Abstract
Scale invariance is a characteristic of neural activity. How this property emerges from neural interactions remains a fundamental question. Here, we studied the relation between scale-invariant brain dynamics and structural connectivity by analyzing human resting-state (rs-) fMRI signals, together with diffusion MRI (dMRI) connectivity and its approximation as an exponentially decaying function of the distance between brain regions. We analyzed the rs-fMRI dynamics using functional connectivity and a recently proposed phenomenological renormalization group (PRG) method that tracks the change of collective activity after successive coarse-graining at different scales. We found that brain dynamics display power-law correlations and power-law scaling as a function of PRG coarse-graining based on functional or structural connectivity. Moreover, we modeled the brain activity using a network of spins interacting through large-scale connectivity and presenting a phase transition between ordered and disordered phases. Within this simple model, we found that the observed scaling features were likely to emerge from critical dynamics and connections exponentially decaying with distance. In conclusion, our study tests the PRG method using large-scale brain activity and theoretical models and suggests that scaling of rs-fMRI activity relates to criticality.
Subject terms: Network models, Neural circuits
Human fMRI and dMRI data are used alongside modelling to test the phenomenological renormalization group (PRG) method, consequentially suggesting that scaling of rsfMRI activity emerges from criticality and exponentially decaying connectivity.
Introduction
Interesting phenomena in biological systems are often collective behaviors emerging from the interactions among their constituents. Growing evidence indicates that large-scale spontaneous brain activity is an emergent phenomenon continuously generating patterned activity at multiple spatiotemporal scales1–3. How patterned activity arise from the brain’s connectivity is a fundamental question in neuroscience. A hallmark of spontaneous, or resting-state (rs), whole-brain activity is scale invariance. Indeed, scale-free (power-law) power spectrum has been reported in human brain dynamics acquired using fMRI4,5, electrocorticography6, LFPs7, and MEG8. Moreover, scale-invariance of propagating clusters of activity (neuronal avalanches) has been observed in human fMRI recordings9, M/EEG fluctuations10,11, and zebrafish whole-brain calcium imaging12. These works add up to a large body of studies showing scale-invariant neuronal avalanches at the microcircuit level13–18.
In physical systems, scale-invariance is observed at critical points. Thus, the observation of power-law statistics in neural activity has contributed to support the idea that spontaneous neural activity operates close to a phase transition19. Several studies have shown that critical neural systems maximize information transmission, storage, and processing12,20–22. Interestingly, it has been shown that brain activity deviates from critical-like behavior in different brain states and neuropsychiatric disorders23–29. For instance, using fMRI30,31 and voltage imaging32, it has been shown that awake resting state displays critical-like dynamics, yielding maximal information capacity and transmission, while anesthesia states depart from criticality. For these reasons it is believed that critical dynamics are a characteristic of healthy, awake spontaneous neural activity.
Because the critical behavior of a physical system is governed by fluctuations that are statistically self-similar, its statistics are re-scaled after gradual elimination of the correlated degrees of freedom. This is achieved through the Renormalization Group (RG) procedure. This method tracks the change of the joint probability distribution of the system variables after successive coarse-graining at different scales. In the case of critical systems at equilibrium, probability distributions are scale-invariant under iterated coarse-graining and represent fixed points of the RG. In the case of neural activity, to account for the (unknown) topology of interactions, a phenomenological renormalization group (PRG) procedure has been proposed in which maximally correlated variables are grouped together33, instead of locally grouping variables that are spatially close as in most applications of RG. This method successfully revealed scaling features in local single-neuron recordings from the mouse hippocampus33,34 and, recently, in other areas of the mouse brain35. The method has been previously tested on theoretical models36, but it remains to be tested at the large scale and combining theoretical models and data.
Moreover, how the scaling properties of brain dynamics relate to structural principles remains unclear. Previous investigations using retrograde tract tracing methods in mice and nonhuman primates’ cortices37–39 have shown that the probability of a connection existing between two given cortical areas declines with distance. An exponential decay with interareal distance, known as exponential distance rule (EDR), has been proposed as a simple, geometrically-constraint wiring principle38–41. Nevertheless, a recent study using fluorescent mapping of neuronal projections found that a power law decay was a marginally better fit than exponential decay42. Special attention has been paid to long-range connections that deviate from simple connection decay as a function of distance, in terms of their impact on wiring-cost43 and on dynamics of oscillator models44,45.
Here, we studied how scale-invariance of brain dynamics relates to structural connectivity. For this, we examined the relation between human rs-fMRI signals and diffusion MRI (dMRI) structural connectivity, focusing on spatial anatomical constrains. Next, we studied the scaling properties of rs-fMRI dynamics using the correlation function and the PRG method. Finally, using a simple spin model with a geometrically-constraint connectivity, we showed that scaling in rs-fMRI signals is suggestive of critical behavior.
Results
Linear prediction of fMRI signals from structural connections
We analyzed fMRI signals in a parcellation of = 1000 regions of interest (ROIs) or nodes. The dataset was composed of 1003 individual scans of 1200 time frames. Structural connectivity was obtained using dMRI and probabilistic tractography in the same parcellation, resulting in a coupling matrix (see “Methods”). The data was obtained from the Human Connectome Project (HCP) public database. In this section, we studied the relation between fMRI dynamics and structural couplings.
We first note that the relation between the connectivity weight between two nodes and the Euclidean spatial distance between those nodes was approximately a power law (Fig. 1a), indicating the presence of long-range connections. To evaluate the importance of long-range connections, we compared the dMRI connectivity to a model connectivity based on EDR, i.e., , where , and and are the positions in 3D space of the centers of ROIs and , respectively. Using least squares and for distances <50 mm, we obtained: 0.106 ± 0.007 mm−1. Notably, the value of is consistent with the extrapolation to human brain based on the relation between the EDR and the brain volume39. We noted that the EDR fit was higher for intra-hemispheric connections than for the long-range inter-hemispheric ones (Fig. 1b): the correlation between EDR and dMRI connectivities was equal to 0.69 and 0.49 for intra- and inter-hemispheric connections, respectively. By construction, dMRI and EDR connectivity matrices are symmetric.
Fig. 1. Linear prediction of fMRI signals using dMRI and EDR coupling matrices.
a dMRI weights as a function of the distance between pairs of nodes (blue) and the exponential approximation (red). The thickness of the blue trace indicates SEM. b Large-scale connectivity matrices: dMRI (left), EDR (right). Connectivity weights are presented in logarithmic scale. Portions of the matrices corresponding to intra- and inter-hemispheric connections are highlighted in red and blue, respectively. c Three example fMRI signals and their corresponding linear predictions using the EDR connectivity, i.e., . The correlation between the actual and the predicted signals is also indicated. d Distributions of signal predictions using the dMRI connectivity (blue), the EDR (red), and a shuffled connectivity that preserves the distribution of dMRI weights but destroys the spatial organization (gray) (n = 1000 × 1003). e Average signal prediction as a function of the strength of the nodes of the EDR connectivity. Each dot represents a node (n = 1000); the blue trace represents the average relation. Error bars indicate SD.
We next tested the linear signal prediction of both connectivity matrices. Let be the data matrix containing the fMRI signals. Assuming linear couplings, we calculated the predicted signals as (i.e., the prediction of each signal given the rest of the network and couplings ). The goodness of the linear prediction was given by the Pearson correlation between and for all nodes and all subjects. Correlation coefficients were remarkably similar for the dMRI and the EDR, with means equal to 0.51 ± 0.01 and 0.49 ± 0.01, respectively (Fig. 1c, d). These average values were significantly higher (p < 0.001, Welch’s t-test) than the one obtained using a shuffled connectivity that preserves the distribution of dMRI weights but destroys their spatial organization (mean correlation: 0.32 ± 0.01). Consistently, we noted that nodes for which the linear prediction was the lowest were those nodes that were weakly connected to the network, i.e., nodes with low node strength, where the strength of node is the sum of the weights of its connections with other nodes in the network, i.e., (Fig. 1e). Finally, we also tested (i) the linear prediction of each signal given the rest of signals from the same hemisphere and intra-hemispheric couplings and (ii) the linear prediction of each signal given the signals from the contralateral hemisphere and inter-hemispheric couplings (Supplementary Fig. S1). We found that, although inter-hemispheric predictions were reduced with respect to intra-hemispheric ones, they remained significant and were practically indistinguishable using the dMRI and the EDR connectivity matrices. Altogether, we concluded that both the dMRI and the EDR connectivity matrices were equally good linear predictors of the fMRI signals.
Correlation function and phenomenological renormalization-group
We next evaluated the relation between functional correlations of pairs of ROIs and their distance. As required for the modelling in the next section, we used binarized fMRI signals (see “Methods”). Briefly, for each scan, the z-scored time-series of each ROI, (), was binarized by imposing a threshold . Thus, at each time frame , the collective activity was described by a binary vector , with 1 if and otherwise. Binarization of time-series has proven to effectively capture and compress fMRI large-scale collective dynamics9,31,46.
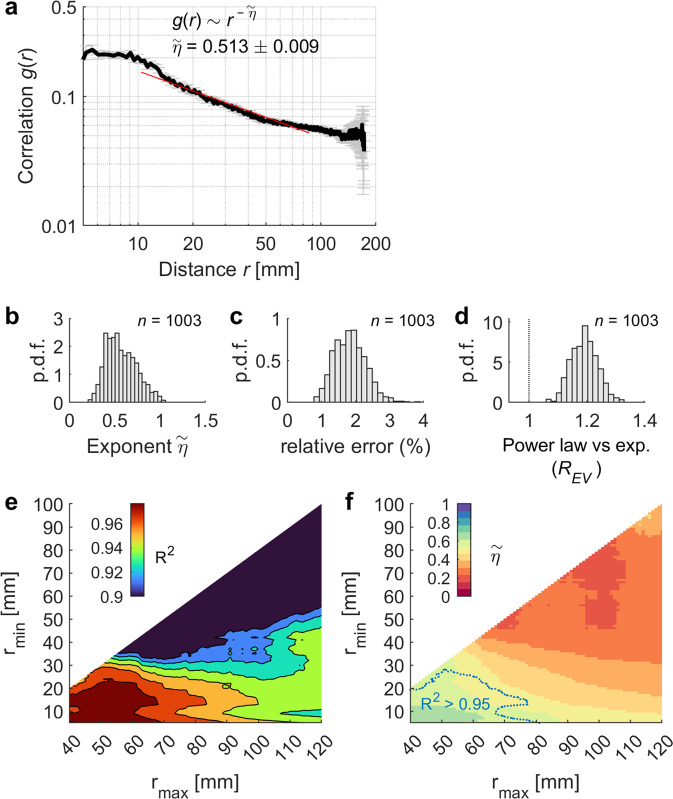
Consistent with previous work47, we found that the average functional correlation, across all ROI pairs and all subjects, was approximately power-law, i.e., , with a power exponent equal to 0.513 ± 0.009 (Fig. 2a–c). For each individual scan, we tested the power-law hypothesis against an exponential alternative by calculating the ratio between explained variances () of least-squares fits using the two competing regression models. We found a ratio of 1.2 of explained variances systematically in favor of the power-law hypothesis (Fig. 2d). The exact value of the exponent depends on the cutoffs used to constrain the power-law fitting, but for a large region in the plane we found a good fitting of the power law (explained variance 0.95) and an exponent around 0.52 (Fig. 2e, f).
Fig. 2. Correlation function.
a Correlation function of fMRI signals as a function of the distance between nodes. Error bars indicate SEM. The correlation function of fMRI signals was approximately power-law, i.e., . The power law was fitted in the distance interval mm. b Distribution of the estimated power exponent for single-subject scans (n = 1003). c Distribution of the relative estimation error of exponent , i.e., , where is the least square estimation error of exponent . Note that the average relative estimation error is <3%. d The power law fit was compared the one obtained using an exponential function by calculating the ratio between the explained variance of the competing regression models. Ratios larger than 1 favor the power law hypothesis. e, f When fitting the power law to in the distance interval , for several combinations of and , we found a large region in the plane with high explained variance (e) yielding power exponents 0.52 (f, the blue dotted line indicates the region for which 0.95).
Power-law correlations are a hallmark of critical systems, but neither a necessary nor a sufficient condition. Recently, a PRG approach has been proposed to identify scale-invariant activity in neural systems33. Within this method, the collective activity is iteratively coarse-grained by grouping maximally correlated variables (see “Methods”). At each coarse-graining step , clusters of size are built, resulting in a system of coarse-grained variables and successively ignoring degrees of freedom. We calculated several observables of the coarse-grained variables and studied their evolution as a function of .
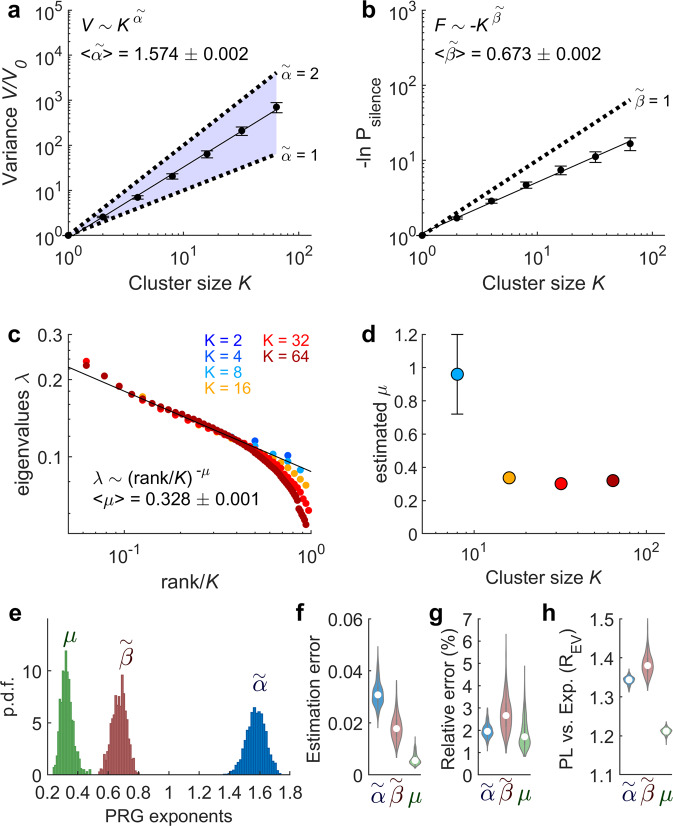
We found that the variance of coarse-grained variables scaled as a power of the cluster size, i.e., , with an average exponent 1.574 ± 0.002 across subjects (Fig. 3a). This exponent lies in the region between linear ( 1) and quadratic growth ( 2), corresponding to uncorrelated and fully-correlated systems, respectively. The distribution of the value of the exponents from individual scans is shown in Fig. 3e. Individual exponents had low uncertainty: on average, the exponent estimation error was equal to 0.03 (Fig. 3f), corresponding to an average relative estimation error equal to 1.99% (Fig. 3g). Moreover, the ratio between explained variances of least-square fits of using a power law vs. an exponential distribution systematically favored the power-law hypothesis ( 1.34; Fig. 3h).
Fig. 3. Phenomenological renormalization-group.
a Variance of coarse-grained variables as a function of cluster size , average over subjects (black points; error bars indicate SD over subjects, n = 1003). The solid black line indicates least squares power law fit, i.e. . Dashed lines indicate linear ( 1) and quadratic ( 2) growths, corresponding to uncorrelated and fully correlated systems, respectively. indicates the average exponent across subjects. b Silence log-probability, , of coarse-grained variables as a function of cluster size, average over subjects (black points; error bars indicate SD over subjects, n = 1003). The solid black line indicates least squares power law fit, i.e. . The dashed line indicates the prediction for uncorrelated variables ( 1). In (a) and (b), the variance and the silence log-probability were normalized by their corresponding values at coarse-graining step (original system). indicates the average exponent across subjects. c Eigenvalues of the covariance matrix as a function of their relative rank, for clusters of different sizes, for one example subject. The solid black line indicates least squares power law fit, i.e. , for . indicates the average exponent across subjects. d Estimated exponent for different cluster sizes. Error bars indicate the estimation error of the exponent (for 8 error bars are smaller than the symbols). e Distribution of exponents , , for single-subject scans (n = 1003). f Least square estimation errors of PRG exponents. g Relative estimation error of exponents; e.g., , where is the least square estimation error of exponent (n = 1003). White circles indicate medians. h The power-law fits of , , and were compared to those obtained using an exponential function by calculating the ratio between the explained variance of the competing regression models (). Ratios >1 favor the power law hypothesis. Violin plots represent the distribution of ratios across subjects (n = 1003). White circles indicate medians.
Another interesting observable is the probability of silence activity , i.e., time frames in which all signals within a cluster are below their activation threshold. Assuming that the probability distribution of the collective activity in a cluster of size is a Boltzmann distribution and that the configuration of complete silence has a null energy, relates to the partition function of the distribution, i.e., . Thus can be associated to an effective free energy 34 The effective free energy scales with the cluster size, i.e., , with an average exponent 0.673 ± 0.002 across subjects (average relative estimation error: 2.27%; 1.38; Fig. 3b, e–h).
The last observable that we studied was the eigenvalue spectra of coarse-grained variables. For this, for each cluster of size , we decomposed the covariance matrix into eigenvectors and studied the behavior of eigenvalues as a function of their relative rank. Consistent with Meshulam et al.33,34, we found that the spectra in clusters of different size collapsed when the rank was normalized, i.e., , and a power-law scaling of the eigenvalues as a function of their rank, i.e., , with an average exponent 0.328 ± 0.001 across subjects, followed by exponential truncation due to finite size effect (average relative estimation error: 1.88%; 1.21; Fig. 3c, e–h). The estimated least-squares exponent stabilized for 8 (Fig. 3d).
We further compared the measured PRG exponents to those obtained from shuffled data for which the correlations between signals were destroyed. To do this, the time frames of the binarized fMRI signals were randomly permuted, independently for each ROI. The PRG method applied to the shuffled data yielded exponents that were significantly different than those obtained using the original data (see Supplementary Fig. S2). Indeed, shuffled data yielded exponents and that approach 1 (as expected for independent signals) and showed no evidence of scaling of the eigen-spectrum. Finally, we examined whether the number of ROIs affect the PRG scaling. For this, we built subsampled systems by randomly selecting a fraction of the ROIs and we applied the PRG method to the subsampled data. We found that PRG exponents from subsampled data converged to those obtained using the full-size original data when the fraction of selected ROIs was larger than (see Supplementary Fig. S3).
In conclusion, using the correlation function and the PRG method we were able to describe the scale invariance of collective fMRI binarized activity by means of four power-law exponents , , , and .
Connectivity-based phenomenological renormalization-group
The above PRG approach was designed to study scale-invariance in neural systems when information about the neural connectivity was unknown33. This method is based on the correlation matrix (functional correlations) of variables at successive coarse-graining. We here extended this method to incorporate information about structural connectivity (see “Methods”) that, in our case, was provided by the dMRI connectivity and its EDR approximation. For this, one needs to coarse-grain the variables and the connectivity at each coarse-graining step. When applying this connectivity-based PRG to the fMRI data, we found that , , and exponents computed based on the dMRI connectivity strongly correlated with exponents computed based on the EDR (correlation > 0.98; Fig. 4a, see also Supplementary Fig. S4). Interestingly, the connectivity-based PRG exponents also strongly correlate with the exponents obtained using the original version of the PRG method, i.e., based on functional correlations (correlation > 0.89, Fig. 4b). In fact, we found that connectivity-based PRG exponents were shifted by a systematic bias from PRG exponents equal to −0.161, 0.095, and 0.106 for exponents , , and , respectively. We concluded that scaling exponents were consistently and reliably estimated using the PRG based on functional correlations or structural connections.
Fig. 4. Connectivity-based phenomenological renormalization group.
a Connectivity-based PRG exponents calculated from the fMRI data assuming structural connections given by dMRI or EDR. Each dot represents an individual scan (n = 1003). : correlation between corresponding exponents computed based on dMRI and EDR. b Comparison between dMRI-based PRG exponents and those obtained using the original PRG method (based on functional correlations, FC). Each dot represents an individual scan (n = 1003). : correlation between corresponding exponents computed based on dMRI and FC.
Whole-brain spin model
To test whether the observed scaling of activity is a signature of criticality, we built a spin model based on large-scale connectivity. The spin model is a canonical model presenting a second-order phase transition between ordered and disordered phases. Using the maximum entropy principle, the spin model can be mapped to binary neural data such as spiking activity (e.g. refs. 48,49) or binarized fMRI data (e.g. refs. 31,46,50). In this model, at each time step, the state of each node is described by a binary variable, i.e., , and the collective activity of the nodes is given by a binary pattern . The probability of each pattern is given by the Boltzmann distribution:
| 1 |
Where represents the energy of the pattern; is the partition function, i.e., ; and is a scaling parameter of the connectivity matrix , equivalent to an inverse temperature, i.e., , which is the free parameter of the model. The connectivity matrix was given by the EDR or the dMRI. Realizations of the spin model were obtained using Monte Carlo Metropolis simulations (see “Methods”). For each configuration , the population activity is defined as the average node value: . The average population activity was obtained by averaging across simulation steps.
For temperatures lower than a critical value, , the spin model is in an ordered phase and presents a spontaneous population activity (i.e., ) that vanishes in the disordered phase for temperatures > (Fig. 5a). A power-law correlation function was observed around the critical point separating the two phases (Fig. 5b; see also Supplementary Fig. S5), with a critical exponent that was close to the one measured in the data for the EDR ( = 0.515 ± 0.013; relative error from the empirical exponent: 0.4%), but slightly different for dMRI ( = 0.310 ± 0.009; 39.6%). When fitting a power law to the model correlation function for the full range of tested temperatures, we found that the power exponent was the closest to the empirical one around the critical point for the two connectivity matrices (Fig. 5c).
Fig. 5. Spin model.
a Population activity as a function of , relative to the critical point . For , the system is disordered and the average population activity is zero. For , the system is ordered and a spontaneous population activity emerges and settles in either a negative or a positive value (depending on the initial conditions). b At the critical point (blue), the correlation function is a power law, with a power exponent close to the one measured in the fMRI data. The correlation function is shown for three example temperatures, also shown in (a) (red: supercritical; blue: critical; purple: subcritical). c Exponent as a function of . d Variance of coarse-grained variables as a function of cluster size , for the three example temperatures. e Exponent as a function of . f Eigenvalues of the covariance matrix as a function of their relative rank, at the critical point, for clusters of different sizes. g Exponent as a function of . In (a), (b), (d) and (f), the connectivity used was the EDR. In (c), (e), and (g): filled symbols indicate explained variance ratios favoring the power law model over the exponential model, i.e., (see also Supplementary Fig. S5); exponent estimation errors are smaller than the symbols; the horizontal line indicates the empirically measured exponent.
We next applied the PRG method to the model activity. We found that, for the EDR and close to the critical point, the scaling of the variance and the covariance eigen-spectrum was similar to the one measured in fMRI data: the critical exponents were equal to 1.62 ± 0.01 and 0.30 ± 0.03 ( 2.9%, 8.5%; Fig. 5d–g). Power-law scaling of the coarse-grained variance was observed for all temperatures, while the eigen-spectrum scaled as a power law of the rank for the supercritical and critical regimes, but it exponentially decays in the subcritical regime (Supplementary Fig. S5). For the dMRI connectivity, the critical exponent was similar to the one measured in the fMRI data, but the critical exponent deviated from the data: 1.53 ± 0.05 ( 2.9%) and 0.26 ± 0.02 ( 21.7%) (Fig. 5d–g). Thus, opposite to the case of the EDR, in the case of the dMRI connectivity the empirical exponents could not be simultaneously fitted using a unique temperature parameter. Similarly, when using the connectivity-based PRG to compare the scaling of the data and the model, we found that the model’s critical exponents were remarkably close to the empirical exponents when the EDR was used both to couple and coarse-grain the spin variables ( 1.1%, 1.0%; see Supplementary Fig. S6c, d), but deviated for the dMRI connectivity ( 11.5%, 11.2%; see Supplementary Fig. S6a, b). We note that, since spin variables are symmetric (−1 or 1), we cannot define a silence probability to be associated to a free energy. Thus, the exponent could not be calculated for the model.
We concluded that, around its critical point, the spin model approximates the fMRI correlations and their scaling features, especially for the EDR, i.e., in the absence of long-range connections.
Discussion
We have shown that rs-fMRI signals present scaling features in the correlation function and as a function of coarse-graining based either on functional or structural connectivity. Notably, we found that the observed scaling can emerge from connections following a simple EDR and critical dynamics, i.e., the scaling exponents observed in the data were strikingly close to those predicted by a critical system of spins interacting through the EDR connectivity. Thus, our results suggest that criticality is the link between the connectome’s structure and scale-invariant brain dynamics.
Previous theoretical work has tested the PRG method in an interacting particle system, the contact process, and the Ising model, both in a regular 2D lattice and with nearest-neighbor interactions36. The scaling of the contact process and Ising models under PRG yields critical exponents that differ between 3.4-27.2% from those measured from collective neuronal activity of mouse hippocampus, which was the target of those models33,36. Interestingly, critical exponents in the contact process were found to be unchanged in the presence of long-range interactions in small-world networks36. Our strategy, however, was different since we modeled the system with a proxy of the underlying brain connectivity, thus conserving both the size of the network and the spatial distribution of the nodes. This allowed us to map the scaling exponents observed in rs-fMRI data to the phases of a model presenting a phase-transition. In our case, using the EDR, the model’s critical exponents were remarkably close to the exponents measured in rs-fMRI signals (both using the PRG or the connectivity-based PRG; see Fig. 5 and Supplementary Fig. S6), suggesting that brain dynamics operate close to a critical point in which order and disorder coexist. This result is in line with previous works showing that rs-fMRI signals display critical-like patterned activity detected using other methods, such as neuronal avalanches9 and maximum entropy models which are equivalent to spin models inferred from data31,50. Since, the EDR has been previously observed in different species (rodents and nonhuman primates), it would be interesting to examine whether large-scale dynamics in these animal models also scale under PRG and relate to criticality.
Overall, our study shows that combination of PRG, connectomes, and models can be useful to distinguish the working regime of the observed neural system. Since different behavioral and pathological brain states deviate from critical dynamics23–32, extending these analyses to different brain states could provide new insights on phase transitions in neural systems. We hypothesize that the PRG exponents change depending on the brain state and could be used as biomarkers in clinical and fundamental research. Indeed, in a recent study, Rocha et al.29 have shown that criticality is lost in the case of stroke lesions, but it is recovered over time as behavior improves. Thus, criticality signatures can represent promising tools for translational research. Furthermore, we note that the PRG method can be formalized using different interaction measures (e.g., synchrony) and in Fourier space33, which makes it suitable to study oscillatory dynamics recorded using LFPs, MEG, or electrocorticography. However, notice that, when applying the PRG method to finite data, both the cluster size’s range on which scaling can be tested and the accuracy of coarse-grained statistics (e.g., covariances) are limited by the amount of data and the tradeoff between the spatial and temporal resolutions of the used recording technique.
The dMRI is a non-invasive method to estimate the large-scale brain connectivity, but it has methodological caveats and limitations51. In particular, it estimates short-range intra-hemispheric connections more reliably than inter-hemispheric ones. On the contrary, at the mesoscale, invasive chemical tracers are considered to be the gold standard for estimating the connecting fibers with high accuracy. This technique has shown that connection strength exponentially declines with distance. Here, we showed that the EDR with a characteristic scale that was consistent with the extrapolation to human brain given its volume39, acheived a good linear prediction of fMRI signals, and yielded an accurate, consistent prediction of scaling exponents of brain activity using a critical whole-brain model. The whole-brain model constraint with the dMRI connectivity did not fit all scaling exponents neither at the critical point nor using a single temperature different from (see Fig. 5 and Supplementary Fig. S6). We concluded that, at least within the framework of our simple model, the EDR-based critical spin model represents a more parsimonious description of the observed rs-fMRI dynamics than the model based on dMRI connectivity (which presents long-range connections departing from the EDR). Future studies could explore the link between scaling features and connectivity using different recording techniques and more realistic models including interacting excitatory and inhibitory neural populations.
In conclusion, we have shown that whole-brain dynamics display scaling properties that emerge from exponentially decaying connections and critical dynamics, which are two features of connectivity and dynamics largely supported by fiber-tracking research and studies of neural activity at different scales and with different techniques.
Methods
Neuroimaging ethics
The Washington University–University of Minnesota (WU-Minn HCP) Consortium obtained full written informed consent from all participants to study procedures and data sharing outlined by HCP, and research procedures and ethical guidelines were followed in accordance with Washington University institutional review board approval.
Functional MRI data
In this study we analyzed publicly available rs-fMRI data from the Human Connectome Project (HCP), from 1003 participants. The participants were scanned on a 3 T connectome-Skyra scanner (Siemens). The rs-fMRI data was acquired for ~15 min, with eyes open and relaxed fixation on a projected bright cross-hair on a dark background. The HCP website (https://www.humanconnectome.org/) provides the details of participants, the acquisition protocol and preprocessing of the functional data. Briefly, the fMRI data was preprocessed using standardized methods using FSL (FMRIB Software Library), FreeSurfer, and the Connectome Workbench software52,53. This preprocessing included correction for spatial and gradient distortions and head motion, intensity normalization and bias field removal, registration to the T1 weighted structural image, transformation to the 2 mm Montreal Neurological Institute (MNI) space, using the FIX artefact removal procedure53,54. The head motion parameters were regressed out and structured artefacts were removed by ICA + FIX processing55,56. Preprocessed timeseries of all grayordinates are in HCP CIFTI grayordinates standard space and available in the surface-based CIFTI file for each participant. A custom-made Matlab script, using the ft_read_cifti function from the Fieldtrip toolbox57, was used to extract the average timeseries of all the grayordinates in each region of the Schaefer parcellation, which are defined in the HCP CIFTI grayordinates standard space. Sequence and imaging parameters: Sequence: Gradient-echo EPI; TR: 720 ms; TE: 33.1 ms; flip angle 52 deg; FOV: 208 × 180 mm (RO x PE); Matrix: 104 × 90 (RO x PE); Slice thickness: 2.0 mm, 72 slices, 2.0 mm isotropic voxels; Multiband: factor 8; Echo spacing: 0.58 ms; BW: 2290 Hz/Px.
Structural connectivity using dMRI
Structural connectivity was estimated from diffusion spectrum and T2-weighted imaging data from 32 participants from the HCP database, scanned over 89 min. Acquisition parameters are described in detail in the HCP website58. The freely available Lead-DBS software package (http://www.lead-dbs.org/) provided the preprocessing which is described in detail in Horn and colleagues59 but, briefly, the data was processed using a generalized q-sampling imaging algorithm implemented in DSI studio (http://dsi-studio.labsolver.org). Segmentation of the T2-weighted anatomical images produced a white-matter mask and co-registering the images to the b0 image of the diffusion data using SPM12. In each HCP participant, 200,000 fibers were sampled within the white-matter mask. Fibers were transformed into MNI space using Lead-DBS60. We used the standardized methods in Lead-DBS to produce the structural connectomes for the Schaefer 1000 parcellation scheme61. The connectivity weight was given by the number of fibers connecting two brain regions. To have values between 0 and 1, we normalized the weights by dividing them by the largest value, i.e., . Diffusion MRI parameters: Sequence: Spin-echo EPI; TR: 5520 ms; TE: 89.5 ms; flip angle: 78 deg; refocusing flip angle: 160 deg; FOV: 210 × 180 (RO x PE); matrix: 168 × 144 (RO x PE); slice thickness: 1.25 mm, 111 slices, 1.25 mm isotropic voxels; Multiband factor: 3; Echo spacing: 0.78 ms; BW: 1488 Hz/Px; Phase partial Fourier: 6/8; b-values: 1000, 2000, and 3000 s/mm2.
Schaefer parcellation
Schaefer and colleagues created a publicly available population atlas of cerebral cortical parcellation based on estimation from a large data set (n = 1489)61. They provide parcellations of 400, 600, 800, and 1000 areas available in surface spaces, as well as MNI152 volumetric space. We used here the Schaefer parcellation with 1000 areas and estimated the Euclidean distances from the MNI152 volumetric space and extracted the timeseries from HCP using the HCP surface space version.
Data binarization
The rs-fMRI time-series were binarized to study the data statistics and to compared them to those predicted by the spin model. For each scan, the z-scored time-series of each ROI, (), was binarized by imposing a threshold . The binarized activity was if and otherwise. Transformation of continuous signals into discrete point processes has proven to effectively capture and compress fMRI large-scale dynamics9. Importantly, the fluctuations that cross the threshold do not merely represent noise, since the resulting point process largely overlaps with deconvoluted fMRI signals using the hemodynamic response function and preserves the topology of the resting state networks9. Furthermore, using maximum entropy models to estimate the probability distribution of binarized activity, it has been shown that binarized rs-fMRI data is poised close at a critical point31.
Correlation function
We calculated the average correlation as a function of the Euclidean distance between ROIs. For this, we calculated the average correlation among pairs of nodes that were separated by distances between and , with = 0.43 mm, i.e.:
| 2 |
where is the number of pairs of ROIs such that , and and denote the distance and the Pearson correlation between ROIs and , respectively. Distances between ROIs range between 4.28 mm and 173.16 mm.
Phenomenological renormalization-group method
We here review the recently proposed PRG approach to study scale-invariance in neural systems33. Within this method, the collective activity is iteratively coarse-grained by grouping together the variables that are maximally correlated.
Let be the binary activity of ROI for , with . The superscript 0 indicates that the data is not coarse-grained. In the first coarse-graining step, we seek for the pair of variables with maximal correlation and sum them:
| 3 |
where . We repeat this procedure for the second maximally correlated pair among the remaining variables, i.e., from the set , and so on until all pairs are used. This process is iterated for coarse-grained variables , resulting in clusters of size . The size of the system is equal to at each coarse-graining step.
Along the coarse-graining procedure, several statistics of are calculated and their change at different coarse-graining steps are examined. A first observable is the variance of coarse-grained variables:
| 4 |
For calcium imaging recordings in the mouse hippocampus, it has been shown that the variance scales with the cluster size, 33 with a power-law exponent that lies between linear ( 1) and quadratic growth ( 2), corresponding to uncorrelated and full-correlated systems, respectively.
A second quantity is the probability of silence, for all . Assuming that the probability distribution of the collective activity in a cluster of size is a Boltzmann distribution and that the configuration of complete silence has null energy, relates to the partition function of the distribution, i.e., . Thus can be associated to a free energy . In calcium imaging recordings in the mouse hippocampus, it has been shown that the free energy scales with the cluster size as 34 with an exponent <1, which is the expected value for independent variables.
A third quantity is the spectrum of the covariance matrix inside a cluster of size . Let denote the eigenvalues of the covariance matrix. The eigenvalues are ordered from the highest eigenvalue, 1, to the lowest, . It has been shown that, at the fixed point of RG, one expects that the eigen-spectrum scales with the relative rank33:
| 5 |
Notice that the eigen-spectrum presents scaling in two senses: the spectra in clusters of different size collapse when the rank is normalized, i.e., , and the eigenvalues have a power-law decay as a function of rank, followed by exponential truncation due to finite size effect33.
Spectral properties of covariance matrices often depend on the ratio between the number of samples and the number of variables. For this reason and following Meshulam et al.34, we studied the eigen-spectrum for cluster sizes for which we have >10 times more samples than variables. In our case, with 1200 time frames per scan we required . Throughout this article, we coarse-grained the activity up to six times and evaluated the PRG power-laws of and for . The eigen-spectrum was computed for and its power-law fit was evaluated for < < 0.4 to avoid the finite-size truncation36.
In a D-dimensional system with translational invariance and a power-law correlation function (as expected for a critical system), the exponent is related to . In this case, the eigenvalues are given by the Fourier transform of the correlation function62:
| 6 |
Since the rank scales as , one has: . However, the exponent does not satisfy this relation neither in the critical spin model nor in the fMRI data. This might indicate that the system is not translational invariant.
Connectivity-based phenomenological renormalization-group method
Given that, in our case, we have information about the structural connectivity, we can extend the PRG method to coarse-grain the collective fMRI activity based on this structural connectivity matrix . Here, the matrix is given by the dMRI or the EDR. Note that in both cases, the connectivity matrix is symmetric. In the connectivity-based PRG (CBPRG) method, we grouped the variables that are maximally connected. In the first coarse-graining step, we seek for the pair of variables with maximal connection , where , and sum them as in Equation (3). This was repeated for the second maximally connected pair among the remaining variables and so on until all pairs are used. Let and be two indices corresponding to two groups formed in the first step from variables and , i.e.:
| 7 |
| 8 |
Where and . A new connectivity matrix of size is defined as follows:
| 9 |
This connectivity matrix was then used to group the variables at step 2. As above, the process was iterated to obtain coarse-grained variables , built by grouping variables based on their connectivity , resulting in clusters of size . Notice that the weights of matrix result from averaging connectivity weights from the original connectivity matrix.
Whole-brain spin model
To relate the observed fMRI statistics to critical dynamics, we built a spin model based on large-scale connectivity. In this model, the state of each node is described by a binary variable or “spin”, i.e., , and the collective activity of the nodes is given by a binary pattern or configuration . The probability of each pattern is given by the Boltzmann distribution:
| 10 |
where represents the energy of the pattern and is given as:
| 11 |
is the partition function, i.e., . Spins interact through the connectivity matrix . is a scaling parameter of the connectivity matrix ; it is equivalent to an inverse temperature, i.e., , which is the free parameter of the model.
For each configuration , the population activity is defined as the average node value: . The average population activity was obtained by averaging across simulation steps. The model presents a second-order phase transition that can be detected by examining the behavior of as a function of the temperature parameter. For temperatures lower than a critical value, , the spin model presents a spontaneous population activity, i.e., (subcritical regime), that vanishes for temperatures > (supercritical regime).
Realizations of the spin model were obtained using Monte Carlo Metropolis simulations. The algorithm starts with an initial random configuration of spins, then flips the spin of a randomly chosen node, and calculates the change in energy induced by the spin flip. If , the spin flip is accepted, otherwise it is accepted with probability . For each tested value of , we ran 5 realizations (with different initial conditions) of simulation steps. The system’s configuration was stored every flip attempts. To avoid dependences on initial conditions, simulations started with extra 500,000 steps without storing the configurations. The population activity, the correlation function, the coarse-grained variance, and the coarse-grained eigen-spectrum were averaged over realizations, for each .
Note that, using the maximum entropy principle, the spin model (and its extensions) can be mathematically map to binary data. This has been done using spiking data at the microcircuit level (e.g. refs. 48,49) but also using binarized fMRI data at the large-scale level (e.g. refs. 31,46,50). Briefly, within the framework of maximum entropy models, to estimate the probability distribution of binary patterns, one seeks for the distribution that matches some statistics of the data and has maximum entropy. It is known that the maximum entropy distribution that preserves the pairwise correlations of the data is , where is the effective connectivity between the variables and 49 The resulting maximum entropy distribution is thus equivalent to the Boltzmann distribution of the present spin model.
Power law fit
We fitted power laws using least squares on log-log scattered data. Note that we were unable to use a maximum likelihood estimation (MLE) approach which applies only to probability distributions. MLE, together with Kolmogorov–Smirnov (KS) statistics63,64 or log-likelihood ratios (LLR) between the candidate heavy-tailed distributions65, are commonly used to study critical power-law behavior in neural systems12,66,67. However, this approach is only applicable to probability densities. For this reason, we here used least squares to fit power laws and we evaluated the goodness of fit by comparing the explained variance of the least-squares fit using the power law and the one obtained using an exponential function. Specifically, we calculated the ratio , where is the explained variance (or coefficient of determination) of the linear regression model (power law) and is the explained variance of the linear regression model (exponential). Ratios > 1 favor the power law hypothesis against the exponential alternative. The estimation error of the power-law exponent was given by the error of the slope of the linear regression model . The relative error was defined as: 100.
Statistics and reproducibility
The goodness of the linear prediction was given by the Pearson correlation between and for all nodes and all subjects (n = 1000 × 1003), where denotes the dMRI or the EDR connectivity matrices and denotes the data signals (Fig. 1d). The distributions of correlation coefficients were compared to the one obtained using a shuffled connectivity. These comparisons were done using a Welch’s t-test, after Fisher z-transformation of the correlation coefficients.
For each subject, shuffled data were built by randomly permuting the time frames of binarized fMRI signals for each ROI separately. Next, the PRG method was applied to the resulting shuffled data (Supplementary Fig. S2). The distributions of the PRG exponents , , from the original data and the shuffled data were compared using Wilcoxon tests (n = 1003 for each distribution).
MATLAB (R2021a) software was used to perform all analyses and to simulated the model. Numerical simulations were performed in a 50-nodes computer cluster (Intel® Xeon® E5-2684 at 2.1 Ghz, 256 GB RAM, 1 TB disk).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
A.P.-A. was supported by a Ramón y Cajal fellowship (RYC2020-029117-I) from FSE/Agencia Estatal de Investigación (AEI), Spanish Ministry of Science and Innovation. A.P.-A. and G.D. were supported by the EU Fet Flagship Human Brain Project SGA3 (945539). G.D. was supported by the Spanish Research Project AWAKENING (PID2019-105772GB-I00/AEI/10.13039/501100011033), financed by the Spanish Ministry of Science, Innovation and Universities (MCIU), State Research Agency (AEI). M.L.K. is supported by the Centre for Eudaimonia and Human Flourishing (funded by the Pettit and Carlsberg Foundations) and Center for Music in the Brain (funded by the Danish National Research Foundation, DNRF117).
Author contributions
A.P.-A. and G.D. designed the research. A.P.-A., M.L.K. and G.D. analyzed/processed data. A.P.-A. designed and performed model analysis and wrote the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Christian Beste and Karli Montague-Cardoso. A peer review file is available.
Data availability
We used a publicly available dataset of fMRI data from the Human Connectome Project (HCP), from 1003 participants selected from the March 2017 public data release from the Human Connectome Project (HCP). The HCP dataset is available at https://www.humanconnectome.org/study/hcp-young-adult. Source data for main figures presented in this study are provided as Supplementary Data 1–5.
Code availability
The codes to perform the PRG analysis and to simulate the model are available at https://github.com/adrianponce/Scaling-of-whole-brain-resting-state-dynamics, 10.5281/zenodo.796210968.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-023-05001-y.
References
- 1.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reason. Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 2.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 3.Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He BJ. Scale-free properties of the functional magnetic resonance imaging signal during rest and task. J. Neurosci. 2011;31:13786–13795. doi: 10.1523/JNEUROSCI.2111-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciuciu P, Varoquaux G, Abry P, Sadaghiani S, Kleinschmidt A. Scale-free and multifractal time dynamics of fMRI signals during rest and task. Front. Physiol. 2012;3:186. doi: 10.3389/fphys.2012.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He BJ, Zempel JM, Snyder AZ, Raichle ME. The temporal structures and functional significance of scale-free brain activity. Neuron. 2010;66:353–369. doi: 10.1016/j.neuron.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milstein J, Mormann F, Fried I, Koch C. Neuronal shot noise and Brownian 1/f2 behavior in the local field potential. PLoS ONE. 2009;4:e4338. doi: 10.1371/journal.pone.0004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dehghani N, Bédard C, Cash SS, Halgren E, Destexhe A. Comparative power spectral analysis of simultaneous electroencephalographic and magnetoencephalographic recordings in humans suggests non-resistive extracellular media: EEG and MEG power spectra. J. Comput. Neurosci. 2010;29:405. doi: 10.1007/s10827-010-0263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo DR. Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front. Physiol. 2012;3:15. doi: 10.3389/fphys.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palva JM, et al. Neuronal long-range temporal correlations and avalanche dynamics are correlated with behavioral scaling laws. Proc. Natl. Acad. Sci. USA. 2013;110:3585–3590. doi: 10.1073/pnas.1216855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shriki O, et al. Neuronal Avalanches in the Resting MEG of the Human Brain. J. Neurosci. 2013;33:7079–7090. doi: 10.1523/JNEUROSCI.4286-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponce-Alvarez A, Jouary A, Privat M, Deco G, Sumbre G. Whole-brain neuronal activity displays crackling noise dynamics. Neuron. 2018;100:1446–1459. doi: 10.1016/j.neuron.2018.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beggs JM, Plenz D. Neuronal avalanches in neocortical circuits. J. Neurosci. 2003;23:11167–11177. doi: 10.1523/JNEUROSCI.23-35-11167.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasquale V, Massobrio P, Bologna LL, Chiappalone M, Martinoia S. Self-organization and neuronal avalanches in networks of dissociated cortical neurons. Neuroscience. 2008;153:1354–1369. doi: 10.1016/j.neuroscience.2008.03.050. [DOI] [PubMed] [Google Scholar]
- 15.Hahn G, et al. Neuronal avalanches in spontaneous activity in vivo. J. Neurophysiol. 2010;104:3312–3322. doi: 10.1152/jn.00953.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn G, et al. Spontaneous cortical activity is transiently poised close to criticality. PLoS Comput. Biol. 2017;13:e1005543. doi: 10.1371/journal.pcbi.1005543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ribeiro TL, et al. Spike avalanches exhibit universal dynamics across the sleep-wake cycle. PLoS One. 2010;5:e14129. doi: 10.1371/journal.pone.0014129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman N, et al. Universal critical dynamics in high resolution neuronal avalanche data. Phys. Rev. Lett. 2012;108:208102. doi: 10.1103/PhysRevLett.108.208102. [DOI] [PubMed] [Google Scholar]
- 19.di Santo S, Villegas P, Burioni R, Muñoz MA. Landau–Ginzburg theory of cortex dynamics: Scale-free avalanches emerge at the edge of synchronization. Proc. Natl. Acad. Sci. USA. 2018;115:e1356–e1365. doi: 10.1073/pnas.1712989115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shew WL, Yang H, Petermann T, Roy R, Plenz D. Neuronal avalanches imply maximum dynamic range in cortical networks at criticality. J. Neurosci. 2009;29:15595–15600. doi: 10.1523/JNEUROSCI.3864-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shew WL, Plenz D. The functional benefits of criticality in the cortex. Neuroscientist. 2013;19:88–100. doi: 10.1177/1073858412445487. [DOI] [PubMed] [Google Scholar]
- 22.Marinazzo D, et al. Information transfer and criticality in the Ising model on the human connectome. PLoS One. 2014;9:e93616. doi: 10.1371/journal.pone.0093616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs JP, Smith JL, Beggs JM. Aberrant neuronal avalanches in cortical tissue removed from juvenile epilepsy patients. J. Clin. Neurophysiol. 2010;27:380–386. doi: 10.1097/WNP.0b013e3181fdf8d3. [DOI] [PubMed] [Google Scholar]
- 24.Meisel C, Storch A, Hallmeyer-Elgner S, Bullmore E, Gross T. Failure of adaptive self-organized criticality during epileptic seizure attacks. PLoS Comput. Biol. 2012;8:e1002312. doi: 10.1371/journal.pcbi.1002312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesse J, Gross T. Self-organized criticality as a fundamental property of neural systems. Front. Syst. Neurosci. 2014;8:166. doi: 10.3389/fnsys.2014.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tinker J, Perez Velazquez JL. Power law scaling in synchronization of brain signals depends on cognitive load. Front. Syst. Neurosci. 2014;8:73. doi: 10.3389/fnsys.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massobrio P, de Arcangelis L, Pasquale V, Jensen HJ, Plenz D. Criticality as a signature of healthy neural systems. Front. Syst. Neurosci. 2015;9:22. doi: 10.3389/fnsys.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cocchi L, Gollo LL, Zalesky A, Breakspear M. Criticality in the brain: a synthesis of neurobiology, models and cognition. Prog. Neurobiol. 2017;158:132–152. doi: 10.1016/j.pneurobio.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Rocha RP, et al. Recovery of neural dynamics criticality in personalized whole-brain models of stroke. Nat. Comm. 2022;13:3683. doi: 10.1038/s41467-022-30892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tagliazucchi E, et al. Large-scale signatures of unconsciousness are consistent with a departure from critical dynamics. J. R. Soc. Interface. 2016;13:20151027. doi: 10.1098/rsif.2015.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponce-Alvarez A, et al. Macroscopic quantities of collective brain activity during wakefulness and anesthesia. Cereb. Cortex. 2022;32:298–311. doi: 10.1093/cercor/bhab209. [DOI] [PubMed] [Google Scholar]
- 32.Fekete T, et al. Critical dynamics, anesthesia and information integration: Lessons from multi-scale criticality analysis of voltage imaging data. NeuroImage. 2018;183:919–933. doi: 10.1016/j.neuroimage.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 33.Meshulam L, Gauthier JL, Brody CD, Tank DW, Bialek W. Coarse graining, fixed points, and scaling in a large population of neurons. Phys. Rev. Lett. 2019;123:178103. doi: 10.1103/PhysRevLett.123.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meshulam, L., Gauthier, J. L., Brody, C. D., Tank, D. W. & Bialek, W. Coarse-graining and hints of scaling in a population of 1000+ neurons. arXiv10.48550/arXiv.1812.11904 (2018).
- 35.Morales, G., di Santo, S. & Muñoz, M. A. Quasiuniversal scaling in mouse-brain neuronal activity stems from edge-of-instability critical dynamics. Proc. Natl. Acad. Sci. USA120, e2208998120 (2023). [DOI] [PMC free article] [PubMed]
- 36.Nicoletti G, Suweis S, Maritan A. Scaling and criticality in a phenomenological renormalization group. Phys. Rev. Res. 2020;2:023144. doi: 10.1103/PhysRevResearch.2.023144. [DOI] [Google Scholar]
- 37.Markov NT, et al. The role of long-range connections on the specificity of the macaque interareal cortical network. Proc. Natl. Acad. Sci. USA. 2013;110:5187–5192. doi: 10.1073/pnas.1218972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ercsey-Ravasz M, et al. A predictive network model of cerebral cortical connectivity based on a distance rule. Neuron. 2013;80:184–197. doi: 10.1016/j.neuron.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theodoni P, et al. Structural attributes and principles of the neocortical connectome in the marmoset monkey. Cereb. Cortex. 2022;32:15–28. doi: 10.1093/cercor/bhab191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiser M, Hilgetag C. Spatial growth of real-world networks. Phys. Rev. E. 2004;69:036103. doi: 10.1103/PhysRevE.69.036103. [DOI] [PubMed] [Google Scholar]
- 41.Horvát S, et al. Spatial embedding and wiring cost constrain the functional layout of the cortical network of rodents and primates. PLoS Biol. 2016;14:e1002512. doi: 10.1371/journal.pbio.1002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knox J, et al. High-resolution data-driven model of the mouse connectome. Netw. Neurosci. 2018;3:217–236. doi: 10.1162/netn_a_00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubinov M, Ypma R, Watson C, Bullmore E. Wiring cost and topological participation of the mouse brain connectome. Proc. Natl. Acad. Sci. USA. 2015;112:10032–10037. doi: 10.1073/pnas.1420315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi H, Mihalas S. Synchronization dependent on spatial structures of a mesoscopic whole-brain network. PLoS Comput. Biol. 2019;15:e1006978. doi: 10.1371/journal.pcbi.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deco G, et al. Rare long-range cortical connections enhance human information processing. Curr Biol. 2021;31:4436–4448. doi: 10.1016/j.cub.2021.07.064. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T, et al. A pairwise maximum entropy model accurately describes resting-state human brain networks. Nat Commun. 2013;4:1370. doi: 10.1038/ncomms2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Expert, P. et al. Self-similar correlation function in brain resting-state functional magnetic resonance imaging. J. R. Soc. Int., 8, 472–479 (2011). [DOI] [PMC free article] [PubMed]
- 48.Schneidman E, Berry MJ, Segev R, Bialek W. Weak pairwise correlations imply strongly correlated network states in a neural population. Nature. 2006;440:1007–1012. doi: 10.1038/nature04701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tkačik G, et al. Searching for collective behavior in a large network of sensory neurons. PLoS Comput. Biol. 2014;10:e1003408. doi: 10.1371/journal.pcbi.1003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ezaki T, et al. Closer to critical resting-state neural dynamics in individuals with higher fluid intelligence. Commun Biol. 2020;3:52. doi: 10.1038/s42003-020-0774-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sotiropoulos SN, Zalesky A. Building connectomes using diffusion MRI: why, how and but. NMR biomed. 2019;32:e3752. doi: 10.1002/nbm.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glasser MF, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith SM, et al. Resting-state fMRI in the Human Connectome Project. NeuroImage. 2013;80:144–168. doi: 10.1016/j.neuroimage.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro Schroder, T., Haak, K. V., Zaragoza Jimenez, N. I., Beckmann, C. F.& Doeller, C. F. Functional topography of the human entorhinal cortex. eLife10.7554/eLife.06738 (2015). [DOI] [PMC free article] [PubMed]
- 55.Griffanti L, et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage. 2014;95:232–247. doi: 10.1016/j.neuroimage.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salimi-Khorshidi G, et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. NeuroImage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oostenveld R, Fries P, Maris E, Schoffelen JM. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011;2011:156869. doi: 10.1155/2011/156869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Setsompop K, et al. Pushing the limits of in vivo diffusion MRI for the Human Connectome Project. NeuroImage. 2013;80:220–233. doi: 10.1016/j.neuroimage.2013.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horn A, Neumann WJ, Degen K, Schneider GH, Kuhn AA. Toward an electrophysiological “sweet spot” for deep brain stimulation in the subthalamic nucleus. Hum. Brain Mapp. 2017;38:3377–3390. doi: 10.1002/hbm.23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horn A, Blankenburg F. Toward a standardized structural-functional group connectome in MNI space. NeuroImage. 2016;124:310–322. doi: 10.1016/j.neuroimage.2015.08.048. [DOI] [PubMed] [Google Scholar]
- 61.Schaefer A, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cerebral Cortex. 2018;28:3095–3114. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vinayak S, Prosen T, Buča B, Seligman TH. Spectral analysis of finite-time correlation matrices near equilibrium phase transitions. EPL. 2014;108:20006. doi: 10.1209/0295-5075/108/20006. [DOI] [Google Scholar]
- 63.Clauset A, Shalizi CR, Newman MEJ. Power-law distributions in empirical data. SIAM Rev. 2009;51:661–703. doi: 10.1137/070710111. [DOI] [Google Scholar]
- 64.Deluca A, Corral Á. Fitting and goodness-of-fit test of non-truncated and truncated power-law distributions. Acta Geophys. 2013;61:1351–1394. doi: 10.2478/s11600-013-0154-9. [DOI] [Google Scholar]
- 65.Alstott J, Bullmore E, Plenz D. Powerlaw: a python package for analysis of heavy-tailed distributions. PLoS One. 2014;9:e85777. doi: 10.1371/journal.pone.0085777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marshall N, et al. Analysis of power laws, shape collapses, and neural complexity: new techniques and MATLAB support via the NCC toolbox. Front. Physiol. 2016;7:250. doi: 10.3389/fphys.2016.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu C. Toward a unified analysis of the brain criticality hypothesis: reviewing several available tools. Front. Neural Circuits. 2022;16:911245. doi: 10.3389/fncir.2022.911245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ponce-Alvarez, A. adrianponce/Scaling-of-whole-brain-resting-state-dynamics: critical scaling of whole-brain resting-state dynamics. Zenodo10.5281/zenodo.7962109 (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
We used a publicly available dataset of fMRI data from the Human Connectome Project (HCP), from 1003 participants selected from the March 2017 public data release from the Human Connectome Project (HCP). The HCP dataset is available at https://www.humanconnectome.org/study/hcp-young-adult. Source data for main figures presented in this study are provided as Supplementary Data 1–5.
The codes to perform the PRG analysis and to simulate the model are available at https://github.com/adrianponce/Scaling-of-whole-brain-resting-state-dynamics, 10.5281/zenodo.796210968.