Abstract
The HIV-1-encoded protein Vpu forms an oligomeric ion channel/pore in membranes and interacts with host proteins to support the virus lifecycle. However, Vpu molecular mechanisms are currently not well understood. Here, we report on the Vpu oligomeric organization under membrane and aqueous conditions and provide insights into how the Vpu environment affects the oligomer formation. For these studies, we designed a maltose-binding protein (MBP)-Vpu chimera protein and produced it in E. coli in soluble form. We analyzed this protein using analytical size-exclusion chromatography (SEC), negative staining electron microscopy (nsEM), and electron paramagnetic resonance (EPR) spectroscopy. Surprisingly, we found that MBP-Vpu formed stable oligomers in solution, seemingly driven by Vpu transmembrane domain self-association. A coarse modeling of nsEM data as well as SEC and EPR data suggests that these oligomers most likely are pentamers, similar to what was reported regarding membrane-bound Vpu. We also noticed reduced MBP-Vpu oligomer stability upon reconstitution of the protein in β-DDM detergent and mixtures of lyso-PC/PG or DHPC/DHPG. In these cases, we observed greater oligomer heterogeneity, with MBP-Vpu oligomeric order generally lower than in solution; however, larger oligomers were also present. Notably, we found that in lyso-PC/PG, above a certain protein concentration, MBP-Vpu assembles into extended structures, which had not been reported for Vpu. Therefore, we captured various Vpu oligomeric forms, which can shed light on Vpu quaternary organization. Our findings could be useful in understanding Vpu organization and function in cellular membranes and could provide information regarding the biophysical properties of single-pass transmembrane proteins.
Keywords: HIV-1 Vpu, Vpu oligomer, MBP-Vpu fusion protein, Negative staining electron microscopy of oligomeric protein, EPR spectroscopy of oligomeric protein
1. Introduction
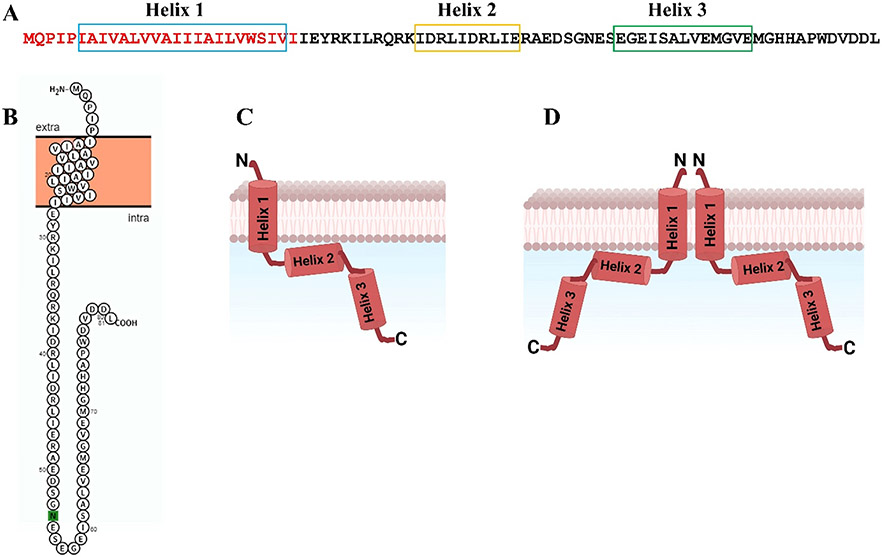
We studied the structure and function of the viral protein U (Vpu) protein encoded by the human immunodeficiency virus 1 (HIV-1). Vpu is among the key HIV-1 proteins aiding virus adaptation and proliferation. Of the four virus subtypes currently known, viz. M, N, O, and P, the M subtype is globally distributed, being the major cause of HIV-1 pandemic spread (Sharp and Hahn, 2011). All HIV-1 subtypes encode the accessory protein Vpu, but it is noteworthy that this protein was found to be most active in the pandemic-causing subtype M, thereby suggesting that Vpu may play an important role in HIV-1 lifecycle and infectivity (Sharp and Hahn, 2011; Gonzalez, 2015; Khan and Geiger, 2021). Vpu is expressed in the infected cells and is located in the membranes of the trans-Golgi network, endoplasmic reticulum (ER), and plasma membrane (Hussain et al., 2007; Gonzalez, 2015). It is a small protein containing 81 amino acids and is organized in the N-terminal domain with a conserved hydrophobic helical region (Helix 1, transmembrane domain [TMD]), which resides in and traverses the cellular lipid membranes, and in a C-terminal domain containing two amphipathic helical regions, Helix 2 and Helix 3, with Helix 2 possibly associated with membrane surfaces (Fig. 1) (Cohen et al., 1988; Strebel et al. 1988; Lewinski et al., 2015; Khan and Geiger, 2021).
Fig. 1. Amino acid sequence of Vpu protein and its topology in lipid membranes:
(A) Amino acid sequence of HIV-1 Vpu. The N-terminus is in red, and the C-terminus is in black; the amino acids forming Helix 1 (TMD), Helix 2, and Helix 3 are enclosed in boxes. (B) Monomer organization predicted by the PROTTER program. (C) Proposed monomer domain organization is shown with the N-terminal Helix 1 traversing the lipid bilayer, Helix 2 interacting with the membrane surface (in correspondence with NMR studies (Ma et al., 2002)), and Helix 3 being highly soluble. (D) Suggested Vpu oligomerization via TMD self-association (only two monomers are shown for clarity).
Vpu is known to oligomerize in lipid environments through its N-terminal Helix 1 (TMD), forming a pentameric ion-conducting channel (Maldarelli et al., 1993; Ewart et al., 1996; Lu et al., 2010). Therefore, based on its monomer structure and self-oligomerization, Vpu is referred to as a member of the viroporins family (Nieva et al., 2012; Gonzalez, 2015). The proteins in this family are encoded by almost all human health-threatening viruses. They support virus adaptation, proliferation and pathogenicity and are targets for drug development (Agirre et al., 2002; Gonzalez and Carrasco, 2003; Fischer and Hsu, 2011; Nieva et al., 2012). In infected cells, Vpu has two main biological functions, performed by its N- and C-terminal domains: (i) modifying signaling pathways, carried out mainly through the C-terminal interactions with host proteins, leading to the downregulation of the CD4 receptor, and (ii) enhancing the release of newly formed virions, usually through viroporin activity encompassed by its N-terminal homo-oligomer (Strebel et al., 1989; Willey et al., 1992; Jia et al., 2014) but also through the counteraction of the tetherin (BST2) host protein (Douglas et al., 2009; Mitchell et al., 2009). These functions greatly facilitate HIV-1 infection by either cell-to-cell or cell-free spread (Klimkait et al., 1990; Sato et al., 1992).
Because of diverse functions of Vpu that are critical to HIV-1 adjustment and proliferation, thorough knowledge of its structure–function relationship at the molecular level could reveal approaches to regulate this protein’s function. Yet, the structural information about the full-length (FL) Vpu in its monomeric and oligomeric forms is very limited. It is also not well understood how Vpu interacts with lipid membranes to form homo-oligomers and participates in interactions via its TMD or C-terminal region with host proteins.
Here, we report on our findings about the oligomerization forms of FL Vpu in solution and lipid-like environments. These studies were possible, owing to our success in producing the Vpu protein in soluble form, which was aided by the design of a fusion construct containing MBP moiety connected at C terminus by an engineered short linker to the FL Vpu (MBP-Vpu chimeric protein). We produced large quantities of highly pure MBP-Vpu protein and conducted structural studies to explore this protein’s oligomerization state. To this end, we applied analytical size exclusion chromatography (SEC), negative staining transmission electron microscopy (nsEM), and continuous wave (CW) and pulse EPR spectroscopy with protein spin labeling.
2. Results
2.1. Design, cloning, and production in soluble form of MBP-Vpu fusion protein
We engineered a construct of MBP fused to the N-terminus of FL Vpu (MBP-Vpu protein) and cloned it in a bacterial expression vector (pET15b), resulting in the MBP-Vpu/pET15b plasmid (Fig. 2).
Fig. 2. Design and cloning of MBP-Vpu protein construct:
(A) The amino acid sequence of the FL MBP-Vpu fusion protein—The His8 tag is in red, MBP is in black, Vpu is in blue, and the linkers, His8-MBP and MBP-Vpu, are underlined. (B) Schematic representation of the MBP-Vpu protein—the color code is as in (A) except the linkers, which here are in orange. (C) MBP-Vpu plasmid map—the gene encoding the MBP-Vpu protein was cloned in the pET15b vector.
The MBP-Vpu chimera also included the His8 affinity tag, which allowed for more efficient Ni-affinity purification than the standard His6 tag (Georgieva et al., 2020). Furthermore, the incorporation of MBP served multiple purposes. First, it increased the solubility of the fusion protein construct, enabling us to produce in E. coli large (milligram-scale) quantities of MBP-Vpu in soluble form. Second, we used MBP as an affinity purification tag. Finally, the increased molecular weight and size of Vpu fusion protein made it suitable for routine imaging by EM. We designed a relatively short linker of just eleven amino acids connecting MBP to Vpu to provide sufficient conformational stability of the MBP-Vpu protein, thereby facilitating its imaging by EM. Such a fusion-construct strategy to study by EM small proteins and their complexes in lipid or buffer environments has grown in popularity and enjoyed success over the last few years (Liu et al., 2019; Nygaard et al., 2020).
We transformed the MBP-Vpu/pET15b plasmid into E. coli cells, and we indeed found the expressed protein in the cytosolic fraction, which enabled us to handle the MBP-Vpu as a soluble protein in the following purification procedures. The calculated molecular weight and molar extinction coefficient (ε) of our MBP-Vpu protein are 52.1 kDa and 78,840 M−1cm−1, respectively (based on ExPasy Prot Param software). We used this extinction coefficient to estimate the protein molar concentration in the subsequent experiments.
By successively applying Ni-affinity and amylose-affinity chromatography, we obtained highly pure MBP-Vpu, as detected by SDS-PAGE and WB (Fig. 3).
Fig. 3. SDS-PAGE and western blotting (WB) of double affinity purified MBP-Vpu.
All detected bands were WB positive, indicating they all are from MBP-Vpu.
SDS-PAGE and WB detected several protein bands, and the result was reproducible in repeated experiments. These bands appeared most likely due to heterogeneous MBP-Vpu oligomers in the SDS environment, which is quite feasible because SDS could be considered as a membrane mimetic (Georgieva et al., 2010). Specifically, we detected bands at ca. 50 kDa, ca. 100 kDa, and ca. 250 kDa, corresponding to the MBP-Vpu monomer, dimer and pentamer, which is in line with the altered and destabilized oligomerization of MBP-Vpu under many protein/detergent (lipid) conditions. The observed prevalence of MBP-Vpu monomers is not surprising because SDS can serve as a detergent/membrane mimetic, but it is also a harsh detergent capable of altering MBP-Vpu’s quaternary structure. The bands above 250 kDa could appear due to even larger oligomers, possibly such as observed in lyso-PC/PG (linear arrays, vide infra), but we cannot rule out unspecific SDS-induced oligomers. It is unlikely that the apparent bands at molecular weight lower than 50 kDa (for MBP-Vpu monomer) are a result of protein degradation because we did not observe such an effect by SEC (also discussed below). They are most likely a result of anomalous protein migration due to a smaller effective size of folded in SDS Vpu monomer. Indeed, artefacts of SDS effects on the oligomerization state and SDS-PAGE migration of other membrane protein Aβ42 were previously observed (Pujol-Pina et al., 2015).
An MBP-FL Vpu construct was previously created and investigated, but mostly in context of in cell experiments than as a tool for in vitro structural studies (Hussain et al., 2007). Here, we focus on in vitro studies to characterize Vpu’s oligomeric structure benefiting from available methods and tools. Also, MBP was utilized previously as a fusion tag in studies of Vpu C-terminal interactions with host proteins, (Jia et al., 2014) adding confidence for its utility in researching this class of viral proteins. The study we report here, however, goes beyond the using MBP just as a fusion tag for expression, since we were able to clearly visualize soluble MBP-Vpu oligomers due to increased protein size, paving the way to characterize Vpu in significant detail by EM and pulse EPR.
2.2. MBP-Vpu forms protein concentration-dependent oligomers in aqueous solution
Literature strongly suggests that Vpu forms oligomers in the native membranes of Golgi, ER, and plasma membrane, as well as in model membrane mimetics (Maldarelli et al., 1993; Ewart et al., 1996; Hussain et al., 2007; Lu et al., 2010; Gonzalez, 2015). Therefore, our initial goal was to produce FL Vpu in soluble form as a fusion construct with MBP and then reconstitute it in a lipid or lipid-like environments for functional and structural investigations. Studying the structure of Vpu within an aqueous non-membrane context was not initially planned. Vpu has highly hydrophobic regions (TMD) and, in general, has not been anticipated to acquire any thought-provoking structure outside membrane. However, surprisingly, our SEC experiments on MBP-Vpu in solution showed that this protein elutes predominantly as a high molecular weight oligomer (Fig. 4A). In some cases, a second much less intensive elution peak corresponding to lower molecular-weight protein was observed as well (Supporting Fig. 1). We collected the SEC fractions corresponding to the main protein elution peak and subjected them to SDS-PAGE and WB. The results from this experiment explicitly confirmed that the elution peak is of MBP-Vpu protein (Fig. 4B). Next, we ran SEC using a control sample containing a mixture of soluble proteins with known molecular weight (SEC protein standard) to estimate the molecular weight of the MBP-Vpu oligomers (Fig. 4A). We found that the molecular weight of the oligomer, which elutes between 8.5 ml and 10.5 ml, is greater than 200 kDa, as the elution peak of this protein occurs before but close to the elution peak of β-amylase from a sweet potato (200 kDa) (Fig. 4A). The position of the elution peak also suggests that the MBP-Vpu oligomers do not elute in the column void volume because the void volume is typically 20–25% of the column bed volume, which is less than 6 ml for the 24 ml column we used. Furthermore, the protein migration could be altered due to 10% glycerol in the buffer. Thus, our SEC results reveal that MBP-Vpu forms stable oligomers in solution with molecular weight greater than 200 kDa. The low-intensity peak at 15–17 ml in the SEC elution profile (Supporting Fig. 1) corresponds to MBP-Vpu monomer, as it occurs between the peaks of albumin from bovine serum (66 kDa) and carbonic anhydrase from bovine erythrocytes (29 kDa) from the SEC standards; the protein in this peak is also WB-positive.
Fig. 4. SEC results confirm that MBP-Vpu forms pentamers in solution:
(A) The SEC data for MBP-Vpu is shown in blue. An elution peak at 8.5 ml to 10.5 ml was observed. The fractions corresponding to this peak were pooled and subjected to SDS-PAGE and WB. The SEC data for mixture of protein standards is shown in red with the protein molecular weight indicated next to each elution peak (β-amylase from a sweet potato – 200 kDa, alcohol dehydrogenase from yeast – 150 kDa, albumin from bovine serum – 66 kDa, carbonic anhydrase from bovine erythrocytes – 29 kDa, and cytochrome c from a horse heart – 12.4 kDa). (B) SDS-PAGE and WB results for the pooled MBP-Vpu fractions as described in (A) are shown—the protein in this elution peak was WB-positive, confirming that it is MBP-Vpu. The samples were not heated before loading them onto the gel.
We sought to find out which region of MBP-Vpu is responsible for the oligomerization in solution. MBP does not self-oligomerize in solution when used as a fusion tag (Hu et al., 2011; Selmke et al., 2018). On the other hand, the TMD region of Vpu is responsible for oligomerization in a lipid environment (Ewart et al., 1996; Lu et al., 2010). Therefore, it may well be that this region contributes to the observed protein self-association in aqueous solution, possibly due to the hydrophobic effects minimizing contacts with water (Lins and Brasseur, 1995). We further examined the Vpu C-terminus (residues 28–81 in FL Vpu) and found that in solution it is predominantly a monomer (Supporting Fig. 2), although it was earlier suggested that this region is important for oligomer stabilization in lipids; (Maldarelli et al., 1993) in addition, self-association in buffer solution was previously observed for the cytodomain of gB fusogen from herpes simplex virus, (Cooper et al., 2018) indicating that juxtamembrane soluble regions of single-pass transmembrane proteins might play a stabilizing role in their oligomerization.
We next utilized nsEM to visualize these MBP-Vpu oligomers in solution. Certainly, our goal to increase the effective molecular weight and size of Vpu by fusing it to MBP was fruitful, and we successfully observed well-defined oligomeric structures for samples containing either 5 μM (Fig. 5A) or 10 μM of MBP-Vpu (Supporting Fig. 3). These protein assemblies were similar in size and were about 20 nm in diameter in the axial view of the oligomer (Fig. 5A). We confirmed the uniformity of protein aggregates by the particle size analysis. Within a set of more than 30 protein particles, we found high homogeneity with an average particle diameter of ca. 19.8 nm (Supporting Fig. 4).
Fig. 5. Representative nsEM images of negatively stained MBP-Vpu in solution:
(A) Data for 5 μM of protein. (B) Data for 0.5 μM of protein. In all images, the black scale bars correspond to 100 nm, and the red bars in magnified views correspond to 20 nm. It is clear from the figure that at the higher protein concentration more uniform oligomers form with typical diameters, for those apparently viewed from the top being ca. 20 nm. (They are emphasized with yellow circles in (A)). The qualitative conclusion is that these oligomers are pentamers (see the inset in the rightmost panel in (A)). The blue arrows in (B) indicate smaller protein oligomers, noticeable only in a low concentration sample of 0.5 μM of MBP-Vpu.
The coarse analysis of the MBP-Vpu oligomers imaged by nsEM, which we performed further, suggests a pentamer as a plausible state (Fig. 5), in agreement with SEC results and available data on Vpu in lipids (Ewart et al., 1996; Lu et al., 2010). Thus, considering that the empty of substrate MBP monomer is about 6.8 nm long based on its crystal structure,(Telmer and Shilton, 2003), when this number is taken together with the possible size of Vpu pentameric pore/self-associate, the length of MBP-Vpu linker, and the size of the MBP monomer, observing pentameric structures of 20 nm size is up to expectations (Supporting Fig. 6). In addition, we found that the oligomer stability was affected by protein dilution because when the protein concentration was lowered to 0.5 μM, oligomers of smaller size compared to those formed at 5 μM protein concentration were also present (Fig. 5B); MBP-Vpu monomers were possibly present as well. This indicates that the MBP-Vpu oligomer assembly is protein (monomer) concentration dependent. However, apparently this dependence could only be observed in low micromolar to nanomolar concentration range. At higher protein concentrations, fully assembled and stable oligomers are populated, i.e., the process could be cooperative.
To corroborate our findings from nsEM experiments, we conducted EPR study on spin-labeled at residue Q36C MBP-Vpu in buffered solution (The residue numbering is based on the Vpu sequence). The EPR results provided further support to the formation of soluble MBP-Vpu oligomers. We recorded CW EPR spectrum with the lineshape (Fig. 6A) indicative of highly restricted mobility of the spin label, and thereby the corresponding protein region (the loop between Vpu TMD and Helix 2) (Georgieva, 2017; Garcia-Rubio, 2020). This result agrees with protein self-association creating specific structural arrangement in this Vpu region which may bury the labeling site. In solution, the Q36C residue is highly likely to be occluded and consequently be refractory to spin labeling, based on our unsuccessful attempts to spin label this site in solution. Furthermore, the DEER data proved beyond any doubt that in solution MBP-Vpu assembles into a distinct and stable oligomer. For the singly spin-labeled protein at position Q36C, we recorded avery clear, well-defined DEER signal due to the dipole–dipole interactions between spin labels (Fig. 6B). The high dipolar signal amplitude of 0.56 proves that the protein is oligomer (Bode et al., 2007; Georgieva et al., 2013; Georgieva et al., 2015) because singly spin-labeled protein monomers do not produce a DEER signal. The presence of two different distances in equal proportion in a presumed pentamer and possibly the existence of a set of spin label conformers are likely the reason for the lack of clear dipolar oscillations for the chosen site.
Fig. 6. EPR data for the spin-labeled MBP-Vpu Q36C residue in solution.
(A) CW EPR spectrum corresponds to relatively slow motion of spin label and thereby the protein region it is attached to. (B), center panel: The baseline-subtracted and normalized time-domain DEER trace for spin-labeled MBP-Vpu. The data value at zero time of 0.56 (DEER “modulation depth”) is indicative of at least three coupled spins. It will be only 0.35 for a dimer labeled with 100% efficiency, therefore most likely the data point to more than three spins based on realistic spin labeling efficiency for this case. (B), right panel: The distances distribution reconstructed from the time-domain DEER data plotted in the center. The distances are inter-spin label distances possible among the spin-labeled sites in the oligomer. The distance distribution reveals two main peaks at about 3 nm and 4.3 nm. This distribution is consistent with the expectations for a symmetric pentamer.
It is currently uncertain whether this oligomer has five-fold symmetry, but more detailed future spin labeling DEER studies could answer this and other questions, helping to characterize these oligomers. If we assume an MBP-Vpu pentamer, as suggested by SEC and nsEM (Figs. 4 and 5, and Supporting Fig. 6, the spin labels in the protein monomers would have nearly a pentagon arrangement. Therefore, in the case of a 5-fold symmetry, one would expect the DEER signal reconstruction to produce two distances, R1 and R2 = 1.618R1, where R1 is the distance between the juxtaposed labels in a pentamer. Experimentally, we obtained the distance distribution having the two maxima at R1 ≈ 3 nm and R2 ≈ 4.3 nm (Fig. 6B). Thus, the experimental ratio of R2 = 1.43R1 is close to the calculated value and is in support of having two distances possibly originating from a protein pentamer. The smaller experimental ratio could be explained based on the location of the spin labeling site in the linker at the N-terminal side of the Helix-2, the orientation of the spin label and possibly more than one spin label conformers, which also contributes to the broad distance distribution. Pentamer asymmetry could be another reason for broader distance distribution. Similar DEER studies on other oligomeric proteins, such as the heptameric mechanosensitive channel MscS, were conducted before (Pliotas et al., 2012) and proved highly suitable for oligomer characterization. The DEER method can provide even further rationale about the oligomeric order. This is because of the DEER capacity to quantify the number of interacting spin labels in a spin-labeled protein oligomer, which is based on the “modulation depth” of the DEER signal, which is the (processed) DEER signal amplitude at zero time (see Fig. 6B) (Milov et al., 1984; Bode et al., 2007; Jeschke et al., 2009; Georgieva et al., 2015). In our experimental setup, the modulation depth for efficiently labeled proteins or synthetic biradicals is always very close to the calculated value of 0.35. For a symmetric oligomer of order N, the modulation depth is 1 — (1 — pf)N-1, wherein f is spin-labeling efficiency. Experimentally, we obtained V ≈ 0.56, which unambiguously indicates oligomer of order higher than dimer and even trimer. However, given the incomplete spin labeling (less than 100% due to relatively inaccessible spin-labeling site) and the bimodal distance distribution, the result is fairly consistent with a protein pentamer.
Taken together, the SEC, nsEM and DEER data provide sufficient arguments in support of conceivably pentameric MBP-Vpu in solution. Indeed, fractional pentamerization of Vpu in solution was observed before,(Hussain et al., 2007) but in that study, the oligomers were only scetchy defined and not characterized any further.
2.3. Lipid and detergent environments as well as protein-to-lipid (detergent) molar ratio affect MBP-Vpu oligomerization and oligomer morphology
After revealing that MBP-Vpu forms stable oligomers in solution, we intended to understand the behavior of these oligomers and the self-assembly of MBP-Vpu in general upon protein reconstitution in detergent or lipids. To this end, we supplemented the buffer of soluble MBP-Vpu with either β-DDM, lyso-PC/PG mixture at 50:50 mol%, or DHPC/DHPG mixture at the same ratio. All these are membrane-substituting mimetics, forming micelles of different sizes and shapes (Fernandez et al., 2002; Koehler et al., 2010; Hutchison et al., 2017). However, we used DHPC/DHPG at 2 mM or less, which is below the critical micelles concentration (CMC) of about 15 mM (concentrations at and above 2 mM produced difficult to interpret nsEM data). Furthermore, we selected these hydrophobic environments due to their extensive use in studies of membrane proteins because they possess appropriate membrane-like properties (le Maire et al., 2000; Fernandez et al., 2002; Georgieva et al., 2013; Georgieva et al., 2014; Georgieva et al., 2015; Kotov et al., 2019). Membrane proteins maintain their activity in lyso-lipids,(Koehler et al., 2010) and the structure of Glycophorin A, also a single-pass membrane protein, was solved in DPC micelles (Mineev et al., 2011). Additionally, the structure of Vpu monomer was previously studied by NMR in DHPC micelles,(Ma et al., 2002) proving these lipid mimetics provide a suitable environment for membrane proteins. In our studies, we also used lipids with negatively charged headgroups (lyso-PG and DHPG) because they are deemed important for interactions with positively charged residue in similar proteins (Nieva et al., 2012).
We conducted analytical SEC experiments on MBP-Vpu reconstituted in 1 mM of β-DDM or 1–2 mM of lyso-PC/PG. In both cases the protein concentration used was 25–50 μM. Interestingly, in both detergent and lipid environments, we detected several SEC elution peaks. In β-DDM, we observed elution peaks at 8.5–10.5 ml, as well as peaks with maxima at 12 ml and close to 16 ml (Fig. 7A). Our experimental conditions of 50 μM protein produced a peak with the highest intensity at 12 ml. A similar SEC profile was observed for protein in lyso-PC/PG. However, in this case, the intensities of the elution peaks for different protein oligomeric states depended also on the protein concentration. Additional shoulders at 7–8.5 ml and 12.5–14.5 ml were apparent, and the peak at 16 ml was not present (Fig. 7B). When compared to the elution profile of protein standards with known molecular weights (Fig. 4A), the rough estimate is that the peak at 8.5–10.5 ml corresponds to protein with a molecular weight of 260 kDa, likely representing MBP-Vpu pentamers. The peaks from 10.5 ml to 12.5 ml correspond to protein with a molecular weight of approximately 200 kDa, suggesting MBP-Vpu tetramers, and the last peak at 16 ml is of an MBP-Vpu monomer. Therefore, our results point to significant restructuring of the MBP-Vpu protein in lipid environments. However, this restructuring is not in the direction of stabilizing a unique protein oligomeric form. Instead, greater oligomer heterogeneity occurred when Vpu transitioned from an aqueous to a detergent or lipid environment. Again, for protein in β-DDM and lyso-PC/PG, we collected the SEC fractions corresponding to different peaks and subjected them to SDS-PAGE and WB analysis (Supporting Fig. 8). Notably, the SDS-PAGE and WB results were like those for protein in solution. Furthermore, heat treatment of the samples before loading them onto the gel had no effect on the result (Supporting Fig. 9).
Fig. 7. SEC data for MBP-Vpu in β-DDM and lyso-PC/PG:
Data for 50 μM of protein in 1 mM of β-DDM; (B) Data for 50 μM and 25 μM of protein in 2 mM of lyso PC/PG. In both environments, multiple elution peaks of MBP-Vpu corresponding to distinct protein oligomeric states were detected.
We performed nsEM to visualize the MBP-Vpu oligomers in β-DDM, lyso-PC/PG, and DHPC/DHPG. We used a range of protein and detergent/lipid concentrations: 5 μM and 1 μM of protein both in 1 mM β-DDM (protein-to-detergent molar ratios, P/Ds, of 1:200 and 1:1000, respectively) (Fig. 8); 5 μM of protein in 1 mM of lyso-PC/PG, 5 μM of protein in 2 mM of lyso-PC/PG, and 0.5 μM of protein in 2 mM of lyso-PC/PG (protein-to-lipid molar ratios, P/Ls, of 1:200, 1:400 and 1:4000, respectively) (Fig. 9); and 1 μM of protein in 1 mM of DHPC/DHPG, 1 μM of protein in 2 mM of DHPC/DHPG, and 5 μM of protein in 2 mM of DHPC/DHPG (P/Ls of 1:1000, 1:2000, and 1:400, respectively) (Fig. 10).
Fig. 8. Representative nsEM images of MBP-Vpu in β-DDM:
(A) data for 5 μM of protein in 1 mM of β-DDM; (B) data for 1 μM of protein in 1 mM of β-DDM; and (C) data for control sample of 1 mM of β-DDM. Heterogeneous oligomers, including linear arrays of MBP-Vpu, are encircled in yellow. The 100-nm bar is in black.
Fig. 9. Representative nsEM images of MBP-Vpu in lyso-PC/PG:
(A) data for 5 μM of protein in 2 mM of lyso-PC/PG; (B) data for 0.5 μM of protein in 2 mM of lyso-PC/PG; and (C) data for 5 μM of protein in 1 mM of lyso-PC/PG; (D) data for control sample of 1 mM of lyso-PC/PG. The 100-nm bar is in black. The formation of linear MBP-Vpu oligomers is clearly visible in (A), whereas larger protein self-assemblies were observed in (B) and (C). The linear MBP-Vpu assemblies are enclosed in yellow, and the smaller oligomers are indicated with blue arrows. The red bar in (A) rightmost panel shows 27 nm linear protein arrays.
Fig. 10. Representative nsEM images of MBP-Vpu in DHPC/DHPG:
(A) Data for 1 μM of protein in 2 mM of lipids; (B) Data for 5 μM of protein in 2 mM of lipids; (C) Data for 1 μM of protein in 1 mM of lipids; (D) Data for control sample of 1 mM of lipids. The aggregates that are similar to MBP-Vpu pentamers in just a buffer are encircled in yellow and the 20-nm bar is in red. The 100-nm bar is in black.
In β-DDM, we found that the MBP-Vpu oligomeric state is highly heterogeneous with aggregation order increasing upon increasing the protein concentration, i.e., decreasing P/D (Fig. 8A vs. Fig. 8B). Remarkably, this indicates that the Vpu oligomers possess reduced stability in a detergent environment compared to the oligomers in an aqueous environment, i.e., the self-association of Vpu monomers in a hydrophobic environment is less efficient. Thus, in the case of 1 μM of protein (P/D of 1:1000), we observed mostly dispersed low-order oligomers and possibly monomers (which are too small to be clearly distinguished by the technique used). At 5 μM of MBP-Vpu (P/D of 1:200), larger protein oligomers persisted (encircled in yellow in Fig. 8A), but they were apparently smaller than pentamers. Moreover, under the P/D of 1:200 condition, elongated linear-like aggregates of MBP-Vpu were also formed (surrounded by a yellow ellipse in Fig. 8A), representing novel structural organization of membrane-bound Vpu, which has not been reported previously.
MBP-Vpu oligomers were detected in lyso-PC/PG as well, but varying patterns of oligomerization were observed for P/Ls of 1:400 and 1:200 vs. 1:4000. Remarkably, for P/L of 1:400, similarly to P/D 1:200 (Fig. 8A), we observed the formation of well-pronounced close to linear MBP-Vpu arrays with a length of up to or even longer than 30 nm (Fig. 9A). To confirm this hydrophobic environment-driven protein organization, we further conducted EM on negatively stained MBP-Vpu linked to 5 nm gold nanoparticles (GNPs) in lyso-PC/PG as well as EM on non-stained MBP-Vpu linked to 1.8 nm GNPs (we reproduced the 5 μM of protein in 2 mM of lyso-PC/PG conditions). In both cases, we could again trace the GNP-MBP-Vpu arrays (Supporting Fig. 10). Thus, in a hydrophobic membrane-mimicking environment, we observed a novel organization of Vpu protein. More heterogeneous MBP-Vpu oligomers were detected for P/Ls of 1:4000 and 1:200 (Fig. 9B, C). Still, along with shorter and more round-shaped protein oligomers, we also observed elongated protein structures as for P/L 1:400.
Large variations in MBP-Vpu oligomers were detected by nsEM in DHPC/DHPG as well (Fig 10). The most homogeneous and well-shaped oligomers were observed for samples of 1 μM or 5 μM of protein in 2 mM of lipids (P/L of 1:2000 and 1:400) (Fig. 10A, B), validating further the finding that the protein concentration plays a role in Vpu oligomer assembly in a lipid environment.
Again, we conducted CW measurement on the spin labeled at residue Q36C MBP-Vpu protein in β-DDM and lyso-PC/PG. In these environments, we observed an altered CW EPR spectra lineshape with narrower lines (Fig. 11A) compared to the spectrum in just buffer (Fig. 6A, Supporting Fig. 7B). Thus, the CW EPR results for protein in detergent and lysolipid clearly point to protein restructuring upon transition from solution to detergent- and lysolipid-bound states, resulting in increased mobility of the spin label and more dynamic properties of the Vpu region to which the spin label is attached. Indeed, this restructuring allowed the spin labeling of the residue Q36C, possibly by making it more exposed and therefore accessible to the spin label to interact. Further, for the spin-labeled protein in detergent β-DDM, we observed a decrease in the DEER modulation depth to 0.24 (Fig. 11B) as compared to 0.56 in solution (Fig. 6B). This reduction in the modulation depth suggests a reduced MBP-Vpu oligomeric order due to the destabilizing effect of the detergent environment, but it could also result from the altered arrangement of MBP-Vpu monomers within the oligomers (Georgieva et al., 2015; Tang et al., 2015a, 2015b). The DEER-derived distance distributions are even broader than for the MBP-Vpu oligomer in solution and also exhibit an extra peak at a distance of ~ 3.6 nm, which also indicates increased oligomer heterogeneity. While the exact origin of the central peak in the distance distribution is unknown, it may be due to a distinct low-order oligomer or a distinct and more confined spin-label conformer. Thus, the EPR results agree with the SEC and nsEM data showing MBP-Vpu restructuring upon transition from an aqueous to a lipid-bound state.
Fig. 11. EPR data for spin-labeled residue Q36C in hydrophobic environment.
(A) CW EPR data for protein in β-DDM (top) and lyso-PC/PG (bottom). Spectral changes indicating faster motion were observed upon transition to a hydrophobic environment compared to these in solution. (B) Baseline-corrected DEER signal (top) and reconstructed distances (bottom) in β-DDM. A broader distance distribution with additional peaks compared to those in solution were obtained. A broader spectrum may be due to a more opened conformation of helix 2.
All in all, the results from experiments using SEC, nsEM, and EPR techniques strongly suggest that a membrane environment modifies the affinity of Vpu monomers’ self-association in proteomicelles due to additional forces of Vpu-lipid interaction. The results also indicate that the Vpu oligomer assembly is likely more efficient in closer to native lipid environment (lyso-PC/PG and DHPC/DHPG) compared to detergent (β-DDM). Previously, such an effect of detergent vs. lipid on the influenza M2 protein self-associating was also identified (Georgieva et al., 2015). We recognize that the membrane mimetics of the current study do not provide perfect membrane properties, e.g., they do not form lipid bilayers, and lack the curvature and lateral pressure characteristic for cellular membranes. But as they well represent lipid local environment these mimetics have provided practical and important platforms to widen studies of membrane proteins; their application is also justified by the particular experimental techniques used (Warschawski et al., 2011; Majeed et al., 2021).
3. Discussion
Vpu protein plays an important role in the HIV-1 life cycle through its function as an ion-conducting channel in host membranes, (Ewart et al., 1996; Schubert et al., 1996) owing to the protein TMD oligomerization (Fig. 1) and interaction of its soluble C-terminal domain, as well as TMD, with host proteins (Maldarelli et al., 1993; Ewart et al., 1996; Hussain et al., 2007; Lu et al., 2010; Gonzalez, 2015). In addition to its involvement in transporting ions across cellular membranes, viral egress, and budding,(Dube et al., 2010; Roy et al., 2014) Vpu was found to suppress cellular signals critical for proper immune response in already infected cells (Langer et al., 2019). In spite of the extensive studies to elucidate the structure–function relationship of this protein, there are still unanswered questions. NMR spectroscopy and X-ray crystallography provided insights into the structure of the isolated C-terminal region, (Lu et al., 2010; Jia et al., 2014) TMD, (Lu et al., 2010) and FL Vpu monomer (Ma et al., 2002). However, currently, there is no complete understanding of the structural basis of Vpu association with host proteins, and details about how this protein interacts with a lipid environment are unknown. Despite the existing hypothesis that pentamer is one of the functional Vpu states,(Lu et al., 2010) the assembled pentamer of FL Vpu has not been visualized. Furthermore, given that Vpu forms pentamer in the membranes of Golgi and intracellular vesicles, but not in the endoplasmic reticulum (ER),(Hussain et al., 2007) it is apparent that this protein functions in multiple quaternary structures. This lack of comprehensive knowledge about the structure–function relationship of Vpu presents an obstacle in understanding how this protein fulfils multiple roles in HIV-1 adaptation and persistence.
Through our studies here, we aimed to contribute to overcoming this deficiency. We utilized a combination of analytical SEC, nsEM, and EPR spectroscopy to capture and characterize multiple oligomerization states of Vpu residing in detergent and lipid environments. Due to the small size of FL Vpu, ca. 9 kDa, conducting EM on this protein, even in its pentameric form, would be difficult due to the current limitations of the method. Therefore, we benefited from the recently developed fusion-construct methodology to visualize small proteins in membranes by linking this protein to a larger and soluble protein, thus increasing the effective protein size/molecular weight (Nygaard et al., 2020). To do so, we generated a fusion construct of MBP linked to the N-terminal of FL Vpu (Fig. 2A and B). The linker of 11 amino acids between the two proteins was wisely selected to minimize the protein flexibility and, at the same time, avoid protein oligomerization hindrance due to the presence of the bulkier MBP.
Interestingly, when we subjected the purified MBP-Vpu protein in buffer, with no lipid or detergent present, to analytical SEC and nsEM, we revealed that the protein forms stable oligomers, probably pentamers in an aqueous environment (Figs. 4, 5 and 6). Altogether, SEC, nsEM, and DEER confirm that above a 5-μM concentration (used in nsEM experiments) and up to a 65-μM concentration (used in SEC and DEER experiments), and likely even higher concentrations, MBP-Vpu exists in stable and possibly unique oligomer whose structure could be solved. Based on prior studies,(Hu et al., 2011; Jin et al., 2017; Selmke et al., 2018) it is well known that in solutions MBP does not form homo oligomers on its own. In the cases of oligomeric arrangement of MBP fused to other proteins, the observed self-association was caused by its part-ners (Coscia et al., 2016). Therefore, we strongly believe that in our case the observed oligomerization is caused by the Vpu moiety of the protein, primarily its TMD.
We forgo discussions of the physiological relevance of a Vpu oligomer (pentamer) in solution, as it is currently unclear to what extent these oligomers may represent a Vpu physiological state. It could be that the assembly of such an oligomer is an isolated special case, where highly hydrophobic TMDs are driven together reducing the contact with aqueous phase. The solubility of the whole complex could be supported by the large MBP content contributing to the pronounced hydrophilic nature to the MBP-Vpu fusion construct. Still, this remains an unique observation, poised to contribute to our knowledge of protein structure and biophysics. Furthermore, of note is the stable oligomer (pentamer) organization of MBP-Vpu in solution, the MBP moieties are well-ordered, being arranged in a circular pattern in axial view on the oligomer (Fig. 5 and Supporting Fig. 4). This protein behavior may suggest that the pentameric form of Vpu could be optimal for oligomer stability and is amino-acid-sequence-defined. Strikingly, the oligomeric order in solution corresponds to that proposed for Vpu channel/pore in lipid membranes. Thus, by means of similarity, the oligomer (pentamer) in solution could contribute a useful model to reflect on the Vpu channel in lipid environments. One, of course, could point out that the forces stabilizing the structure of the Vpu oligomer in solution and in a lipid environment are different; therefore, distinct folds of the FL protein monomer within the oligomers (pentamers) in these two environments cannot be ruled out. Further studies to determine the high-resolution structure of Vpu oligomer in solution would be beneficial to better understand our observations.
We further studied the oligomerization profile of MBP-Vpu reconstituted in detergent and lipid-like environments, β-DDM, lyso-PC/PG, and DHPC/DHPG. A large protein oligomer polymorphism was detected by both SEC and nsEM in all three hydrophobic environments (Figs. 7, 8, 9, and 10). Moreover, it seems that the transition from hydrophilic to hydrophobic more membrane-like environments led to substantial protein restructuring, including destabilization of the stable oligomers in solution. These results were further supported by the CW ERP and DEER results. The CW EPR spectrum of spin-labeled MBP-Vpu in detergent and in lyso-PC/PG indicated increased protein-region dynamics compared to those for a protein in buffer (Figs. 6 and 11 and Supporting Fig. 7). The observed decrease in the DEER modulation depth for spin-labeled MBP-Vpu in β-DDM vs. MBP-Vpu in buffer provides one more unambiguous piece of evidence for protein restructuring and the altered oligomer organization/oligomeric order upon transition from aqueous to hydrophobic environments. Indeed, the oligomeric helical proteins in lipid bilayers participate in complex and even competing contacts, including helix–helix and helix (amino acid)-lipid interactions (Stangl and Schneider, 2015). Hence, it seems that the Vpu monomer-monomer stabilizing contacts in the aqueous environment are now provided by readily available protein–lipid hydrophobic interactions.
The most striking result about MBP-Vpu in a lipid environment is the observation of MBP-Vpu linear oligomers (arrays) when the protein was reconstituted in lyso-PC/PG at P/L 1/400 (Fig. 8). Such Vpu structural organization has not been reported before and linking it to a specific function is currently quite imprecise. It is plausible that this Vpu organization affects membrane morphology by destabilizing effects. Indeed, viroporins were found to participate in membrane remodeling; (Campbell and Monje-Galvan, 2022) therefore, it might be interesting to further explore the nature and roles of these array-like arrangements of Vpu in lipids.
Concluding remarks:
We conducted in vitro studies to elucidate the oligomerization properties of HIV-1 Vpu protein. To this end, we generated an MBP-Vpu construct, which helped to produce the protein in soluble form and to observe protein oligomerization in aqueous and membrane environments. We found that in buffer, MBP-Vpu forms well-defined stable oligomers. We, thus, propose that these oligomers could provide the targets for high-resolution structure determination. This might shed light into the Vpu oligomerization in lipid membranes, but could also help uncover yet uncharacterized soluble Vpu forms, the notion particularly plausible because soluble Vpu oligomers have been spotted in cells (Maldarelli et al., 1993). We also detected multiple forms of MBP-Vpu self-assembly in detergent and lipid environments, which are channel-like oligomers but assemble into linear arrays. The observed in vitro Vpu oligomerization states might represent functional states of Vpu under physiological conditions. Our study provides significant mechanistic insights about the Vpu quaternary structure and the effect of the environment on Vpu oligomer stability and it sets the stage for structure characterization in detail and gaining insights into protein-lipid interactions. Further studies aiming at elucidating the properties and specific lipid-Vpu interactions in membrane bilayers would provide even deeper understanding of this protein structure and function.
4. Materials and methods
4.1. Design and cloning of MBP-Vpu fusion construct
We generated a fusion construct of MBP followed by a short linker and FL HIV-1 Vpu for optimized structural studies, creating an MBP-Vpu fusion protein. In addition, this construct had an Hisx8 (His8) affinity tag at its N-terminal to facilitate protein purification. We also introduced a single cysteine residue at position Q36C (residue numbering is for the WT Vpu sequence). The DNA encoding this fusion protein was commercially synthesized (GenScript Inc) and cloned into a pET15b bacterial expression vector at NcoI/BamHI cloning sites. We then used the obtained MBP-Vpu/pET15b vector for protein expression. We used the ExPasy ProtParam software (Duvaud et al., 2021) to characterize the MBP-Vpu protein properties, e.g., molecular weight and extinction co-efficient, and the PROTTER version 1.0 program (Omasits et al., 2014) to predict the transmembrane topology of MBP-Vpu.
4.2. Expression and purification of MBP-Vpu construct
First, we transfected a pET15b/MBP-Vpu plasmid into E.coli BL21 (DE3)-competent cells (Lucigen), following the manufacturer’s protocol. We plated the transfected cells on an LB/agar/ampicillin (Amp) plate (100 μg/mL Amp) and incubated them overnight at 37 °C. Then, we selected a single colony and inoculated it in a flask containing 200 ml of LB broth Lennox (Sigma-Aldrich), supplemented with 100 μg/mL of Amp (Gold Biotechnology), and we grew bacterial stock solution in an incubator-shaker (Innova 43/43R) overnight for 17–18 h at 37 °C. The next day, we inoculated 30 ml of the overnight cell culture in a 3-L Fernbach flask containing 2 L of LB broth and 100 μg/mL of Amp, and we grew it in an incubator-shaker at 37 °C at 200 RPM for 2 h 30 min–3 h, until the absorbance at 600 nm (OD600) of the inoculated LB medium reached 0.70–0.80. Afterward, we reduced the temperature in the incubator-shaker from 37 °C to 18 °C and added IPTG (Isopropyl-β-D-1-thiogalactopyranoside) (Gold Biotechnology) to the bacterial solution to a final concentration of 1 mM to induce the protein expression under the control of the T7 promoter. The protein expression proceeded overnight at 18 °C. The next day, we harvested the cells by spinning them down in an Avanti J-15R centrifuge (Beckman coulter; JA-4.750 rotor) at 4,100 RPM (1,880g) for 15 min at 4 °C. We discarded the supernatant and collected and resuspended cell pellets in the resuspension buffer containing 20 mM of HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (Sigma) at pH 7.4 and 200 mM of NaCl (Sigma)]. Then, we added TCEP (tris (2-carboxyethyl) phosphine) (Gold Biotechnology), chicken egg lysozyme (Roche), and PMSF (phenyl-methylsulphonyl fluoride) (Gold Biotechnology) to the resuspended cell solution to final concentrations of 200 μM, 0.5–0.6 mg/mL, and 1 mM, respectively. We then subjected this solution to sonication with a sonicator (U.S. solid ultrasonic processor) to break the cells open, and we separated the cell debris by centrifugation at 7,000 RPM (5,380g) for 15 min in the same centrifuge (JA-10.100 rotor) at 4 °C. We collected the supernatant containing the soluble fraction of cell lysate and subjected it to double-affinity chromatography (Routzahn and Waugh, 2002) to purify the MBP-Vpu protein.
We utilized double-affinity chromatography, i.e., Nickel (Ni2+, Ni)-affinity chromatography, followed by amylose affinity chromatography to obtain highly pure MBP-Vpu protein. For Ni-affinity purification, we incubated Ni-NTA agarose resin (Qiagen) (1.5 ml/1 L cell culture) under constant agitation with soluble cell lysate in the resuspension buffer at 4 °C for 1–1.5 h. Afterward, we transferred Ni-NTA agarose resin bound with protein to a gravity column and discarded flow-through containing the unbound material. Then, to elute most of the weakly bound protein impurities, we washed the column with 10 resin volumes of buffer A containing 50 mM of sodium phosphate buffer, pH 7.4; 150 mM of NaCl, and 5 % (w/v) glycerol supplemented with 40 mM of imidazole (Im) (Sigma). Subsequently, we added 2 resin volumes of buffer A supplemented with 300 mM of Im to the column for the elution of the target protein, i.e., MBP-Vpu protein. Later, we concentrated the protein at 3,900 RPM (1,700g) in 10-kDa MWCO ultra centrifugal filters (Amicon®) at 4 °C. We measured the protein concentration using nanodrop spectrophotometer (Thermo Scientific) and then subjected it to an amylose purification technique after removing Im by washing it with an exchange Buffer B made of 20 mM of Tris, pH 7.4; 200 mM of NaCl; and 1 mM of EDTA and 5% glycerol. For the amylose affinity chromatography, pre-equillibrated with buffer B amylose resin was incubated with Ni-affinity purified MBP-Vpu protein at 4 °C for 1.5–2 h. Afterward, we transferred the amylose resin bound with protein to a gravity column and discarded the flow-through containing the unbound material. Then we added buffer B containing 20 mM of Tris, pH 7.4; 200 mM of NaCl; and 1 mM of EDTA and 5% glycerol to the gravity column to wash out the unbound impurities. Next, we introduced buffer B supplemented with 25 mM of maltose (Sigma) into the column for the elution of the target MBP-Vpu protein. We collected the flow-through and concentrated the protein at 3,900 RPM (1,700g) in 10-kDa MWCO ultra centrifugal filters (Amicon®) at 4 °C. We measured protein concentration with the nanodrop spectrophotometer using the calculated extinction coefficient of 78,840 M−1 cm−1. We used this highly pure protein in further experiments.
4.3. Sodium dodecyl sulfate gel electrophoresis (SDS-PAGE) and western blotting (WB)
We assessed the protein purity by using SDS-PAGE and WB. All the samples for SDS-PAGE and WB constituted MBP-Vpu protein, dithiothreitol (DTT; Sigma), and a loading buffer (BioRad). We loaded the samples and marker (precision plus protein dual-color standard; BioRad) onto 4–20% Criterion™ TGX™ precast gels (BioRad) immersed in Tris/glycine/SDS buffer (BioRad) and then conducted electrophoresis in a midi Criterion™ vertical electrophoresis cell (BioRad) at 170 V. For blotting, we transferred the protein from the gel onto a 0.2-μm nitro-cellulose membrane (BioRad) using a Criterion™ blotter (BioRad) overnight at 10 V and 4 °C. We visualized protein bands on the gel by Coomassie Blue dye staining and washing with de-stain solution. We utilized colorimetric detection to visualize the MBP-Vpu band(s) on the membrane after WB transfer. We used primary mouse anti-histidine tag antibodies (BioRad) and secondary goat anti-mouse IgG antibodies conjugated to Alkaline Phosphatase (BioRad).
4.4. Preparation of lipid and detergent stock solutions
Lipids stock solutions were used for reconstituting MBP-Vpu as follows. The mixtures of lyso-lipids, 14:0 lyso-PC (1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine) and 14:0 lyso-PG (1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt)) were prepared in 1:1 M ratio in chloroform, methanol, H2O. The organic solvents were evaporated under the gentle stream of N2 gas flowing into the vials until the lipids got dry; two more hours of N2 flow followed to remove the residual organic solvent from the lipid films. Thereafter, the films were rehydrated in ultrapure water for 1–2 h at 4 °C to produce 50 mM lyso-lipid stock solutions. In the same manner we prepared 40 mM stocks of DHPC (6:0 PC) and DHPG (6:0 PG) lipids in the molar ratio of 1:1 in ultrapure water. We used all these stock solutions to reconstitute MBP-Vpu protein at the desired protein-to-lipid molar ratios. We also prepared 200 mM aqueous stock solution of β-DDM (n-Dodecyl-β-D-Maltoside) to use throughout the experiments. The detergents and lipids used form micelles, but the lyso lipids and DHPC/DHPG are closer in their head group region to native membrane than β-DDM although their fatty tails, while locally adequate, cannot support longer range ordering of bilayers. This is a small price to pay for providing more uniform ensembles for structural study. All lipids were purchased from Avanti® Polar Lipids Inc., β-DDM came from Anatrace.
4.5. Assessing the MBP-Vpu oligomerization using analytical SEC
We used SEC to assess the oligomerization state of the purified MBP-Vpu protein. Because no other protein impurities were present, the SEC elution profile directly reports on the MBP-Vpu oligomerization state through the position of the protein elution peaks. For these experiments, we used a Superdex™ 200 increase 10/300 GL size-exclusion column (GE Healthcare) plugged into an AKTÄ explorer 100 (Amersham Bio-sciences) protein purifier system. For all SEC experiments, we exchanged the protein buffer with 50 mM of NaPi (sodium phosphate), pH 7.4, and 150 mM of NaCl (Buffer C); then we added 5% glycerol to the protein sample without lipids, or we reconstituted the protein in either 2 mM of lyso-PC/PG (50:50 mol% of 14:0 lyso-PC and 14:0 lyso-PG) or 1 mM of β-DDM. To characterize the protein in buffer with no lipids, we pre-equilibrated the SEC column with Buffer C supplemented with 10% glycerol. We added the 10% glycerol because in buffer with no glycerol, the protein elution was hindered by interaction of its hydrophobic regions with the column beads. When studying protein in a detergent or lipid environment, we pre-equilibrated the column with Buffer C supplemented with either 1 mM of β-DDM or 2 mM of lyso PC/PG. Then, we injected the protein (in the corresponding buffer with or without detergent or lipid) in the column and eluted it by running an SEC program with a 0.5-ml/min flow rate. We collected fractions with a volume of 0.5 ml. In the case of protein in Buffer C with no lipid or detergent, the concentration of injected MBP-Vpu was 50 μM or 25 μM. In the case of protein in Buffer C (no glycerol) with 1 mM of β-DDM, the concentration of injected MBP-Vpu was 50 μM. In the case of Buffer C (no glycerol) with 2 mM of lyso PC/PG, the concentration of the injected MBP-Vpu was 100 μM, 50 μM, 25 μM, or 15 μM. We combined, concentrated, and analyzed the fractions corresponding to protein elution peaks using SDS-PAGE and WB as described above.
We conducted SEC on the SEC protein standards (molecular weights 12.4–200 kDa; Sigma), including β-amylase from sweet potato (200 kDa), alcohol dehydrogenase from yeast (150 kDa), albumin from bovine serum (66 kDa), carbonic anhydrase from bovine erythrocytes (29 kDa), and cytochrome c from horse heart (12.4 kDa). First, we mixed 0.4 mg of β-amylase, 0.6 mg of alcohol dehydrogenase, 1.25 mg of albumin, 0.4 mg of carbonic anhydrase, and 0.25 mg of cytochrome C in 500uL of buffer C and loaded it on the SEC column, pre-equilibrated with buffer C supplemented with 10% glycerol. We adjusted the quantity of each protein standard to produce peaks (280 nm absorbance) of similar intensity. Later, we referenced the positions of elution peaks of MBP-Vpu in buffer, in lipid (lyso-PC/PG), and in detergent (β-DDM) with the elution peaks of standard proteins.
4.6. Preparation of samples for negative staining transmission electron microscopy (nsEM) experiments
To prepare the protein for nsEM, we first exchanged the protein buffer with a buffer containing 25 mM of Tris, pH 7.4; 150 mM of NaCl; and 100 μM of TCEP. Then, we prepared multiple samples of MBP-Vpu protein with and without β-DDM or lipids (14:0 lyso PC/PG, DHPC/DHPG). These samples can be categorized as four types: (i) protein in just buffer with no lipid/detergent added at protein concentrations of 0.5 μM, 1 μM, and 5 μM; (ii) protein reconstituted in β-DDM at 1 μM and 5 μM of protein in 1 mM of β-DDM as well as a control of 1 mM of β-DDM; (iii) protein reconstituted in lyso PC/PG at 0.5 μM and 5 μM of protein in 2 mM of lyso-PC/PG, 5 of μM protein in 1 mM of lyso-PC/PG plus a sample of 5 μM of protein labeled with 5-nm Gold Nanoparticles (5 nm Ni-NTA-Nanogold®, Nanoprobes) (GNPs) in 2 mM of lyso-PC/PG with 0.04 μM, and a control of 1 mM of lyso-PC/PG; and (iv) protein reconstituted in DHPC/DHPG (06:0 PC/06:0 PG) at 0.5 μM, 5 μM, and 10 μM of protein in 2 mM of DHPC/DHPG, 1 μM of protein in 0.5 mM of DHPC/DHPG, 1 μM of protein in 1 mM of DHPC/DHPG, and 5 μM of protein in 38 mM of DHPC/DHPG plus control samples of 1 mM of DHPC/DHPG and 38 mM of DHPC/DHPG. Indeed, the low DHPC/DHPG concentrations at 1–2 mM are much below the critical micelle concentration of these lipids, i.e., about 15 mM, but we found these concentrations more suited for nsEM imaging. The protein was hardly visible in 38 mM of DHPC/DHPG; therefore, we did not continue the experiment at this lipid concentration. We imaged each of these samples using nsEM. Additionally, we labeled 5 μM of MBP-Vpu with 1.8-nm GNPs (Ni-NTA-Nanogold®, Nanoprobes) and reconstituted it in 2 mM of lyso-PC/PG. We conducted the protein reconstitution by mixing the soluble protein with detergent or lipid solution, in a manner similar to previously described procedures for reconstitution of lyophilized Vpu (Ma et al., 2002).
4.7. Negative staining and electron microscopy imaging
We loaded a 10 μL sample onto a carbon-coated copper grid and allowed it to settle for 1 min or 1 min 30 sec at room temperature (RT). Then, we gently removed the solution on the grid using filter paper and the settled on the grid protein or protein-lipid/detergent was stained by adding 10 μL of 1.5% uranyl acetate (UA) and incubated it for 1 min or 1 min 30 sec. We again gently cleared the remaining UA solution using filter paper. We air-dried stained protein or protein-lipid/detergent samples for 30 min to 1 h at RT and then used them for EM imaging.
Afterward, we collected digital micrographs with a transmission electron microscope (TEM Hitachi H-7650) equipped with a fluorescent screen with a visual field diameter of 160 mm and 0.204-nm resolution (×20,000 final magnification) and operated them at an accelerating voltage of 60 kV and 10-μA emission current. We scanned micrographs free of drift and astigmatism with 1,024 × 1,024 pixels. We visualized, contrast-adjusted, and analyzed the final images using ImageJ software (Schneider et al., 2012).
4.8. Protein spin labeling and EPR spectroscopy
We labeled the single cysteine residue in MBP-Vpu (Q36C, numbering in Vpu sequence), located in the linker between Vpu TMD and Helix 2, with an MTS spin label. The residue was inaccessible for labeling in detergent- or lipid-free MBP-Vpu. Therefore, we reconstituted the protein in 3 mM of β-DDM prior to the spin labeling procedure. We conducted the spin labeling at the protein-to-lipid molar ratio of 1:30 and incubated it at 22 °C for 4 h and overnight at 4 °C, similarly to previous studies (Georgieva et al., 2013; Georgieva et al., 2015). We removed the unreacted spin label using dialysis for ca. 24 h in a large volume of spin label-free buffer supplemented with 1 mM of β-DDM and changed the buffer 4 times. Thereafter, we subjected some of the spin-labeled protein to β-DDM removal by washing out the detergent in a concentrator using detergent-free buffer.
We studied the spin-labeled MBP-Vpu with EPR spectroscopy, using continuous wave (CW) and pulse (DEER) methods (Borbat and Freed, 2007; Jeschke, 2012a, 2012b)). We prepared samples of about 60–85 μM of protein in buffer solution and in 1 mM of β-DDM or 2 mM of lyso-PC/PG. The samples for the DEER experiment contained 18% (w/v) glycerol as a cryoprotectant. We recorded the CW EPR spectra using an X-band Bruker EMXmicro spectrometer at RT and a modulation amplitude of 2 gauss. We carried out DEER measurements at 60 K using a home-built Ku-band pulse ESR spectrometer operating at approximately 17.3 GHz (Borbat et al., 1997; Borbat et al., 2013). A four-pulse DEER experiment (Jeschke and Polyhach, 2007) used for spin-echo detection a π/2-π-π pulse sequence with 32-ns π-pulses applied at the low-field edge of the nitroxide spectrum; we set a 16-ns pump π-pulse at a frequency 70 MHz lower to pump on the spectrum center peak. A 32-step phase cycle (Gamliel and Freed, 1990) was necessary to suppress unwanted echo contributions to the signal except spurious dipolar pathways, of which there were three in the 4-pulse DEER(Borbat and Freed, 2014a, 2014b). We eliminated nuclear electron spin-echo envelope modulation (ESEEM) caused by surrounding protons by summing the data traces from four measurements, which had initial pulse separations and the start of detection advanced by 9.5 ns in subsequent measurements, i.e., by the quarter period of the 26.2-MHz nuclear Zeeman frequency for protons at 0.615 T corresponding to the 17.3-GHz frequency (Tang et al., 2015a, 2015b).
The solution and detergent samples had phase memory times, Tms, of about 1.0 μs, allowing us to record the data with up to a 2-μs evolution time (tm). The dipolar signals all easily reached their asymptotic backgrounds. We averaged the DEER data for 48 h to facilitate effective reconstruction by Tikhonov regularization (Chiang et al., 2005a, 2005b) or singular-value decomposition (SVD) methods (Srivastava and Freed, 2017; Srivastava and Freed, 2019).
We preprocessed the time domain DEER data, V(t), using standard approaches (Borbat and Freed, 2007; Jeschke and Polyhach, 2007; Jeschke, 2012a, 2012b; Borbat and Freed, 2014a, 2014b) and reconstructed it into distance distributions, P(r)s, using an SVD application on the ACERT web site. The DEER signal, V(t), had the decay caused by intermolecular dipole–dipole couplings removed by fitting the latter points (about half of the record) of ln[V(t)] to the straight line, extrapolating it to zero time, and subtracting it from ln[V(t)] so that the antilog yields u(t) = d(t) + 1. Here, d(t) is the dipolar evolution part of the DEER signal. Then, we normalized u(t) as u(t) / u(0). It serves as a typical form of DEER data presentation (cf. Fig. 7), whereas v(t) = d(t) / u(0) gives background-free “dipolar” data, which we converted by SVD to a distance distribution between spins in the oligomers.
Dipolar signal amplitude (“modulation depth”) is given as v(0). This would be accurate to the extent the background or V(∞), the asymptotic value of V(t), is known. This depth, tabulated by calculations for typical pulse sequence setup and verified over years of experiments, is a measure of a fraction of spins in pairs or oligomers (Milov et al., 1984; Bode et al., 2007; Georgieva et al., 2015). We used this method to estimate oligomeric order.
Supplementary Material
Acknowledgements
This work was supported by start-up funds from the Department of Chemistry and Biochemistry (to ERG). The College of Arts & Sciences Microscopy (CASM) is acknowledged for providing nsEM resources. The DEER experiments were conducted at ACERT Biomedical Resource, Cornell University. ACERT is supported by NIH/NIGMS grant 1R24GM146107.
Footnotes
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Elka R. Georgieva serves on the Deputy Editorial Board of JSB.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jsb.2023.107943.
Data availability
Data will be made available on request.
References
- Agirre A, Barco A, Carrasco L, Nieva JL, 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem 277 (43), 40434–40441. [DOI] [PubMed] [Google Scholar]
- Bode BE, Margraf D, Plackmeyer J, Durner G, Prisner TF, Schiemann O, 2007. Counting the monomers in nanometer-si zed oligomers by pulsed electron - Electron double resonance. J. Am. Chem. Soc 129 (21), 6736–6745. [DOI] [PubMed] [Google Scholar]
- Borbat PP, Crepeau RH, Freed JH, 1997. Multifrequency two-dimensional Fourier transform ESR: an X/Ku-band spectrometer. J. Magn. Reson 127 (2), 155–167. [DOI] [PubMed] [Google Scholar]
- Borbat PP, Freed JH, 2007. Measuring distances by pulsed dipolar ESR spectroscopy: spin-labeled histidine kinases. Methods Enzymol. 423, 52–116. [DOI] [PubMed] [Google Scholar]
- Borbat PP, Freed JH, 2014a. Pulse Dipolar Electron Spin Resonance: Distance Measurements. Structural Information from Spin Labels and Intrinsic Paramagnetic Centres in the Biosciences. Timmel CR Harmer JR. 152, 1–82. [Google Scholar]
- Borbat PP, Freed JH, 2014b. Pulse Dipolar ESR: Distance Measurements. Struct. Bond Heidelberg Springer. 152, 1–82. [Google Scholar]
- Borbat PP, Georgieva ER, Freed JH, 2013. Improved Sensitivity for Long-Distance Measurements in Biomolecules: Five-Pulse Double Electron-Electron Resonance. J. Phys. Chem. Lett 4 (1), 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell O, Monje-Galvan V, 2022. Protein-driven membrane remodeling: Molecular perspectives from Flaviviridae infections. Biophys. J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang YW, Borbat PP, Freed JH, 2005a. The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson 172 (2), 279–295. [DOI] [PubMed] [Google Scholar]
- Chiang YW, Borbat PP and Freed JH (2005a). “Maximum entropy: a complement to Tikhonov regularization for determination of pair distance distributions by pulsed ESR.” Journal of magnetic resonance (San Diego, Calif.: 1997) 177(2): 184–196. [DOI] [PubMed] [Google Scholar]
- Cohen EA, Terwilliger EF, Sodroski JG, Haseltine WA, 1988. Identification of a protein encoded by the vpu gene of HIV-1. Nature 334 (6182), 532–534. [DOI] [PubMed] [Google Scholar]
- Cooper RS, Georgieva ER, Borbat PP, Freed JH, Heldwein EE, 2018. Structural basis for membrane anchoring and fusion regulation of the herpes simplex virus fusogen gB. Nat. Struct. Mol. Biol 25 (5), 416–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscia F, Estrozi LF, Hans F, Malet H, Noirclerc-Savoye M, Schoehn G, Petosa C, 2016. Fusion to a homo-oligomeric scaffold allows cryo-EM analysis of a small protein. Sci. Rep 6, 30909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas JL, Viswanathan K, McCarroll MN, Gustin JK, Fruh K, Moses AV, 2009. Vpu directs the degradation of the human immunodeficiency virus restriction factor BST-2/Tetherin via a {beta}TrCP-dependent mechanism. J. Virol 83 (16), 7931–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube M, Bego MG, Paquay C, Cohen EA, 2010. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology 7, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C, 2021. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49 (W1), W216–W227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart GD, Sutherland T, Gage PW, Cox GB, 1996. The Vpu protein of human immunodeficiency virus type 1 forms cation-selective ion channels. J. Virol 70 (10), 7108–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez C, Hilty C, Wider G, Wuthrich K, 2002. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. PNAS 99 (21), 13533–13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer WB, Hsu HJ, 2011. Viral channel forming proteins - modeling the target. BBA 1808 (2), 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamliel D, Freed JH, 1990. THEORY OF 2-DIMENSIONAL ESR WITH NUCLEAR MODULATION. J. Magn. Reson 89 (1), 60–93. [Google Scholar]
- Garcia-Rubio I, 2020. EPR of site-directed spin-labeled proteins: A powerful tool to study structural flexibility. Arch. Biochem. Biophys 684, 108323. [DOI] [PubMed] [Google Scholar]
- Georgieva ER, 2017. Nanoscale lipid membrane mimetics in spin-labeling and electron paramagnetic resonance spectroscopy studies of protein structure and function. Nanotechnol. Rev 6 (1), 75–92. [Google Scholar]
- Georgieva ER, Borbat PP, Fanouraki C, Freed JH, 2020. High-yield production in E. coli and characterization of full-length functional p13II protein from human T-cell leukemia virus type 1. Protein Expr. Purif 173, 105659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O, 2013. Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol 20 (2), 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva ER, Borbat PP, Norman HD, Freed JH, 2015. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Sci. Rep 5, 11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D, 2010. The lipid binding domain of wild type and mutant alpha-synuclein: compactness and interconversion between the broken and extended helix forms. J. Biol. Chem 285 (36), 28261–28274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva ER, Xiao S, Borbat PP, Freed JH, Eliezer D, 2014. Tau binds to lipid membrane surfaces via short amphipathic helices located in its microtubule-binding repeats. Biophys. J 107 (6), 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, 2015. Vpu Protein: The Viroporin Encoded by HIV-1. Viruses 7 (8), 4352–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez ME, Carrasco L, 2003. Viroporins. FEBS Lett. 552 (1), 28–34. [DOI] [PubMed] [Google Scholar]
- Hu J, Qin H, Gao FP, Cross TA, 2011. A systematic assessment of mature MBP in membrane protein production: overexpression, membrane targeting and purification. Protein Expr. Purif 80 (1), 34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A, Das SR, Tanwar C, Jameel S, 2007. Oligomerization of the human immunodeficiency virus type 1 (HIV-1) Vpu protein–a genetic, biochemical and biophysical analysis. Virol. J 4, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison JM, Lu Z, Li GC, Travis B, Mittal R, Deatherage CL, Sanders CR, 2017. Dodecyl-beta-melibioside Detergent Micelles as a Medium for Membrane Proteins. Biochemistry 56 (41), 5481–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G (2012). DEER Distance Measurements on Proteins. Annual Review of Physical Chemistry, Vol 63. Johnson MA and Martinez TJ. 63: 419–446. [DOI] [PubMed] [Google Scholar]
- Jeschke G, 2012b. DEER distance measurements on proteins. Annu. Rev. Phys. Chem 63, 419–446. [DOI] [PubMed] [Google Scholar]
- Jeschke G, Polyhach Y, 2007. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. PCCP 9 (16), 1895–1910. [DOI] [PubMed] [Google Scholar]
- Jeschke G, Sajid M, Schulte M, Godt A, 2009. Three-spin correlations in double electron–electron resonance. PCCP 11 (31), 6580–6591. [DOI] [PubMed] [Google Scholar]
- Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, Xiong Y, 2014. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 3, e02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Chuenchor W, Jiang J, Cheng J, Li Y, Fang K, Huang M, Smith P, Xiao TS, 2017. Design of an expression system to enhance MBP-mediated crystallization. Sci. Rep 7, 40991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Geiger JD, 2021. Role of Viral Protein U (Vpu) in HIV-1 Infection and Pathogenesis. Viruses 13 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimkait T, Strebel K, Hoggan MD, Martin MA, Orenstein JM, 1990. The human immunodeficiency virus type 1-specific protein vpu is required for efficient virus maturation and release. J. Virol 64 (2), 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler J, Sulistijo ES, Sakakura M, Kim HJ, Ellis CD, Sanders CR, 2010. Lysophospholipid micelles sustain the stability and catalytic activity of diacylglycerol kinase in the absence of lipids. Biochemistry 49 (33), 7089–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov V, Bartels K, Veith K, Josts I, Subhramanyam UKT, Gunther C, Labahn J, Marlovits TC, Moraes I, Tidow H, Low C, Garcia-Alai MM, 2019. High-throughput stability screening for detergent-solubilized membrane proteins. Sci. Rep 9 (1), 10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer S, Hammer C, Hopfensperger K, Klein L, Hotter D, De Jesus PD, Herbert KM, Pache L, Smith N, van der Merwe JA, Chanda SK, Fellay J, Kirchhoff F, Sauter D, 2019. HIV-1 Vpu is a potent transcriptional suppressor of NF-kappaB-elicited antiviral immune responses. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M, Champeil P, Moller JV, 2000. Interaction of membrane proteins and lipids with solubilizing detergents. BBA 1508 (1–2), 86–111. [DOI] [PubMed] [Google Scholar]
- Lewinski MK, Jafari M, Zhang H, Opella SJ, Guatelli J, 2015. Membrane Anchoring by a C-terminal Tryptophan Enables HIV-1 Vpu to Displace Bone Marrow Stromal Antigen 2 (BST2) from Sites of Viral Assembly. J. Biol. Chem 290 (17), 10919–10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lins L, Brasseur R, 1995. The hydrophobic effect in protein folding. FASEB J. 9 (7), 535–540. [DOI] [PubMed] [Google Scholar]
- Liu Y, Huynh DT, Yeates TO, 2019. A 3.8 A resolution cryo-EM structure of a small protein bound to an imaging scaffold. Nat. Commun 10 (1), 1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Sharpe S, Ghirlando R, Yau WM, Tycko R, 2010. Oligomerization state and supramolecular structure of the HIV-1 Vpu protein transmembrane segment in phospholipid bilayers. Protein Sci. 19 (10), 1877–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Marassi FM, Jones DH, Straus SK, Bour S, Strebel K, Schubert U, Oblatt-Montal M, Montal M, Opella SJ, 2002. Expression, purification, and activities of full-length and truncated versions of the integral membrane protein Vpu from HIV-1. Protein Sci. 11 (3), 546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed S, Ahmad AB, Sehar U, Georgieva ER, 2021. Lipid Membrane Mimetics in Functional and Structural Studies of Integral Membrane Proteins. Membranes (Basel) 11 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Chen MY, Willey RL, Strebel K, 1993. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J. Virol 67 (8), 5056–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milov AD, Ponomarev AB, Tsvetkov YD, 1984. Modulation beats of signal of double electron-electron resonance in spin echo for biradical systems. J. Struct. Chem 25 (5), 710–713. [Google Scholar]
- Mineev KS, Bocharov EV, Volynsky PE, Goncharuk MV, Tkach EN, Ermolyuk YS, Schulga AA, Chupin VV, Maslennikov IV, Efremov RG, Arseniev AS, 2011. Dimeric structure of the transmembrane domain of glycophorin a in lipidic and detergent environments. Acta Nat. 3 (2), 90–98. [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC, 2009. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 5 (5), e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieva JL, Madan V, Carrasco L, 2012. Viroporins: structure and biological functions. Nat. Rev. Microbiol 10 (8), 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygaard R, Kim J, Mancia F, 2020. Cryo-electron microscopy analysis of small membrane proteins. Curr. Opin. Struct. Biol 64, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B, 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30 (6), 884–886. [DOI] [PubMed] [Google Scholar]
- Pliotas C, Ward R, Branigan E, Rasmussen A, Hagelueken G, Huang HX, Black SS, Booth IR, Schiemann O, Naismith JH, 2012. Conformational state of the MscS mechanosensitive channel in solution revealed by pulsed electron-electron double resonance (PELDOR) spectroscopy. PNAS 109 (40), E2675–E2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol-Pina R, Vilaprinyo-Pascual S, Mazzucato R, Arcella A, Vilaseca M, Orozco M, Carulla N, 2015. SDS-PAGE analysis of Abeta oligomers is disserving research into Alzheimer s disease: appealing for ESI-IM-MS. Sci. Rep 5, 14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routzahn KM, Waugh DS, 2002. Differential effects of supplementary affinity tags on the solubility of MBP fusion proteins. J. Struct. Funct. Genomics 2 (2), 83–92. [DOI] [PubMed] [Google Scholar]
- Roy N, Pacini G, Berlioz-Torrent C, Janvier K, 2014. Mechanisms underlying HIV-1 Vpu-mediated viral egress. Front. Microbiol 5, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Orenstein J, Dimitrov D, Martin M, 1992. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology 186 (2), 712–724. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9 (7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Ferrer-Montiel AV, Oblatt-Montal M, Henklein P, Strebel K, Montal M, 1996. Identification of an ion channel activity of the Vpu transmembrane domain and its involvement in the regulation of virus release from HIV-1-infected cells. FEBS Lett. 398 (1), 12–18. [DOI] [PubMed] [Google Scholar]
- Selmke B, Borbat PP, Nickolaus C, Varadarajan R, Freed JH, Trommer WE, 2018. Open and Closed Form of Maltose Binding Protein in Its Native and Molten Globule State As Studied by Electron Paramagnetic Resonance Spectroscopy. Biochemistry 57 (38), 5507–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH, 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med 1 (1), a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Freed JH, 2017. Singular Value Decomposition Method to Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance. J. Phys. Chem. Lett 8 (22), 5648–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Freed JH, 2019. Singular Value Decomposition Method To Determine Distance Distributions in Pulsed Dipolar Electron Spin Resonance: II. Estimating Uncertainty. J. Phys. Chem. A 123 (1), 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangl M, Schneider D, 2015. Functional competition within a membrane: Lipid recognition vs. transmembrane helix oligomerization. BBA 1848 (9), 1886–1896. [DOI] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Maldarelli F, Martin MA, 1989. Molecular and biochemical analyses of human immunodeficiency virus type 1 vpu protein. J. Virol 63 (9), 3784–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebel K, Klimkait T, Martin MA, 1988. A novel gene of HIV-1, vpu, and its 16-kilodalton product. Science 241 (4870), 1221–1223. [DOI] [PubMed] [Google Scholar]
- Tang S, Henne WM, Borbat PP, Buchkovich NJ, Freed JH, Mao Y, Fromme JC, Emr SD, 2015. Structural basis for activation, assembly and membrane binding of ESCRT-III Snf7 filaments. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telmer PG, Shilton BH, 2003. Insights into the conformational equilibria of maltose-binding protein by analysis of high affinity mutants. J. Biol. Chem 278 (36), 34555–34567. [DOI] [PubMed] [Google Scholar]
- Warschawski DE, Arnold AA, Beaugrand M, Gravel A, Chartrand E, Marcotte I, 2011. Choosing membrane mimetics for NMR structural studies of transmembrane proteins. BBA 1808 (8), 1957–1974. [DOI] [PubMed] [Google Scholar]
- Willey RL, Maldarelli F, Martin MA, Strebel K, 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol 66 (12), 7193–7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.