Abstract
Background:
Inhaled corticosteroids (CS) are a backbone of asthma treatment, improving quality of life, exacerbation rates and mortality. Though effective for most, a subset of asthma patients experience CS resistant disease despite receipt of high medication doses.
Objective:
Our goal was to investigate the transcriptomic response of bronchial epithelial cells (BEC) to inhaled corticosteroids.
Methods:
Independent component analysis was performed on datasets detailing the transcriptional response of BECs to CS treatment. The expression of these CS response components was examined in two patient cohorts and investigated in relation to clinical parameters. Supervised learning was used to predict BEC CS responses using peripheral blood gene expression.
Results:
We identified a signature of CS response that was closely correlated with CS use in asthma patients. Participants could be separated based on CS response genes into groups with high and low signature expression. Patients with low expression of CS-response genes, particularly those with a severe asthma diagnosis, showed worse lung function and quality of life. These individuals demonstrated enrichment for T lymphocyte infiltration in endobronchial brushings. Supervised machine learning identified a 7 gene signature from peripheral blood that reliably identified patients with poor CS response expression in BECs.
Conclusion:
Loss of CS transcriptional responses within bronchial epithelium was related to impaired lung function and poor quality of life, particularly in severe asthma. These individuals were identified using minimally invasive blood sampling, suggesting these findings may enable earlier triage to alternative treatments.
Clinical Implications:
The specific transcriptional changes in BECs and blood identified here may guide early use of additional therapies.
Keywords: Asthma, Corticosteroids, Systems Biology, severe asthma, transcriptomics
Capsule Summary:
Severe asthma patients have reduced corticosteroid-linked transcriptional changes in the bronchial epithelium, show enrichment for T lymphocyte immune processes, and can be identified using a peripheral blood gene signature.
Graphical Abstract
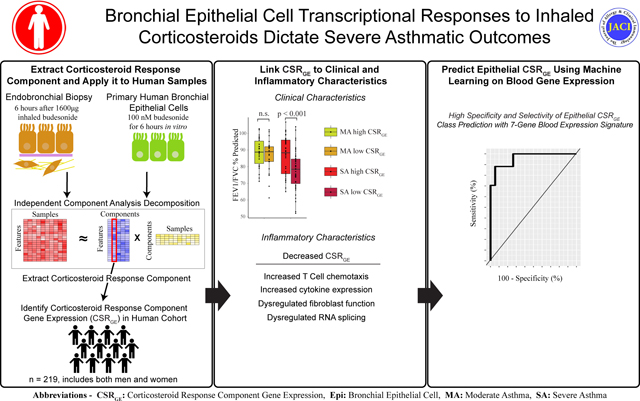
Introduction
Asthma is a common disease, effecting more than 300 million people worldwide(1, 2). Inhaled corticosteroids (CS) are the current standard of care, improving quality of life, exacerbation rates and mortality(3). Response is not universal, however, and 5–10% of patients experience poor control despite high dose CS therapy(4). This variation in therapeutic efficacy led to investigations describing heterogeneity amongst asthma patients at the clinical, immunological and molecular levels(2, 5, 6). Though prior work illustrated activation of specific pathways in CS refractory patients, the interplay between CS response in bronchial epithelial cells (BECs) and asthma phenotype remains incompletely understood(5, 7–9).
CS signaling is mediated by the glucocorticoid receptor (GR), which, following ligand binding, translocates to the nucleus to activate target gene expression(10). The GR may also associate with other transcription factors, promoting or suppressing expression of their target genes(11). CS efficacy in the context of asthma has been largely attributed to downregulation of T cell cytokine production and induction of eosinophil apoptosis(12, 13). Though downregulation of immune cell inflammatory cytokines may be crucial for CS response in asthma, growing work illustrates a role for airway epithelium in immune cell activation and propagation of inflammation(14, 15). Prior work clearly demonstrated transcriptional hallmarks of GR activation in human BECs(16). Murine models of asthma also show that epithelial GR expression is required for CS-mediated alleviation of airway hyperresponsiveness and inflammation(17). Together, these data suggest that interplay between BECs and immune cells is crucial for asthma pathogenesis.
Here, we use independent component analysis (ICA) on transcriptomic datasets of CS treatment in heathy volunteers and cell lines to extract the transcriptomic CS response signal in epithelial cells. We then evaluate the expression of this CS response signal in two large, well described asthma patient cohorts, linking poor CS response to clinical traits including impaired lung function and inadequate disease control. We describe a subset of asthma patients with low expression of CS response genes despite self-reported medication adherence, suggesting their disease may be intrinsically recalcitrant to this therapeutic modality. Importantly, patients with low CS response expression in their BECs could be identified using a peripheral blood gene expression signature, offering hope that minimally-invasive monitoring may be one day used to guide therapeutic choices.
Methods
For full methods, see online supplementary methods.
Human Participants
Endobronchial brushings from the Immune Modulation in Severe Asthma (IMSA; GSE158752) and Severe Asthma Research Program (SARP; dbGaP Study Accession: phs001446.v2.p1) cohorts were used as previously described(18). Gene expression counts for all data were normalized using the variance stabilizing transform from the DESeq2 suite(19). Batch effects within and between endobronchial brush data from the two studies were controlled for using the COMBAT algorithm(20), creating a combined epithelial dataset of the two cohorts.
Peripheral blood was also extracted from a subset of the IMSA patients. Transcriptomic data was prepared from the whole peripheral blood using the same methods as the endobronchial brushing and the data are available at GSE207751.
Corticosteroid Signal Source Datasets
Two microarray datasets were downloaded from GEO and used in our analysis. The first dataset, GSE83233, consisted of paired endobronchial biopsies obtained from healthy volunteers from before and after inhaled budesonide(21). The second dataset, GSE161805 consisted of primary human bronchial epithelial cells (pHBECs) from six different donors under control conditions, budesonide, formoterol, or budesonide and formoterol combined(22). For our analysis of GSE161805, only the control and budesonide groups were used.
Corticosteroid component extraction with independent component analysis
Joint Approximate Diagonalization of Eigenmatrices (JADE) was used to perform independent component analysis (ICA) on the GEO datasets(23). Each component produced by ICA is a vector containing a score for each gene which denotes the amount that gene contributes to the component.
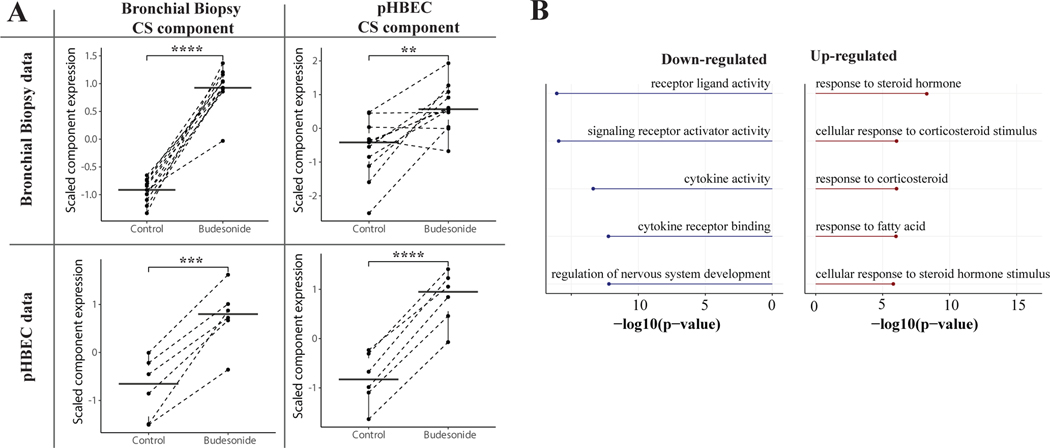
After extracting the components, paired T-tests were performed to compare the relative expression of each component in the pre-CS samples against the post-CS samples. In each case, the expression of one of the components was highly significant between the pre- and post-CS samples, and thus deemed as the CS components (Fig 1A). Because the expression of the two CS components in the combined IMSA+SARP epithelial dataset were similar (Fig 2A), we averaged their expression levels to create a set of one CS component expression value per patient.
Figure 1: Independent Component Analysis Extracts the Corticosteroid Response Signal.
(A) Corticosteroid components were extracted from both the bronchial biopsy data and the pHBEC data. Each component was then expressed in each of the two datasets. The corticosteroid components expression levels were significantly different for both components in their native dataset as well as the other dataset. (B) Gene ontology of the top positively and negatively contributing genes for the average of the two corticosteroid components.
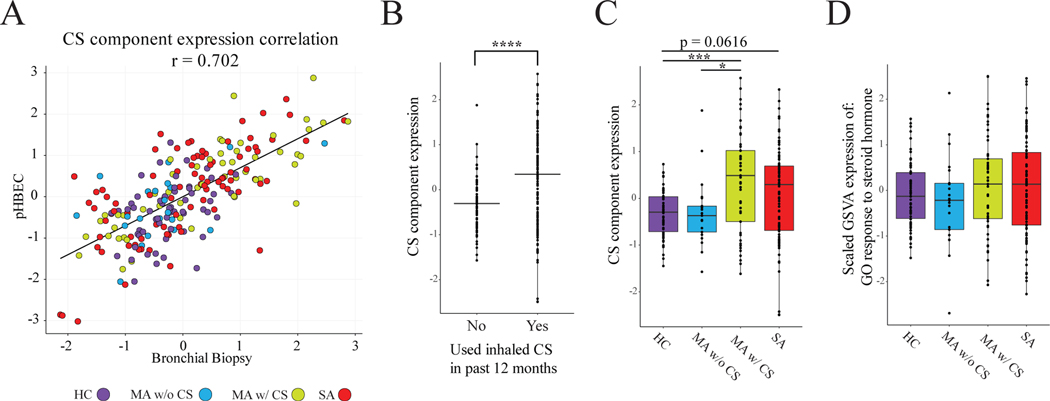
Figure 2: The CS Component is Correlated With CS Usage and Response to Corticosteroid.
(A) The two CS components extracted from GSE161805 (pHBEC) and GSE83233 (Bronchial Biopsy) are highly correlated (r = 0.702, p < 0.00001) when expressed in the bronchial epithelial cells of the merged IMSA+SARP dataset. (B) There is a significant difference in CS component expression between patients who had used inhaled CS in the past 12 months and those who had not (Student’s t-test). (C) Comparison of CS component expression between healthy controls, MA-, MA+ and SA patients. Moderate asthma patients with CS have a significantly higher CS component expression level than moderate asthma patients without CS and healthy controls (ANOVA with Tukey post-hoc test). (D) There is no significant difference between the four groups of GSVA expression of the GO term Response to Steroid Hormone (ANOVA, p = 0.506).
Computational and Machine Learning Methods
Packages used for computational analysis include ClusterProfiler (24) for gene ontology analysis, GSVA (25) to quantify the expression of the GO term Response to Steroid Hormone, ComplexHeatmap (26) for visualization, Deseq2 (19) for differential expression, and WGCNA to identify groups of coexpressed genes (referred to as modules) (27).
Patients in the IMSA cohort that had transcriptomic data for both blood and epithelial cells (n = 36) were used for machine learning. For estimation of epithelial CSRGE from blood transcriptomics, elastic net regularized regression analysis was performed with glmnet (28). For supervised modeling, the mixOmics package (29) was used to perform sparse-partial least square discriminant analysis training.
Statistical Analysis
All statistical analysis was performed using the R computing environment (Version 4.1.0) unless otherwise noted(30, 31). Statistical testing and methodology are described within figure legends or above within context specific methodologies. For all figures, significance levels are denoted as *p< 0.05, **p< 0.01, ***p< 0.001, ****p < 0.0001.
For information on other computational methods, see the online supplementary material.
Results
Extraction of Corticosteroid Signal
We used Independent Component analysis (ICA) to extract a corticosteroid (CS)-response component from two separate datasets (one CS-response component from each dataset; see methods). The first (GSE83233) consisted of paired endobronchial biopsies before and after inhaled budesonide and the second (GSE161805) of primary human bronchial epithelial cells (pHBEC) cultured with and without budesonide.
We found that many known steroid and inflammation-related genes were among the top genes contributing to the CS response component in both datasets, and that there was a general overall correlation of gene contribution scores in the CS components (r = 0.186, p < 0.00001, Fig S1, Table S1). To test if the CS-response component from one dataset could be used to examine gene expression patterns in the other, we cross interrogated their expression in control and budesonide treated samples (Fig 1A). Expression of the CS response component derived from endobronchial biopsies was significantly higher in budesonide treatment conditions compared to control in both the original dataset and in pHBECs. A similar expression pattern was observed for the CS response component originating from pHBECs. These results show that a transcriptional signal for CS response can be extracted with ICA and validated across external datasets. Averaging the scores from the two CS components and performing gene ontology on the top 1% of positively and negatively-contributing genes confirmed that these components are highly enriched for multiple CS-related ontologies (Fig 1B, Table S2).
Expression of CS Signals in Combined IMSA and SARP Dataset
Satisfied that our CS response components could be validated between exposure data sets, we next investigated their utility in assessing asthma patient cohorts. To this end, we examined bronchial epithelial cell (BEC) expression of the extracted CS components in a pooled data set (clinical information in Table S3) from the Immune Modulation in Severe Asthma (IMSA) and Severe Asthma Research Program (SARP) cohorts(18, 32). We calculated an expression value for both of the previously identified CS components in each patient in the IMSA+SARP dataset and found that expression of the two components was highly correlated (Fig 2A; r = 0.70205, p< 0.0001). For clarity in presentation moving forward, we averaged their expression levels to create a single value of CS-response gene expression (CSRGE) for each cohort participant. Unsurprisingly, CSRGE was higher in patients who had used inhaled CS in the past 12 months than in those who had not (Fig 2B).
We next split the moderate asthma patients into two groups: those who had used inhaled CS in the past 12 months (MA+, n = 53) and those who had not (MA-, n = 21). We found that there was no significant difference in CSRGE between MA- and HC (Figure 2C). While CSRGE differed between MA+ and MA-, MA+ and SA (who by definition were taking ICS) showed comparable levels. To further support the relationship between CS exposure and CSRGE values, we next created modules of coexpressed genes using Weighted Gene Co-expression Network Analysis (WGCNA)(27). WGCNA identified a module that was strongly correlated with CSRGE (Fig S2A) and included many of the top contributing genes to the combined CS component (Fig S2B). Gene ontology analysis on this module showed enrichment for CS response-related terms (Fig S2C).
To further test the utility of our CSRGE in determining transcriptional response to CS, we compared its performance to gene set variation analysis (GSVA), which scores individuals using known pathways(25). For the purpose of this comparison, we selected the top result from gene ontology analysis of the combined CS component: GO response to steroid hormone(33). While there was a strong correlation between the expression of the GSVA response to steroid hormone ontology and CSRGE (r = 0.548, p< 0.001), GSVA was less effective at separating CS+ from CS- patients (Fig. 2D). These data suggest that CSRGE is more effective than currently available GO libraries at determining the transcriptomic response of patients to CS treatment.
CSRGE and Clinical Traits in Asthma
Although CSRGE expression identified asthma patients receiving CSs in aggregate, we noticed high variability in CSRGE amongst MA+ and SA patients, with many having CSRGE values at or below the levels of HCs (Figure 2C). To investigate the nature of this variability in CSRGE, we first correlated CSRGE in MA+ and SA patients with various clinical traits. We found that CSRGE had a strong positive correlation with FEV1% predicted and FEV1/FVC % predicted in SA but not MA+ patients (Fig 3A&B), suggesting that appropriate CS response may protect lung function in SA. In both MA+ and SA patients there was a correlation between CSRGE and worsening Asthma Control Questionaire-6 (ACQ6) score, although the negative correlation was stronger in SA patients (Fig 3C). SA patients are older on average than those with MA. As age is known to impact lung gene expression(34), we next determined whether a relationship exists with CSRGE value. We found a positive correlation between CSRGE and age in MA+ patients but not in SA patients (Fig 3D), suggesting that age has minimal relationship to any differences seen in CSRGE value in SA. Overall, these results suggest that age independent, high CSRGE values were linked with improved clinical outcomes in SA but not MA.
Figure 3: Correlations Between CS-Response Gene Expression and Clinical Traits for Asthma Patients Taking CS.
A-D: Correlation between CSRGE and various clinical traits, split between severe asthma (red) and moderate asthma with CS (yellow).
Corticosteroid Response Dictates Clinical Outcomes in Severe Asthma
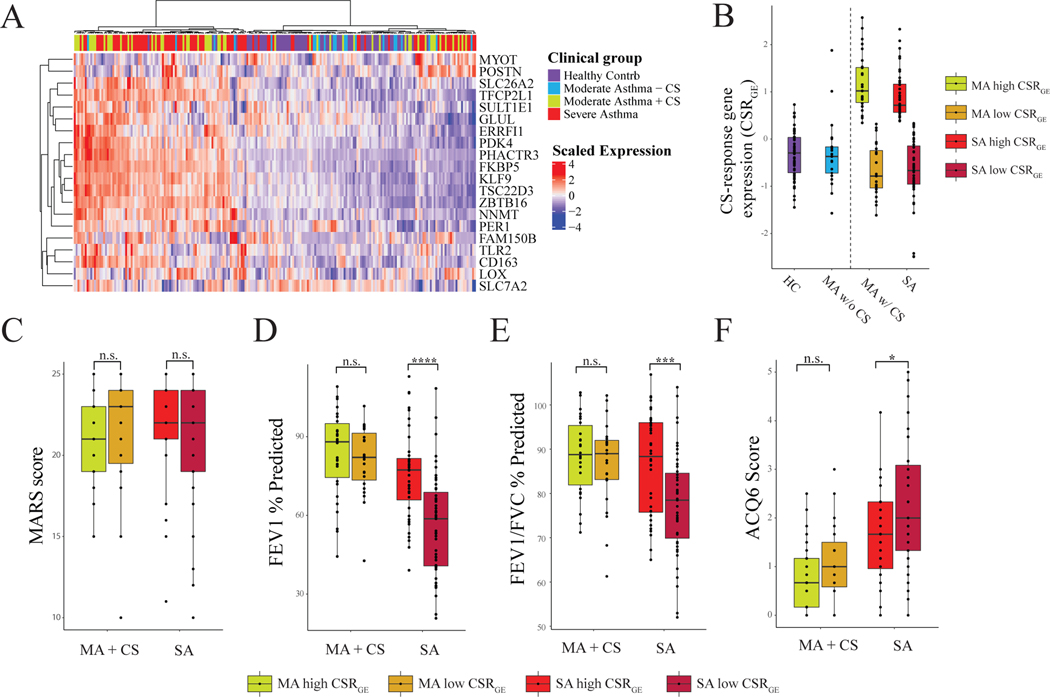
The current ERS/ATS definition of SA includes treatment with high dose inhaled CS, thereby suggesting poor responsiveness to therapy as an intrinsic feature(4). Our data suggest, however, that transcriptional response to CS may in fact vary amongst SA patients and this may have important implications for clinical outcomes. We clustered cohort participants by the top 20 genes in our combined CS component, effectively identifying HCs and MA- patients (Figure 4A). Though the majority of MA+ and SA patients were effectively delineated from those not receiving CSs, there were some individuals whose gene expression profile was similar but not identical to those not on therapy.
Figure 4: Heterogeneity in CSRGE Dictates Clinical Outcomes in SA.
(A) Heatmap showing clustering of patients for the combined IMSA+SARP dataset by the top 20 contributing genes to the two corticosteroid components (determined by taking the absolute value of the average contribution of each gene to the two components). (B) CSRGE for each diagnosis group, with Severe Asthma and Moderate Asthma with Corticosteroids split into high- and low- CSRGE groups. (C-F): Comparisons of clinical traits between high- and low- CSRGE groups. Statistics are performed with student’s t-test comparing high- and low- groups within each diagnosis group.
To further examine differences in clinical outcomes between high- and low-CSRGE groups, we split both the MA+ and SA patients into high- and low- CSRGE groups by the median CSRGE expression of the patients in the MA+ and SA groups (Fig 4B, Table S4 & S5). As medication adherence may complicate our assessment of CS response, we next investigated prevalence of CS associated comorbidities. We found no difference between MA-, MA+ and SA groups in prevalence of coronary artery disease, osteoporosis or diabetes mellitus. Importantly, we also found no difference in Medication Adherence Report Scale (MARS) score, a validated self-reporting metric for medication compliance, between CSRGE-high or -low patients with either MA+ or SA (Figure 4C)(35). There were, however, large differences in FEV1 % predicted and FEV1/FVC% predicted between high- and low- CSRGE groups in SA, but not MA+ (Figure 4D&E). There was also a significant difference in ACQ6 between high- and low-CSRGE groups in SA, but not MA+ (Figure 4F). Intriguingly, no difference in peripheral blood cell count, including eosinophils, fraction of exhaled nitric oxide (FeNO) or BMI was found between high- and low-CSRGE groups in either SA or MA+ (Figure S3), suggesting currently available biomarkers for evaluation of asthma patients cannot discern BEC CS response.
Transcriptomic Differences in CS Response Between Severe and Moderate Asthma Patients
These differences in clinical traits between CSRGE-high and -low asthma patients suggest that therapeutic response may result in, or be attributed to, broader changes to molecular phenotype. To investigate potential genes and pathways to explain this phenomenon, we performed differential expression between MA+ and SA patients, as well as between high- and low-CSRGE patients. We then plotted the log-fold change from the two differential expression comparisons together and split the plot into quadrants (Fig 5, Table S6). Gene ontology was then performed on each quadrant (Fig 5 and S4). Unsurprisingly, the high- CSRGE-specific quadrant (top-middle; yellow-green; Fig 5 and S4) was enriched for several CS-response related ontologies. Interestingly, the SA-specific quadrant (middle-left, green, Fig 5 and S4) was enriched for several terms related to bone morphogenic protein (BMP) signaling and prostaglandin pathways. The MA+ high-CSRGE quadrant (upper-right; light blue; Fig5 and S4), i.e. the gene expression for the patients most responsive to treatment, was enriched for several neurological ontologies. Both the low-CSRGE-specific quadrant (bottom- middle; orange; Fig 5 and S4) and the SA low-CSRGE quadrant (bottom-left; pink; Fig 5 and S4) were enriched for T-cell and lymphocyte related ontologies and cytokines. The bottom SA and low- CSRGE quadrant was also enriched for several ontologies related to peptidase activities.
Figure 5: Comparison of Differentially Expressed Genes by Asthma Severity and CS-Response Gene Expression.
Log-fold changes in differential expression between high- vs low- CSRGE groups (y-axis) and Moderate-with-CS vs Severe Asthma patients (x-axis). Hashed lines separating the plot into quadrants are drawn at +/− 1 on each axis. Summary terms for Gene Ontology analysis (Figure S4) are included in each hashed area.
The SA high- CSRGE quadrant (top-left; dark blue; Fig 5 and S4) expressed many genes associated with activation of fibroblasts and other mesenchymal cells, including MHY11, DES, ACTG2, and surfactant genes(36, 37), suggesting that activation and proliferation of mesenchymal cells could play a role in the pathophysiology of severe asthma. We further note that the contrast between CSRGE-high samples with upregulated mesenchymal gene expression and CSRGE-low samples with upregulated cytokine expression is similar to the contrast between myofibroblastic and inflammatory subpopulations of cancer-associated-fibroblasts(38–40).
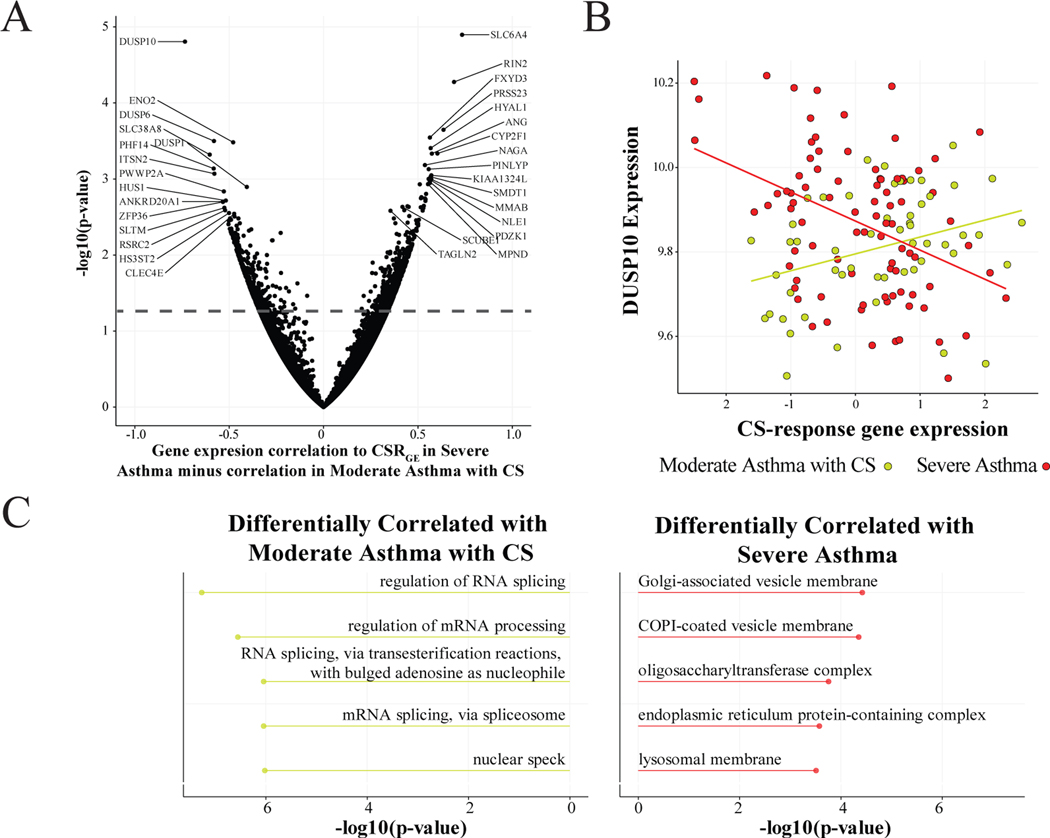
Although differential expression can identify genes with different mean expression between groups, there is also the possibility of a gene being correlated with CSRGE in one direction for SA and in the opposite direction for MA+. Such a gene might not be differentially expressed but could still be important in explaining the mechanism behind CS response in SA. Thus, we performed differential correlation analysis between MA+ and SA (Fig 6A, Table S7). Interestingly, among the genes with the lowest ratio of SA vs CSRGE correlation to MA+ vs CSRGE correlation were DUSP10 (Fig 6B) and DUSP6, which both negatively regulate members of the MAP kinase superfamily. Gene ontology on the top differentially correlated genes found that genes differentially corelated with MA+ CSRGE are enriched for multiple ontologies related to RNA splicing. This suggests that dysregulation of RNA splicing in SA may be responsible for blunted response to CS treatment.
Figure 6: Differential Correlation Analysis by Asthma Severity.
(A) Differential gene correlation between MA-with-CS and SA for CSRGE. (B) Expression levels of DUSP10 compared to CSRGE for Severe Asthma and Moderate-Asthma-with-CS. DUSP10 is negatively correlated with CSRGE in Severe Asthma (r = −0.404), but positively correlated with CSRGE in Moderate-Asthma-with-CS (r = 0.330). (C) Gene ontology for the top differentially correlated genes for Moderate-Asthma-with-CS and Severe Asthma. Top genes are selected as those with p-value < 0.05.
Predicting Lung CS Response Using Peripheral Blood
Though bronchial epithelial sampling is an invaluable tool for understanding asthma pathobiology, the resources required and risk imposed by the procedure are prohibitive for regular clinical evaluation of asthma patients. Prior work from our group showed that peripheral blood cell composition may reflect BEC transcriptional phenotype. We next determined whether peripheral blood transcriptional profile can be used to predict BEC CSRGE. Elastic net modeling on IMSA patients that had matched epithelial and peripheral blood (n = 36) showed strong correlation between measured BEC CSRGE values and those predicted by blood gene expression from matched IMSA cohort participants (Figure 7A). As CSRGE response class showed important relationship to both lung function and quality of life, we developed supervised learning models for class prediction. BEC gene expression could be reliably used to predict CSRGE response class using 55 genes (Figure 7B). We repeated this process using blood gene expression (Figure 7C), with the goal of identifying a minimal set of genes required to meet or exceed the sensitivity of our BEC model. CSRGE class prediction using supervised learning with blood gene expression was highly effective (Figure 7D). A 7-gene model was able to achieve a test sensitivity of 88%, comparable to performance of the original BEC data set. Of these 7 genes, 6 were down-regulated and 1 was up-regulated in CSRGE-high patients (Figure 7E). These data suggest that peripheral blood can be used to predict BEC expression of corticosteroid responsiveness genes.
Figure 7: Prediction of BEC CSRGE Using Peripheral Blood.
(A) Elastic Net (EN) predicted BEC CSRGE based on blood gene expression versus measured BEC CSRGE in IMSA. Grayed area indicates the 95% confidence bounds around a linear regression model comparing the two. Spearman’s rho and p-value are indicated in plot area. (B) Stacked bar plot of leave-one-out CSRGE response class prediction using BEC transcriptional data. P-value of chi square testing = 4.03e-22. (C) Stacked bar plot of leave-one-out CSRGE response class prediction using blood transcriptional data. P-value of chi square testing = 6.17e-05. (D) Receiver operating characteristics (ROC) curve of a sparse-partial least squares discriminant analysis (sPLS-DA) model for CSRGE class prediction. ROC curves were calculated using 5 fold-validation. Area under the curve (AUC) value is 0.9598 based on comparison of predicted scores of one class vs the other. Wilcoxon test of predicted scores < 1e-10. (E) Boxplot for expression of genes included the in the blood prediction model. Upper and lower hinges correspond to the first and third quartiles. The upper whisker extends from the hinge to 1.5 * Inter-quartile range (IQR) and the lower whisker extends from the hinge 1.5 * IQR of the hinge. Data beyond the end of the whiskers are plotted individually.
Discussion
Inhaled CSs have become ubiquitous in the treatment of persistent asthma symptoms. Morbidity and mortality from asthma have been significantly impacted and as a result these can be life changing medicines for many(1, 3). For some, however, disease control is never accomplished, even with dose escalation into ranges that characterize a patient as having severe disease. Prior studies have uncovered heterogeneity in immune cell profile and BEC transcriptional profile that are associated with SA using clustering techniques on patient cohort data(5, 6, 41). Here, we use a different approach to assessing heterogeneity amongst asthma patients, first defining CS response using publicly available data sets that specifically test their effect on BEC transcription. We then identify heterogeneity in CS response that dictates important clinical outcomes such as lung function and asthma control within the SA population.
Correlation between contributors to the CS components from pHBECs and endobronchial biopsies was highly significant but only showed a modest coefficient in statistical comparison of contribution (Figure S1), likely owed to technical and biological considerations. ICA utilizes every gene entry in the transcriptome for component assembly, meaning that many genes included in that calculation had very small component loading values(28). Correspondence between the data sets improved dramatically when considering top contributors to each component. It is important to note that pHBECs represent a more homogenous cell population than endobronchial biopsy samples which contain sub-epithelial stroma and embedded immune cells. It is also important to consider that the experiments in these external datasets were measured 6 hours after exposure to CS, and thus may not contain information regarding long-term repeated exposure to CS.
The lack of significant difference in CSRGE between MA+ and SA patients (Figure 2B) was surprising, as we expected to see a blunted CS response in the SA patients who, by clinical definition, are poorly responsive to CS treatment. Heterogeneity in these individuals instead suggests that CS dose escalation may remain an effective treatment modality for some moderate asthma patients. As CSRGE-low SA patients show decrements in lung function, use of alternative treatment modalities in those that show ineffective CS response may have significant impact on quality of life and objective measures of lung health. The detection of non-responsiveness thereby becomes a critical consideration. Whether CSRGE-low MA+ patients will progress to SA designation, and along the way develop worsening lung function and asthma control, remains to be seen.
Severe asthma patients in CSRGE-low and CSRGE-high groups could not be discerned using peripheral blood cell count or FeNO, two commonly used measures in clinical evaluation of asthma patients. Prior study has associated high levels of Type-2 inflammation, driven by cytokines such as IL-4, IL-5 and IL-13, with CS responsiveness in many but not all patients(42). Here we see that the most commonly used biomarkers of Type-2 inflammation (blood eosinophilia and FeNO) were not significantly different between those with BEC transcriptional response to CS and those without, suggesting that additional biomarkers may be needed to guide expedient patient triage to alternative therapies such as targeted biologics.
As FeNO has been linked to CS-responsiveness (and low adherence(43)) in asthma, a lack of difference between CSRGE-low and CSRGE-high groups also supports the notion that compliance alone does not explain transcriptional changes in BECs(44). Cohort participants identified by CSRGE levels and disease severity were transcriptionally distinct, which would also not be expected if medication adherence was the principal driver. Rather, we would have expected low CSRGE SA patients to resemble MA patients not on ICS. Although we did not have directly observed therapy or pharmacy data available, we did not find difference in self-reported adherence using the MARS scale(35), a scale validated for use across multiple disease states and care contexts, including asthma (35, 45–47).
Both moderate and SA patients with low CSRGE shared upregulation of IFNG, CXCL9 and CXCL10. These cytokines have been previously reported by our group and others in association with CS-resistant inflammation in asthma(6, 8, 9). Interferon-γ is a predominantly T cell derived cytokine; although production from macrophages has been noted, it is unlikely that this signal is being driven by bronchial epithelial cells present in endobronchial brushings. These data suggest that nested T cells may be a critical factor in driving CS resistance. Our recent data confirmed the presence of CD4 and CD8 T cells bearing surface markers of tissue residency within the airways of CS resistant asthma patients(6). Epithelial derived factors that may contribute to tissue resident memory (TRM) cell generation and maintenance have yet to be elucidated, but may offer novel avenues of treatment in asthma and other pathologies.
CS-resistant severe asthma was characterized by expression of CYP1B1, CCL26, IDO1, SLC26A4 and CPA4 in our study. The high expression of these genes could explain the poor prognosis of SA patients with low CSRGE: CYP1B1 and IDO1 are involved in the metabolism of tryptophan, which has been implicated in the pathology of asthma(48, 49). Furthermore, CYP1B1 is involved in the metabolism of steroid hormones and arachidonic acid(50). Curiously, CCL26, CYP1B1, and SLC26A4 are upregulated by IL4, whereas IDO1 expression in suppressed by IL4(51). IDO1 is, however, strongly induced by IFN-γ, suggesting a complex inflammatory microenvironment featuring both T1 and T2 signaling in the airways of these individuals(52, 53). We also found that severe asthma was enriched for terms related to BMP signaling, which has previously been implicated in asthma by a study that found increased BMP-4 and BMP-6 in asthma sputum supernatants with increased neutrophils (54). These data open the possibility for differential signaling within the airway microenvironment and may point to the importance of temporal regulation.
Mesenchymal genes were highly expressed in CSRGE-high patients, whereas cytokine-related genes were highly expressed in CSRGE-low patients. We noticed that this contrast is similar to the contrast between myofibroblastic and inflammatory cancer-associated fibroblasts, suggesting that differentiation of fibroblasts into heterogeneous subpopulations during activation could contribute to variation in treatment response among SA patients. Although further in vivo experiments are needed to test this hypothesis, these data suggest that experimental treatments focused on reverting cancer-associated fibroblasts to their quiescent state(55, 56) could also be beneficial in severe asthma.
In addition to their impact on transcription, corticosteroids are also known to exert post-transcriptional and translational effects (57). We identified several genes related to post-translational effects that had differential correlation of CSRGE and gene expression between severe asthma and moderate asthma. Among these was ZFP36 (also known as tristetraprolin), which is known to regulate immune response and inflammation through post-translational degradation of AU-rich element mRNAs, including several cytokines such as TNFα (58–60). Therefore, dysregulation of CS-induced post-transcriptional modification may play a role in resistance to CS in some severe asthma patients.
The ability to identify CS unresponsive patients using peripheral blood assessment may be a valuable tool in evaluation of asthma patients. A 7 gene test is tenable for use in a clinical laboratory setting and may offer value to individuals and health systems for expediting the triage of asthma patients to other treatments such as targeted biologic therapies. Some of the genes identified by predictive modeling may also give insight into biological processes governing systemic immune response of steroid-resistant asthma patients. TAS2R38 codes for a bitter taste receptor expressed by neutrophils and mast cells, as well as ciliated airway epithelium(61). Agonists of the TAS2R family have been shown to curtail allergen-induced inflammatory responses and mitigate airway remodeling in animal models of asthma. Peptidoglycan recognition protein (Pglyrp) 1 is a pattern-recognition protein that mediates antibacterial host defense(62). Mice lacking Pglyrp1 on hematopoietic cells have shown a reduction in HDM-induced eosinophilic and lymphocytic airway inflammation(63).
Using publicly available datasets, we report a transcriptional signal for CS response in BECs that is heterogeneously expressed in asthma patients and linked to lung function and symptom burden in severe disease. Those with poor CS response show hallmarks of T lymphocyte infiltration within their airways and are able to be identified using a blood gene expression signature, suggesting a systemic immune process. Further study may allow us to identify these individuals and restore CS sensitivity or tailor treatment to the specific drivers of their disease.
Supplementary Material
Acknowledgments
We thank Kathryn Scholl and John Trudeau for processing of the samples. Thank you to the co-investigators, staff at Washington University for their recruitment of participants and preparation of epithelial brushing samples. The following companies provided financial support for study activities at the Coordinating and Clinical Centers beyond the third year of patient follow-up: AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi-Genzyme-Regeneron, and TEVA.
Funding Support:
NIH grants P01AI106684 (to A.R. and S.E.W.), R01HL113956 (to A.R.), R01AI048927 (to A.R.), U10HL109152 (S.E.W.), U10HL109172 (to E.I. and B.D.L), U10HL109168 (N.N.J.), U10HL109250 (S.C.E), U10HL109164 (W.C.M), U10HL109257 (M.C), and U10HL109146 (to P.G.W. and S.A.C.)
Declaration of Interests
Sally Wenzel is a consultant for AstraZeneca, Novartis, Knopp, Glaxo Smith-Kline and Sanofi-Genzyme. She is also involved in clinical trials being run by AstraZeneca and an investigator-initiated study with Regeneron. Sally Wenzel and Anuradha Ray have research support from Pieris Pharmaceuticals. Matthew Camiolo is an independent consultant to Pieris Pharmaceuticals. The following companies provided financial support for study activities at the Coordinating and Clinical Centers beyond the third year of patient follow-up: AstraZeneca, Boehringer-Ingelheim, Genentech, GlaxoSmithKline, Sanofi-Genzyme-Regeneron, and TEVA. These companies had no role in study design or data analysis, and the only restriction on the funds was that they be used to support the SARP initiative.
Abbreviations:
- ICA
independent component analysis
- pHBEC
primary human bronchial epithelial cell
- BEC
bronchial epithelial cell
- CS
corticosteroid
- CSRGE
Corticosteroid response component gene expression
- GR
glucocorticoid receptor
- FeNO
exhaled nitric oxide
- IMSA
Immune Modulation in Severe Asthma
- SARP
Severe Asthma Research Program
- FDR
false discovery rate
- ROC
Receiver operating characteristic
- MA
moderate asthma
- SA
severe asthma
- MA+
moderate asthma with inhaled corticosteroid
- MA-
moderate asthma without inhaled corticosteroids
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Pavord ID, Mathieson N, Scowcroft A, Pedersini R, Isherwood G, Price D. The impact of poor asthma control among asthma patients treated with inhaled corticosteroids plus long-acting β(2)-agonists in the United Kingdom: a cross-sectional analysis. NPJ Primary Care Respiratory Medicine. 2017;27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray A, Camiolo M, Fitzpatrick A, Gauthier M, Wenzel SE. Are We Meeting the Promise of Endotypes and Precision Medicine in Asthma? Physiological reviews. 2020;100(3):983–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akinbami LJ, Sullivan SD, Campbell JD, Grundmeier RW, Hartert TV, Lee TA, et al. Asthma Outcomes: Healthcare Utilization and Costs. The Journal of allergy and clinical immunology. 2012;129(3 0):S49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. The European respiratory journal. 2014;43(2):343–73. [DOI] [PubMed] [Google Scholar]
- 5.Camiolo MJ, Zhou X, Wei Q, Trejo Bittar HE, Kaminski N, Ray A, et al. Machine learning implicates the IL-18 signaling axis in severe asthma. JCI Insight. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camiolo M, Zhou X., Oriss TB, Yan Q, Gorry M, Horne W, Trudeau JB, Chen W, Kolls JK, Ray P, Weisel FJ, Weisel NM, Aghaeepour N, Nadeau K, Wenzel SE, Ray A High Dimensional Profiling Clusters Asthma Severity by Lymphoid and Non-lymphoid Statu. Cell Reports. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camiolo MJ, Kale SL, Oriss TB, Gauthier M, Ray A. Immune responses and exacerbations in severe asthma. Current Opinion in Immunology. 2021;72:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raundhal M, Morse C, Khare A, Oriss TB, Milosevic J, Trudeau J, et al. High IFN-γ and low SLPI mark severe asthma in mice and humans. J Clin Invest. 2015;125(8):3037–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gauthier M, Chakraborty K, Oriss TB, Raundhal M, Das S, Chen J, et al. Severe asthma in humans and mouse model suggests a CXCL10 signature underlies corticosteroid-resistant Th1 bias. JCI Insight. 2017;2(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol. 2017;18(3):159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vockley CM, D’Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, et al. Direct GR Binding Sites Potentiate Clusters of TF Binding across the Human Genome. Cell. 2016;166(5):1269–81.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Druilhe A, Létuvé S, Pretolani M. Glucocorticoid-induced apoptosis in human eosinophils: mechanisms of action. Apoptosis. 2003;8(5):481–95. [DOI] [PubMed] [Google Scholar]
- 13.Brinkmann V, Kristofic C. Regulation by corticosteroids of Th1 and Th2 cytokine production in human CD4+ effector T cells generated from CD45RO- and CD45RO+ subsets. The Journal of Immunology. 1995;155(7):3322–8. [PubMed] [Google Scholar]
- 14.Levine SJ, Larivee P, Logun C, Angus CW, Shelhamer JH. Corticosteroids differentially regulate secretion of IL-6, IL-8, and G-CSF by a human bronchial epithelial cell line. American Journal of Physiology-Lung Cellular and Molecular Physiology. 1993;265(4):L360–L8. [DOI] [PubMed] [Google Scholar]
- 15.Frey A, Lunding LP, Ehlers JC, Weckmann M, Zissler UM, Wegmann M. More Than Just a Barrier: The Immune Functions of the Airway Epithelium in Asthma Pathogenesis. Front Immunol. 2020;11:761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loke T-K, Mallett KH, Ratoff J, O’Connor BJ, Ying S, Meng Q, et al. Systemic glucocorticoid reduces bronchial mucosal activation of activator protein 1 components in glucocorticoid-sensitive but not glucocorticoid-resistant asthmatic patients. Journal of Allergy and Clinical Immunology. 2006;118(2):368–75. [DOI] [PubMed] [Google Scholar]
- 17.Klaßen C, Karabinskaya A, Dejager L, Vettorazzi S, Van Moorleghem J, Lühder F, et al. Airway Epithelial Cells Are Crucial Targets of Glucocorticoids in a Mouse Model of Allergic Asthma. The Journal of Immunology. 2017;199(1):48. [DOI] [PubMed] [Google Scholar]
- 18.Nagasaki T, Schuyler AJ, Zhao J, Samovich SN, Yamada K, Deng Y, et al. 15LO1 dictates glutathione redox changes in asthmatic airway epithelium to worsen type 2 inflammation. J Clin Invest. 2022;132(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England). 2012;28(6):882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leigh R, Mostafa MM, King EM, Rider CF, Shah S, Dumonceaux C, et al. An inhaled dose of budesonide induces genes involved in transcription and signaling in the human airways: enhancement of anti- and proinflammatory effector genes. Pharmacology Research & Perspectives. 2016;4(4):e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mostafa MM, Rider CF, Shah S, Traves SL, Gordon PMK, Miller-Larsson A, et al. Glucocorticoid-driven transcriptomes in human airway epithelial cells: commonalities, differences and functional insight from cell lines and primary cells. BMC Medical Genomics. 2019;12(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miettinen J, Nordhausen K, Taskinen S. Blind Source Separation Based on Joint Diagonalization in R: The Packages JADE and BSSasymp. Journal of Statistical Software. 2017;76(2):1–31.36568334 [Google Scholar]
- 24.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS: A Journal of Integrative Biology. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNASeq data. BMC Bioinformatics. 2013;14(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–9. [DOI] [PubMed] [Google Scholar]
- 27.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedman JH, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. Journal of Statistical Software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 29.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS computational biology. 2017;13(11):e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Core Team R. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: (2013). Supplementary Figure S. 2015;2. [Google Scholar]
- 31.Team RC. R: A language and environment for statistical computing. 2013. [Google Scholar]
- 32.Kasela S, Ortega VE, Martorella M, Garudadri S, Nguyen J, Ampleford E, et al. Genetic and non-genetic factors affecting the expression of COVID-19-relevant genes in the large airway epithelium. Genome Med. 2021;13(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Izzotti A, Calin GA, Steele VE, Croce CM, De Flora S. Relationships of microRNA expression in mouse lung with age and exposure to cigarette smoke and light. FASEB J. 2009;23(9):3243–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fialko L, Garety PA, Kuipers E, Dunn G, Bebbington PE, Fowler D, et al. A large-scale validation study of the Medication Adherence Rating Scale (MARS). Schizophr Res. 2008;100(1–3):53–9. [DOI] [PubMed] [Google Scholar]
- 36.Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, et al. Cross-tissue organization of the fibroblast lineage. Nature. 2021;593(7860):575–9. [DOI] [PubMed] [Google Scholar]
- 37.Helms EJ, Berry MW, Chaw RC, DuFort CC, Sun D, Onate MK, et al. Mesenchymal Lineage Heterogeneity Underlies Nonredundant Functions of Pancreatic Cancer-Associated Fibroblasts. Cancer Discov. 2022;12(2):484–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, et al. IL1-Induced JAK/STAT Signaling Is Antagonized by TGFbeta to Shape CAF Heterogeneity in Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2019;9(2):282–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov. 2019;9(8):1102–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. American journal of respiratory and critical care medicine. 2014;190(12):1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters MC, Kerr S, Dunican EM, Woodruff PG, Fajt ML, Levy BD, et al. Refractory airway type 2 inflammation in a large subgroup of asthmatic patients treated with inhaled corticosteroids. The Journal of allergy and clinical immunology. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNicholl DM, Stevenson M, McGarvey LP, Heaney LG. The utility of fractional exhaled nitric oxide suppression in the identification of nonadherence in difficult asthma. American journal of respiratory and critical care medicine. 2012;186(11):1102–8. [DOI] [PubMed] [Google Scholar]
- 44.Silkoff PE, Laviolette M, Singh D, FitzGerald JM, Kelsen S, Backer V, et al. Identification of airway mucosal type 2 inflammation by using clinical biomarkers in asthmatic patients. The Journal of allergy and clinical immunology. 2017;140(3):710–9. [DOI] [PubMed] [Google Scholar]
- 45.Chan AHY, Horne R, Hankins M, Chisari C. The Medication Adherence Report Scale: A measurement tool for eliciting patients’ reports of nonadherence. British Journal of Clinical Pharmacology. 2020;86(7):1281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smits D, Brigis G, Pavare J, Maurina B, Barengo NC. Factors related to good asthma control using different medical adherence scales in Latvian asthma patients: an observational study. NPJ Primary Care Respiratory Medicine. 2017;27(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen JL, Mann DM, Wisnivesky JP, Horne R, Leventhal H, Musumeci-Szabó TJ, et al. Assessing the validity of self-reported medication adherence among inner-city asthmatic adults: the Medication Adherence Report Scale for Asthma. Annals of Allergy, Asthma & Immunology. 2009;103(4):325–31. [DOI] [PubMed] [Google Scholar]
- 48.Gostner JM, Becker K, Kofler H, Strasser B, Fuchs D. Tryptophan Metabolism in Allergic Disorders. Int Arch Allergy Immunol. 2016;169(4):203–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hayashi T, Beck L, Rossetto C, Gong X, Takikawa O, Takabayashi K, et al. Inhibition of experimental asthma by indoleamine 2,3-dioxygenase. J Clin Invest. 2004;114(2):270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Divanovic S, Dalli J, Jorge-Nebert LF, Flick LM, Gálvez-Peralta M, Boespflug ND, et al. Contributions of the three CYP1 monooxygenases to pro-inflammatory and inflammation-resolution lipid mediator pathways. J Immunol. 2013;191(6):3347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kagami S, Saeki H, Komine M, Kakinuma T, Tsunemi Y, Nakamura K, et al. Interleukin-4 and interleukin-13 enhance CCL26 production in a human keratinocyte cell line, HaCaT cells. Clin Exp Immunol. 2005;141(3):459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odemuyiwa SO, Ghahary A, Li Y, Puttagunta L, Lee JE, Musat-Marcu S, et al. Cutting edge: human eosinophils regulate T cell subset selection through indoleamine 2,3-dioxygenase. J Immunol. 2004;173(10):5909–13. [DOI] [PubMed] [Google Scholar]
- 53.Hastie AT, Steele C, Dunaway CW, Moore WC, Rector BM, Ampleford E, et al. Complex association patterns for inflammatory mediators in induced sputum from subjects with asthma. Clin Exp Allergy. 2018;48(7):787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hastie AT, Moore WC, Meyers DA, Vestal PL, Li H, Peters SP, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. The Journal of allergy and clinical immunology. 2010;125(5):1028–36 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99–115. [DOI] [PubMed] [Google Scholar]
- 56.Steele NG, Biffi G, Kemp SB, Zhang Y, Drouillard D, Syu L, et al. Inhibition of Hedgehog Signaling Alters Fibroblast Composition in Pancreatic Cancer. Clin Cancer Res. 2021;27(7):2023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Post-transcriptional Stellato C. and nongenomic effects of glucocorticoids. Proc Am Thorac Soc. 2004;1(3):255–63. [DOI] [PubMed] [Google Scholar]
- 58.Brahma PK, Zhang H, Murray BS, Shu FJ, Sidell N, Seli E, et al. The mRNA-binding protein Zfp36 is upregulated by beta-adrenergic stimulation and represses IL-6 production in 3T3-L1 adipocytes. Obesity (Silver Spring). 2012;20(1):40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Makita S, Takatori H, Nakajima H. Post-Transcriptional Regulation of Immune Responses and Inflammatory Diseases by RNA-Binding ZFP36 Family Proteins. Front Immunol. 2021;12:711633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newton R, Shah S, Altonsy MO, Gerber AN. Glucocorticoid and cytokine crosstalk: Feedback, feedforward, and co-regulatory interactions determine repression or resistance. J Biol Chem. 2017;292(17):7163–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nayak AP, Shah SD, Michael JV, Deshpande DA. Bitter Taste Receptors for Asthma Therapeutics. Front Physiol. 2019;10:884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, et al. Peptidoglycan Recognition Proteins Are a New Class of Human Bactericidal Proteins. Journal of Biological Chemistry. 2006;281(9):5895–907. [DOI] [PubMed] [Google Scholar]
- 63.Park SY, Jing X, Gupta D, Dziarski R. Peptidoglycan recognition protein 1 enhances experimental asthma by promoting Th2 and Th17 and limiting regulatory T cell and plasmacytoid dendritic cell responses. J Immunol. 2013;190(7):3480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.