molecular docking and SPR analysis indicate that Rk3 targets PI3K/AKT to regulate autophagy and apoptosis to inhibit HCC proliferation.
CRediT author statement
Linlin Qu: Methodology, Validation, Data curation, Investigation, Visualization, Writing - Original draft preparation, Conceptualization; Yannan Liu: Validation, Methodology, Supervision, Project administration; Jianjun Deng: Supervision, Funding acquisition, Visualization, Writing – Reviewing and Editing; Xiaoxuan Ma: Data curation, Investigation, Writing - Reviewing and Editing; Daidi Fan: Resources, Project administration, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was financially supported by the National Key R&D Program of China (Grant No.: 2021YFC2101500), the National Natural Science Foundation of China (Grant Nos.: 22078264, 21978235, 22108224, and 21978236), the Natural Science Basic Research Program of Shaanxi, China (Grant Nos.: 2023-JC-JQ-17 and 2023-JC-QN-0109), the Xi'an Science and Technology Project, China (Project No.: 20191422315KYPT014JC016), and Key Research and Development Program of Shaanxi, China (Grant No.: 2022ZDLSF05-12).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2023.03.006.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R., Zhang S., Zeng H., et al. Cancer incidence and mortality in China, 2016. J. Natl. Cancer Cent. 2022;2:1–9. doi: 10.1016/j.jncc.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai Z., Wang X., Peng R., et al. Induction of IL-6Rα by ATF3 enhances IL-6 mediated sorafenib and regorafenib resistance in hepatocellular carcinoma. Cancer Lett. 2022;524:161–171. doi: 10.1016/j.canlet.2021.10.024. [DOI] [PubMed] [Google Scholar]
- 4.Xu J., Ji L., Ruan Y., et al. UBQLN1 mediates sorafenib resistance through regulating mitochondrial biogenesis and ROS homeostasis by targeting PGC1β in hepatocellular carcinoma. Signal Transduct. Target. Ther. 2021;6:190. doi: 10.1038/s41392-021-00594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G. Tossetta, D. Marzioni, Natural and synthetic compounds in ovarian cancer: A focus on NRF2/KEAP1 pathway, Pharmacol. Res. 183 (2022), 106365. [DOI] [PubMed]
- 6.Zhong R., Farag M., Chen M., et al. Recent advances in the biosynthesis, structure–activity relationships, formulations, pharmacology, and clinical trials of fisetin. eFood. 2022;3 [Google Scholar]
- 7.Hai Y., Zhang Y., Liang Y., et al. Advance on the absorption, metabolism, and efficacy exertion of quercetin and its important derivatives. Food Front. 2020;1:420–434. [Google Scholar]
- 8.Higbee J., Solverson P., Zhu M., et al. The emerging role of dark berry polyphenols in human health and nutrition. Food Front. 2022;3:3–27. [Google Scholar]
- 9.Ren J. Bringing to fore the role of peptides, polyphenols, and polysaccharides in health: the research profile of Jiaoyan Ren. Food Front. 2021;2:29–31. [Google Scholar]
- 10.Liu Y., Fan D. Ginsenoside Rg5 induces G2/M phase arrest, apoptosis and autophagy via regulating ROS-mediated MAPK pathways against human gastric cancer. Biochem. Pharmacol. 2019;168:285–304. doi: 10.1016/j.bcp.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y., Zhu C., Yang H., et al. Protective effect of ginsenoside Rg5 against kidney injury via inhibition of NLRP3 inflammasome activation and the MAPK signaling pathway in high-fat diet/streptozotocin-induced diabetic mice. Pharmacol. Res. 2020;155 doi: 10.1016/j.phrs.2020.104746. [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Lu X., Hu Y., et al. Chemical constituents of Panax ginseng and Panax notoginseng explain why they differ in therapeutic efficacy. Pharmacol. Res. 2020;161 doi: 10.1016/j.phrs.2020.105263. [DOI] [PubMed] [Google Scholar]
- 13.Chen H., Yang H., Fan D., et al. The anticancer activity and mechanisms of ginsenosides: An updated review. eFood. 2020;1:226–241. [Google Scholar]
- 14.Qu L., Zhu Y., Liu Y., et al. Protective effects of ginsenoside Rk3 against chronic alcohol-induced liver injury in mice through inhibition of inflammation, oxidative stress, and apoptosis. Food Chem. Toxicol. 2019;126:277–284. doi: 10.1016/j.fct.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 15.Wei B., Duan Z., Zhu C., et al. Anti-anemia effects of ginsenoside Rk3 and ginsenoside Rh4 on mice with ribavirin-induced anemia. Food Funct. 2018;9:2447–2455. doi: 10.1039/c8fo00368h. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Deng J., Fan D. Ginsenoside Rk3 ameliorates high-fat-diet/streptozocin induced type 2 diabetes mellitus in mice via the AMPK/Akt signaling pathway. Food Funct. 2019;10:2538–2551. doi: 10.1039/c9fo00095j. [DOI] [PubMed] [Google Scholar]
- 17.Duan Z., Deng J., Dong Y., et al. Anticancer effects of ginsenoside Rk3 on non-small cell lung cancer cells: In vitro and in vivo. Food Funct. 2017;8:3723–3736. doi: 10.1039/c7fo00385d. [DOI] [PubMed] [Google Scholar]
- 18.Noorolyai S., Shajari N., Baghbani E., et al. The relation between PI3K/AKT signalling pathway and cancer. Gene. 2019;698:120–128. doi: 10.1016/j.gene.2019.02.076. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y., Zhang Y., Qin X., et al. PI3K/AKT/mTOR pathway-related long non-coding RNAs: Roles and mechanisms in hepatocellular carcinoma. Pharmacol. Res. 2020;160 doi: 10.1016/j.phrs.2020.105195. [DOI] [PubMed] [Google Scholar]
- 20.Hennessy B., Smith D., Ram P., et al. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 21.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Capodanno A., Camerini A., Orlandini C., et al. Dysregulated PI3K/Akt/PTEN pathway is a marker of a short disease-free survival in node-negative breast carcinoma. Hum. Pathol. 2009;40:1408–1417. doi: 10.1016/j.humpath.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Vivanco I., Chen Z.C., Tanos B., et al. A kinase-independent function of AKT promotes cancer cell survival. eLife. 2014;3 doi: 10.7554/eLife.03751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanidas I., Polytarchou C., Hatziapostolou M., et al. Phosphoproteomics screen reveals Akt isoform-specific signals linking RNA processing to lung cancer. Mol. Cell. 2014;53:577–590. doi: 10.1016/j.molcel.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Q., Cai W., Zheng Y., et al. ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene. 2014;33:1828–1839. doi: 10.1038/onc.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue S., Li J., Lee S., et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19:393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue S., Zhou Y., Zhang J., et al. Anemoside B4 exerts anti-cancer effect by inducing apoptosis and autophagy through inhibiton of PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Am. J. Transl. Res. 2019;11:2580–2589. [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon M., Nam T. A polysaccharide of the marine Alga Capsosiphon fulvescens induces apoptosis in AGS gastric cancer cells via an IGF-IR-mediated PI3K/Akt pathway. Cell Biol. Int. 2007;31:768–775. doi: 10.1016/j.cellbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Hopkins A. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 30.Nottingham E., Mazzio E., Surapaneni S., et al. Synergistic effects of methyl 2-cyano-3, 11-dioxo-18beta-olean-1,-12-dien-30-oate and erlotinib on erlotinib-resistant non-small cell lung cancer cells. J. Pharm. Anal. 2021;11:799–807. doi: 10.1016/j.jpha.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z., Mao L., Yu B., et al. GB7 acetate, a galbulimima alkaloid from Galbulimima belgraveana, possesses anticancer effects in colorectal cancer cells. J. Pharm. Anal. 2022;12:339–349. doi: 10.1016/j.jpha.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Hernández M., González R., de la Rosa A., et al. Molecular characterization of autophagic and apoptotic signaling induced by sorafenib in liver cancer cells. J. Cell Physiol. 2018;234:692–708. doi: 10.1002/jcp.26855. [DOI] [PubMed] [Google Scholar]
- 33.Uehara T., Pogribny I., Rusyn I. The DEN and CCl4-induced mouse model of fibrosis and inflammation-associated hepatocellular carcinoma. Curr. Protoc. Pharmacol. 2014;66 doi: 10.1002/0471141755.ph1430s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J., Li L., Yin J., et al. Tumor-targeting intravenous lipid emulsion of paclitaxel: Characteristics, stability, toxicity, and toxicokinetics. J. Pharm. Anal. 2022;12:901–912. doi: 10.1016/j.jpha.2022.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X., Lin Y., Wu Y., et al. Identification of potential anti-pneumonia pharmacological components of Glycyrrhizae Radix et Rhizoma after the treatment with Gan An He Ji oral liquid. J. Pharm. Anal. 2022;12:839–851. doi: 10.1016/j.jpha.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Wang Y., Wu Y., et al. PKM2-mediated neuronal hyperglycolysis enhances the risk of Parkinson’s disease in diabetic rats. J. Pharm. Anal. 2023;13:187–200. doi: 10.1016/j.jpha.2022.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye W., Li L., Feng Z., et al. Sensitive detection of alkaline phosphatase based on terminal deoxynucleotidyl transferase and endonuclease IV-assisted exponential signal amplification. J. Pharm. Anal. 2022;12:692–697. doi: 10.1016/j.jpha.2021.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Shen Y., Wang S., et al. PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D., Morris J., Cook H., et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P., Markiel A., Ozier O., et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang D., Sherman B., Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Meng Y., Chen J., Liu Y., et al. A highly efficient protein corona-based proteomic analysis strategy for the discovery of pharmacodynamic biomarkers. J. Pharm. Anal. 2022;12:879–888. doi: 10.1016/j.jpha.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandrashekar D., Bashel B., Balasubramanya S., et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerbel R. Antiangiogenic therapy: A universal chemosensitization strategy for cancer? Science. 2006;312:1171–1175. doi: 10.1126/science.1125950. [DOI] [PubMed] [Google Scholar]
- 45.Harkus U., Wankell M., Palamuthusingam P., et al. Immune checkpoint inhibitors in HCC: Cellular, molecular and systemic data. Semin. Cancer Biol. 2022;86:799–815. doi: 10.1016/j.semcancer.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 46.Singal A., Zhang E., Narasimman M., et al. HCC surveillance improves early detection, curative treatment receipt, and survival in patients with cirrhosis: A meta-analysis. J. Hepatol. 2022;77:128–139. doi: 10.1016/j.jhep.2022.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai J., Li Y., Zhang G. Cell cycle regulation and anticancer drug discovery. Cancer Biol. Med. 2017;14:348–362. doi: 10.20892/j.issn.2095-3941.2017.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Hong, M. Almutairi, S. Li, et al., Wogonin inhibits cell cycle progression by activating the glycogen synthase kinase-3 beta in hepatocellular carcinoma, Phytomedicine 68 (2020), 153174. [DOI] [PubMed]
- 49.Ingham M., Schwartz G. Cell-cycle therapeutics come of age. J. Clin. Oncol. 2017;35:2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ko K., Mak L., Cheung K., et al. Hepatocellular carcinoma: Recent advances and emerging medical therapies. F1000Res. 2020;9 doi: 10.12688/f1000research.24543.1. F1000 Faculty Rev-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Offermanns S., Rosenthal W. Springer Science & Business Media; Berlin: 2008. Encyclopedia of molecular pharmacology. [Google Scholar]
- 52.Wan Y., Wang J., Xu J., et al. Panax ginseng and its ginsenosides: Potential candidates for the prevention and treatment of chemotherapy-induced side effects. J. Ginseng Res. 2021;45:617–630. doi: 10.1016/j.jgr.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Y. Liu, X. Wang, D. He, et al., Protection against chemotherapy-and radiotherapy-induced side effects: A review based on the mechanisms and therapeutic opportunities of phytochemicals, Phytomedicine 80 (2021), 153402. [DOI] [PubMed]
- 54.Varghese J., Balasubramanian B., Velayuthaprabhu S., et al. Therapeutic effects of vitamin D and cancer: An overview. Food Front. 2021;2:417–425. [Google Scholar]
- 55.Yao Z., Wang L., Cai D., et al. Warangalone induces apoptosis in HeLa cells via mitochondria-mediated endogenous pathway. eFood. 2021;2:259–270. [Google Scholar]
- 56.Zhang Q., Luo P., Zheng L., et al. 18beta-glycyrrhetinic acid induces ROS-mediated apoptosis to ameliorate hepatic fibrosis by targeting PRDX1/2 in activated HSCs. J. Pharm. Anal. 2022;12:570–582. doi: 10.1016/j.jpha.2022.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao C., Lin G., Wu D., et al. The algal polysaccharide ulvan suppresses growth of hepatoma cells. Food Front. 2020;1:83–101. [Google Scholar]
- 58.Cheng S., Chen N., Kuo H., et al. Prodigiosin stimulates endoplasmic reticulum stress and induces autophagic cell death in glioblastoma cells. Apoptosis. 2018;23:314–328. doi: 10.1007/s10495-018-1456-9. [DOI] [PubMed] [Google Scholar]
- 59.Zhang H., Caprioli G., Hussain H., et al. A multifaceted review on dihydromyricetin resources, extraction, bioavailability, biotransformation, bioactivities, and food applications with future perspectives to maximize its value. eFood. 2021;2:164–184. [Google Scholar]
- 60.Yonekawa T., Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105–117. doi: 10.1042/bse0550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y., Mao X., Chen W., et al. A discovery of clinically approved formula FBRP for repositioning to treat HCC by inhibiting PI3K/AKT/NF-κB activation. Mol. Ther. Nucleic Acids. 2020;19:890–904. doi: 10.1016/j.omtn.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L., Hou Y., Yuan J., et al. Twist promotes reprogramming of glucose metabolism in breast cancer cells through PI3K/AKT and p53 signaling pathways. Oncotarget. 2015;6:25755–25769. doi: 10.18632/oncotarget.4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fu Q., Liu Y., Fan Y., et al. Alpha-enolase promotes cell glycolysis, growth, migration, and invasion in non-small cell lung cancer through FAK-mediated PI3K/AKT pathway. J. Hematol. Oncol. 2015;8 doi: 10.1186/s13045-015-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sophia J., Kowshik J., Dwivedi A., et al. Nimbolide, a neem limonoid inhibits cytoprotective autophagy to activate apoptosis via modulation of the PI3K/Akt/GSK-3β signalling pathway in oral cancer. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-1126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Braicu C., Zanoaga O., Zimta A., et al. Natural compounds modulate the crosstalk between apoptosis-and autophagy-regulated signaling pathways: Controlling the uncontrolled expansion of tumor cells. Semin. Cancer Biol. 2022;80:218–236. doi: 10.1016/j.semcancer.2020.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.