Abstract
MYC is a transcription factor frequently overexpressed in cancer. To determine how MYC drives the neoplastic phenotype, we performed transcriptomic analysis using a panel of MYC-driven autochthonous transgenic mouse models. We found that MYC elicited gene expression changes mostly in a tissue and lineage specific manner across B-cell lymphoma, T-cell acute lymphoblastic lymphoma, hepatocellular carcinoma, renal cell carcinoma, and lung adenocarcinoma. However, despite these gene expression changes being mostly tissue-specific, we uncovered a convergence on a common pattern of upregulation of embryonic stem cell gene programs and downregulation of tissue-of-origin gene programs across MYC-driven cancers. These changes are representative of lineage dedifferentiation, that may be facilitated by epigenetic alterations that occur during tumorigenesis. Moreover, while several cellular processes are represented among embryonic stem cell genes, ribosome biogenesis is most specifically associated with MYC expression in human primary cancers. Altogether, MYC’s capability to drive tumorigenesis in diverse tissue types appears to be related to its ability to both drive a core signature of embryonic genes that includes ribosomal biogenesis genes as well as promote tissue and lineage specific dedifferentiation.
Keywords: MYC, oncogene, dedifferentiation, gene signature, transcriptomics, pan-cancer, tissue-lineage, embryonic stem cells, TCGA
Introduction
The MYC proto-oncogene, when mutated or overexpressed, has been implicated in the pathogenesis of most types of human cancer (1). MYC activation, in tandem with other genetic and epigenetic insults, results in initiation of tumorigenesis (2). Cancers driven by MYC become dependent on its sustained oncogenic activity, a phenomenon known as oncogene addiction (3-5). Hence, despite the genomic complexity of cancers, inactivation of MYC alone can cause reversal of the neoplastic phenotype (6-8).
MYC orchestrates a transcriptional program of genes involved in proliferation, survival, self-renewal, ribosome biogenesis, mitochondrial biogenesis, glucose and glutamine metabolism, nucleotide biosynthesis, lipogenesis, angiogenesis, and immune evasion (4,5,9-11). Genes regulated by MYC in cancer could present druggable targets. Thus, uncovering bona fide MYC-regulated genes could have promising therapeutic implications for MYC-driven cancers (12).
MYC acts as a transcriptional amplifier that can also specifically induce or suppress gene expression (13-17). Moreover, MYC and its proximal network are generally involved in human cancer (18). Clearly, MYC regulates the expression of many genes and many reports have identified MYC gene signatures (19-24). However, it is less clear if MYC regulates any common set of genes required for tumorigenesis. Alternatively, MYC may upregulate genes in a particular cancer that are needed for the growth and survival of a particular cellular lineage. MYC’s transcriptional network has been suggested to be cell type and context dependent (25). Therefore, the gene expression changes elicited by MYC that are generally important for tumorigenesis versus for pathogenesis of a specific cancer still remain elusive.
Many studies have used experimental cancer models or human clinical specimens to identify genes regulated by MYC. A recent study identified a prognostic MYC gene signature through microarray analysis of a transgenic murine model of MYC-induced lung adenocarcinoma (24). Other studies have probed the transcriptional landscape of MYC using human clinical specimens. One such study derived a prognostic MYC gene signature in human epithelial ovarian cancer and neuroblastoma (23). Furthermore, a pan-cancer analysis of The Cancer Genome Atlas (TCGA) data identified multi-cancer MYC-associated pathways (18).
Here, we examined how MYC regulates gene expression across different types of cancers using five genetically engineered mouse models (GEMMs) including tetracycline (Tet)-regulated conditional transgenic mouse models (26) of MYC-induced renal cell carcinoma (RCC) (27), hepatocellular carcinoma (HCC) (7), lung adenocarcinoma (LAC) (28), T-cell acute lymphoblastic lymphoma (T-ALL) (6), and a transgenic model of B-cell lymphoma (BCL) (29). In all these models, MYC can be overexpressed in a tissue-specific manner, allowing us to interrogate tissue-specific changes in gene expression brought about by MYC. Our findings point to large heterogeneity in gene expression changes among different MYC-driven cancer types. Despite this heterogeneity, we find that the gene expression changes across different MYC-driven cancers manifest a common profile of tissue dedifferentiation, represented by alterations in embryonic stem cell-like gene programs, particularly ribosome biogenesis genes, as well as tissue-of-origin gene programs.
Results
Differentially Expressed Genes in MYC-Induced Cancer
We used conditional transgenic mouse models with tissue-specific inducible MYC expression for RCC, HCC, LAC, and T-ALL (6,7,27,28) (Supplementary Fig. 1) under the Tet System (26,30). For BCL, we leveraged RNA-seq data from the transgenic model of Eμ-Myc lymphomagenesis (15,29).
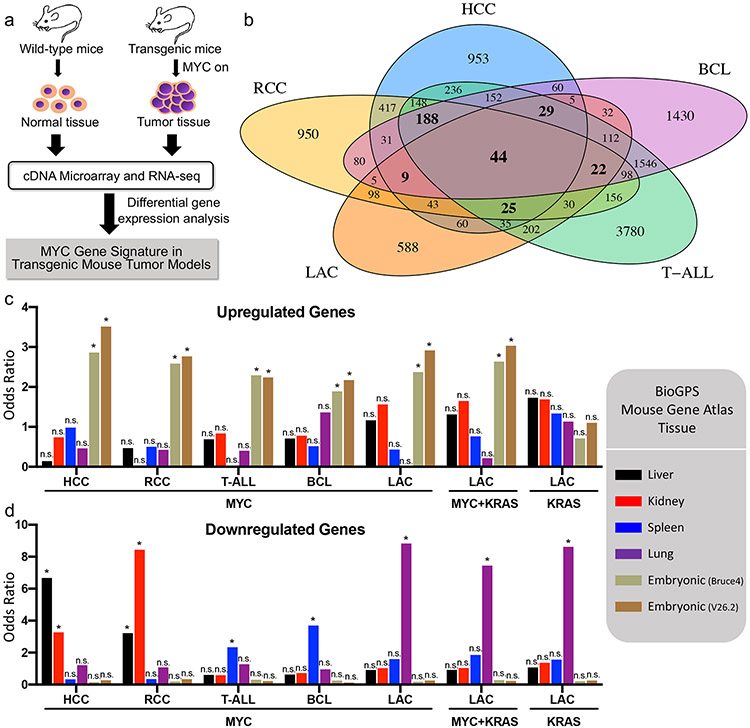
We performed differential gene expression analysis on these five transgenic mouse tumor models (Table 1), comparing MYC-induced tumor tissue to control normal tissue (Fig. 1a). Genes were considered differentially expressed (DE) if they changed at least 2-fold with adjusted p-value < 0.05. This analysis unveiled thousands of DE genes (Supplementary Table 1), only 44 of which were common to all five transgenic mouse models (Fig. 1b, Supplementary Fig. 2). Thus, there is large heterogeneity in differentially expressed genes among the five MYC-driven transgenic mouse tumor models.
Table 1:
Transgenic mouse models used in gene expression profiling in this study
| Tumor Type | Mouse Background |
Mouse Genotype | Number of Samples | |
|---|---|---|---|---|
| Hepatocellular carcinoma (HCC)1 | FVB/N | LAP-tTA/TetO-MYC | n = 4 liver | n = 6 tumor |
| Renal cell carcinoma (RCC)1 | FVB/N | GGT-tTA/TetO-MYC | n = 3 kidney | n = 3 tumor |
| Lung adenocarcinoma (LAC)1 | FVB/N | CCSP-rtTA/TetO-MYC | n = 2 lung | n = 5 tumor |
| FVB/N | CCSP-rtTA/TetO-KRASG12D | n = 5 tumor | ||
| FVB/N | CCSP-rtTA/TetO-MYC/TetO-KRASG12D | n = 4 tumor | ||
| T-cell acute lymphoblastic lymphoma (T-ALL)2 | FVB/N | EμSRα-tTA/TetO-MYC | n = 3 spleen | n = 3 tumor |
| B-cell lymphoma (BCL)3 | C57/Bl6 | Eμ-Myc | n = 4 spleen | n = 3 tumor |
Figure 1. MYC overexpression selectively dysregulates embryonic stem cell genes and tissue-lineage genes in tumorigenesis.
a, Workflow for identifying genes differentially expressed (DE) in MYC-induced transgenic mouse tumor models. b, Venn diagram of the DE genes among five MYC transgenic mouse models. c, Enrichment of mouse tissue-lineage genes among DE upregulated genes from transgenic mouse tumors. Mouse tissue-lineage genes are genes highly expressed in mouse liver, kidney, spleen, lung, or embryonic stem cell tissue based on BioGPS Mouse Gene Atlas data. For LAC, KRASG12D-induced (KRAS) and MYC/KRASG12D co-induced (MYC+KRAS) tumors were also included in addition to the MYC-induced tumors. d, Same as c except enrichment of mouse tissue-lineage genes was assessed among DE downregulated genes from transgenic mouse tumors. *p < 0.000595 (Bonferroni correction for 84 tests, α = 0.05, one-sided Fisher's exact test), n.s. = not significant.
Tissue Lineage Specificity of MYC Cancer Gene Signatures
Tissue enrichment analysis was performed using differential gene expression data for each of the five transgenic mouse models to identify whether specific tissue-lineages were enriched among the differentially expressed genes. Tissue-lineage genes, which consist of genes abundantly expressed in a given tissue type, were derived from the BioGPS Mouse Gene Atlas (31,32). We specifically investigated gene sets of liver, kidney, lung, and spleen tissue, which were the tissue-of-origin of the mouse tumors. Additionally, we explored embryonic stem cell genes, which are commonly overexpressed in cancer (33). We identified that, across MYC-driven cancers, the upregulated genes were enriched in embryonic stem cell (ESC) genes (Fig. 1c) while downregulated genes for a given cancer type were enriched in genes abundantly expressed in the respective tissue-of-origin (Fig. 1d, Supplementary Table 2). However, in KRAS-driven LAC, although lung tissue genes were downregulated, ESC genes were not upregulated unlike in MYC-driven cancers, demonstrating that different oncogenes alter different gene programs to promote tumorigenesis. Taken together, the results suggest that MYC-driven cancers repress corresponding tissue-lineage genes and promote ESC gene profiles.
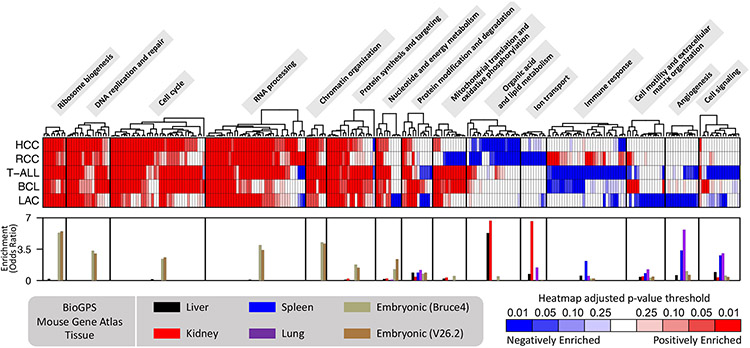
We identified, using gene set enrichment analysis (GSEA), the top gene ontology (GO) terms associated with the differential gene expression profiles in each MYC-induced transgenic mouse tumor model (Fig. 2, Supplementary Table 3). Related GO terms were grouped into pathways (Supplementary Table 4). Across models, programs related to cell cycle control, DNA replication, chromatin organization, protein synthesis, RNA processing, and ribosome biogenesis were consistently upregulated by MYC activation. This is congruent with long-standing knowledge about MYC’s role in regulating cell proliferation (9) and with reports suggesting a cell-type independent function of MYC is to drive biomass accumulation (20). We also observed tissue-specific pathway enrichment patterns, such as downregulation of lipid metabolism genes in HCC and RCC, downregulation of ion transport genes in RCC, downregulation of immune response genes in T-ALL and BCL, and downregulation of angiogenesis genes and extracellular matrix (ECM) organization genes in LAC. However, we were careful to note that the corresponding tissue-of-origin also showed enrichment for these pathways (Supplementary Table 5). For example, genes abundantly expressed in normal kidney tissue exhibit enrichment for ion transport and genes abundantly expressed in normal lung tissue exhibit enrichment for angiogenesis. Furthermore, embryonic stem cell genes showed enrichment for the MYC-upregulated pathways. Thus, these pathway enrichment results for the differential gene expression profiles are likely just a consequence of the normal tissue gene expression profile becoming dedifferentiated, lending further support for the importance of considering dedifferentiation in interpreting gene expression changes in MYC-driven cancers.
Figure 2. Pathways associated with genes differentially expressed in MYC-driven tumorigenesis reflect tissue dedifferentiation.
Gene ontology (GO) gene set enrichment analysis (GSEA) of genes, ranked by log2 fold change, was performed for each of the five MYC-induced transgenic mouse tumor models. Heatmap shows the GSEA adjusted p-values for the top GO terms among the different models. Each column of the heatmap represents a distinct GO term and similar GO terms were grouped together into pathways. The bar graphs at the bottom depict pathway enrichment among lung, liver, kidney, spleen, and embryonic stem cell tissue-lineage genes (BioGPS Mouse Gene Atlas); each bar shows the median enrichment across the GO terms in the pathway.
Epigenetic Changes Underlying MYC-Driven Differential Gene Expression in Cancer
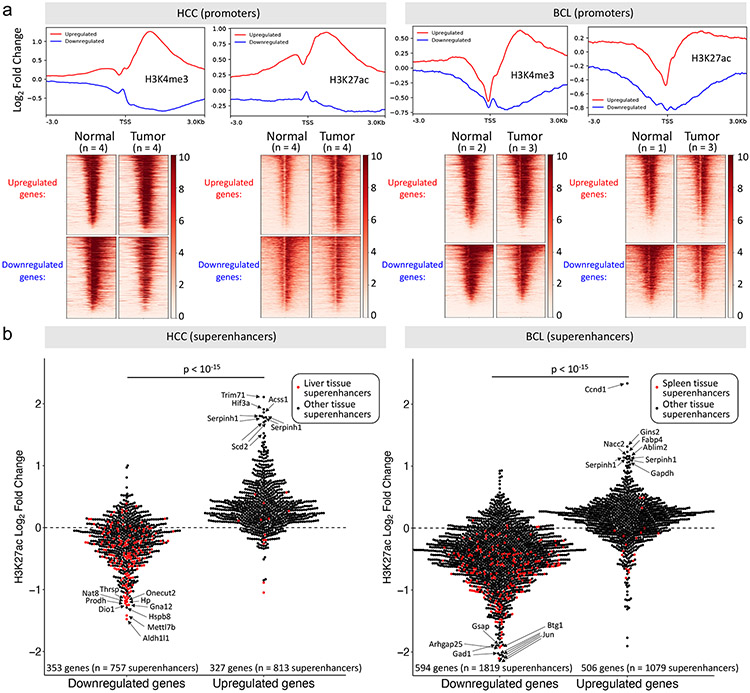
To investigate the mechanisms underlying the gene expression changes in MYC-induced tumorigenesis, we explored chromatin immunoprecipitation sequencing (ChIP-seq) data from the HCC and BCL transgenic mouse models (15,34). Comparing tumor to normal tissue, promoters of upregulated genes in each model became enriched in H3K4me3 and H3K27ac, histone marks of active transcription, while promoters of downregulated genes became depleted of these marks (Fig. 3a). Next, we looked at superenhancers (35) associated with the differentially expressed genes (Supplementary Table 6). We found tissue-specific changes in superenhancer activity in that, for each tumor model, most superenhancers associated with the upregulated genes became enriched in H3K27ac, an active enhancer mark, while most superenhancers associated with the downregulated genes became depleted in H3K27ac (Fig. 3b, Supplementary Fig. 3). Of note, liver tissue superenhancers tended to be associated with the downregulated genes, not the upregulated genes, in HCC and the same was observed for spleen tissue superenhancers with regards to BCL. This agrees with our gene expression analyses showing the high tissue-of-origin specificity of downregulated genes. These results collectively show that MYC-driven tumors have an altered epigenetic state that is consistent with gene expression changes.
Figure 3. Gene expression changes in MYC-induced tumorigenesis are associated with epigenetic changes.
a, Metagene plots and heatmaps showing changes in H3K4me3 and H3K27ac in the promoters of genes upregulated and downregulated in MYC-induced mouse HCC (left) and BCL (right) (GEO accession numbers: GSE76042 and GSE51004, respectively). Log2 fold changes reflect ChIP-Seq signal changes in tumor relative to normal tissue, both normalized to an input sample. b, ChIP-Seq H3K27ac signal changes in tumor relative to normal for superenhancers (obtained from dbSUPER) associated with genes upregulated and downregulated in the MYC-induced mouse HCC (left) and BCL (right) model. P-values were determined by two-sided Welch’s t-test.
Identification of a Gene Signature Associated with MYC Expression in Human Cancers
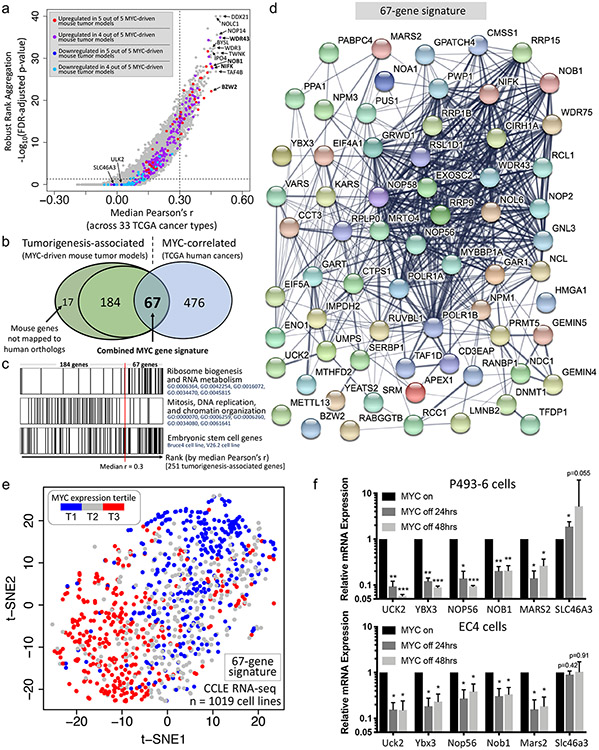
Human TCGA pan-cancer RNA-seq data of 9354 patient primary cancer samples across 33 cancer types were used to identify genes associated with MYC expression. We performed pairwise Pearson correlation between each gene and MYC gene expression values and performed robust rank aggregation (RRA) (36) using the Pearson correlation coefficients (Pearson’s r) for each gene across the 33 cancer types (Fig. 4a, Supplementary Table 7). We considered genes with median Pearson’s r > 0.30 and RRA adjusted p-value < 0.05 to be MYC-correlated genes. These genes are the ones most likely to be specific to MYC’s activity whereas genes with lesser correlation may be involved in many cancers in general, not just those with high MYC expression. We considered tumorigenesis-associated genes to be those upregulated in at least 4 out of 5 MYC-driven mouse tumor models. These genes are likely to be important in MYC-induced tumorigenesis as they have been demonstrated upregulation across multiple MYC-driven mouse tumor models. These genes are also generally upregulated in TCGA human cancers (Supplementary Fig. 4). We filtered the 251 tumorigenesis-associated genes that had a human ortholog to only include genes that are also MYC-correlated, resulting in a final set of 67 genes (Fig. 4b, Supplementary Table 8). These 67 genes, unlike the other 184 tumorigenesis-associated genes that aren’t as strongly correlated with MYC expression, are enriched in ribosome biogenesis and RNA metabolism genes but do not exhibit enrichment for DNA replication, cell division, and chromatin organization pathways which were highly enriched among the other 184 genes (Fig. 4c, Supplementary Table 9). Both the 67-gene signature and the remaining 184 tumorigenesis-associated genes are enriched in embryonic stem cell genes. Together, these results suggest that while embryonic stem cell genes are involved in multiple pathways and are upregulated in MYC-driven cancers, some pathways like ribosome biogenesis tend to be more specific for MYC activity.
Figure 4. A tumorigenesis gene signature that is highly associated with MYC expression in primary human cancers.
a, One-sided volcano plot representing the pairwise Pearson correlation between each gene’s expression and MYC expression across 33 cancer types in The Cancer Genome Atlas (TCGA)’s RNA-seq dataset (n = 9354 primary human cancers). Robust rank aggregation (RRA) was performed across the 33 cancer types by ranking each gene by its Pearson correlation with MYC expression within each cancer type. b, Venn diagram showing the overlap between tumorigenesis-associated genes (genes considered upregulated in at least 4 out of 5 MYC-driven mouse tumor models) and MYC-correlated genes (genes with median Pearson’s r > 0.30 and RRA adjusted p-value < 0.05). This 67-gene overlap constitutes the combined MYC gene signature. c, Barcode plot showing the representation of two distinct pathways (based on gene ontology) and embryonic stem cell genes (based on BioGPS Mouse Gene Atlas data) among the tumorigenesis-associated genes when the genes are ordered from low correlation with MYC expression (left) to high correlation with MYC expression (right). d, Protein-protein interaction (PPI) network of genes from the 67-gene MYC gene signature. e, t-distributed stochastic neighbor embedding (t-SNE) plot showing the clustering of cancer cell lines (from CCLE RNA-seq data) based on this study’s 67-gene signature. MYC expression tertiles are colored. f, Changes in mRNA expression upon MYC inactivation of five selected signature genes plus SLC46A3 in P493-6 cells and EC4 cells, a cell line derived from a transgenic mouse HCC tumor. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed one-sample Student's t test (n = 3 independent RT-qPCR experiments).
The 67-gene signature forms a densely interconnected protein-protein interaction network (Fig. 4d), representative of the fact that most of the genes are related in function. We validated this gene signature using cancer cell line RNA-seq data from the Cancer Cell Line Encyclopedia (CCLE) (37). Clustering analysis on CCLE expression data shows that these 67 genes could separate cancer cells with high MYC expression from those with low MYC expression fairly well (Fig. 4e). Moreover, several signature genes such as NOB1, RSL1D1, and MARS2 were among the genes most correlated with MYC expression (Pearson’s r > 0.475) across the CCLE cancer cell lines (Supplementary Table 10). We further validated five signature genes: UCK2, YBX3, NOP56, NOB1, and MARS2 plus the MYC-downregulated gene, SLC46A3, by RT-qPCR in two cell lines which possess tetracycline-regulated MYC overexpression: P493-6 cells, a transformed human B cell line, and EC4 cells, a tumor-derived mouse HCC cell line. RT-qPCR confirmed MYC inactivation (Supplementary Fig. 5) and confirmed that all these genes, barring SLC46A3, were significantly downregulated by MYC inactivation, while SLC46A3 either showed no change or slight upregulation (Fig. 4f). Immunohistochemistry on mouse tissue further validates that Apex1, a signature gene that is also a member of the embryonic stem cell gene sets, and Mki67, a tumorigenesis-associated gene, decrease in expression at the protein-level upon MYC inactivation in both MYC-driven T-ALL (Fig. 5a,b, Supplementary Fig. 6) and MYC-driven HCC (Fig. 5c,d, Supplementary Fig. 6). Additionally, liquid chromatography with tandem mass spectrometry (LC-MS/MS)-based proteomics on mouse tissue from the MYC-driven HCC transgenic mouse model (38) reveals that embryonic signature genes, including the aforementioned Apex1, are upregulated while Adh1, which is a specific hepatocyte marker as it encodes the main alcohol dehydrogenase responsible for ethanol oxidation in the liver, is downregulated in MYC-driven HCC compared to normal liver tissue (Fig. 5e, Supplementary Fig. 7). These proteomic results provide additional validation of identified signature genes and, importantly, provide further support for tissue dedifferentiation in MYC-driven cancers. Finally, a patient survival meta-analysis (39) across 33 cancer types in TCGA suggests that the 67 gene signature is associated with poor prognosis (Supplementary Fig. 8; see Methods).
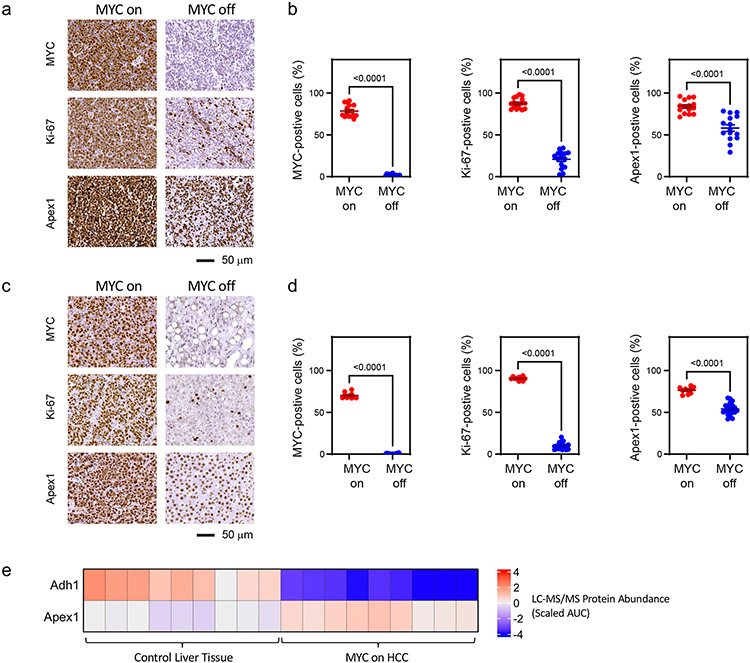
Figure 5. Examples of in situ gene expression changes upon alterations in MYC status.
Immunohistochemical analysis of thymic tissue (a,b) from EμSRα-tTA/TetO-MYC mice (MYC on: n = 3, MYC off: n = 3) and liver tissue (c,d) from LAP-tTA/TetO-MYC mice (MYC on: n = 2, MYC off: n = 4). Tissues were evaluated for expression of MYC, Ki-67, and Apex1. a,c) Representative images of immuno-stained tissue sections. b,d) Quantified frequency of MYC-, Ki-67-, and Apex-1-positive cells. Five regions of interest were analyzed from each tissue sample. P-values were calculated using the two-sided Mann-Whitney U test. e, Liquid chromatography with tandem mass spectrometry (LC-MS/MS)-based proteomic analysis showing protein abundance changes for Adh1 and Apex1 in LAP-tTA/TetO-MYC mice (MYC on: n = 3 samples, Control liver tissue: n = 3 samples) (n = 3 technical replicates per sample). AUC: Area Under the Curve.
Discussion
MYC and its associated network of interacting gene products are important to the regulation of gene transcription and have been implicated in the pathogenesis of most types of human cancer (18). Here we have performed a multi-cancer transcriptomic analysis on transgenic mouse models to reveal that although the genes differentially expressed in MYC-driven tumorigenesis depend on the tissue in which MYC is overexpressed, a common gene expression pattern of tissue dedifferentiation emerges across different types of MYC-driven tumors.
Our results are the first to look across multiple MYC-driven conditional transgenic mouse models. We identified MYC-driven differential gene expression profiles in BCL (3843 DE genes), T-ALL (6803 DE genes), HCC (2435 DE genes), RCC (2344 DE genes) and LAC (1339 DE genes) using transgenic mouse models. We noted a very limited degree of overlap in DE genes among the different models, suggesting that the DE genes are highly tissue-specific. This could be one reason why, while many previous reports have defined MYC gene signatures (19-24), often the gene lists are dissimilar. Moreover, our work agrees with prior reports that MYC functions as an amplifier of gene expression (13,14), by selectively affecting particular sets of genes (15-17). However, we note that even though MYC overexpression can influence total RNA production, the transcriptional effect of MYC is, in general, selective and specific. We found that across multiple tissue types, certain groups of genes are differentially affected by MYC overexpression and the specific genes affected by MYC are tissue-dependent.
We had wondered whether the disparate differential gene expression profiles represented different pathways or different genes within the same pathway. We hypothesized that because MYC overexpression can drive tumorigenesis in various tissue types, the disparate differential gene expression profiles should converge on a common oncogenic theme. Indeed, we found that MYC appears to commonly upregulate embryonic stem cell-like gene programs and downregulate tissue-of-origin gene programs in tumorigenesis, alterations that are representative of tissue dedifferentiation. Our results have implications for how MYC initiates and maintains a cancer state.
We therefore propose a model whereby MYC overexpression causes a new tissue state to form which allows for tissue-specific dedifferentiation gene expression changes (Fig. 6). These changes depend on the initial tissue type and lineage. For example, the downregulated genes for a given cancer type tend to be enriched in genes that are highly expressed in the tissue type from which the cancer originated and not other tissue types. We suggest that the gene expression profile formed in the new tumor tissue state is likely the result of both direct effects of MYC on gene expression in its capacity as a transcription factor as well as epigenetic changes. These epigenetic changes, specifically in enhancer and promoter activity markers, that occur in MYC-driven tumorigenesis are corroborated by our analysis of ChIP-seq data. MYC is known to epigenetically alter gene expression by directly recruiting proteins that catalyze histone modifications (40,41) and by reprogramming cell state (42,43). In breast cancer, it has been shown that MYC-driven epigenetic reprogramming represses luminal lineage-specifying transcription factors and induces a stem cell-like state (44). Our results implicate that MYC may function in a similar way across multiple other tissue types.
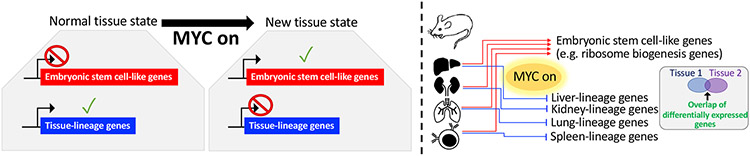
Figure 6. Proposed model for how MYC overexpression results in tissue-specific selective gene expression changes in tumorigenesis.
Graphical representation showing that MYC overexpression results in a tissue state which involves higher expression of embryonic stem cell-like genes and lower expression of tissue-lineage genes. This gene expression program change is likely the result of both direct effects of MYC on gene expression in its capacity as a transcription factor as well as epigenetic changes.
Genes that more broadly seem to be regulated by MYC across cancers appear to mostly be related to embryonic stem cell genes. The upregulation of embryonic stem cell genes in cancer has been suggested previously (33,45) and stem cell pluripotency depends on higher rates of rRNA synthesis and ribosome biogenesis (46-48) like MYC-driven cancers (49,50). MYC has been shown to exhibit stem-cell specific gene expression even in primitive metazoans such as the Hydra (51). MYC is reported to function in self-renewal and maintenance of pluripotency in mouse embryonic stem cells (52) as well as in promoting embryonic stem cell expression programs in mouse fibroblasts (53). We realized that even genes broadly differentially expressed across MYC-driven cancers in transgenic mouse models may not necessarily be specific to MYC-driven cancers. To identify genes that are more likely to be specific to MYC-driven cancers, we explored the TCGA dataset and narrowed our MYC gene signature to a set of 67 genes that were at least moderately correlated with MYC expression in primary human cancers. These 67 genes, unlike the genes with weaker correlation with MYC, were significantly enriched in ribosome biogenesis genes suggesting that ribosome biogenesis is most specific to MYC-driven cancers. In contrast, other pathways, such as DNA replication and cell division, might not depend specifically on MYC activity but rather are features of many cancers in general. Thus, these 67 genes warrant further investigation as potential vulnerabilities in MYC-driven cancers.
Our observations are consistent with a multitude of reports that suggest that the MYC oncogene is most capable of eliciting tumorigenesis in immature tissue lineages (54). For example, experimental MYC overexpression in embryonic liver elicits rapid onset of the immature pediatric associated cancer, hepatoblastoma; whereas, in adult liver, elicits the longer latency onset of adult hepatocellular carcinoma (7,55). Similarly, the inactivation of MYC appears to elicit differentiation of lymphoma, leukemia, osteosarcoma, and hepatocellular carcinoma (6-8,56). Our results support a model of tumorigenesis that MYC oncogene activation and differentiative context are intrinsically tied together: changing MYC activation status can change the differentiative context of a cell and a particular differentiative state of a cell is permissive for tumorigenesis (56). That model, in turn, is consistent with our proposed explanation for the pattern of differential expression of lineage-specific genes with a core signature of embryonic genes across MYC-driven tumors of various tissue origins: each tissue progresses from its unique initial state toward a common dedifferentiated end state in tumorigenesis. Thus, MYC appears to hijack gene expression programs of less mature, embryonic cellular lineages, thereby resulting in dedifferentiation; and correspondingly, suppression of MYC can result in cellular maturation and differentiation.
Our findings further support the idea that the transition to the tumor tissue state involves specific epigenetic and transcriptional profile changes, which are dictated by both MYC overexpression and the initial tissue state. In part, this could be a direct consequence of MYC’s regulation of epigenetic programs, whether through miR-17-92 which controls the expression of certain chromatin-modifying genes (57), physical interaction with chromatin-remodeling complexes (58-60), direct regulation of DNA methyltransferases (61), or other mechanisms of regulation of chromatin regulatory genes (57,62-64). Since tumorigenesis is a multi-step process (65), it is probable that a step-wise succession of feedback loops involving gene expression changes gives rise to new cellular states. As such, future work should explore the progression of the dedifferentiation gene expression trajectory over a tumorigenesis time course. Future studies should additionally explore the effect of other members of the MYC proximal network that shield a more nuanced insight into how gene expression is regulated in a specific manner across cancer types (18). Finally, further identification of how MYC regulates these key regulatory nodal points may uncover therapeutic targets.
Materials and methods
Code Availability
For transparency of methods, all the code and procedures that can be used to reproduce the results in this paper, starting from the raw data, are available at https://github.com/Yenaled/felsher and the relevant data from all public datasets used in this study (e.g. TCGA, CCLE, dbSUPER, etc.) are also accessible via this repository.
Mouse Microarray and RNA-Seq Data Analysis and Availability
All mouse microarray and RNA-Seq data used in this study are publicly available on the Gene Expression Omnibus (GEO) repository. Microarrays were performed by the Stanford Functional Genomics Facility using Illumina mouse microarray platforms. The microarray data is available at GEO accession number GSE143254. The microarray data was further processed to remove poorly detected probes (detection p-value > 0.01 across all samples) followed by quantile-normalization then log2-transformation using the R package, lumi (version 2.38.0). All animal studies, which were performed to generate immunohistochemistry and the microarray datasets, were approved by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). For the conditional mouse models, oncogene activation was induced after weaning age (4-6 weeks of age). Male and female mice were used in roughly equal proportion and were not distinguished in this study. Eμ-Myc RNA-seq data (15) and T-ALL RNA-seq data (66) from previous studies were obtained from GEO (accession numbers GSE51008 and GSE106078, respectively). For RNA-seq, kallisto (version 0.44.0) (67) was used to pseudoalign raw reads to the GRCm38 (mm10) mouse with 100 bootstraps. The R libraries, limma (version 3.42.0) (68) and sleuth (version 0.30.0) (69), were used for differential gene expression analysis of microarray and RNA-seq data, respectively.
Analysis of TCGA and CCLE RNA-Seq Data
Human RNA-seq data from The Cancer Genome Atlas (TCGA) was downloaded from the UCSC Xena Toil project (70), which provides the log2-transformed RSEM expected counts (71) normalized using DESeq2 (72). The R library, ComBat (73), was used for batch effect correction in TCGA data using biospecimen batch identifier data obtained from the National Cancer Institute's Genomic Data Commons (GDC) data portal. RNA-seq TPM data from the Cancer Cell Line Encyclopedia (CCLE) was obtained from the Broad Institute and this data was further normalized using the DESeq2 method followed by log2-transformation.
Gene List Analysis
Enrichment analysis, providing odds ratios and Fisher’s exact test p-values to test the representation of gene sets given a list of genes, was performed using Enrichr (74). For gene ontology (GO) terms, the Biological Process category is used. Gene set enrichment analysis (GSEA) (75) was performed on the log2 fold change differential gene expression profiles of each of the five transgenic mouse tumor models using the fgsea (version 1.12.0) R package (76). Protein-protein interaction network analysis was performed using the STRING database (77), with the settings "meaning of network edges": "confidence", "active interaction sources": all selected, "minimum required interaction score": 0.400, and "max number of interactors to show": "query proteins only".
ChIP-Seq Analysis
MYC and histone ChIP-Seq data were processed using the ENCODE ChIP-Seq pipeline (78) (with Macs2 as the peak caller) and further analyzed using deepTools (79). For ChIP-Seq analysis of mouse superenhancer regions, mouse superenhancer genomic coordinates and associated gene symbols were obtained from dbSUPER (35) and the coordinates were converted to mm10 coordinates using the UCSC Genome Browser tool: liftOver.
Cell Culture
P493-6 cells were maintained in RPMI 1640 medium (with L-glutamine) supplemented with 10% FBS and Antibiotic-Antimycotic. EC4 cells were maintained in maintained in Dulbecco's Modified Eagle Medium (DMEM) (with 4.5g/L D-glucose), supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine, 1% sodium pyruvate, 1% nonessential amino acids, and Antibiotic-Antimycotic. For these cell lines, MYC inactivation was achieved by adding doxycycline to the culture medium at a concentration of 20 ng/mL. Cells were not routinely authenticated or monitored for mycoplasma. Source of cells - P493-6: Laboratory of D. Eick; EC4: Derived from tumor of HCC mouse model.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA was isolated from cells by the RNeasy Mini Kit (QIAGEN) and reverse-transcribed into cDNA using the SuperScript III First-Strand Synthesis kit (ThermoFisher) following manufacturer's protocol. Quantitative PCR was performed in 384-well plates on the QuantStudio 12K Flex Real-Time PCR System, using SYBR Green I dye as fluorophore to detect amplicons. Reactions were carried out in 20-μL volumes that contained 0.5 μL cDNA, 0.25 μM forward and reverse primers, and SYBR Green PCR Master Mix (Applied Biosystems). The following amplification cycle settings were used: 50 °C for 2 minutes; 95 °C for 10 minutes; and 40 cycles of 95 °C for 15 seconds, 60 °C for 1 minute, and 72 °C for 30 seconds. The 2−ΔΔCT method was used to plot relative mRNA levels with UBC serving as a reference gene. Primers used in this study are shown in Supplementary Table 11.
Survival Meta-analysis
TCGA patient overall survival data was obtained from the TCGA Pan-Cancer Clinical Data Resource (TCGA-CDR) dataset (80). The z-scores from the Cox proportional hazards regression model for each gene were obtained using the coxph function of the survival (version 3.1-12) package in R and were aggregated across all TCGA cancer types into meta-z scores via Stouffer's method (unweighted) akin to the methods described in PRECOG (39). The z-scores quantify prognostic significance of genes with positive z-scores suggesting poor prognosis and negative z-scores suggesting good prognosis. The z-scores and meta z-scores for all genes are supplied in Supplementary Table 12.
Immunohistochemistry
Thymus tissue was collected from EμSRα-tTA/TetO-MYC mice that were untreated (MYC on, n = 3), or treated with 0.1mg/ml doxycycline in drinking water for 8 days upon apparent signs of disease (MYC off, n = 3). Liver tissue was collected from LAP-tTA/TetO-MYC mice that were untreated (MYC on, n = 2), or treated with 0.1mg/ml doxycycline in drinking water for 2 weeks upon apparent signs of disease (MYC off, n = 4). Tissue was fixed in 10% formalin at room temperature for two days, incubated in 70% ethanol overnight, and embedded in paraffin. Tissue was cut in 5 μm sections. After deparaffinization, antigen retrieval was performed in 95°C-heated epitope retrieval buffer (10% in water, Dako Target Retrieval Solution (10x), Agilent) for 45 minutes. Slides were washed with PBS, incubated in 3% hydrogen peroxide (Sigma Aldrich) in phosphate-buffered saline (PBS) and washed in PBS before blocking with protein block (Serum-Free Protein Block, Agilent). Slides were stained with primary antibodies in DAKO diluent at 4°C overnight. Slides were washed in PBS and incubated with biotinylated secondary antibodies in DAKO diluent for 30 minutes at room temperature if primary antibody was unlabeled. The following antibodies were used in this study: c-MYC (1:500, clone D84C12, Cell Signaling Technology, Cat# 5605), Ki-67-biotin (1:100, clone SolA15, Invitrogen, Cat# 13-5698-82), Apex1 (1:100, clone E5Y2C, Cell Signaling Technology, Cat# 10519), biotinylated anti-rabbit (1:300, Vectastain ABC kit, Vector Laboratories, Cat# BA-1000). DAB staining (ImmPACT DAB Peroxidase Substrate Kit, Vector Laboratories) was performed and nuclei were counterstained with hematoxylin (Hematoxylin QS, Vector Laboratories). Slides were mounted in Permount (Fisher Chemical) mounting medium and slide scans were acquired using Aperio AT2 scanner (Leica Biosystems). Thresholding for positive cell detection was based on DAB intensity in corresponding normal tissue for each target. QuPath software was used for image quantifications. Five regions of interest were analyzed from each tissue sample. Statistical analysis by Mann-Whitney test was done in Prism (Version 9.3.1, GraphPad Software, LLC).
Proteomics
The methods and analysis of liquid chromatography with tandem mass spectrometry (LC-MS/MS) proteomics on MYC-driven HCC (n = 3 biological replicates; n = 3 technical replicates) and control liver tissue (n = 3 biological replicates; n = 3 technical replicates) was described previously (38). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (81) partner repository with the dataset identifier PXD031478.
Statistics
Statistical tests for investigating gene sets or for differential gene expression analysis were performed using appropriate established tools in the literature. For normally distributed log-scale data, the Welch’s t-test was performed when the variances were unequal between two groups, otherwise, the standard Student’s t-test was performed. When variation within a group of data is reported in a figure, the standard deviation is used. No statistical methods were used to predetermine sample sizes, no randomization was performed, and no blinding was performed. For cell culture experiments, three independent experiments were performed. All false discovery rate (FDR) adjusted p-values were determined by the Benjamini-Hochberg procedure (82).
Supplementary Material
Acknowledgments
We thank Andrew J. Gentles and Felsher laboratory members for their advice and guidance. We thank Stanford Research Computing Center for computational resources, including the Sherlock cluster. We are grateful for the public data from TCGA and from the Broad Institute’s CCLE. This work was funded by National Institutes of Health (NIH) grants: R35 CA253180, R01 CA089305, R01 CA170378, and U01 CA188383, with additional support as follows. D.K.S. - UCLA-Caltech Medical Scientist Training Program (NIH NIGMS training grant T32 GM008042). A.D. - Lymphoma Research Foundation. R.D. - NIH grant CA222676 from the National Cancer Institute (NCI), American College of Gastroenterology Junior Faculty Career Development Grant. A.M.G. - Stanford Cancer Translational Nanotechnology Training T32 Training Grant CA196585 (NCI). D.F.L - Tumor Biology Training Grant (NIH 5T32CA009151-38), Stanford University (NCI), Burroughs Wellcome Fund Postdoctoral Enrichment Award, Research Supplement Award (NCI), 3U01CA188383-03S1, and (K01) CA234453 (NCI).
Footnotes
The authors declare no competing financial interests.
Declaration of Interests
The authors have no conflicts of interest to disclose.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012. Mar 30;149(1):22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014. Jun 2;4(6):a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002. Jul 5;297(5578):63–4. [DOI] [PubMed] [Google Scholar]
- 4.Felsher DW. MYC Inactivation Elicits Oncogene Addiction through Both Tumor Cell-Intrinsic and Host-Dependent Mechanisms. Genes Cancer. 2010. Jun;1(6):597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Casey SC, Felsher DW. Inactivation of MYC reverses tumorigenesis. J Intern Med. 2014. Jul;276(1):52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. [DOI] [PubMed] [Google Scholar]
- 7.Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004. Oct 28;431(7012):1112–7. [DOI] [PubMed] [Google Scholar]
- 8.Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002. Jul 5;297(5578):102–4. [DOI] [PubMed] [Google Scholar]
- 9.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med. 2013. Aug;3(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouw AM, Margulis K, Liu NS, Raman SJ, Mancuso A, Toal GG, et al. The MYC Oncogene Cooperates with Sterol-Regulated Element-Binding Protein to Regulate Lipogenesis Essential for Neoplastic Growth. Cell Metab. 2019. Sep 3;30(3):556–572.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002. Oct 1;16(19):2530–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018. Dec 23;3(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012. Sep 28;151(1):68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012. Sep 28;151(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabò A, Kress TR, Pelizzola M, de Pretis S, Gorski MM, Tesi A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014. Jul 24;511(7510):488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walz S, Lorenzin F, Morton J, Wiese KE, von Eyss B, Herold S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014. Jul 24;511(7510):483–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science. 2018;360(6390):800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, et al. Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst. 2018. Mar 28;6(3):282–300.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4(10):R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One. 2011;6(10):e26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C-H, Sahoo D, Arvanitis C, Bradon N, Dill DL, Felsher DW. Combined Analysis of Murine and Human Microarrays and ChIP Analysis Reveals Genes Associated with the Ability of MYC To Maintain Tumorigenesis. Cheung VG, editor. PLoS Genet. 2008. Jun 6;4(6):e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006. Jan 6;439(7074):353–7. [DOI] [PubMed] [Google Scholar]
- 23.Jung M, Russell AJ, Liu B, George J, Liu PY, Liu T, et al. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res. 2017. Feb 15;77(4):971–81. [DOI] [PubMed] [Google Scholar]
- 24.Ciribilli Y, Borlak J. Oncogenomics of c-Myc transgenic mice reveal novel regulators of extracellular signaling, angiogenesis and invasion with clinical significance for human lung adenocarcinoma. Oncotarget. 2017. Nov 24;8(60):101808–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006. Aug;16(4):253–64. [DOI] [PubMed] [Google Scholar]
- 26.Yeh ES, Vernon-Grey A, Martin H, Chodosh LA. Tetracycline-regulated mouse models of cancer. Cold Spring Harb Protoc. 2014. Oct 1;2014(10):pdb.top069823. [DOI] [PubMed] [Google Scholar]
- 27.Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, et al. MYC oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci U S A. 2015. May 26;112(21):6539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J, et al. Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS One. 2008. May 7;3(5):e2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985. Dec 1;318(6046):533–8. [DOI] [PubMed] [Google Scholar]
- 30.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992. Jun 15;89(12):5547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004. Apr 20;101(16):6062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008. May;40(5):499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kress TR, Pellanda P, Pellegrinet L, Bianchi V, Nicoli P, Doni M, et al. Identification of MYC-Dependent Transcriptional Programs in Oncogene-Addicted Liver Tumors. Cancer Res. 2016;76(12):3463–72. [DOI] [PubMed] [Google Scholar]
- 35.Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res. 2016. Jan 4;44(D1):D164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019. May;569(7757):503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnan MS, Rajan Kd A, Park J, Arjunan V, Garcia Marques FJ, Bermudez A, et al. Genomic Analysis of Vascular Invasion in HCC Reveals Molecular Drivers and Predictive Biomarkers. Hepatology. 2021;73(6):2342–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015. Aug 20;21(8):938–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001. Aug 15;15(16):2069–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS One. 2008;3(11):e3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006; [DOI] [PubMed] [Google Scholar]
- 43.Farrell AS, Joly MM, Allen-Petersen BL, Worth PJ, Lanciault C, Sauer D, et al. MYC regulates ductal-neuroendocrine lineage plasticity in pancreatic ductal adenocarcinoma associated with poor outcome and chemoresistance. Nat Commun. 2017;8(1):1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poli V, Fagnocchi L, Fasciani A, Cherubini A, Mazzoleni S, Ferrillo S, et al. MYC-driven epigenetic reprogramming favors the onset of tumorigenesis by inducing a stem cell-like state. Nat Commun. 2018. Dec 9;9(1):1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008. Apr 10;2(4):333–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brombin A, Joly J-S, Jamen F. New tricks for an old dog: ribosome biogenesis contributes to stem cell homeostasis. Curr Opin Genet Dev. 2015. Oct;34:61–70. [DOI] [PubMed] [Google Scholar]
- 47.Turi Z, Lacey M, Mistrik M, Moudry P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY). 2019. Apr 26;11(8):2512–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe-Susaki K, Takada H, Enomoto K, Miwata K, Ishimine H, Intoh A, et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 2014. Dec;32(12):3099–111. [DOI] [PubMed] [Google Scholar]
- 49.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008. Dec 18;456(7224):971–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010. Apr 1;10(4):301–9. [DOI] [PubMed] [Google Scholar]
- 51.Hartl M, Mitterstiller AM, Valovka T, Breuker K, Hobmayer B, Bister K. Stem cell-specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan Hydra. Proc Natl Acad Sci U S A. 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, et al. Myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009. Jan 23;136(2):364–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene — the grand orchestrator of cancer growth and immune evasion. Nature Reviews Clinical Oncology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beer S, Zetterberg A, Ihrie RA, McTaggart RA, Yang Q, Bradon N, et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Biol. 2004;2(11):e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsher DW. Cancer revoked: Oncogenes as therapeutic targets. Nature Reviews Cancer. 2003. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Choi PS, Casey SC, Dill DL, Felsher DW. MYC through miR-17-92 suppresses specific target genes to maintain survival, autonomous proliferation, and a neoplastic state. Cancer Cell. 2014. Aug 11;26(2):262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stojanova A, Tu WB, Ponzielli R, Kotlyar M, Chan P-K, Boutros PC, et al. MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation. Cell Cycle. 2016;15(13):1693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soshnikova NV, Tatarskiy EV, Tatarskiy VV, Klimenko NS, Shtil AA, Nikiforov MA, et al. PHF10 subunit of PBAF complex mediates transcriptional activation by MYC. Oncogene. 2021. Oct 21;40(42):6071–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng SWG, Davies KP, Yung E, Beltran RJ, Yu J, Kalpana GV. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet. 1999. May;22(1):102–5. [DOI] [PubMed] [Google Scholar]
- 61.Poole CJ, Zheng W, Lodh A, Yevtodiyenko A, Liefwalker D, Li H, et al. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget. 2017. Sep 29;8(44):76898–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tesi A, de Pretis S, Furlan M, Filipuzzi M, Morelli MJ, Andronache A, et al. An early Myc-dependent transcriptional program orchestrates cell growth during B-cell activation. EMBO Rep. 2019. Sep 23;20(9):e47987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sabò A, Amati B. Genome recognition by MYC. Cold Spring Harb Perspect Med. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kieffer-Kwon K-R, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, et al. Myc Regulates Chromatin Decompaction and Nuclear Architecture during B Cell Activation. Mol Cell. 2017. Aug 17;67(4):566–578.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Land H, Parada LF, Weinberg RA. Cellular oncogenes and multistep carcinogenesis. Science. 1983. Nov 18;222(4625):771–8. [DOI] [PubMed] [Google Scholar]
- 66.Swaminathan S, Hansen AS, Heftdal LD, Dhanasekaran R, Deutzmann A, Fernandez WDM, et al. MYC functions as a switch for natural killer cell-mediated immune surveillance of lymphoid malignancies. Nat Commun. 2020;11(1):2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–7. [DOI] [PubMed] [Google Scholar]
- 68.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 69.Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017. Jul;14(7):687–90. [DOI] [PubMed] [Google Scholar]
- 70.Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, et al. Toil enables reproducible, open source, big biomedical data analyses. Nat Biotechnol. 2017;35(4):314–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011. Aug 4;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007. Jan;8(1):118–27. [DOI] [PubMed] [Google Scholar]
- 74.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005. Oct 25;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019. Jan 1;60012. [Google Scholar]
- 77.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019. Jan 8;47(D1):D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012. Sep;22(9):1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018;173(2):400–416.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50(D1):D543–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B. 1995. Jan;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.