Abstract
Microwave-assisted reaction of 3,5-bis((E)-ylidene)-1-phosphonate-4-piperidones 3a‒g with azomethine ylide (produced through interaction of isatins 4 and sarcosine 5) cycloaddition afforded the corresponding (dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl)phosphonates 6a‒l in excellent yields (80–95%). Structure of the synthesized agents was evidenced by single crystal X-ray studies of 6d, 6i and 6l. Some of the synthesized agents revealed promising anti-SARS-CoV-2 properties in the viral infected Vero-E6 cell technique with noticeable selectivity indices. Compounds 6g and 6b are the most promising agents synthesized (R = 4-BrC6H4, Ph; R' = H, Cl, respectively) with considerable selectivity index values. Mpro-SARS-CoV-2 inhibitory properties supported the anti-SARS-CoV-2 observations of the potent analogs synthesized. Molecular docking studies (PDB ID: 7C8U) are consistent with the Mpro inhibitory properties. The presumed mode of action was supported by both experimentally investigated Mpro-SARS-CoV-2 inhibitory properties and explained by docking observations.
Keywords: Spiroindole, Phosphonate, Azomethine ylide, SARS-CoV-2, COVID-19
Graphical abstract
Spiroindoles bearing phosphonate group were regioselectively synthesized exhibiting promising Mpro-SARS-CoV-2 properties.
1. Introduction
The calamitous global pandemic due to the COVID-19 (coronavirus disease 2019) is one of the most widespread in recorded history. The SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) put the entire world under social and economic stress due to its high infectivity. An aggravating factor was the limited access to urgent medical treatment for a large proportion of the world population. Initial discovery of the disease was associated with local fish and wild animals market in Wuhan, China, towards the end of 2019 when it started spreading rapidly worldwide effectively impacting all countries and nations. Lack of principal knowledge concerning the cell biology of the new virus with high prevalence and fatality rate compelled the entire scientific society to explore for promising pathways to save human life and avoid socio-economic disaster [1].
SARS-CoV-2 is an RNA zoonotic virus belonging to the Coronaviridae family, order: Nidovirales, genus: Betacoronavirus. It is commonly found in bats but, for unclear reasons, became infectious with the ability to transfer from human to human thereby spreading worldwide resulting in a global pandemic [2,3]. It is a positive-sense single-stranded RNA virus (ssRNA(+)) with genetic material capable of performing the function of messenger RNA (mRNA) [[4], [5], [6]].
The number of infected patients and mortality increased dramatically after the first alarm was raised (pandemic status was declared by the World health organization (WHO) in March 2019) [7]. About 761.4 million infected patients and 6.887 million deaths were confirmed by WHO [8]. Symptoms due to SARS-CoV-2 infection are similar to many other diseases (for example flu) and include; cough, headache, fever, diarrhea, breathing difficulty, and loss of taste and/or smell. For severe cases, supplemental oxygen and intensive care hospitalization are needed due to respiratory problems that can seriously impact human organ function and possibly lead to death. Several waves of SARS-CoV-2 mutants were observed, namely: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), Epsilon (B.1.427 and B.1.429), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), Mu (B.1.621, B.1.621.1) and Zeta (P.2). The Omicron (B.1.1.529) variant was first identified in Botswana in Nov. 2021 and rapidly spread to all countries worldwide [[9], [10], [11], [12]]. The Omicron variant is highly efficiently transmitted between humans. Due to the continuous virus mutation and limitation of effective therapeutics, it is estimated that the epidemic disease is away from its end [13].
Some proteins have been mentioned as having a critical role in SARS-CoV-2 infection making them potential targets for optimizing therapeutic agents (Fig. 1 ) [14]. Coronavirus main protease (Mpro, also called 3CLpro) plays an essential role in viral maturation, replication and transcription. Thus, the inhibition of Mpro is an attractive target for therapeutics against COVID-19 [15,16]. By the end of 2021, FDA (Food and Drug Administration) approved (under the emergency use authorization) Paxlovid as anti-SARS-CoV-2 therapeutic for mild and moderate COVID-19 patients. It is a combination of two effective agents, Nirmatrelvir (3CL protease inhibitor) and Ritonavir (protease inhibitor originally developed for HIV/AIDS). Paxlovid can reduce the risk of death or the hospitalization period if administrated within a few days of symptoms due to infection appearing [[17], [18], [19]]. PF-00835231 is an Mpro inhibitor (Ki = 0.27 nM) with potential inhibitory properties against SARS-CoV-2 [EC50 (Vero-E6 cell) = up to 0.23 μM] reported by Pfizer as a repurposing therapeutic (Fig. 2 ) [20]. 2-Substituted indolealkylamines were mentioned as potential anti-SARS-CoV-2 candidates with viral main protease (Mpro) inhibitory properties [21]. Many indole-containing compounds have also been mentioned by the in-silico studies as Mpro inhibitory properties but lack of experimental bio-evidence has hindered their applicability [15,[22], [23], [24], [25]].
Fig. 1.
Some of the proteins involved in the SARS-CoV-2 infection, that make them potential targets for developing therapeutical agents.
Fig. 2.
Repurposed drugs against COVID-19.
The scientific community pulled all stops to investigate and develop effective diagnostic and therapeutical agents in addition to vaccines capable of preventing infection [26]. Vaccination was considered as an integral part for controlling the long-term COVID-19 pandemic as the protective neutralizing antibodies produced in the human body that can either completely protect from infection or control the severe symptoms of the infectious virus. A number of vaccines with substantial efficacy against different variants of SARS-CoV-2 were developed and administered [27]. The most well-known are BNT162b2 by Pfizer/BioNTech, mRNA-1273 by Moderna, and AZD1222 by University of Oxford & AstraZeneca [28]. Large scale vaccination was pursued for controlling the infection as herd immunity was risky due to no certain evidence being available for acquired immunity in recovered COVID-19 patients [29]. Although convalescent plasma was considered as a therapeutic tool for severely infected patients, the feasibility of this approach was questionable [30].
Drug repurposing has been the main technique for identifying potential therapeutics against COVID-19. Investigating new drugs usually needs lots of research investigations requiring effort, time and money. However, redirecting already known drugs towards a new disease is an attractive shortcut towards the desired target. Many repurposed therapeutics have been identified with potential potency for mild and moderate infections [31]. However, none up to our knowledge has efficacy for severe conditions. Arbidol (an indolyl scaffold) is a broad spectrum antiviral agent (anti-influenza, hepatitis “HBV, HCV”, Ebola, Lassa, and chikungunya) with hemagglutinin esterase inhibitory properties. It was successfully repurposed against COVID-19 [[32], [33], [34], [35], [36], [37], [38]]. Obatoclax which was subjected to phase II clinical trial as an anticancer agent (leukemia, lymphoma, and lung) revealed potent anti-SARSCoV-2 properties in addition to the inhibitory properties against S protein-mediated virus entry suggesting its potenial applicibility against COVID-19 [39]. Melatonin which is a natural product found in plants and animals with antioxidant and anti-inflammatory properties was mentioned as a promising agent useful for preventing and treating COVID-19 with safety profile. Reports supported that treatment of COVID-19 patients with melatonin alone or in combination with another therapeutic drug either shortened the hospitalization period or reduced the severity of the viral infection (Fig. 2) [[40], [41], [42], [43], [44], [45], [46], [47]].
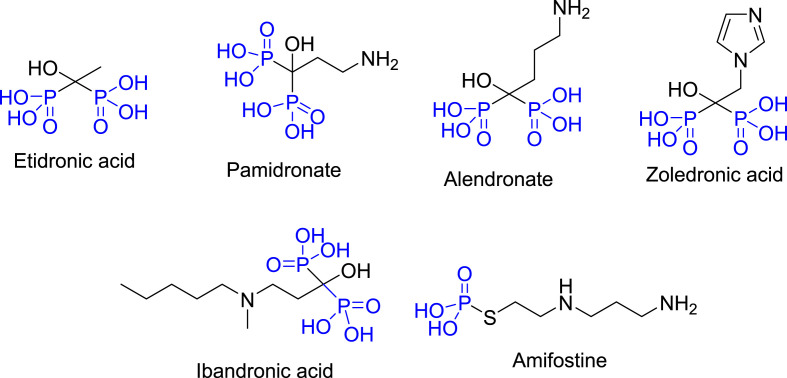
The current study is directed towards construction of new spiroindole-containing compounds bearing a phosphonate moiety. Interest of this scaffold is attributed to the previously described anti-COVID-19 properties of indole-containing heterocycles especially those considered repurposed drug (Arbidol) and the potent clinically supported ones (PF-00835231, Obatoclax and Melatonin). The potent anti-SARS-CoV-2 properties of indole and spiroindole-containing analogs also encouraged the current study [48,49]. Interest in insertion of the phosphoryl group into the heterocyclic system is the ability of the group to improve the physicochemical properties as the electron-rich phosphoryl residue that may increase the bioavailability of the targeted agents [50]. Previous reports describing the antiviral properties of phosphonate-containing compounds also support the rationality of the current design study [51,52]. Many therapeutics incorporating the phosphonate group are well known and include Etidronate [53], Pamidronate [54], Alendronate [55], Zoledronic acid [56], Ibandronic acid [57] (FDA approved in 1977, 1991, 1995, 2001, 2003 for osteoporosis) and Amifostine (FDA approved in 1995 to reduce kidney toxicity “nephrotoxic” effect upon repeated treatment of ovarian cancer with cisplatin) [58,59] (Fig. 3 ).
Fig. 3.
Drugs with phosphonate group.
2. Results and discussion
2.1. Chemical synthesis
The N-diethyl phosphonate-4-piperidones 3a‒g were synthesized in good to excellent yields (71–87%) through dehydrohalogenation reaction of diethylchlorophosphate 2 with 3,5-bis(ylidene)-4-piperidones 1a‒g in DMF (N,N-dimethylformamide) in presence of a sufficient amount of TEA (triethylamine) at 0 °C. The single signal integrated to two protons at δ H = 7.67–7.89 evidenced the E-configuration [[60], [61], [62], [63]]. Azomethine cycloaddition (obtained through condensation of sarcosine 5 with isatins 4) with the appropriate N-diethyl phosphonate-4-piperidones 3a‒g in ethanolic solution under microwave condition (60 Watt, 60 °C) gave the targeted spiroindoles 6a‒l in excellent yields (80–95%) as the only isolable products (Scheme 1 ). The spectral data (IR, 1H, 13C NMR, 1H, 1H-Cosy, and HSQC) evidenced the chemical structure (Supplementary information file Fig. S1‒S59).
Scheme 1.
Synthetic route toward spiroindoles 6a‒l.
2.2. X-ray studies
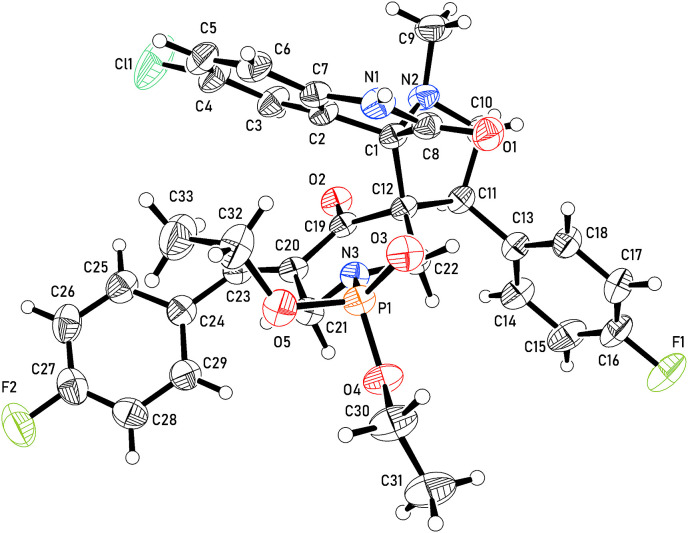
The molecule of 6d is shown in Fig. 4 . In the diethyl phosphonate group (P1, O3 – O5, C30 – C33); one ethoxy group is disordered with two components. The P–O–C–C bonds are in trans conformation (except for one disorder component which is gauche). The pyrrolidine ring (C1, C10 – C12, N2) is in envelope conformation with the nitrogen located 0.574(4) Å from the least squares plane through the other atoms. The pyrrolidine ring is linked to three other ring systems which are oriented perpendicular to its plane, namely: fluorophenyl (C13–C18, F1), chloro-indol-2-one (C1 – C8, O1, N1, Cl1) and piperidone (C12, C19 – C22, O2, N3) rings. The piperidone ring is essentially planar except for atom N3 which deviates by 0.580(2) Å from the plane through the rest of the atoms. The fluorophenylmethylene group (C23–C29, F2) is twisted by about 40° from the plane of the piperidone ring.
Fig. 4.
An ortep representation of the asymmetric unit of the crystal structure of 6d showing 50% probability atomic displacement ellipsoids for one disorder component.
The molecule of 6i is shown in Fig. 5 . In the diethyl phosphonate group (P1, O3 – O5, C33 – C36), one P–O–C–C bond is in trans conformation and the second is gauche. The pyrrolidine ring (C2, C10 – C12, N2) is in envelope conformation with the nitrogen located 0.594 (3) Å from the least squares plane through the other atoms. The pyrrolidine ring is linked to three other ring systems which are oriented perpendicular to its plane, namely: methylphenyl (C13–C19), indol-2-one (C1 – C8, O1, N1) and piperidone (C11, C21 – C24, O2, N3) rings. The piperidone ring is essentially planar except for atom N3 which deviates by 0.462(2) Å from the plane through the rest of the atoms. The tolylmethylene group (C25–C32) is disordered with two components and is twisted by about 35° from the plane of the piperidone ring.
Fig. 5.
An ortep representation of the asymmetric unit of the crystal structure of 6i showing 50% probability atomic displacement ellipsoids for one disorder component.
There are two unique molecules in the crystal of 6l (Fig. 6 ). In both molecules, the thiophene ([C13 – C16, S1] for the first molecule and [C42 – C45, S3] for the second molecule), (thienyl)methylene ([C21 – C25, S2] for the first molecule and [C50 – C54, S4] for the second molecule), and the diethyl phosphonate ([P1, O3 – O5, C26 – C29] for the first molecule and [P2, O8 – O10, C55 – C58] for the second molecule) are disordered with two components. The P–O–C–C bonds are in trans conformation for the major components whereas one torsion of each minor component of the molecule is gauche. The pyrrolidine rings ([C2, C10 – C12, N2] for the first molecule and [C31, C39 – C40, N5] for the second molecule) are in envelope conformation with the nitrogen atoms N2 and N5 being located 0.595 (4) Å and 0.596 (5) Å from the least squares plane through the other atoms of the respective rings. Each pyrrolidine ring is linked to three ring systems which are oriented perpendicular to its plane. These are thiophene, chloroindol-2-one ([C1 – C8, O1, N1, Cl1] for the first molecule and [C30 – C37, O6, N4, Cl2] for the second molecule) and piperidone ([C11, C17 – C20, O2, N3] for the first molecule and [C40, C46 – C49, O7, N6] for the second molecule) rings. The piperidone rings are essentially planar other than for atoms N3 and N6 which are located 0.503(3) Å and 0.541(3) Å from the planes through the rest of the atoms of their respective rings. The (thienyl)methylene group is nearly co-planar with the piperidone ring which is linked to.
Fig. 6.
An ortep representation of the asymmetric unit of the crystal structure of 6l showing 50% probability atomic displacement ellipsoids for one disorder component.
The molecules of 6d, 6i and 6l have a common core comprising pyrrolidine, indol-2-one and piperidone rings. This unit is relatively rigid with the same geometry in all three compounds. Any variation in the biological properties of these molecules, therefore can be reasonably attributed to the conformational flexibility of the pendant groups as well as their derivatization.
2.3. Anti-SARS-CoV-2 properties
The standard viral infected Vero-E6 cell technique was used to assess the anti-SARS-CoV-2 properties of the synthesized spiroindoles 6a‒l [[64], [65], [66]]. The results (Table 1 , Fig. 7 ), indicate that some of the synthesized spiroindoles are more potent SARS-CoV-2 inhibitor than the standard references used (Favipiravir, Hydroxychloroquine and Chloroquine) with promising selectivity indices (SI). Of the synthesized agents, compound 6g (R = 4-BrC6H4, R' = H) is the most promising, with IC50 = 8.88 μM, CC50 = 20.33 μM, and SI = 2.29. Compound 6b (R = Ph, R' = Cl) also has comparable anti-SARS-CoV-2 properties (IC50 = 10.39 μM, CC50 = 22.0 μM, SI = 2.12) but with lower efficacy than 6g. Although compound 6i (R = 4-H3CC6H4, R' = H) exhibits a higher SI value relative to the other synthesized analogs (IC50 = 35.21 μM, CC50 = 87.01 μM, SI = 2.47), it has lower efficacy than 6g and 6b. Compound 6f (R = 4-ClC6H4, R' = Cl) displays higher anti-SARS-CoV-2 activity but with a lower SI value (IC50 = 7.26 μM, CC50 = 10.0 μM, SI = 1.38) than those of 6g, 6b and 6i. Similar observations are also made for compound 6d (R = 4-FC6H4, R' = Cl) and 6j (R = 4-H3CC6H4, R' = Cl), The values for 6d, 6j are IC50 = 13.53, 21.66 μM; CC50 = 18.83, 40.10 μM; and SI = 1.39, 1.85 respectively.
Table 1.
Anti-SARS-CoV-2 properties of the synthesized spiroindoles 6a‒l and standard references.
| Compd. | IC50 (μM) | CC50 (μM) | Selectivity index (SI) |
|---|---|---|---|
| 6a | 88.61 | 73.07 | 0.82 |
| 6b | 10.39 | 22.0 | 2.12 |
| 6c | 85.84 | 126.0 | 1.47 |
| 6d | 13.53 | 18.83 | 1.39 |
| 6e | 340 | 9.834 | 0.03 |
| 6f | 7.26 | 10.0 | 1.38 |
| 6g | 8.88 | 20.33 | 2.29 |
| 6h | 46.74 | 38.33 | 0.82 |
| 6i | 35.21 | 87.01 | 2.47 |
| 6j | 21.66 | 40.10 | 1.85 |
| 6k | 106.1 | 49.92 | 0.47 |
| 6l | 578.4 | 23.71 | 0.04 |
| Favipiravir [65] | 1382 | 5262 | 3.8 |
| Hydroxychloroquine [61] | 36.92 | 356.4 | 9.7 |
| Chloroquine [61] | 24.98 | 377.7 | 15.1 |
Fig. 7.
Dose-response curves for the spiroindole 6a‒l against SARS-CoV-2.
SAR (structure-activity relationship) deduced from anti-SARS-CoV-2 activity indicate that the chloroindolyl-containing compounds have higher efficacy than the unsubstituted analogs (compound 6l which contains thienyl heterocycle is an exception). This rule seems to apply to the synthesized compounds derived from (un)substitutd benzylidene-4-piperidones in general as supported by comparison of pairs 6a/6b (IC50 = 88.61, 10.39 μM, respectively), 6c/6d (IC50 = 85.84, 13.53 μM, respectively), 6e/6f (IC50 = 340, 7.26 μM, respectively) and 6i/6j (IC50 = 35.21, 21.66 μM, respectively). These observations support the role of indolyl heterocyclic substitution in the anti-SARS-CoV-2 properties.
2.4. Mpro-SARS-CoV-2 properties
Mpro is one of the essential proteins for replication and gene expression of the SARS-CoV-2 virus. Inhibition of Mpro is considered one of the most potential pathways for treating infected COVID-19 patients [67]. The Mpro inhibitory properties of the most effective agents synthesized 6g, 6b and 6d were determined by an appropriate Kit assay [68]. From the results (Table 2 ), compound 6b has the most potent against Mpro-SARS-CoV-2 and is comparable to Tipranavir (IC50 = 9.605, 7.38 μM, respectively). Tipranavir (Fig. 8 ) is a non-peptidic protease inhibitor useable for therapy and prevention of HIV (human immunodeficiency virus) [[69], [70], [71]]. Spiroindole 6g also shows promising Mpro-SARS-CoV-2 inhibitory properties (IC50 = 15.59 μM). Considerable protease inhibitory properties are also shown by spiroindole 6d (IC50 = 42.82 μM). It is notable that the protease (Mpro) inhibitory properties are comparable to the anti-SARS-CoV-2 inhibitory properties presented in Table 1 (IC50 = 10.39, 13.53, 8.88 μM for compounds 6b, 6d and 6g, respectively). The slight differences in the results are attributed to differences in the experimental techniques applied (in-vitro and biochemical assays).
Table 2.
IC50 values of Mpro-SARS-CoV-2 for the synthesized agents (6b, 6d and 6g) and standard reference Tipranavir.
| Entry | Compd. | IC50 (μM ± SE) |
|---|---|---|
| 1 | 6b | 9.605 ± 0.66 |
| 2 | 6d | 42.82 ± 2.53 |
| 3 | 6g | 15.59 ± 1.02 |
| 4 | Tipranavir | 7.38 ± 0.42 |
Fig. 8.
Tipranavir, protease inhibitor useable for therapy and prevention of HIV.
2.5. Molecular modeling
Various molecular modeling techniques have been developed over the last decades and are used intensively in medicinal chemical studies for identifying and optimizing the effective hits/leads and determining the parameters controlling biological properties [72,73]. The effective Mpro-SARS-CoV-2 agents identified (6b, 6d and 6g) were used for molecular modeling (docking) studies utilizing PDB ID: 7C8U [74] by Discovery Studio 2.5 software (RMS: 0.088). The standard CDOCKER technique was adopted after protein and ligand optimization (force field: CHARMm, Partial charge: MMFF94, radius of the active site employed: 8.0368 Å) [61]. The results (Fig. 9 , Table 3 ), reveal that compound 6b is involved in hydrogen bonding interaction of indolyl NH with GLU166 and indolyl Cl with CYS145. The amino acids concerned interact with the co-crystallized ligand in the protein active site (supplementary information file Fig. S60). The high CDOCKER interaction energy score (−52.58 kcal mol−1) is consistent with the higher Mpro-SARS-CoV-2 inhibitory properties (IC50 = 9.605 μM) compared to the other tested analogs. Similar observations have been made for compound 6g revealing hydrogen bonding interaction of the phosphonate P O with GLU166 and acceptable docking interaction energy score (−48.823 kcal mol−1) supporting its Mpro-SARS-CoV-2 inhibitory properties (IC50 = 15.59 μM). Compound 6d shows hydrogen bonding interaction of piperidinyl nitrogen with GLN189 and phosphonate P O with GLN189. Although, GLN189 is one of the amino acids of the protein active site, it is not one of the lead amino acids revealing hydrogen bonding interaction with the co-crystallized ligand. These observations explain the low Mpro-SARS-CoV-2 inhibitory properties (IC50 = 42.82 μM) of 6d relative to the other tested agents. Thus, the docking studies support the observed Mpro-SARS-CoV-2 inhibitory properties and explain the potency of the tested agents based on the presumed mode of action.
Fig. 9.
Docking poses of the spiroindoles (6b, 6d and 6g) in the active site of PDB ID: 7C8U.
Table 3.
Interaction CDOCKER energy scores and hydrogen bonding interactions of the synthesized spiroindoles (6b, 6d and 6g) in the active site of PDB ID: 7C8U.
| Compd. | CDOCKER interaction energy (‒Kcal mol−1) | Hydrogen bonding interactions |
|---|---|---|
| 6b | 52.58 | Phosphonate P O … GLN189, Indolyl NH … GLU166, Indolyl Cl…CYS145 |
| 6d | 49.017 | Piperidinyl N … GLN189, Phosphonate P O … GLN189 |
| 6g | 48.823 | Phosphonate P O … GLU166, Phosphonate OEt … GLN189 |
3. Conclusion
Spiroindoles 6a‒l bearing the phosphonate group were obtained in excellent yields (80–95%) through microwave-assisted azomethine ylide (produced through interaction of isatins 4 and sarcosine 5) cycloaddition of N-diethyl phosphonate-3,5-bis(ylidene)-4-piperidones 3a‒g. X-ray diffraction studies evidenced the stereochemical configuration. Some of the synthesized spiroindoles revealed potent anti-SARS-CoV-2 properties in the viral infected Vero-E6 cell technique. Mpro inhibitory properties were evidenced for the synthesized agents with potent bio-properties by the appropriate Kit assay. Molecular modeling studies (PDB ID: 7C8U) supported the Mpro-SARS-CoV-2 inhibitory properties. The observations provide a compelling case for the compounds to be considered in future studies for optimizing potent anti-SARS-CoV-2 agents.
4. Experimental
Melting points were determined on a capillary point apparatus (Stuart SMP3) equipped with a digital thermometer. IR spectra (KBr) were recorded on a Shimadzu FT-IR 8400S spectrophotometer. Reactions were monitored using thin layer chromatography (TLC) on 0.2 mm silica gel F254 plates (Merck) utilizing various solvents for elution. The chemical structures of the synthesized compounds were characterized by nuclear magnetic resonance spectra (1H NMR, 13C NMR) and determined on a Bruker NMR spectrometer (500 MHz, 125 MHz for 1H and 13C, respectively). 13C NMR spectra are fully decoupled. Chemical shifts were reported in parts per million (ppm) using the deuterated solvent peak or tetramethylsilane as an internal standard. Microwave oven used is a Milestone Italy (model: StartSynth, Reactor: Pack2B Basic Single Vessel Kit).
4.1. Chemical synthesis
4.1.1. Synthesis of diethyl [3,5-di((E)-ylidene)-4-oxopiperidin-1-yl]phosphonates 3a‒g (general procedure)
To a stirring solution of 3,5-di(E)-ylidene)piperidin-4-ones 1a‒g (5 mmol) in DMF (10 ml) containing TEA (triethylamine, 0.8 ml, 6 mmol) in an ice bath (0 °C), diethylchlorophosphate 2 (0.8 ml, 6 mmol) in DMF (10 ml) was added dropwise (within 10 min). The reaction mixture was stirred at the mentioned conditions for 2 h and stored at room temperature (20–25 °C) overnight. After the completion of the reaction (TLC), it was poured into ice-cold water (200 ml) containing NaCl (1.0 g). The separated solid was collected, washed with tap water, dried and crystallized from a suitable solvent affording the corresponding 3a‒g.
4.1.1.1. Diethyl [3,5-di((E)-benzylidene)-4-oxopiperidin-1-yl]phosphonate (3a)
Obtained from the reaction of 1a and 2, pale-yellow microcrystals from cyclohexane, mp 134–135 °C (127 °C [75]) and yield 71% (1.45 g). IR: ν max/cm−1 1670, 1612, 1585, 1485, 1261, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 1.05 (t, J = 7.1 Hz, 6H, 2 CH3), 3.75–3.84 (m, 4H, 2 OCH2), 4.44 (d, J = 9.2 Hz, 4H, 2 NCH2), 7.45–7.51 (m, 10H, arom. H), 7.73 (s, 2H, 2 olefinic CH). 13C NMR (DMSO‑d 6) δ (ppm): 15.67, 15.72 (CH3), 45.51, 45.54 (NCH2), 61.80, 61.84 (OCH2), 128.7, 129.4, 130.3, 133.01, 133.04, 134.3, 135.5 (arom. C + olefinic C), 185.8 (C O). Anal. Calcd. for C23H26NO4P (411.44): C, 67.14; H, 6.37; N, 3.40. Found: C, 67.26; H, 6.17; N, 3.54.
4.1.1.2. Diethyl [3,5-bis((E)-4-fluorobenzylidene)-4-oxopiperidin-1-yl]phosphonate (3b)
Obtained from the reaction of 1b and 2, yellow microcrystals from cyclohexane, mp 112–114 °C and yield 85% (1.9 g). IR: ν max/cm−1 1670, 1601, 1574, 1508, 1443, 1238, 1049. 1H NMR (DMSO‑d 6) δ (ppm): 1.07 (t, J = 7.0 Hz, 6H, 2 CH3), 3.77–3.84 (m, 4H, 2 OCH2), 4.42 (dd, J = 1.8, 9.2 Hz, 4H, 2 NCH2), 7.33–7.37 (m, 4H, arom. H), 7.57–7.60 (m, 4H, arom. H), 7.71 (s, 2H, 2 olefinic CH). 13C NMR (DMSO‑d 6) δ (ppm): 15.76, 15.81 (CH3), 45.50, 45.53 (NCH2), 61.91, 61.95 (OCH2), 115.8, 116.0, 130.89, 130.91, 132.79, 132.86, 134.4, 161.5, 163.5 (arom. C + olefinic C), 185.7 (C O). Anal. Calcd. for C23H24F2NO4P (447.42): C, 61.74; H, 5.41; N, 3.13. Found: C, 61.90; H, 5.12; N, 2.93.
4.1.1.3. Diethyl [3,5-bis((E)-4-chlorobenzylidene)-4-oxopiperidin-1-yl]phosphonate (3c)
Obtained from the reaction of 1c and 2, pale yellow microcrystals from cyclohexane, mp 136–137 °C (132 °C [75]) and yield 75% (1.8 g). IR: ν max/cm−1 1678, 1616, 1582, 1489, 1269, 1018. 1H NMR (DMSO‑d 6) δ (ppm): 1.05 (t, J = 7.1 Hz, 6H, 2 CH3), 3.73–3.85 (m, 4H, 2 OCH2), 4.40 (dd, J = 1.9, 9.3 Hz, 4H, 2 NCH2), 7.53 (d, J = 8.9 Hz, 4H, arom. H), 7.56 (d, J = 8.9 Hz, 4H, arom. H), 7.68 (s, 2H, 2 olefinic CH). 13C NMR (DMSO‑d 6) δ (ppm): 15.79, 15.84 (CH3), 45.53, 45.56 (NCH2), 61.95, 61.99 (OCH2), 128.9, 132.2, 133.2, 133.57, 133.61, 134.2, 134.3 (arom. C + olefinic C), 185.7 (C O). Anal. Calcd. for C23H24Cl2NO4P (480.32): C, 57.51; H, 5.04; N, 2.92. Found: C, 57.65; H, 5.10; N, 2.81.
4.1.1.4. Diethyl [3,5-bis((E)-4-bromobenzylidene)-4-oxopiperidin-1-yl]phosphonate (3d)
Obtained from the reaction of 1d and 2, pale yellow microcrystals from n-butanol, mp 141–142 °C and yield 74% (2.1 g). IR: ν max/cm−1 1670, 1612, 1585, 1485, 1261, 1022. 1H NMR (DMSO‑d 6) δ (ppm): 1.06 (t, J = 7.0 Hz, 6H, 2 CH3), 3.74–3.86 (m, 4H, 2 OCH2), 4.40 (d, J = 9.3 Hz, 4H, 2 NCH2), 7.47 (d, J = 8.4 Hz, 4H, arom. H), 7.67 (s, 2H, 2 olefinic CH), 7.71 (d, J = 8.3 Hz, 4H, arom. H). 13C NMR (DMSO‑d 6) δ (ppm): 15.7 (CH3), 45.4 (NCH2), 61.9 (OCH2), 123.0, 131.7, 132.2, 133.4, 133.57, 133.60, 134.3 (arom. C + olefinic C), 185.6 (C O). Anal. Calcd. for C23H24Br2NO4P (569.23): C, 48.53; H, 4.25; N, 2.46. Found: C, 48.68; H, 4.37; N, 2.52.
4.1.1.5. Diethyl [3,5-bis((E)-4-methoxybenzylidene)-4-oxopiperidin-1-yl]phosphonate (3e)
Obtained from the reaction of 1e and 2, yellow microcrystals from methanol, mp 158–160 °C (144 °C [75]) and yield 81% (1.9 g). IR: ν max/cm−1 1667, 1605, 1578, 1508, 1261, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 1.08 (t, J = 7.1 Hz, 6H, 2 CH3), 3.77–3.86 (m, 10H, 2 OCH2 + 2 OCH3), 4.42 (d, J = 9.0 Hz, 4H, 2 NCH2), 7.07 (d, J = 8.9 Hz, 4H, arom. H), 7.48 (d, J = 8.9 Hz, 4H, arom. H), 7.67 (s, 2H, 2 olefinic CH). 13C NMR (DMSO‑d 6) δ (ppm): 15.8, 15.9 (CH3), 45.66, 45.69 (NCH2), 55.3 (OCH3), 61.83, 61.87 (OCH2), 114.4, 127.0, 131.0, 131.1, 132.5, 135.2, 160.3 (arom. C + olefinic C), 185.5 (C O). Anal. Calcd. for C25H30NO6P (471.49): C, 63.69; H, 6.41; N, 2.97. Found: C, 63.76; H, 6.55; N, 3.05.
4.1.1.6. Diethyl [3,5-bis((E)-4-methylbenzylidene)-4-oxopiperidin-1-yl]phosphonate (3f)
Obtained from the reaction of 1f and 2, pale yellow microcrystals from cyclohexane, mp 158–160 °C (151 °C [75]) and yield 87% (1.9 g). IR: ν max/cm−1 1670, 1609, 1582, 1508, 1261, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 1.05 (t, J = 7.0 Hz, 6H, 2 CH3), 2.36 (s, 6H, 2 ArCH3), 3.74–3.83 (m, 4H, 2 OCH2), 4.42 (d, J = 9.0 Hz, 4H, 2 NCH2), 7.31 (d, J = 8.1 Hz, 4H, arom. H), 7.40 (d, J = 8.1 Hz, 4H, arom. H), 7.68 (s, 2H, 2 olefinic CH). 13C NMR (DMSO‑d 6) δ (ppm): 15.79, 15.84 (CH3), 21.0 (ArCH3), 45.6, 45.7 (NCH2), 61.8, 61.9 (OCH2), 129.5, 130.5, 131.6, 132.3, 132.4, 135.5, 139.5 (arom. C + olefinic C), 185.7 (C O). Anal. Calcd. for C25H30NO4P (439.49): C, 68.32; H, 6.88; N, 3.19. Found: C, 68.50; H, 6.99; N, 3.35.
4.1.1.7. Diethyl [(3E,5E)-4-oxo-3,5-bis(thiophen-2-ylmethylene)piperidin-1-yl]phosphonate (3g)
Obtained from the reaction of 1g and 2, buff microcrystals from cyclohexane, mp 123–125 °C and yield 85% (1.8 g). IR: ν max/cm−1 1659, 1593, 1558, 1504, 1443, 1261, 1022. 1H NMR (DMSO‑d 6) δ (ppm): 1.16 (t, J = 7.0 Hz, 6H, 2 CH3), 3.86–3.95 (m, 4H, 2 OCH2), 4.47 (d, J = 9.3 Hz, 4H, 2 NCH2), 7.29 (dd, J = 3.7, 5.0 Hz, 2H, arom. H), 7.63 (d, J = 3.4 Hz, 2H, arom. H), 7.89 (s, 2H, 2 olefinic CH), 7.97 (d, J = 5.1 Hz, 2H, arom. H). 13C NMR (DMSO‑d 6) δ (ppm): 15.9, 16.0 (CH3), 45.22, 45.25 (NCH2), 62.06, 62.10 (OCH2), 127.8, 128.6, 129.61, 129.64, 132.4, 134.6, 137.5 (arom. C + olefinic C), 184.7 (C O). Anal. Calcd. for C19H22NO4PS2 (423.48): C, 53.89; H, 5.24; N, 3.31. Found: C, 53.98; H, 5.38; N, 3.38.
4.1.2. Synthesis of dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl)phosphonates 6a‒l (general procedure)
A mixture of equimolar amounts of the appropriate diethyl [3,5-di((E)-ylidene)-4-oxopiperidin-1-yl]phosphonates 3a‒g (1.25 mmol) and the corresponding isatins 4a,b with sarcosine 5 in ethanol (10 ml) was heated in the microwave reactor at 60 °C (60 Watt) for 60–90 min (hold time). After the completion of the reaction (TLC), the reaction mixture was allowed to cool at room temperature, and the solvent was evaporated under reduced pressure. The separated solid upon triturating the residual material with methanol (5 ml) was collected and crystallized from a suitable solvent affording the corresponding 6a‒l.
4.1.2.1. Diethyl (E)-(5″-benzylidene-1′-methyl-2,4″-dioxo-4′-phenyldispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl)phosphonate (6a)
Obtained from the reaction of 3a, 4a and 5, reaction time 90 min as pale yellow microcrystals from ethanol (90%), mp 181–182 °C and yield 92% (0.67 g). IR: ν max/cm−1 3175, 1713, 1674, 1585, 1470, 1227, 1018. 1H NMR (DMSO‑d 6) δ (ppm): 0.84 (t, J = 7.0 Hz, 3H, CH3), 0.98 (t, J = 7.1 Hz, 3H, CH3), 1.96 (s, 3H, NCH3), 2.14 (d, J = 13.5 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.28–3.32 (m, 2H, upfield H of pyrrolidinyl H2C-5' + upfield H of piperidinyl H2C-6″), 3.48–3.88 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.71 (t, J = 9.1 Hz, 1H, pyrrolidinyl H-4′), 6.70 (d, J = 7.7 Hz, 1H, arom. H), 6.88–6.92 (m, 2H, arom. H), 7.11–7.15 (m, 3H, arom. H), 7.26 (t, J = 7.3 Hz, 1H, arom. H), 7.33–7.40 (m, 5H, 4 arom. H + olefinic CH), 7.43–7.46 (m, 3H, arom. H), 10.53 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.56, 15.62, 15.80, 15.81 (CH3), 33.8 (NCH3), 45.1 (pyrrolidinyl HC-4′), 46.1 (piperidinyl H2C-6″), 47.4 (piperidinyl H2C-2″), 57.1 (pyrrolidinyl H2C-5′), 61.84, 61.86, 61.88, 61.91, 61.95, 62.03 [spiro-C-3' (C-3″) + OCH2], 75.1 [spiro-C-3 (C-2′)], 109.1, 120.9, 125.4, 126.8, 127.0, 128.3, 128.5, 129.0, 129.2, 129.6, 129.8, 131.6, 131.7, 134.2, 137.9, 138.2, 143.7 (arom. C + olefinic C), 175.2, 197.2 (C O). Anal. Calcd. for C33H36N3O5P (585.64): C, 67.68; H, 6.20; N, 7.18. Found: C, 67.85; H, 6.31; N, 7.41.
4.1.2.2. Diethyl (E)-(5″-benzylidene-5-chloro-1′-methyl-2,4″-dioxo-4′-phenyldispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl)phosphonate (6b)
Obtained from the reaction of 3a, 4b and 5, reaction time 90 min as buff microcrystals from ethanol (90%), mp 220–221 °C and yield 93% (0.72 g). IR: ν max/cm−1 3175, 3140, 1717, 1682, 1601, 1470, 1238, 1022. 1H NMR (DMSO‑d 6) δ (ppm): 0.81 (t, J = 7.0 Hz, 3H, CH3), 1.00 (t, J = 7.0 Hz, 3H, CH3), 1.97 (s, 3H, NCH3), 2.16 (d, J = 13.6 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.32 (d, J = 8.9 Hz, 1H, upfield H of pyrrolidinyl H2C-5′), 3.49–3.52 (m, 2H, upfield H of piperidinyl H2C-6'' + downfield H of pyrrolidinyl H2C-5′), 3.65–3.90 (m, 6H, downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.65 (t, J = 8.9 Hz, 1H, pyrrolidinyl H-4′), 6.68 (d, J = 8.3 Hz, 1H, arom. H), 6.82 (s, 1H, arom. H), 7.18–7.21 (m, 3H, arom. H), 7.27 (t, J = 7.3 Hz, 1H, arom. H), 7.34–7.46 (m, 8H, 7 arom. H + olefinic CH), 10.70 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.47, 15.52, 15.74, 15.79 (CH3), 33.9 (NCH3), 45.8 (pyrrolidinyl HC-4′), 46.2 (piperidinyl H2C-6″), 48.0 (piperidinyl H2C-2″), 57.4 (pyrrolidinyl H2C-5′), 61.87, 61.92, 61.96, 62.26, 62.34 [spiro-C-3' (C-3″) + OCH2], 75.2 [spiro-C-3 (C-2′)], 110.6, 125.0, 126.7, 127.0, 127.5, 128.3, 128.5, 128.7, 129.3, 129.5, 129.8, 131.7, 131.8, 134.0, 137.9, 142.6 (arom. C + olefinic C), 174.8, 197.2 (C O). Anal. Calcd. for C33H35ClN3O5P (620.08): C, 63.92; H, 5.69; N, 6.78. Found: C, 63.86; H, 5.78; N, 6.86.
4.1.2.3. Diethyl (E)-[5''-(4-fluorobenzylidene)-4'-(4-fluorophenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6c)
Obtained from the reaction of 3b, 4a and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 193–194 °C and yield 94% (0.73 g). IR: ν max/cm−1 3186, 1705, 1682, 1601, 1508, 1223, 1030. 1H NMR (DMSO‑d 6) δ (ppm): 0.85 (t, J = 7.1 Hz, 3H, CH3), 1.00 (t, J = 7.0 Hz, 3H, CH3), 1.94 (s, 3H, NCH3), 2.17 (d, J = 13.4 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.27–3.28 (m, 1H, upfield H of pyrrolidinyl H2C-5′) 3.32 (d, J = 8.4 Hz, 1H, upfield H of piperidinyl H2C-6″), 3.49–3.81 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.66 (dd, J = 8.1, 10.0 Hz, 1H, pyrrolidinyl H-4′), 6.69 (d, J = 7.7 Hz, 1H, arom. H), 6.88–6.90 (m, 2H, arom. H), 7.11–7.27 (m, 7H, arom. H), 7.41 (s, 1H, olefinic CH), 7.50 (dd, J = 5.6, 8.5 Hz, 2H, arom. H), 10.56 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.6, 15.7, 15.8, 15.9 (CH3), 33.9 (NCH3), 45.2 (pyrrolidinyl HC-4′), 45.4 (piperidinyl H2C-6″), 47.4 (piperidinyl H2C-2″), 57.6 (pyrrolidinyl H2C-5′), 61.7, 61.8, 61.91, 61.95, 61.99 [spiro-C-3' (C-3″) + OCH2], 75.3 [spiro-C-3 (C-2′)], 109.2, 115.0, 115.1, 115.6, 115.8, 121.0, 125.2, 126.9, 129.1, 130.69, 130.72, 131.4, 131.76, 131.82, 132.08, 132.15, 134.39, 134.41, 136.8, 143.6, 160.3, 161.3, 162.2, 163.2 (arom. C + olefinic C), 175.3, 197.2 (C O). Anal. Calcd. for C33H34F2N3O5P (621.62): C, 63.76; H, 5.51; N, 6.76. Found: C, 63.52; H, 5.34; N, 6.57.
4.1.2.4. Diethyl (E)-[5-chloro-5''-(4-fluorobenzylidene)-4'-(4-fluorophenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6d)
Obtained from the reaction of 3b, 4b and 5, reaction time 60 min as pale yellow microcrystals from n-butanol, mp 234–235 °C and yield 93% (0.76 g). IR: ν max/cm−1 3183, 1713, 1682, 1601, 1508, 1227, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 0.82 (t, J = 7.0 Hz, 3H, CH3), 1.01 (t, J = 7.0 Hz, 3H, CH3), 1.94 (s, 3H, NCH3), 2.18 (d, J = 13.5 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.34 (t, J = 8.5 Hz, 1H, upfield H of pyrrolidinyl H2C-5′), 3.44–3.82 (m, 8H, upfield H of piperidinyl H2C-6'' + downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.61 (dd, J = 8.1, 9.9 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 8.4 Hz, 1H, arom. H), 6.79 (d, J = 2.2 Hz, 1H, arom. H), 7.18 (dt, J = 1.9, 8.9 Hz, 3H, arom. H), 7.23–7.30 (m, 4H, arom. H), 7.39 (s, 1H, olefinic CH), 7.49 (dd, J = 5.5, 8.5 Hz, 2H, arom. H), 10.72 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.59, 15.64, 15.86, 15.90 (CH3), 33.9 (NCH3), 45.6 (pyrrolidinyl HC-4′), 45.9 (piperidinyl H2C-6″), 48.0 (piperidinyl H2C-2″), 57.9 (pyrrolidinyl H2C-5′), 62.00, 62.05, 62.07, 62.10, 62.15 [spiro-C-3' (C-3″) + OCH2], 75.4 [spiro-C-3 (C-2′)], 110.7, 115.0, 115.2, 115.7, 115.9, 125.0, 126.7, 127.5, 128.9, 130.54, 130.57, 131.7, 131.8, 131.9, 132.1, 132.2, 134.19, 134.21, 136.9, 142.6, 160.3, 161.4, 162.3, 163.3 (arom. C + olefinic C), 175.0, 197.1 (C O). Anal. Calcd. for C33H33ClF2N3O5P (656.06): C, 60.42; H, 5.07; N, 6.41. Found: C, 60.29; H, 5.13; N, 6.28.
4.1.2.5. Diethyl (E)-[5''-(4-chlorobenzylidene)-4'-(4-chlorophenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6e)
Obtained from the reaction of 3c, 4a and 5, reaction time 60 min as pale yellow microcrystals from benzene ‒ light petroleum as 1:2 v/v, mp 143–145 °C and yield 91% (0.74 g). IR: ν max/cm−1 3260, 1713, 1682, 1605, 1489, 1227, 1015. 1H NMR (DMSO‑d 6) δ (ppm): 0.84 (t, J = 7.1 Hz, 3H, CH3), 0.99 (t, J = 7.0 Hz, 3H, CH3), 1.92 (s, 3H, NCH3), 2.18 (d, J = 13.5 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.27–3.33 (m, 2H, upfield H of pyrrolidinyl H2C-5' + upfield H of piperidinyl H2C-6″), 3.46–3.81 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.64 (t, J = 9.0 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 7.8 Hz, 1H, arom. H), 6.87–6.91 (m, 2H, arom. H), 7.10–7.15 (m, 3H, arom. H), 7.39 (d, J = 7.0 Hz, 2H, arom. H), 7.41 (s, 1H, olefinic CH), 7.46 (d, J = 8.6 Hz, 2H, arom. H), 7.48 (d, J = 8.3 Hz, 2H, arom. H), 10.58 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.6, 15.7, 15.84, 15.88 (CH3), 33.9 (NCH3), 45.2 (pyrrolidinyl HC-4′), 45.6 (piperidinyl H2C-6″), 47.4 (piperidinyl H2C-2″), 57.4 (pyrrolidinyl H2C-5′), 61.8, 61.88, 61.95, 61.98, 62.00, 62.03 [spiro-C-3' (C-3″) + OCH2], 75.3 [spiro-C-3 (C-2′)], 109.3, 121.0, 125.2, 126.9, 128.3, 128.7, 129.1, 131.4, 131.7, 131.8, 132.15, 132.23, 133.0, 134.0, 136.5, 137.3, 143.6 (arom. C + olefinic C), 175.3, 197.1 (C O). Anal. Calcd. for C33H34Cl2N3O5P (654.52): C, 60.56; H, 5.24; N, 6.42. Found: C, 60.66; H, 5.42; N, 6.61.
4.1.2.6. Diethyl (E)-[5-chloro-5''-(4-chlorobenzylidene)-4'-(4-chlorophenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6f)
Obtained from the reaction of 3c, 4b and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 229–230 °C and yield 88% (0.76 g). IR: ν max/cm−1 3175, 3140, 1713, 1682, 1605, 1493, 1238, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 0.84 (t, J = 7.1 Hz, 3H, CH3), 1.02 (t, J = 7.0 Hz, 3H, CH3), 1.95 (s, 3H, NCH3), 2.22 (d, J = 13.5 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.35 (t, J = 8.4 Hz, 1H, upfield H of pyrrolidinyl H2C-5′), 3.45–3.81 (m, 8H, upfield H of piperidinyl H2C-6'' + downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.61 (dd, J = 8.1, 9.9 Hz, 1H, pyrrolidinyl H-4′), 6.69 (d, J = 8.3 Hz, 1H, arom. H), 6.79 (d, J = 2.2 Hz, 1H, arom. H), 7.19–7.22 (m, 3H, arom. H), 7.37–7.52 (m, 7H, arom. H + olefinic CH), 10.74 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.62, 15.67, 15.85, 15.90 (CH3), 33.9 (NCH3), 45.7 (pyrrolidinyl HC-4′), 45.8 (piperidinyl H2C-6″), 47.9 (piperidinyl H2C-2″), 57.7 (pyrrolidinyl H2C-5′), 62.0, 62.08, 62.13, 62.15, 62.22 [spiro-C-3' (C-3″) + OCH2), 75.4 [spiro-C-3 (C-2′)], 110.8, 125.0, 126.8, 127.4, 128.3, 128.8, 128.9, 131.4, 131.8, 131.9, 132.3, 132.4, 132.9, 134.2, 136.5, 137.1, 142.6 (arom. C + olefinic C), 175.9, 197.0 (C O). Anal. Calcd. for C33H33Cl3N3O5P (688.97): C, 57.53; H, 4.83; N, 6.10. Found: C, 57.69; H, 5.15; N, 6.21.
4.1.2.7. Diethyl (E)-[5''-(4-bromobenzylidene)-4'-(4-bromophenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6g)
Obtained from the reaction of 3d, 4a and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 137–139 °C and yield 85% (0.79 g). IR: ν max/cm−1 3167, 1709, 1686, 1616, 1597, 1238, 1022. 1H NMR (DMSO‑d 6) δ (ppm): 0.85 (t, J = 7.0 Hz, 3H, CH3), 1.01 (t, J = 7.1 Hz, 3H, CH3), 1.93 (s, 3H, NCH3), 2.20 (d, J = 13.4 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.28–3.32 (m, 1H, upfield H of pyrrolidinyl H2C-5′) 3.49–3.80 (m, 8H, upfield H of piperidinyl H2C-6'' + downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.63 (dd, J = 8.1, 9.9 Hz, 1H, pyrrolidinyl H-4′), 6.68 (d, J = 7.8 Hz, 1H, arom. H), 6.86–6.90 (m, 2H, arom. H), 7.08 (dd, J = 2.1, 6.5 Hz, 2H, arom. H), 7.13 (dt, J = 2.3, 7.3 Hz, 1H, arom. H), 7.36 (s, 1H, olefinic CH), 7.42 (d, J = 8.2 Hz, 2H, arom. H), 7.55 (d, J = 8.2 Hz, 2H, arom. H), 7.61 (dd, J = 2.0, 6.5 Hz, 2H, arom. H), 10.58 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.6, 15.7, 15.8, 15.9 (CH3), 33.9 (NCH3), 45.2 (pyrrolidinyl HC-4′), 45.6 (piperidinyl H2C-6″), 47.4 (piperidinyl H2C-2″), 57.3 (pyrrolidinyl H2C-5′), 61.8, 61.9, 61.95, 61.99, 62.0 [spiro-C-3' (C-3″) + OCH2], 75.3 [spiro-C-3 (C-2′)], 109.3, 120.2, 121.0, 122.8, 125.1, 126.9, 129.1, 131.2, 131.58, 131.63, 132.2, 133.4, 136.6, 137.7, 143.6 (arom. C + olefinic C), 175.3, 197.1 (C O). Anal. Calcd. for C33H34Br2N3O5P (743.43): C, 53.32; H, 4.61; N, 5.65. Found: C, 53.51; H, 4.77; N, 5.73.
4.1.2.8. Diethyl (E)-[5''-(4-methoxybenzylidene)-4'-(4-methoxyphenyl)-1′-methyl-2,4″-dioxodispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6h)
Obtained from the reaction of 3e, 4a and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 196–198 °C and yield 93% (0.75 g). IR: ν max/cm−1 3171, 1705, 1682, 1593, 1512, 1258, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 0.90 (t, J = 7.1 Hz, 3H, CH3), 1.01 (t, J = 7.0 Hz, 3H, CH3), 1.92 (s, 3H, NCH3), 2.13 (d, J = 13.5 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.23–3.28 (m, 2H, upfield H of pyrrolidinyl H2C-5' + upfield H of piperidinyl H2C-6″), 3.52–3.82 (m, 13H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2 + 2OCH3), 4.62 (t, J = 9.1 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 7.7 Hz, 1H, arom. H), 6.81–6.86 (m, 2H, arom. H), 6.89 (d, J = 9.0 Hz, 2H, arom. H), 6.96 (d, J = 9.0 Hz, 2H, arom. H), 7.11 (d, J = 8.8 Hz, 2H, arom. H), 7.34–7.36 (m, 3H, arom. H), 7.42 (s, 1H, olefinic CH), 10.52 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.72, 15.77, 15.86, 15.90 (CH3), 33.9 (NCH3), 45.2 (pyrrolidinyl HC-4′), 45.4 (piperidinyl H2C-6″), 47.1 (piperidinyl H2C-2″), 55.0, 55.3 (OCH3), 57.4 (pyrrolidinyl H2C-5′), 61.65, 61.73, 61.88, 61.93 [spiro-C-3' (C-3″) + OCH2], 75.2 [spiro-C-3 (C-2′)], 109.1, 113.7, 114.2, 120.8, 125.5, 126.6, 126.8, 128.3, 128.9, 129.3, 129.4, 130.1, 130.9, 132.0, 137.8, 143.7, 158.2, 160.1 (arom. C + olefinic C), 175.3, 197.1 (C O). Anal. Calcd. for C35H40N3O7P (645.69): C, 65.11; H, 6.24; N, 6.51. Found: C, 65.19; H, 6.28; N, 6.68.
4.1.2.9. Diethyl (E)-[1′-methyl-5''-(4-methylbenzylidene)-2,4″-dioxo-4'-(p-tolyl)dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6i)
Obtained from the reaction of 3f, 4a and 5, reaction time 70 min as pale yellow microcrystals from methanol, mp 226–227 °C and yield 95% (0.73 g). IR: ν max/cm−1 3179, 1713, 1678, 1620, 1589, 1261, 1022. 1H NMR (DMSO‑d 6) δ (ppm): 0.87 (t, J = 7.1 Hz, 3H, CH3), 0.99 (t, J = 7.1 Hz, 3H, CH3), 1.93 (s, 3H, NCH3), 2.12 (d, J = 13.4 Hz, 1H, upfield H of piperidinyl H2C-2″), 2.27 (s, 3H, ArCH3), 2.28 (s, 3H, ArCH3), 3.23–3.29 (m, 2H, upfield H of pyrrolidinyl H2C-5' + upfield H of piperidinyl H2C-6″), 3.50–3.84 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.63 (t, J = 9.2 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 8.4 Hz, 1H, arom. H), 6.85–6.87 (m, 2H, arom. H), 7.01 (d, J = 8.3 Hz, 2H, arom. H), 7.09–7.14 (m, 3H, arom. H), 7.19 (d, J = 8.1 Hz, 2H, arom. H), 7.31 (d, J = 8.3 Hz, 2H, arom. H), 7.39 (s, 1H, olefinic CH), 10.51 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.68, 15.73, 15.8, 15.9 (CH3), 20.7, 20.9 (ArCH3), 33.9 (NCH3), 45.1 (pyrrolidinyl HC-4′), 45.7 (piperidinyl H2C-6″), 47.3 (piperidinyl H2C-2″), 57.2 (pyrrolidinyl H2C-5′), 61.8, 61.90, 61.94, 61.96 [spiro-C-3' (C-3″), + OCH2], 75.1 [spiro-C-3 (C-2′)], 109.1, 120.9, 125.5, 126.8, 128.9, 129.0, 129.2, 129.7, 129.9, 130.7, 130.8, 131.4, 135.1, 136.0, 137.9, 139.3, 143.7 (arom. C + olefinic C), 175.3, 197.2 (C O). Anal. Calcd. for C35H40N3O5P (613.69): C, 68.50; H, 6.57; N, 6.85. Found: C, 68.71; H, 6.70; N, 6.91.
4.1.2.10. Diethyl (E)-[5-chloro-1′-methyl-5''-(4-methylbenzylidene)-2,4″-dioxo-4'-(p-tolyl)dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6j)
Obtained from the reaction of 3f, 4b and 5, reaction time 60 min as pale yellow microcrystals from ethyl acetate, mp 227–228 °C and yield 93% (0.75 g). IR: ν max/cm−1 3175, 3140, 1717, 1682, 1597, 1512, 1238, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 0.86 (t, J = 7.1 Hz, 3H, CH3), 1.02 (t, J = 7.0 Hz, 3H, CH3), 1.95 (s, 3H, NCH3), 2.15 (d, J = 13.6 Hz, 1H, upfield H of piperidinyl H2C-2″), 2.29 (s, 3H, ArCH3), 2.30 (s, 3H, ArCH3), 3.28 (t, J = 8.4 Hz, 1H, upfield H of pyrrolidinyl H2C-5′), 3.47–3.86 (m, 8H, upfield H of piperidinyl H2C-6'' + downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.60 (dd, J = 8.0, 10.1 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 8.3 Hz, 1H, arom. H), 6.80 (d, J = 2.3 Hz, 1H, arom. H), 7.09 (d, J = 7.9 Hz, 2H, arom. H), 7.16 (d, J = 7.8 Hz, 2H, arom. H), 7.18 (dd, J = 2.2, 8.3 Hz, 1H, arom. H), 7.23 (d, J = 7.9 Hz, 2H, arom. H), 7.32 (d, J = 7.7 Hz, 2H, arom. H), 7.38 (s, 1H, olefinic CH), 10.68 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.64, 15.69, 15.84, 15.89 (CH3), 20.7, 20.9 (ArCH3), 34.0 (NCH3), 45.9 (pyrrolidinyl HC-4' + piperidinyl H2C-6″), 47.9 (piperidinyl H2C-2″), 57.5 (pyrrolidinyl H2C-5′), 62.00, 62.02, 62.07, 62.19, 62.27 [spiro-C-3' (C-3″) + OCH2), 75.3 [spiro-C-3 (C-2′)], 110.6, 125.0, 126.7, 127.7, 128.8, 129.0, 129.3, 129.8, 129.9, 130.9, 131.0, 131.2, 134.9, 136.1, 138.1, 139.5, 142.6 (arom. C + olefinic C), 174.9, 197.3 (C O). Anal. Calcd. for C35H39ClN3O5P (648.14): C, 64.86; H, 6.07; N, 6.48. Found: C, 64.97; H, 6.19; N, 6.66.
4.1.2.11. Diethyl (E)-[1′-methyl-2,4″-dioxo-4'-(thiophen-2-yl)-5''-(thiophen-2-ylmethylene)dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6k)
Obtained from the reaction of 3g, 4a and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 206–208 °C and yield 83% (0.62 g). IR: ν max/cm−1 3186, 1709, 1674, 1616, 1570, 1238, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 1.06 (t, J = 7.0 Hz, 3H, CH3), 1.10 (t, J = 7.0 Hz, 3H, CH3), 1.92 (s, 3H, NCH3), 2.40 (d, J = 13.4 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.28 (d, J = 16.1 Hz, 1H, upfield H of piperidinyl H2C-6″) 3.39 (t, J = 8.4 Hz, 1H, upfield H of pyrrolidinyl H2C-5′), 3.64–3.97 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.89 (dd, J = 8.1, 9.9 Hz, 1H, pyrrolidinyl H-4′), 6.68–6.73 (m, 2H, arom. H), 6.80 (dd, J = 1.4, 7.7 Hz, 1H, arom. H), 7.02 (dd, J = 3.4, 5.2 Hz, 1H, arom. H), 7.08 (dt, J = 1.4, 7.6 Hz, 1H, arom. H), 7.12 (dd, J = 1.2, 3.6 Hz, 1H, arom. H), 7.24 (dd, J = 3.7, 5.1 Hz, 1H, arom. H), 7.41 (dd, J = 1.2, 5.2 Hz, 1H, arom. H), 7.54 (d, J = 3.7 Hz, 1H, arom. H), 7.83 (d, J = 2.0 Hz, 1H, olefinic CH), 7.91 (d, J = 5.1 Hz, 1H, arom. H), 10.56 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.93, 15.97, 15.99, 16.01 (CH3), 33.7 (NCH3), 40.7 (pyrrolidinyl HC-4′), 45.4 (piperidinyl H2C-6″), 46.0 (piperidinyl H2C-2″), 58.3 (pyrrolidinyl H2C-5′), 61.06, 61.14, 61.95, 62.0, 62.05, 62.09 [spiro-C-3' (C-3″) + OCH2], 75.1 [spiro-C-3 (C-2′)], 109.2, 120.7, 125.0, 125.1, 126.4, 126.76, 126.85, 127.07, 127.2, 128.4, 129.0, 130.3, 133.1, 135.2, 137.1, 140.9, 143.8 (arom. C + olefinic C), 175.0, 195.6 (C O). Anal. Calcd. for C29H32N3O5PS2 (597.68): C, 58.28; H, 5.40; N, 7.03. Found: C, 58.42; H, 5.46; N, 6.84.
4.1.2.12. Diethyl (E)-[5-chloro-1′-methyl-2,4″-dioxo-4'-(thiophen-2-yl)-5''-(thiophen-2-ylmethylene)dispiro[indoline-3,2′-pyrrolidine-3′,3″-piperidin]-1″-yl]phosphonate (6l)
Obtained from the reaction of 3g, 4b and 5, reaction time 60 min as pale yellow microcrystals from methanol, mp 223–225 °C and yield 80% (0.63 g). IR: ν max/cm−1 3179, 3140, 1717, 1678, 1616, 1578, 1242, 1026. 1H NMR (DMSO‑d 6) δ (ppm): 1.06 (t, J = 7.0 Hz, 3H, CH3), 1.10 (t, J = 7.0 Hz, 3H, CH3), 1.93 (s, 3H, NCH3), 2.42 (d, J = 13.6 Hz, 1H, upfield H of piperidinyl H2C-2″), 3.41 (t, J = 8.4 Hz, 1H, upfield H of pyrrolidinyl H2C-5′) 3.45 (d, J = 15.3 Hz, 1H, upfield H of piperidinyl H2C-6″), 3.64–3.95 (m, 7H, downfield H of pyrrolidinyl H2C-5' + downfield H of piperidinyl H2C-2'' + downfield H of piperidinyl H2C-6'' + 2 OCH2), 4.85 (t, J = 9.0 Hz, 1H, pyrrolidinyl H-4′), 6.67 (d, J = 8.3 Hz, 1H, arom. H), 6.72 (d, J = 2.3 Hz, 1H, arom. H), 7.01 (dd, J = 3.4, 5.1 Hz, 1H, arom. H), 7.11 (d, J = 3.5 Hz, 1H, arom. H), 7.13 (dd, J = 2.3, 8.3 Hz, 1H, arom. H), 7.25 (dd, J = 3.7, 5.1 Hz, 1H, arom. H), 7.41 (dd, J = 1.3, 5.1 Hz, 1H, arom. H), 7.54 (d, J = 3.3 Hz, 1H, arom. H), 7.82 (s, 1H, olefinic CH), 7.93 (d, J = 5.1 Hz, 1H, arom. H), 10.70 (s, 1H, NH). 13C NMR (DMSO‑d 6) δ (ppm): 15.94, 15.98, 15.99, 16.02 (CH3), 33.8 (NCH3), 40.7 (piperidinyl H2C-2″), 45.8 (pyrrolidinyl HC-4′), 46.3 (piperidinyl H2C-6″), 58.6 (pyrrolidinyl H2C-5′), 61.4, 61.5 [spiro-C-3' (C-3″)], 62.03, 62.08, 62.15, 62.20 (OCH2), 75.2 [spiro-C-3 (C-2′)], 110.6, 124.96, 125.1, 126.5, 126.86, 126.95, 127.1, 127.2, 127.3, 128.5, 128.8, 130.5, 133.3, 135.2, 137.0, 140.7, 142.7 (arom. C + olefinic C), 174.7, 195.6 (C O). Anal. Calcd. for C29H31ClN3O5PS2 (632.13): C, 55.10; H, 4.94; N, 6.65. Found: C, 55.29; H, 4.99; N, 6.72.
4.2. X-ray, biological and computational studies
Described in the supplementary information.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported financially by National Research Centre, Egypt, project ID: 13060103.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2023.115563.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
No data was used for the research described in the article.
References
- 1.Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19) A Review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. https://doi:10.1001/jama.2020.6019 [DOI] [PubMed] [Google Scholar]
- 2.Zheng L., Zhang L., Huang J., Nandakumar K.S., Liu S., Cheng K. Potential treatment methods targeting 2019-nCoV infection. Eur. J. Med. Chem. 2020;205 doi: 10.1016/j.ejmech.2020.112687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buonaguro L., Tagliamonte M., Tornesello M.L., Buonaguro F.M. SARS-CoV-2 RNA polymerase as target for antiviral therapy. J. Transl. Med. 2020;18:185. doi: 10.1186/s12967-020-02355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durmuş S., Ülgen K.Ö. Comparative interactomics for virus-human protein-protein interactions: DNA viruses versus RNA viruses. FEBS Open Bio. 2017;7:96–107. doi: 10.1002/2211-5463.12167. https://doi:10.1002/2211-5463.12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. RNA-dependent RNA polymerase as a target for COVID-19 drug discovery. SLAS Discovery. 2020;25:1141–1151. doi: 10.1177/24725552209421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I., Malik Y.S., Sah R., Rabaan A.A., Panwar P.K., Singh K.P., Michalak I., Chaicumpa W., Martinez-Pulgarin D.F., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Trav. Med. Infect. Dis. 2020;37 doi: 10.1016/j.tmaid.2020.101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.https://covid19.who.int/
- 9.Jayabal K., Elumalai D., Leelakrishnan S., Bhattacharya S., Rengarajan V., Kannan T., Chuang S.-C. Green and regioselective approach for the synthesis of 3-substituted indole based 1,2-dihydropyridine and azaxanthone derivatives as a potential lead for SARS-CoV-2 and delta plus mutant virus: DFT and docking studies. ACS Omega. 2022;7:43856–43876. doi: 10.1021/acsomega.2c04990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barhoumi T., Alghanem B., Shaibah H., Mansour F.A., Alamri H.S., Akiel M.A., Alroqi F., Boudjelal M. SARS-CoV-2 coronavirus spike protein-induced apoptosis, inflammatory, and oxidative stress responses in THP-1-like-macrophages: potential role of angiotensin-converting enzyme inhibitor (perindopril) Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.728896. https://doi:10.3389/fimmu.2021.728896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.https://www.who.int/activities/tracking-SARS-CoV-2-variants/
- 12.https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
- 13.Araf Y., Akter F., Tang Y.‐d., Fatemi R., Parvez S.A., Zheng C., Hossain G. Omicron variant of SARS‐CoV‐2: genomics, transmissibility, and responses to current COVID‐19 vaccines. J. Med. Virol. 2022;94:1825–1832. doi: 10.1002/jmv.27588. https://doi:10.1002/jmv.27588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yapasert R., Khaw-on P., Banjerdpongchai R. Coronavirus infection-associated cell death signaling and potential therapeutic targets. Molecules. 2021;26:7459. doi: 10.3390/molecules26247459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohseni M., Bahrami H., Farajmand B., Hosseini F.S., Amanlou M., Salehabadi H. Indole alkaloids as potential candidates against COVID-19: an in silico study. J. Mol. Model. 2022;28:144. doi: 10.1007/s00894-022-05137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj V., Lee J.-H., Shim J.-J., Lee J. Antiviral activities of 4H-chromen-4-one scaffold-containing flavonoids against SARS–CoV–2 using computational and in vitro approaches. J. Mol. Liquids. 2022;353 doi: 10.1016/j.molliq.2022.118775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.https://www.drugs.com/history/paxlovid.html
- 18.https://www.drugs.com/history/paxlovid.html
- 19.https://go.drugbank.com/unearth/q?utf8=%E2%9C%93&searcher=drugs&query=Paxlovid
- 20.Agost-Beltrán L., de la Hoz-Rodríguez S., Bou-Iserte L., Rodríguez S., Fernández-de-la-Pradilla A., González F.V. Advances in the development of SARS-CoV-2 Mpro inhibitors. Molecules. 2022;27:2523. doi: 10.3390/molecules27082523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chauhan J., Cecon E., Labani N., Gbahou F., Real F., Bomsel M., Dubey K.D., Das R., Dam J., Jockers R., Sen S. Development of indolealkylamine derivatives as potential multi-target agents for COVID-19 treatment. Eur. J. Med. Chem. 2023;249 doi: 10.1016/j.ejmech.2023.115152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori S.-i., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., Das D., Anson B.J., Lendy E.K., Takamatsu Y., Takamune N., Kishimoto N., Murayama K., Hasegawa K., Li M., Davis D.A., Kodama E.N., Yarchoan R., Wlodawer A., Misumi S., Mesecar A.D., Ghosh A.K., Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Com. 2021;12:668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerjee R., Perera L., Tillekeratne L.M.V. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today. 2021;26:804–816. doi: 10.1016/j.drudis.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almaraz-Girón M.A., Calderón-Jaimes E., Carrillo A.S., Díaz-Cervantes E., Alonso E.C., Islas-Jácome A., Domínguez-Ortiz A., Castañón-Alonso S.L. Search for non-protein protease inhibitors constituted with an indole and acetylene core. Molecules. 2021;26:3817. doi: 10.3390/molecules26133817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hassam M., Bashir M.A., Shafi S., Zahra N.-u.-A., Khan K., Jalal K., Siddiqui H., Uddin R. Identification of potent compounds against SARs-CoV-2: an in-silico based drug searching against Mpro. Comput. Biol. Med. 2022;151 doi: 10.1016/j.compbiomed.2022.106284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., Prashar V., Gupta G.D., Panicker L., Kumar M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He M., Huang Y., Wang Y., Liu J., Han M., Xiao Y., Zhang N., Gui H., Qiu H., Cao L., Jia W., Huang S. Metabolomics-based investigation of SARS-CoV-2 vaccination (Sinovac) reveals an immune-dependent metabolite biomarker. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.954801. https://doi:10.3389/fimmu.2022.954801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mishra S.K., Tripathi T. One year update on the COVID-19 pandemic: where are we now? Acta Trop. 2021;214 doi: 10.1016/j.actatropica.2020.105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A.K., Raghavaiah J., Shahabi D., Yadav M., Anson B.J., Lendy E.K., Hattori S.-i., Higashi-Kuwata N., Mitsuya H., Mesecar A.D. Indole chloropyridinyl ester-derived SARS-CoV-2 3CLpro Inhibitors: enzyme inhibition, antiviral efficacy, structure‒activity relationship, and X-ray structural studies. J. Med. Chem. 2021;64:14702–14714. doi: 10.1021/acs.jmedchem.1c01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trivedi N., Verma A., Kumar D. Possible treatment and strategies for COVID-19: review and assessment. Eur. Rev. Med. Pharmacol. Sci. 2020;24:12593–12608. doi: 10.26355/eurrev_202012_24057. [DOI] [PubMed] [Google Scholar]
- 31.Chen H., Cheng F., Li J. iDrug: integration of drug repositioning and drug-target prediction via cross-network embedding. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aktas A., Tuzun B., Kafa A.H.T., Sayin K., Ataseven H. How do arbidol and its analogs inhibit the SARS-CoV-2? Bratislava Med. J. 2020;121:705–711. doi: 10.4149/BLL_2020_115. https://doi:10.4149/BLL_2020_115 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka H., Miyagi S., Yoshida Y., Lamb J.S., Chick C.N., Luhata L.P., Shibata M., Tanaka E., Suzuki Y., Usuki T. Synthesis and biological evaluation of umifenovir analogues as Anti-SARS-CoV-2 agents. ChemistrySelect. 2022;7 doi: 10.1002/slct.202202097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81 doi: 10.1016/j.jinf.2020.03.060. e21–e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng L., Li C., Zeng Q., Liu X., Li X., Zhang H., Hong Z., Xia J. Arbidol combined with LPV/r versus LPV/r alone against corona virus disease 2019: a retrospective cohort study. J. Infect. 2020;81 doi: 10.1016/j.jinf.2020.03.002. e1–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Xie Z., Lin W., Cai W., Wen C., Guan Y., Mo X., Wang J., Wang Y., Peng P., Chen X., Hong W., Xiao G., Liu J., Zhang L., Hu F., Li F., Zhang F., Deng X., Li L. Efficacy and safety of Lopinavir/Ritonavir or Arbidol in adult patients with mild/moderate COVID-19: an exploratory randomized controlled trial. Méd. 2020;1:105–113. doi: 10.1016/j.medj.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu P., Huang J., Fan Z., Huang W., Qi M., Lin X., Song W., Yi L. Arbidol/IFN-α2b therapy for patients with corona virus disease 2019: a retrospective multicenter cohort study. Microb. Infect. 2020;22:200–205. doi: 10.1016/j.micinf.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei S., Xu S., Pan Y.-H. Efficacy of arbidol in COVID-19 patients: a retrospective study. World J. Clin. Cases. 2021;9:7350–7357. doi: 10.12998/wjcc.v9.i25.7350. https://doi:10.12998/wjcc.v9.i25.7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao B., Le-Trilling V.T.K., Wang K., Mennerich D., Hu J., Zhao Z., Zheng J., Deng Y., Katschinski B., Xu S., Zhang G., Cai X., Hu Y., Wang J., Lu M., Huang A., Tang N., Trilling M., Lin Y. Obatoclax inhibits SARS-CoV-2 entry by altered endosomal acidification and impaired cathepsin and furin activity in vitro. Emerg. Microb. Infect. 2022;11:483–497. doi: 10.1080/22221751.2022.2026739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiter R.J., Sharma R., Simko F., Dominguez-Rodriguez A., Tesarik J., Neel R.L., Slominski A.T., Kleszczynski K., Martin-Gimenez V.M., Manucha W., Cardinali D.P. Melatonin: highlighting its use as a potential treatment for SARS-CoV-2 infection. Cell. Mol. Life Sci. 2022;79:143. doi: 10.1007/s00018-021-04102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R., Wang X., Ni L., Di X., Ma B., Niu S., Liu C., Reiter R.J. COVID-19: melatonin as a potential adjuvant treatment. Life Sci. 2020;250 doi: 10.1016/j.lfs.2020.117583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cecon E., Fernandois D., Renault N., Coelho C.F.F., Wenzel J., Bedart C., Izabelle C., Gallet S., Le Poder S., Klonjkowski B., Schwaninger M., Prevot V., Dam J., Jockers R. Melatonin drugs inhibit SARS-CoV-2 entry into the brain and virus-induced damage of cerebral small vessels. Cell. Mol. Life Sci. 2022;79:361. doi: 10.1007/s00018-022-04390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehrzadi S., Karimi M.Y., Fatemi A., Reiter R.J., Hosseinzadeh A. SARS-CoV-2 and other coronaviruses negatively influence mitochondrial quality control: beneficial effects of melatonin. Pharmacol. Ther. 2021;224 doi: 10.1016/j.pharmthera.2021.107825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mousavi S.A., Heydari K., Mehravaran H., Saeedi M., Alizadeh‐Navaei R., Hedayatizadeh‐Omran A., Shamshirian A. Melatonin effects on sleep quality and outcomes ofCOVID‐19 patients: an open‐label, randomized, controlled trial. J. Med. Virol. 2022;94:263–271. doi: 10.1002/jmv.27312. https://doi:10.1002/jmv.27312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasan Z.T., Al Atrakji M.Q.Y.M.A., Mehuaiden A.K. The effect of melatonin on thrombosis, sepsis and mortality rate in COVID-19 patients. Int. J. Infect. Dis. 2022;114:79–84. doi: 10.1016/j.ijid.2021.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rommasi F., Nasiri M.J., Mirsaeidi M. Immunomodulatory agents for COVID-19 treatment: possible mechanism of action and immunopathology features. Mol. Cell. Biochem. 2022;477:711–726. doi: 10.1007/s11010-021-04325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Begum R., Mamun-Or-Rashid A.N.M., Lucy T.T., Pramanik K., Sil B.K., Mukerjee N., Tagde P., Yagi M., Yonei Y. Potential therapeutic approach of melatonin against omicron and some other variants of SARS-CoV-2. Molecules. 2022;27:6934. doi: 10.3390/molecules27206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fawazy N.G., Panda S.S., Mostafa A., Kariuki B.M., Bekheit M.S., Moatasim Y., Kutkat O., Fayad W., El-Manawaty M.A., Soliman A.A.F., El-Shiekh R.A., Srour A.M., Barghash R.F., Girgis A.S. Development of spiro-3-indolin-2-one containing compounds of antiproliferative and ant-SARS-CoV-2 properties. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-17883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Girgis A.S., Panda S.S., Srour A.M., Abdelnaser A., Nasr S., Moatasim Y., Kutkat O., El Taweel A., Kandeil A., Mostafa A., Ali M.A., Fawzy N.G., Bekheit M.S., Shalaby E.M., Gigli L., Fayad W., Soliman A.A.F. 3-Alkenyl-2-oxindoles: synthesis, antiproliferative and antiviral properties against SARS-CoV-2. Bioorg. Chem. 2021;114 doi: 10.1016/j.bioorg.2021.105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Modranka J., Drogosz-Stachowicz J., Pietrzak A., Janecka A., Janecki T. Synthesis and structure-activity relationship study of novel 3-diethoxyphosphorylfuroquinoline-4,9-diones with potent antitumor efficacy. Eur. J. Med. Chem. 2021;219 doi: 10.1016/j.ejmech.2021.113429. [DOI] [PubMed] [Google Scholar]
- 51.Babushkina A.A., Dogadina A.V., Egorov D.M., Piterskaia J.L., Shtro A.A., Nikolaeva Y.V., Galochkina A.V., Kornev A.A., Boitsov V.M. Efficient synthesis and evaluation of antiviral and antitumor activity of novel 3-phosphonylated thiazolo[3,2-a]oxopyrimidines. Med. Chem. Res. 2021;30:2203–2215. doi: 10.1007/s00044-021-02801-x. [DOI] [Google Scholar]
- 52.Bessières M., Plebanek E., Chatterjee P., Shrivastava-Ranjan P., Flint M., Spiropoulou C.F., Warszycki D., Bojarski A.J., Roy V., Agrofoglio L.A. Design, synthesis and biological evaluation of 2-substituted-6-[(4-substituted-1-piperidyl)methyl]-1H-benzimidazoles as inhibitors of ebola virus infection. Eur. J. Med. Chem. 2021;214 doi: 10.1016/j.ejmech.2021.113211. [DOI] [PubMed] [Google Scholar]
- 53.https://www.drugs.com/mtm/etidronate.html
- 54.https://www.drugs.com/mtm/pamidronate.html
- 55.https://www.drugs.com/mtm/alendronate.html
- 56.https://www.drugs.com/mtm/zoledronic-acid.html
- 57.https://www.drugs.com/cdi/ibandronate-tablets.html
- 58.https://www.drugs.com/mtm/amifostine.html
- 59.https://www.cancer.gov/about-cancer/treatment/drugs/amifostine
- 60.Youssef M.A., Panda S.S., Aboshouk D.R., Said M.F., El Taweel A., GabAllah M., Fayad W., Soliman A.A.F., Mostafa A., Fawzy N.G., Girgis A.S. Novel curcumin mimics: design, synthesis, biological properties and computational studies of piperidone-piperazine conjugates. ChemistrySelect. 2022;7 doi: 10.1002/slct.202201406. [DOI] [Google Scholar]
- 61.Srour A.M., Panda S.S., Mostafa A., Fayad W., El-Manawaty M.A., Soliman A.A.F., Moatasim Y., El Taweel A., Abdelhameed M.F., Bekheit M.S., Ali M.A., Girgis A.S. Synthesis of aspirin-curcumin mimic conjugates of potential antitumor and anti-SARS-CoV-2 properties. Bioorg. Chem. 2021;117 doi: 10.1016/j.bioorg.2021.105466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fawzy N.G., Panda S.S., Fayad W., Shalaby E.M., Srour A.M., Girgis A.S. Synthesis, human topoisomerase IIα inhibitory properties and molecular modeling studies of anti-proliferative curcumin mimics. RSC Adv. 2019;9:33761–33774. doi: 10.1039/c9ra05661k. https://doi:10.1039/c9ra05661k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fawzy N.G., Panda S.S., Fayad W., El-Manawaty M.A., Srour A.M., Girgis A.S. Novel curcumin inspired antineoplastic 1-sulfonyl-4-piperidones: design, synthesis and molecular modeling studies. Anti Cancer Agents Med. Chem. 2019;19:1069–1078. doi: 10.2174/1871520619666190408131639. https://doi:10.2174/1871520619666190408131639 [DOI] [PubMed] [Google Scholar]
- 64.Wyman K.A., Girgis A.S., Surapaneni P.S., Moore J.M., Abo Shama N.M., Mahmoud S.H., Mostafa A., Barghash R.F., Juan Z., Dobaria R.D., Almalki A.J., Ibrahim T.S., Panda S.S. Synthesis of potential antiviral agents for SARS-CoV-2 using molecular hybridization approach. Molecules. 2022;27:5923. doi: 10.3390/molecules27185923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seliem I.A., Girgis A.S., Moatasim Y., Kandeil A., Mostafa A., Ali M.A., Bekheit M.S., Panda S.S. New pyrazine conjugates: synthesis, computational studies, and antiviral properties against SARS-CoV-2. ChemMedChem. 2021;16:3418–3427. doi: 10.1002/cmdc.202100476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seliem I.A., Panda S.S., Girgis A.S., Moatasim Y., Kandeil A., Mostafa A., Ali M.A., Nossier E.S., Rasslan F., Srour A.M., Sakhuja R., Ibrahim T.S., Abdel-samii Z.K.M., Al-Mahmoudy A.M.M. New quinoline-triazole conjugates: synthesis, and antiviral properties against SARS-CoV-2. Bioorg. Chem. 2021;114 doi: 10.1016/j.bioorg.2021.105117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lokhande K.B., Doiphode S., Vyas R., Swamy K.V. Molecular docking and simulation studies on SARS-CoV-2 Mpro reveals Mitoxantrone, Leucovorin, Birinapant, and Dynasore as potent drugs against COVID-19. J. Biomol. Struct. Dyn. 2021;39:7294–7305. doi: 10.1080/07391102.2020.1805019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.3CL Protease (SARS-CoV-2) Assay Kit, BPS Bioscience, Catalog #79955-1, 6042 Cornerstone Court West, Ste. B, San Diego CA 92121, San Diego CA 92121. www.bpsbioscience.com.
- 69.https://pubmed.ncbi.nlm.nih.gov/31643708/
- 70.Plosker G.L., Figgitt D.P. Tipranavir, Drugs. 2003;63:1611–1618. doi: 10.2165/00003495-200363150-00009. https://doi:10.2165/00003495-200363150-00009 [DOI] [PubMed] [Google Scholar]
- 71.Orman J.S., Perry C.M. Tipranavir: a review of its use in the management of HIV infection. Drugs. 2008;68:1435–1463. doi: 10.2165/00003495-200868100-00006. https://doi:10.2165/00003495-200868100-00006 [DOI] [PubMed] [Google Scholar]
- 72.Girgis A.S., Panda S.S., Aziz M.N., Steel P.J., Hall C.D., Katritzky A.R. Rational design, synthesis, and 2D-QSAR study of anti-oncological alkaloids against hepatoma and cervical carcinoma. RSC Adv. 2015;5:28554–28569. https://doi:10.1039/c4ra16663a [Google Scholar]
- 73.Nofal Z.M., Srour A.M., El-Eraky W.I., Saleh D.O., Girgis A.S. Rational design, synthesis and QSAR study of vasorelaxant active 3-pyridinecarbonitriles incorporating 1H-benzimidazol-2-yl function. Eur. J. Med. Chem. 2013;63:14–21. doi: 10.1016/j.ejmech.2013.01.042. [DOI] [PubMed] [Google Scholar]
- 74.https://www.rcsb.org/structure/7C8U
- 75.Das S., Das U., Selvakumar P., Sharma R.K., Balzarini J., De Clercq E., Molnár J., Serly J., Baráth Z., Schatte G., Bandy B., Gorecki D.K.J., Dimmock J.R. 3,5-Bis(benzylidene)-4-oxo-1-phosphonopiperidines and related diethyl esters: potent cytotoxins with multi-drug-resistance reverting properties. ChemMedChem. 2009;4:1831–1840. doi: 10.1002/cmdc.200900288. https://doi:10.1002/cmdc.200900288 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.