Circular RNAs (circRNAs) are single-stranded RNAs that are covalently closed and lack free ends. Our current understanding of the functions and regulation of circRNAs in immune systems is still in the nascent stages, but several groups have recently investigated the immunogenicity of engineered circRNAs. However, which factors determine engineered circRNA immunogenicity still remains unclear. Here, we review and compare these investigations on engineered circRNA immunogenicity. We discuss the possible factors that may alter the immunogenic potential of a circRNA, such as its generation, purification, primary sequence, secondary structure, and RNA modifications. We also suggest approaches for designing a circRNA with immunostimulatory or immune-evasive properties and recommend future areas for the field to investigate. Elucidating the factors underlying engineered circRNA immunogenicity is an important step towards understanding endogenous circRNA immunity and increasing the therapeutic potential of circRNAs.
Introduction
CircRNAs are single-stranded, covalently closed, and lack free ends. Originally thought to be byproducts of aberrant splicing (Nigro et al., 1991), recent advances in RNA sequencing combined with specialized bioinformatic pipelines have led to the identification of thousands of circRNAs with a variety of roles. CircRNAs have a variety of functions, including sponging microRNAs (Hansen et al., 2013; Memczak et al., 2013), serving as templates for translation (Legnini et al., 2017), regulating the cell cycle via protein scaffolding (Du et al., 2016), and driving the phase separation of proteins (Chen et al., 2022). While relatively little is known about the roles of circRNAs in innate immunity, several studies suggest that endogenous circRNAs are immunosuppressive in nature. Upon viral infection, the RNA-binding protein nuclear factor 90 (NF90) and its isoform NF110 dissociate from circRNA–protein complexes in the cytoplasm to bind to viral mRNAs and inhibit viral replication (Li et al., 2017). In long-term hematopoietic stem cells, the endogenous circRNA, cia-cyclic guanosine monophosphate–adenosine monophosphate synthase (cia-cGAS), is an antagonist to the DNA sensor cGAS and can inhibit cGAS-mediated interferon (IFN) production (Xia et al., 2018). Additionally, many endogenous circRNAs have secondary structures with short double-stranded regions that suppress the activation of protein kinase R (PKR), an essential factor in the anti-viral response (Liu et al., 2019). These investigations focused on cellular circRNAs, but whether the engineered circRNAs produced exogenously and then delivered into cells are immunostimulatory or not has been a debated question in recent years (Chen et al., 2017, 2019; Wesselhoeft et al., 2018, 2019; Liu et al., 2022). A clear understanding of the immune effects from engineered circRNAs would offer insight into fundamental principles of how cells differentiate between self and nonself RNAs, and further provide a foundation for translating circRNAs into therapies.
Immunogenic potential of engineered circRNAs
The investigations by Chen et al. (2017, 2019), Wesselhoeft et al. (2018, 2019), and Liu et al. (2022) are the most recent and relevant studies on engineered circRNA immunogenicity and the main comparative focus of this perspective (Figure 1). Chen et al. (2017) first showed that transfection of engineered circRNAs stimulated the expression of several immune genes, most notably retinoic acid-inducible gene-I (RIG-I). They synthesized the circRNAs via a permuted intron–exon (PIE) splicing strategy using the autocatalytic group I intron of the thymidylate synthase (td) gene from the T4 phage. Meanwhile, circRNAs with the same sequence but produced by the endogenous human ZKSCAN1 introns, which direct back-splicing by the spliceosome, were not immunogenic. Taken together, the authors concluded that circRNA immunogenicity depends on the introns that program circRNA biogenesis. Chen et al. (2019) later went on to show that the RNA modification N6-methyladenosine (m6A), likely added during the formation of circRNAs programmed by endogenous introns, is a
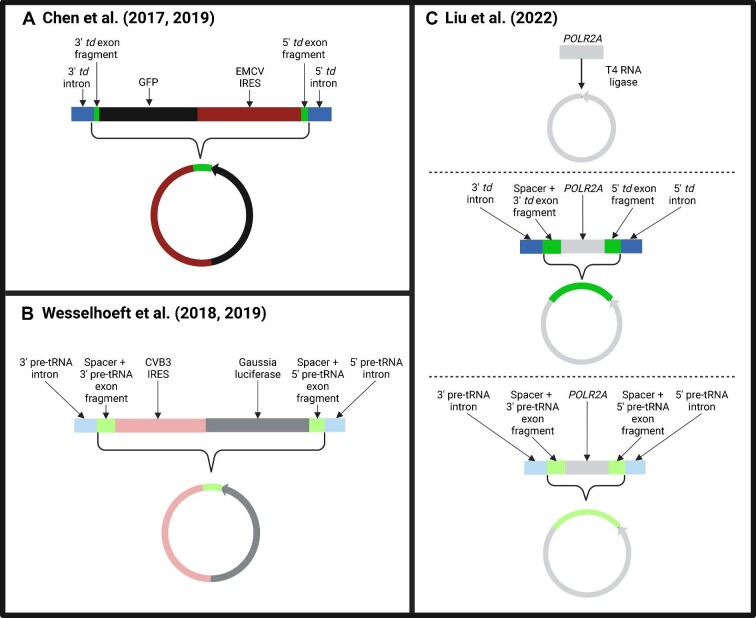
Figure 1.
Main circRNA used in each study with corresponding linear precursor. (A) Chen et al. (2017, 2019) used the permuted td introns (dark blue) surrounding a GFP mRNA (black) and EMCV IRES (dark red). The circular version contains the GFP coding sequence, the EMCV IRES, and a retained fragment of the td exon (dark green). (B) Wesselhoeft et al. (2018, 2019) used the permuted pre-tRNA introns (light blue) with a CVB3 IRES (light red) and Gaussia luciferase mRNA (dark gray). Both the linear precursor and final circular product also contained a spacer sequence and pre-tRNA exon fragments (light green). (C) Liu et al. (2022) circularized the POLR2A sequence (light gray) using either direct ligation (top), the permuted td introns (dark blue) with a spacer and td exon fragments (dark green) (middle), or the permuted pre-tRNA introns (light blue) with a spacer and exon fragments (light green) (bottom).
molecular marker for ‘self’ on these circRNAs. While RIG-I binds to both m6A-modified and unmodified circRNAs, only the unmodified circRNAs were able to activate RIG-I to induce mitochondrial anti-viral signaling protein (MAVS) filamentation, interferon regulatory factor 3 (IRF3) dimerization, and IFN production. Wesselhoeft et al. (2018, 2019) presented a different picture of the effects of delivering engineered circRNAs into cells. While testing circRNAs generated by PIE of the pre-tRNA group I intron from the Anabaena bacteria, Wesselhoeft et al. (2019) found that these exogenous circRNAs were not immunogenic and could evade detection by Toll-like receptors. Most recently, Liu et al. (2022) reported that circRNAs produced via group I introns, such as the ones that Wesselhoeft et al. (2018, 2019) and Chen et al. (2017, 2019) used, stimulated immune responses. The autocatalytic splicing programmed by the group I intron leads to the retention of a portion of the PIE exon in the final circRNA. Liu et al. (2022) postulated that these exon remnants can form double-stranded secondary structures that activate PKR. Meanwhile, circRNAs synthesized by T4 RNA ligase that lack the retained exon sequences suppressed PKR activation and did not provoke an innate immune response.
Of note, each of these groups used a different approach to evaluate immunogenicity. Chen et al. (2017, 2019) primarily used reverse transcription–quantitative polymerase chain reaction (RT–qPCR) to measure the expression of several innate immune genes such as RIG-I, PKR, melanoma-differentiation-associated gene 5 (MDA5), 2′-5′-oligoadentylate synthase 1 (OAS1), and OAS-like protein (OASL). These gene expression results were further normalized by the levels of exogenously synthesized circRNAs or linear RNA detected inside the cell to account for differing transfection efficiencies and cellular stabilities. Liu et al. (2022) also probed immunogenicity by measuring immune gene and protein expression by RT–qPCR and western blot analyses. Compared with Chen et al. (2017, 2019), the panel of innate immune genes used by Liu et al. (2022) was slightly different [RIG-I, IFNβ, tumor necrosis factor alpha (TNFα), and interleukin-6 (IL-6)], and the gene expression was not normalized by levels of in vitro synthesized RNA in the cell. Lastly, Wesselhoeft et al. (2019) evaluated circRNA immunogenicity by measuring cell viability and levels of cytokines and chemokines in a cell culture medium 24 h after transfection with their engineered circRNAs. Each of these assays has specific scopes and limitations that the field should keep in mind when evaluating the investigation’s conclusions.
Another difference among the studies is the type of linear RNA that was compared with the engineered circRNA: Wesselhoeft et al. (2019) compared the immunogenicity of their circRNA to its 5′ triphosphate linear counterpart, whereas Chen et al. (2017, 2019) compared their circRNA to its corresponding 5′ hydroxyl linear RNA. Liu et al. (2022) compared the immunogenicity of their circRNAs to both the 5′ triphosphate and 5′ hydroxyl linear RNAs with the same sequence. These studies all acknowledge that 5′ triphosphate linear RNAs are more immunostimulatory than circRNAs; knowing the type of linear RNA that each study used to compare the circRNA’s immunogenicity is important in evaluating their conclusions.
Given the differing conclusions from these studies, which properties of circRNAs contribute to their immunoge-nicity are unclear. Possible factors may include their generation, purification, primary sequence, secondary structure, and RNA modifications, which are discussed in more detail below.
Effects of circRNA generation and purification on circRNA immunogenicity
There are several chemical and enzymatic strategies to generate engineered circRNAs in vitro [reviewed in Obi and Chen (2021)]. One such method is to in vitro transcribe an exonic sequence flanked by permuted autocatalytic splicing introns from either the phage T4 td gene or the Anabaena pre-tRNA. Other approaches include direct ligation of the linear RNA precursor by DNA or RNA ligases with or without the assistance of a short oligonucleotide splint. Chen et al. (2017, 2019) produced a circRNA encoding the green fluorescent protein (GFP) sequence with the permuted td introns and found it to be highly immunogenic (Figure 1). The circRNA used by Wesselhoeft et al. (2018, 2019) containing the Gaussia luciferase sequence and circularized via the PIE Anabaena pre-tRNA group I intron was found to be immuno-evasive. Wesselhoeft et al. (2019) suggested that the strong immune response previously reported by Chen et al. (2017) was a result of 5′ triphosphate linear RNA contaminants generated during the circRNA production process. Chen et al. (2017) used RNase R digestion and phosphatase to treat their circRNAs but Wesselhoeft et al. (2019) reported that high-performance liquid chromatography (HPLC) was essential to remove linear byproducts of the splicing reaction. Chen et al. (2019) later employed the same purification process as Wesselhoeft et al. (2018, 2019) performed to obtain the circRNA but still found the purified circRNA to be immunostimulatory. Additionally, Chen et al. (2019) found that gel purification of the circRNA, which should eliminate contaminating linear RNA components, continued to lead to the induction of innate immune gene expression. More recently, Liu et al. (2022) generated circRNAs containing a POLR2A exonic sequence using three different approaches with similar efficiencies: (i) PIE splicing directed by the td group I intron from T4 phage, (ii) PIE splicing directed by the pre-tRNA group I intron from Anabaena bacteria, and (iii) direct ligation of the linear RNA with T4 RNA ligase without an oligonucleotide splint. They discovered that the circRNAs produced by PIE splicing were immunogenic but the circ-RNAs produced by direct ligation were not. This difference in immunogenicity was attributed to remnants introduced by the group I intron-directed autocatalytic splicing rather than issues with purification. Similar to Chen et al. (2019), Liu et al. (2022) found that the immunogenicity of circRNAs generated by autocatalytic splicing was not diminished after purification either by HPLC or RNase R digestion. However, Liu et al. (2022) also noted that extremely low (femtogram) levels of contaminating 5′ triphosphate linear RNAs that are undetectable via common circRNA detection methods (e.g. HPLC or gel extraction) may contribute to the immune response. Taken together, the studies above demonstrate that the immunogenicity of engineered circRNAs may depend on the precise strategy used to produce them.
Effects of primary sequence and secondary structure on circRNA immunogenicity
The PIE strategy incorporates portions of the original phage or Anabaena exons into the back-splice junction of the circularized RNA. Thus, for circRNAs generated via the PIE strategy, the final circRNA sequence contains both the exonic sequence of choice and the exon fragments from the PIE construct. Chen et al. (2017, 2019) and Wesselhoeft et al. (2018, 2019) both used an autocatalytic splicing intron (from the td gene and pre-tRNA, respectively) to generate their engineered circRNAs (Figure 1). As a result, both of their final circularized products contained exon fragments from the respective PIE constructs. Wesselhoeft et al. (2018) also included short spacer sequences that improved the efficiency of circRNA generation. Liu et al. (2022) later adopted the same circRNA generation strategy as Wesselhoeft et al. (2018, 2019) but used a different exonic sequence (POLR2A). The remnants in the circRNAs produced by Liu et al. (2022), composed of the PIE exon fragments plus the spacer, were 74 or 186 extra nucleotides for circRNAs generated via the td gene or pre-tRNA introns, respectively. The number of nucleotides retained in the circRNAs produced by Chen et al. (2017, 2019) differed from Liu et al. (2022), even though both groups directed circularization via the td gene intron; the retained sequence was only 48 nucleotides in Chen et al. (2017, 2019) (Figure 1).
Since circRNAs that include the additional sequences from autocatalytic splicing stimulated immune signaling, the primary sequence of the circRNA and the corresponding secondary structures may be critical factors in driving circRNA immunogenicity. Remnant exon fragments and spacer sequences form long double-stranded RNA duplexes that activate PKR (Liu et al., 2022). However, a circRNA with the same POLR2A exonic sequence but generated via direct ligation with 1–3 guanine nucleotides introduced at the splice junction was found to have minimized immunogenicity. This was a result of short RNA duplexes that formed within the exonic POLR2A sequence that prevented PKR activation. Additionally, changes in just the exonic sequence in the circRNA but not the remnants also seem to affect the degree of immunogenicity. Liu et al. (2022) showed that using a reverse complement of the original exonic sequence, which would alter the sequence but preserve the overall RNA secondary structure, affected the expression of several immune genes such as RIG-I, IFNβ, TNFα, and IL-6. Furthermore, replacing the GFP and encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) exon of a circ-RNA with a mCherry coding sequence led to a >10-fold decrease in RIG-I induction (Chen et al., 2017). Thus, the presence of an IRES, which has known double-stranded structures, on circRNA may be one feature that drives immunogenicity. Both the exonic sequence of a circRNA and remnant fragments from autocatalytic splicing can form secondary structures that alter circ-RNA immunogenicity (Liu et al., 2022). However, the degree to which circRNA primary sequence, size, and/or secondary structure affect circRNA immunogenicity remains to be elucidated.
Effects of RNA modification on circRNA immunogenicity
RNA modifications including 5-methylcytidine, pseudouridine, and m6A have been shown to decrease the immunogenicity of in vitro transcribed linear mRNAs (Kariko et al., 2005). RNA modifications may have a similar effect on circRNA immunogenicity. Chen et al. (2019) reported that the m6A modification marks self circRNAs and abrogates circRNA immunity. Endogenous circRNAs are associated with m6A writers and readers, including the YTH domain-containing family protein 2 (YTHDF2) reader protein, and have m6A modifications at 3′ of the back-splice junction (Chen et al., 2019). Furthermore, adding m6A modifications onto engineered circRNAs that are known to be immunostimulatory can abrogate the immune response. Complementarily, mutation of all adenosines or just the adenosines in the canonical m6A motif in a circRNA significantly increases its ability to stimulate immune genes. Chen et al. (2019) also reported that other RNA modifications such as pseudouridine and inosine can decrease activation of innate immune signaling. Wesselhoeft et al. (2019) found that pseudouridine and m6A nucleoside modifications do not further decrease the cytokine release profiles in comparison to an unmodified and non-immunogenic circRNA. It is unclear whether the immune effects from the circRNA modifications in the studies are different due to the type of measurement or the circRNA tested. Since RNA modification sites are dependent on the primary sequence of the circRNA, differences in conclusions about the roles of RNA modifications on circRNAs could also be attributed to the different exons used for testing.
Conclusion
Whether engineered circRNAs that are delivered into cells are immunostimulatory or not is still an open question. Three groups thus far have addressed this topic and highlighted exciting potential therapeutic applications for engineered circRNAs, such as being platforms for the expression of therapeutic proteins (Wesselhoeft et al., 2019), acting as vaccine adjuvants or inducing anti-tumor immunity (Chen et al., 2019), and suppressing PKR activity in autoimmune diseases (Liu et al., 2022). Figure 2 summarizes the primary findings of the investigations. Because three groups used different strategies to generate their circRNAs, select the primary sequences of their circRNAs, and measure the immunogenicity of their circRNAs, it is difficult to evaluate whether these differences lead to conflicting conclusions or whether engineered circRNAs are, as a class, immunogenic or not.
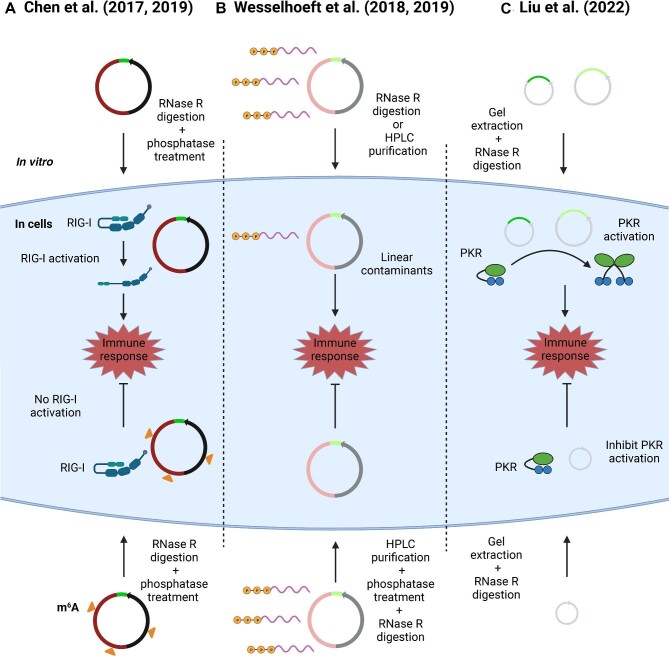
Figure 2.
Immunogenicity of engineered circRNAs. (A) Chen et al. (2017, 2019) found that engineered circRNAs induce a strong immune response via RIG-I but m6A-modified circRNAs would abrogate the immune response. (B) Wesselhoeft et al. (2018, 2019) showed that engineered circRNAs are not immunogenic but linear contaminants with 5′ triphosphates as a result of incomplete purification could stimulate cytokine release. (C) Liu et al. (2022) discovered that remnants included in the circRNA by PIE autocatalytic splicing could activate immune response via PKR. CircRNAs generated without the remnant fragments suppressed PKR activation.
This perspective presents circRNA features, such as primary sequence, secondary structure, and RNA modifications, that may play a role in determining circRNA immunogenicity. The results of these studies (Chen et al., 2017, 2019; Wesselhoeft et al., 2018, 2019; Liu et al., 2022) suggest that researchers could incorporate the PIE remnant primary sequence and double-stranded secondary structures to generate an immunogenic circRNA. For generating immune-evasive circRNAs, researchers could use direct ligation and include RNA modifications. However, interpreting data and drawing strong conclusions from these studies is difficult, because they differed in how to design and produce their circRNAs and how to probe for immune stimulation. Future investigations where the circRNAs from these studies are generated, purified, delivered into cells, and evaluated as immunogenic or not in the same way are warranted. This type of standardized approach is needed to enable direct comparisons and to elucidate the likely synergistic and additive effects of the different elements of a circRNA that dictate its immunogenicity.
[We thank our fellow lab members for helpful discussions and Amy Xue for illustrating the featured image using Procreate. J.T. and Y.G.C. are supported by the NIH R35GM142687. Y.G.C. is also supported by the Rita Allen Foundation and the Paul G. Allen Frontiers Group. Figures were created with BioRender.com.]
Contributor Information
Justin Tai, Department of Immunobiology, Yale University School of Medicine, New Haven, CT, USA.
Y Grace Chen, Department of Immunobiology, Yale University School of Medicine, New Haven, CT, USA.
References
- Chen S., Cao X., Zhang J.et al. (2022). circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Adv. Sci. 9, 2103817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.G., Kim M.V., Chen X.et al. (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.G., Chen R., Ahmad S.et al. (2019). N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W.W., Yang W., Liu E.et al. (2016). Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H.et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Kariko K., Buckstein M., Ni H.et al. (2005). Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175. [DOI] [PubMed] [Google Scholar]
- Legnini I., Timoteo G.D., Rossi F.et al. (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu C.-X., Xue W.et al. (2017). Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227.e7. [DOI] [PubMed] [Google Scholar]
- Liu C.-X., Li X., Nan F.et al. (2019). Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880. [DOI] [PubMed] [Google Scholar]
- Liu C.-X., Guo S.-K., Nan F.et al. (2022). RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434. [DOI] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A.et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Nigro J.M., Cho K.R., Fearon E.R.et al. (1991). Scrambled exons. Cell 64, 607–613. [DOI] [PubMed] [Google Scholar]
- Obi P., Chen Y.G. (2021). The design and synthesis of circular RNAs. Methods 196, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft R.A., Kowalski P.S., Anderson D.G. (2018). Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselhoeft R.A., Kowalski P.S., Parker-Hale F.C.et al. (2019). RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Ye B.et al. (2018). A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701. [DOI] [PubMed] [Google Scholar]