Abstract
The tertiary structure of the 3′-cleaved product of the genomic hepatitis delta virus (HDV) ribozyme was solved by X-ray crystallographic analysis. In this structure, three single-stranded regions (SSrA, -B and -C) interact intricately with one another via hydrogen bonds between nucleotide bases, phosphate oxygens and 2′-OHs to form a nested double pseudoknot structure. Among these interactions, two Watson–Crick (W–C) base pairs, 726G–710C and 727G–709C, that form between SSrA and SSrC (P1.1) seem to be especially important for compact folding. To characterize the importance of these base pairs, ribozymes were subjected to in vitro selection from a pool of RNA molecules randomly substituted at positions 709, 710, 726 and 727. The results establish the importance of the two W–C base pairs for activity, although some mutants are active with one G–C base pair. In addition, the kinetic parameters were analyzed in all 16 combinations with two canonical base pairs. Comparison of variant ribozymes with the wild-type ribozyme reveals that the difference in reaction rates for these variants (ΔΔG‡) is not simply accounted for by the differences in the stability of P1.1 (ΔΔG037). The role played by Mg2+ ions in formation of the P1.1 structure is also discussed.
INTRODUCTION
Human hepatitis delta virus (HDV) is a satellite virus of human hepatitis B virus (1). The genome of HDV is a single-stranded circular RNA molecule (∼1700 nt) that is thought to replicate by a rolling circle mechanism (1,2). During replication both genomic and anti-genomic strands of HDV RNA undergo a self-cleavage reaction (ribozyme) (3–6). The HDV ribozyme motif belongs to a group of small ribozymes, such as hammerhead, hairpin and Neurospora VS ribozymes, in which the cleavage reaction yields products with 2′,3′-cyclic phosphates and 5′-OH termini in the presence of divalent metal ions, usually Mg2+ (3,4). The rate of cleavage is very fast (7,8) and the self-cleavage reaction occurs in human cells (9).
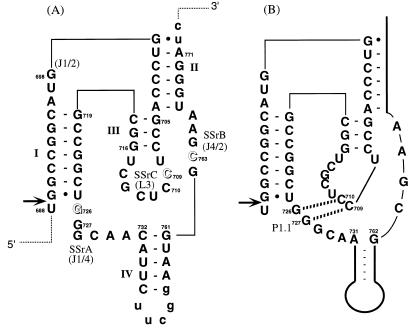
The secondary structure of the HDV ribozyme is quite different from other small ribozymes. Among several proposed secondary structure models, the pseudoknot structure (10) has been generally accepted based on its agreement with experimental results. The pseudoknot consists of four stems (I–IV) and three single-stranded regions (SSrA, -B and -C) as shown in Figure 1A. Stem I includes 7 base pairs in the intact molecule and includes the 5′-cleavage site; a single nucleotide is sufficient on the 5′-side for activity. Stems II and III are required to retain the structure of the catalytic core and stem IV is not essential for ribozyme activity (11). Bases essential for ribozyme activity were previously identified in the three single-stranded regions by site-directed mutagenesis (12,13) followed by in vitro selection from a partly randomized sequence pool of HDV ribozyme molecules (14). It was found that 726G, 709C and 763C cannot be substituted by any other bases.
Figure 1.
Pseudoknot secondary structure of genomic HDV ribozyme. (A) Numbering is based on that of Makino et al. (36). To truncate the original stem IV, the wild-type sequence, from nucleotides 736 to 758, was changed to include a UUCGG sequence. Lowercase letters at the 3′ end indicate vector-derived sequences. Single-stranded regions are marked as SSrA (nucleotides 726–731), SSrB (nucleotides 762–766) and SSrC (nucleotides 708–715) as reported previously (12). Names in parentheses are those used by other groups (2,15). Arrows and open letters indicate self-cleavage site and essential residues for activity, respectively. (B) W–C base pairs identified by X-ray analysis. The dashed lines indicate hydrogen bonds.
The tertiary structure of the 3′-cleaved fragment of genomic HDV ribozyme was elegantly solved by X-ray crystallographic analysis (15). This tertiary structure is very similar to the previously proposed computer graphics model (7). However, the whole structure assumes a more compact folding, that is a nested double pseudoknot structure, and the crystal structure provides information on several previously unrecognized hydrogen bond interactions. In particular, two Watson–Crick (W–C) base pairs between SSrA and SSrC, 726G–710C and 727G–709C (P1.1 in Fig. 1B) are of significant interest because they were not predicted from previous work, including earlier models and many mutagenesis studies (7,12–14,16). However, it was anticipated that these base pairs are necessary to maintain a compact structure and to assemble the essential bases in the catalytic core. Recently, Wadkins et al. (17) analyzed the functional importance of P1.1 using point mutagenesis.
In this paper, to survey all possible combinations of P1.1 base pairs, the importance of the two W–C base pairs at 726–710 and 727–709 for genomic HDV ribozyme activity are clarified by using an in vitro selection procedure. To further characterize these base pairs, the kinetic parameters are compared for all variants of the two canonical base pairs and other variants identified by in vitro selection.
MATERIALS AND METHODS
Synthesis of oligonucleotides and cis-acting HDV ribozymes
G0 DNA PCR template for in vitro selection and PCR primers were synthesized using an automated DNA/RNA synthesizer (model 392; Applied Biosystems). DNA phosphoramidites were purchased from Glen Research. 4N G0 ssDNA for in vitro selection was as follows: 5′-TGTAATACGACTCAC- TATAGGCTCGAGCCTGATGGCCGGCATGGTCCCAGC-CTNNTCGCTGGCGCCGGCTNNGCAACATTCTTCGGC-GAATGGGATCC-3′ (T7 promoter region is underlined and N denotes AGC and T).
The PCR primers were as follows:
(+)T7, 5′-TGTAATACGACTCACTATA-3′;
primer 1, 5′-GGATCCCATTCGCCATTCCG-3′;
primer 2 for first PCR, 5′-GGCCGGCATGGTCCCAGCCT-3′;
primer 3 for second PCR, 5′-TGTAATACGACTCACTATA-GGCCTGATGGCCGGCATGGTCCCAGCCT-3′;
(+) primer and (–) primer for construction of rest of variants were 5′-TATAGGCTCGAGCCTGATGGCCGGCATGGTC-CCAGCCTN1N2TCGCTGGCGCCGGCT-3′ and 5′-TCTAG-AGGATCCCATTCGCCATTCCGAAGAATGTTGCN3N4A-GCCGGCGCCAGCGA-3′, respectively (Nn denotes proper substituted base).
In vitro selection of active variants
For the G0 DNA pool, 4N G0 ssDNA (860 pmol) was converted to dsDNA by PCR (Gene Taq, Nippon Gene) using the primer (+)T7 (860 pmol). The PCR product was used as a template for transcription in vitro by T7 RNA polymerase (T7 Ampliscribe Kit, Epicentre Technologies, USA). After incubation for 3 h at 37°C, an equal volume of stop solution (50 mM EDTA, 9 M urea, 0.1% xylene cyanol, 0.1% bromophenol blue) was added. The mixture was heated at 90°C for 2 min, snap-cooled on ice, and separated on an 8% polyacrylamide gel that contained 7 M urea. After visualization by UV shadowing, the appropriate band was excised from the gel and eluted with elution buffer [0.3 M sodium acetate, 1 mM EDTA (pH 8.0)] and the RNA was recovered from precipitation with ethanol. The 3′-processed G1 RNA pool was reverse-transcribed by avian myeloblastosis virus reverse transcriptase (40 U, Seikagaku Co. Ltd) at 42°C for 1 h. The resulting cDNA was amplified twice by PCR: the first PCR was with primers 1 and 2 (94°C for 1 min, 55°C for 1 min and 72°C for 1 min, 15 cycles). The product was purified by agarose gel electrophoresis and extracted using Quantum Prep Freeze’N Squeeze DNA Gel Extraction Spin column (Bio-Rad). Using this PCR product as a template a second PCR was carried out using primers 2 and 3 (PCR conditions were the same as for the first PCR). The selected DNA pool was then subjected to another cycle of selection.
Sequence analysis of isolated variants
The G3 DNA pool was subcloned into the TA cloning vector (Invitrogen) according to the supplier’s protocol and transformed into a bacterial host (Inv αF′One Shot™ Competent Cell). The transformants were examined by colony PCR screening (14). Plasmids were prepared from an overnight culture and purified with GFX™ Micro Plasmid Prep Kit (Pharmacia Biotech) and sequenced using a BigDye Terminator Cycle Sequencing Kit on a 377 automatic sequencer (Applied Biosystems).
Site-directed mutagenesis
Several mutants not obtained by in vitro selection were prepared by PCR mutagenesis; systematic substitutions were incorporated into (+) and/or (–) primers and the appropriate plasmid was used as a PCR template. The mutated PCR products were digested by XhoI and BamHI, inserted into pUCT7 (14) ligated by T4 DNA ligase (Takara) and transformed into Escherichia coli MV1184. Mutants were characterized by the procedure described above.
Preparation of cis-acting ribozyme and assay of cleavage activity
Plasmid DNA linearized with BamHI was used for transcription in vitro. The reaction mixture (30 µl) for transcription contained 40 mM Tris–HCl (pH 8.0), 8 mM MgCl2, 2 mM spermidine, 5 mM dithiothreitol, 2 mM ribonucleotides, 0.5 mCi/ml [α-32P]CTP, 3 µg of linear plasmid DNA and 150 U of T7 RNA polymerase (Takara). After 30 min at 37°C, an equal volume of stop solution was added, and the mixture was heat-denatured and fractionated by electrophoresis on 8% PAGE containing 7 M urea. The transcript RNA was located by autoradiography and the uncleaved precursor RNA was excised from the gel, extracted with elution buffer, and recovered by ethanol precipitation.
Cleavage reactions containing ∼5–50 nM RNA were conducted in 40 mM Tris–HCl (pH 7.4) and 10 mM MgCl2 at 37°C. The labeled cis-acting ribozyme in 40 mM Tris–HCl (pH 7.4) was denatured at 90°C for 2 min, slowly cooled down over 1 h, and preincubated at 37°C for 10 min. The reaction was started by adding prewarmed MgCl2 solution. At appropriate times, aliquots of the reaction mixture were removed and the reaction was stopped by adding an equal volume of stop solution on ice. After electrophoretic fractionation on 8% PAGE containing 7 M urea, the precursor and 3′-cleaved product were identified using a bioimaging analyzer (BAS2000; Fuji Film). Cleavage activity was determined from the rate at which the cleaved product was formed. The cleaved fraction was calculated as (counts3′product)/(countsprecursor + counts3′product). The first-order rate constant (k) and end point (EP) were obtained by fitting data to the equation: cleaved yield (%) = [EP]·(1 – e–kt). In the case of poor fit to the monophasic equation, kinetic parameters were calculated from the arbitary assumed biphasic first-order equation as a trial; cleavage yield (%) = [EP1]·(1 – e–k1t) + [EP2]·(1 – e–k2t) (18).
RESULTS AND DISCUSSION
Identification of active 4N variants by in vitro selection
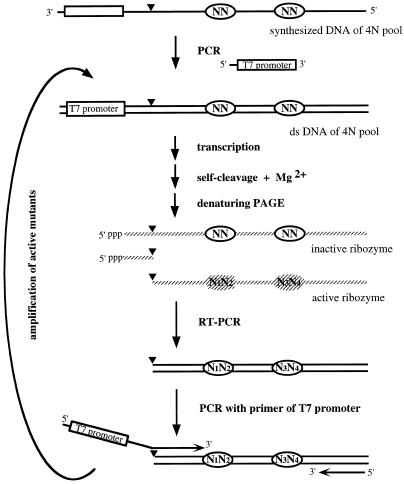
Short pseudoknot base pairs, P1.1, at 726N–710N/727N–709N (Fig. 1B) were observed in the crystal structure of the 3′-fragment of genomic HDV ribozyme (15), but this work did not determine whether these base pairs are necessary for ribozyme activity. Recently, Wadkins et al. (17) prepared 18 mutants by using point mutagenesis at the P1.1 on genomic and anti-genomic HDV ribozymes and reported the importance of P1.1. To study this point more thoroughly, in vitro selection was carried out using a pool of variants at these four positions on the cis-acting HDV ribozyme. The scheme of this procedure is shown in Figure 2. The variants for in vitro selection were generated from a chemically synthesized ssDNA pool of cis-acting HDV ribozyme (CdS4) containing random mutations at the positions 709, 710, 726 and 727 (random mixture of A, G, C and T at each position). The ssDNA pool was converted to dsDNA (G0 DNA) by PCR and transcribed with T7 RNA polymerase in vitro (G0 RNA). Active ribozymes were self-processed during the transcription reaction due to the presence of Mg2+ ions in the reaction buffer. The 3′-fragment was purified by denaturing PAGE, converted to cDNA by RT–PCR and then reamplified using primer 3 (containing the region of the promoter of T7 RNA polymerase) and 5′-processed fragment. A new RNA pool (G1) was prepared with this PCR product (G1 DNA) by in vitro transcription and used for the next cycle of selection.
Figure 2.
Scheme of the in vitro selection of active HDV ribozymes from random sequences at 4N positions. Solid strand, DNA; gray strand; RNA; N, randomized bases.
This cycle was repeated through three generations (G3) and the fraction of active molecules (during transcription for 3 h) in the RNA pool of each generation was monitored. The activity increased with each generation as follows: G0 = 16%, G1 = 39%, G2 = 59% and G3 = 87%. This result clearly indicates that the population of active ribozymes increases during each selection cycle. Theoretically, there are molecules with 256 different sequences in the starting G0 pool. If two W–C base pairs are necessary for the ribozyme to have cleavage activity, it is estimated that 6% of the G0 pool RNA should show cleavage activity (4 × 4 = 16/256). If one W–C base pair is sufficient for activity, 44% of the pool should be active [112/256; 112 = 256 – (16–4)2]. But the actual fraction of the G0 pool with activity was 16%, which is approximately equal to the fraction of the molecules having at least one G–C base pair [12% = 31/256; 31 = 256 – (16–1)2].
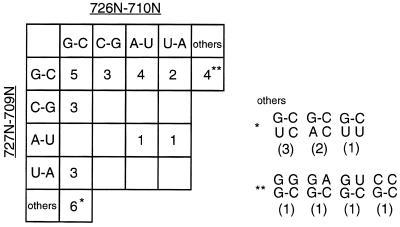
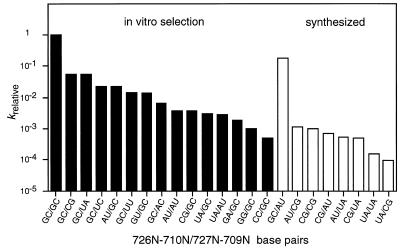
The sequences of the active ribozyme variants were determined by cloning and sequencing G3 DNA. Thirty-two clones were isolated with the correct length insert and all of these were active ribozymes. The nucleotide sequences of the cloned molecules at the randomized positions (709, 710, 726 and 727) and the number of clones of each sequence are summarized in Figure 3. More than two-thirds of the clones possessed two canonical base pairs. Thirty of the 32 clones had a 726G–710C and/or a 727G–709C base pair. This result suggests that at least one G–C base pair, but not a C–G base pair, is sufficient for cleavage activity, although the reaction efficiency varies with the exact sequence. The rate of cleavage of these mutants was 0.1- to 0.001-fold slower than the wild-type, which has two G–C base pairs (Fig. 4). When we cloned the G2 pool, inactive mutants were also obtained, in which no canonical base pairs existed in these positions (i.e. 726N–710N/727N–709N: A·C/U·C, C·A/C·A, G·A/G·G, A·G/G·G; data not shown). This is probably due to a slight contamination of uncleaved molecules at the step of separation and recovering on PAGE. Therefore, the results suggest that catalytic activity requires at least one base pair in this region. The active variants include those mutated at only one position (labeled * and ** in Fig. 3), which agrees with previous point mutagenesis data. In the cis-acting ribozyme, 709C and 726G are essential, but 710C and 727G can be substituted with other bases (7,12–14).
Figure 3.
Combination of base pairs at 4N positions of active clones obtained by in vitro selection. The numbers of clones of each sequence combination are indicated.
Figure 4.
Relative rate of cleavage (krelative) of variants at 4N positions. Black bars indicate variants isolated by in vitro selection and white bars indicate variants synthesized by site-directed mutagenesis.
The cloned variants isolated from the G3 pool did not include all the possible variants with base pairing potential (open boxes in Fig. 3). Therefore, to better understand the relationship between stability of short psuedoknot base pairs and catalytic activity, the eight sequence variants missing from the representative clones were synthesized and their activity level determined (Fig. 4). The G–C/A–U variant shows a high catalytic activity (2.0 min–1; Table 1 and Fig. 4), and despite the presence of one G–C base pair it was unexpectedly not obtained by in vitro selection. We might obtain this variant by screening more clones. The other seven synthesized variants had relatively low catalytic activity in comparison with the variants obtained by in vitro selection. This result indicates that the in vitro selection procedure works to select ones with higher activities, and base pairing is critical for catalytic activity. In other words, the 4N positions tested here (709, 710, 726 and 727) are very important for the assembly of catalytic elements, and the formation of base pairs, P1.1, is necessary to form the active structure, but 4N bases are not directly implicated in the catalytic reaction.
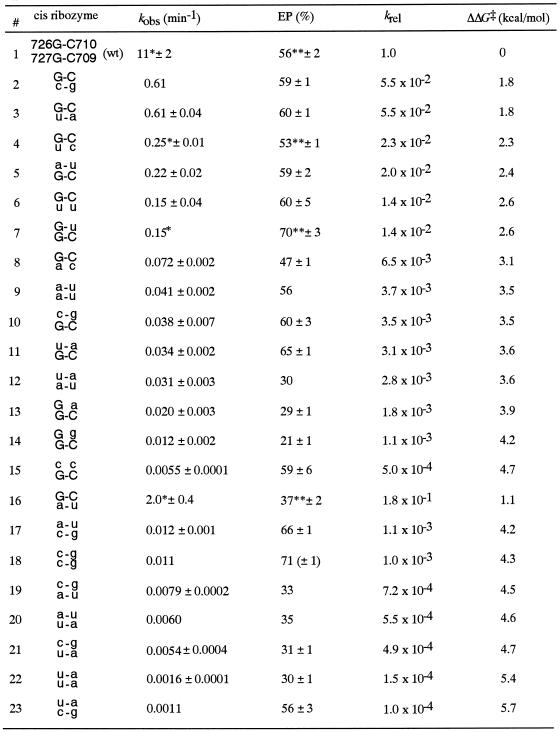
Table 1. Kinetic parameters of all 4N variants.
krel:kobs(variant)/kobs(wt). ΔΔG‡, the difference of free energy of transition-state stabilization, was calculated using the equation ΔΔG‡ = –RTln(krel), where T = 310.15 K (37°C) and R = 1.987 cal K–1·mol–1.
Non-wild-type residues are given in lowercase letters. Numbers 1–15 were obtained from the in vitro selection. Numbers 16–23 were constructed by site-directed mutagenesis. Kinetic parameters are the average of at least two experimental data points. All kobs and EP values were calculated by monophasic equation (see Materials and Methods) except * and **.
*Values of faster reaction calculated from biphasic equation.
**[EP1] + [EP2] from biphasic equation (see Materials and Methods).
Reaction kinetics and sequence-dependent stability
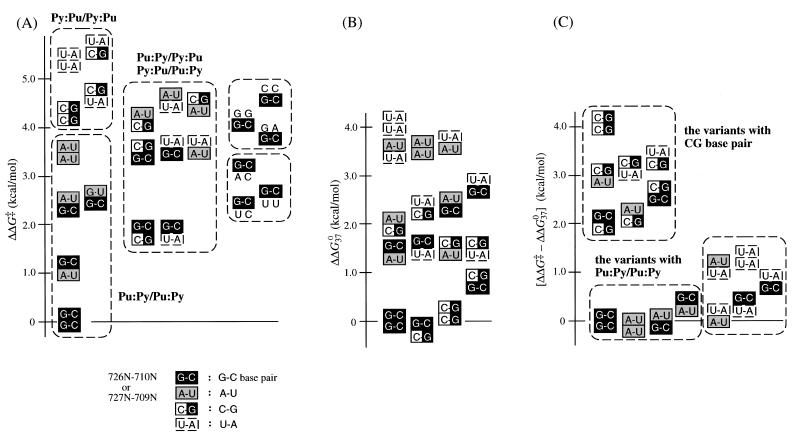
Kinetic parameters of all variants obtained are listed in Table 1. The relative activity of each variant (krel) was used to obtain the change in the apparent free energy of the transition-state stabilization (ΔΔG‡) (19,20). To compare the ΔΔG‡ values of variants more easily, values were ordered, as shown in Figure 5A. From this analysis, the reaction activities can be roughly classified as follows: Pu–Py/Pu–Py ≥ Pu–Py/Py–Pu, Py–Pu/Pu–Py ≥ Py–Pu/Py–Pu. In general, the variants with a 726G–710C base pair had ΔΔG‡ values of 1.1−3.1 kcal/mol, and the variants with a 727G–709C base pair had values of 2.4−4.7 kcal/mol and were less active. This tendency is probably due to cross stacking of 689G with 726G at the position of helical crossover between stem I and P1.1 (15). However, even with stacking between 689G and 726G/A, the cleavage does not occur if there are no base pairs in the 4N region. As a result of this stacking, including base-pair formation (P1.1), the scissile phosphate is brought close to the catalytic core, and HDV ribozyme takes on a favorable conformation for the cleavage reaction that follows.
Figure 5.
(A) Comparison of the difference of apparent free energy of transition-state stabilization (ΔΔG‡). (B) Comparison of the stability of P1.1 compared to wild-type by ΔΔG037. ΔΔG037 = [ΔΔG037(726N–710N/727N–709N) – ΔG037(G–C/G–C)] + [ΔG037(689G·725U/726N–710N) – ΔG037(G·U/G–C)]. (C) Comparison of the difference between ΔΔG‡ and ΔΔG037.
To compare the value of ΔΔG‡ with the stability of the base pairs at P1.1, the approximate values of the difference of free energy between the variant at P1.1 and the wild-type G–C/G–C, ΔΔG037, were calculated from the nearest-neighbor analysis (21–23) and analyzed as shown in Figure 5A (see Fig. 5B). To obtain the values of ΔΔG037, two factors of ΔG037 were considered. The first factor is the thermodynamic value of P1.1 of each variant, which is basically ΔG037 for two adjacent W–C base pairs. In the case of terminal A–U base pairs, the penalty of a terminal A–U base pair (0.45 kcal/mol) was added to the value (22). The second factor is the coaxial stacking effect between the adjacent stem I and P1.1. For adjacent helices, the ΔG037 value for coaxial stacking of helices is approximated as the ΔG037 for the equivalent nearest-neighbor base-pair combination in an intact helix (21). For the coaxial stacking effect, the thermodynamic parameter for the 726N–710N base pair adjacent to the 689G·725U wobble pair was used (23). Finally, the value of ΔΔG037, the sum of each free energy difference between the variant and the wild-type, is shown in Figure 5B.
Comparing Figure 5A and B, it is apparent that the variants with a C–G base pair are among the most stable variants with a W–C base pair (ΔΔG037 ≈ –0.2–2.3 kcal/mol), but these variants are less efficient (ΔΔG‡ ≈ 1.8–5.7 kcal/mol). For example, the G–C/C–G variant is more stable than wild-type (ΔΔG037 = –0.2 kcal/mol), but less active (1.8 kcal/mol of ΔΔG‡). In contrast, in the case of anti-genomic ribozyme, the variant G–C/C–G has higher activity than the wild-type (17). This variant has ΔΔG‡ value of –0.2 kcal/mol, which is coincident with the ΔΔG037 value of –0.2 kcal/mol from our calculation. This result has been explained as follows: since the anti-genomic ribozyme lacks 3 nt (729C,730A,731A) in SSrA, the base pair 727C–709G might stack with stem IV (2) and stabilize a more rigid active structure than the genomic one. The short pseudoknot base pairs P1.1 in the genomic HDV ribozyme seems to have greater flexibility than the anti-genomic variant by using CAA as an adjustable region. The variant with U–A/C–G (ΔΔG037 = 2.3 kcal/mol) has the lowest activity (ΔΔG‡ = 5.7 kcal/mol) of all the variants. As observed in the crystallographic structure, P1.1 gathers the catalytic elements in the catalytic core and the essential 763C is located adjacent to 709C. In the case of variants with a stable C–G base pair, the alteration of the P1.1 structure seems to be more divergent from the wild-type and more energy is required to adjust its position for the catalytic reaction.
The value of ΔΔG‡ measures the energetic penalty for the removal of a functional group involved in the stabilization of the transition state (19,20). If the structural change is negligible during the course of the reaction after the formation of P1.1, the value of ΔΔG‡ should be close to the change in the free energy of the stability of P1.1. It is noteworthy that comparison of Figure 5A and B indicates that the values of ΔΔG‡ do not fit with the values of the relative base pair stability predicted from nearest-neighbor analysis. The range of the values ΔΔG‡ and ΔΔG037 are different and the order of the variants also differ for each parameter. To consider this result systematically for all variants, the difference between the values of ΔΔG‡ and ΔΔG037 were calculated and are presented in Figure 5C. In variants with a Pu–Py/Pu–Py arrangement (similar to wild-type), the values of ΔΔG‡ are close to those of ΔΔG037, as predicted from the nearest-neighbor rules, with only a small range of differences (–0.1–0.3 kcal/mol; Fig. 5C). The other variants (except those with a C–G base pair) have differences within 0.1–1.3 (U–A/U–A) kcal/mol. In contrast, in the variants with a C–G base pair, the difference is larger and in the range of 2.0–4.1 kcal/mol. The variant C–G/C–G has the largest difference value (4.1 kcal/mol; Fig. 5C). It is likely that this large difference is due to interference in the formation of the active conformation, resulting from the interaction of 709G or/and 710G with other proximate bases, as described above. The observed differences between ΔΔG‡ and ΔΔG037 suggest that differences in reaction rate for these variants are not simply accounted for by the differences in the stability of P1.1, but involve other factors such as competition and conformational change. In the mutants, it is possible that the short pseudoknot base pairs, P1.1, act to gather catalytic elements, and then conformational changes occur that allow these elements to assume their proper positions for catalysis.
Contribution of Mg2+ ions to P1.1 formation
In a number of RNAs, it is thought that the binding of cations is important for formation of tertiary structure [studies of tRNA reviewed in (24), the cooperative folding of group I introns (25–27), crystal structure of P4–P6 domain from Tetrahymena group I intron (28), etc.]. Thermodynamic studies of two different pseudoknot sequences elucidated the role played by Mg2+ ions in strongly promoting pseudoknot formation (29,30). At the loop–stem junction, Puglisi et al. (31) have pointed out that a close juxtaposition of loop phosphates would create both a high electrostatic field and the opportunity for ion chelation. The modest ion selectivity and ion binding affinities of pseudoknot structures suggest the existence of a divalent ion binding region intermediate in character between completely delocalized and strongly chelated ions (32). In previous chemical probing studies on HDV ribozyme, it was observed that 709C and 710C were easily modified in the absence of Mg2+ ions by dimethyl sulfate (DMS), which reacts with N3 of C, but these residues are protected in the presence of Mg2+ ions (33). These positions correspond to the 5′-strand of P1.1 in the crystal structure (15). This suggests that Mg2+ ions strengthen the formation of P1.1.
Differences in the pattern of DMS modification in the presence or absence of Mg2+ ions were also detected at the bases near the junctions of stems, such as 698G (JI–II), 705G and 716G (stem III), 719G (stem I), 726G, 729C, 730A and 731A (SSrA). This result suggests that these regions are not tightly packed in the absence of Mg2+ ions, but they become more tightly packed as a result of coaxial stacking by Mg2+ ions as proposed by Puglisi et al. (31). Accordingly, it is concluded that the Mg2+ ions contribute to the formation of the short pseudoknot base pairs, P1.1, and to the stabilization of the tertiary structure of HDV ribozyme. In addition, phosphate modifications by thiophosphate substitution (34) and by ethylnitrosourea (35) showed that there are more or less interaction sites near the junctions of stems and other sites. For example, critical interactions appear at the phosphate oxygens of 688/689, 689/690 and 709/710 by thiophosphate modification interference analysis (34). In particular, the crystal structure demonstrates that 709C/710C phosphate oxygen interacts with the 2′-OH of 708U and the 4-NH2 of 763C, and 763C/764G phosphate oxygen interacts with the 2′-OH of 763C, respectively (15). Although the precise Mg2+ binding sites are not yet known, it is likely that Mg2+ ions bind in a binding pocket, assist in assembling catalytic elements, and stabilize the newly formed base pairs P1.1 and the active structure. In the presence of Mg2+ ions (in the cleavage reaction condition), the active structure contains the additional pseudoknot base pairs P1.1. Accordingly, as the secondary structure of genomic HDV ribozyme, Figure 1B is more suitable than Figure 1A.
CONCLUSIONS
The active structure of genomic HDV ribozyme has been characterized in detail, with particular attention to the role of two W–C base pairs between SSrA and SSrC, 726G–710C/727G–709C. The functional importance of these base pairs was addressed by performing in vitro selection for active molecules from a pool of sequence variants at these positions. All possible variants with the potential of forming base pairs at these sites were analyzed. The results are summarized as follows: (i) short pseudoknot base pairs (726G–710C/727G–709C) are important for ribozyme activity and cleavage can occur with only one base pair in the cis-acting HDV ribozyme; (ii) all W–C base pairs for the short pseudoknot permit cleavage, although the sequence variants show large differences in catalytic efficiency; (iii) sequence variants which contain the 726G–710C base pair are more active (faster cleavage) than other variants with two W–C base pairs because, as a result of the cross stacking between +1G and 726G, the cleavage site is pushed into the catalytic core and places the scissile bond in a favorable location; and (iv) Mg2+ ions are necessary for strengthening the formation of the short pseudoknot base pairs.
Acknowledgments
ACKNOWLEDGEMENT
We thank Dr M. Been for providing a preprint.
REFERENCES
- 1.Lazinski D.W. and Taylor,J.M. (1995) RNA, 1, 225–233. [PMC free article] [PubMed] [Google Scholar]
- 2.Been M.D. and Wickham,G.S. (1997) Eur. J. Biochem., 247, 741–753. [DOI] [PubMed] [Google Scholar]
- 3.Sharmeen L., Kuo,M.Y.P., Dinter-Gottlieb,G. and Taylor,J. (1988) J. Virol., 62, 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo M.Y.P., Sharmeen,L., Dinter-Gottlieb,G. and Taylor,J. (1988) J. Virol., 62, 4439–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H.N., Lin,Y.J., Lin,F.P., Makino,S., Chang,M.F. and Lai,M.M.C. (1989) Proc. Natl Acad. Sci. USA., 86, 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai M.M.C. (1995) Annu. Rev. Biochem., 64, 259–286. [DOI] [PubMed] [Google Scholar]
- 7.Tanner N.K., Schaff,S., Thill,G., Petit-Koskas,E., Crain-Denoyelle,A.M. and Westhof,E. (1994) Curr. Biol., 4, 488–498. [DOI] [PubMed] [Google Scholar]
- 8.Thill G., Vasseur,M. and Tanner,N.K. (1993) Biochemistry, 32, 4254–4262. [DOI] [PubMed] [Google Scholar]
- 9.Jeng K.S., Daniel,A. and Lai,M.M.C. (1996) J. Virol., 70, 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perrotta A.T. and Been,M.D. (1991) Nature, 350, 434–436. [DOI] [PubMed] [Google Scholar]
- 11.Fauzi H., Chiba,A., Nishikawa,F., Roy,M., Kawakami,J. and Nishikawa,S. (1998) Anal. Chim. Acta, 365, 309–317. [Google Scholar]
- 12.Kumar P.K.R., Suh,Y.A., Miyashiro,H., Nishikawa,F., Kawakami,J., Taira,K. and Nishikawa,S. (1992) Nucleic Acids Res., 20, 3919–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suh Y.A., Kumar,P.K.R., Kawakami,J., Nishikawa,F., Taira,K. and Nishikawa,S. (1993) FEBS Lett., 326, 158–162. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami J., Kumar,P.K.R., Suh,Y.-A., Nishikawa,F., Kawakami,K., Taira,K. and Nishikawa,S. (1993) Eur. J. Biochem., 217, 29–36. [DOI] [PubMed] [Google Scholar]
- 15.Ferré-D’Amaré A.R., Zhou,K. and Doudna,J.A. (1998) Nature, 395, 567–574. [DOI] [PubMed] [Google Scholar]
- 16.Perrotta A.T. and Been,M.D. (1996) Nucleic Acids Res., 24, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadkins T.S., Perrota,A.T., Ferré-D’Amaré,A.R., Doudna,J.A. and Been,M.D. (1999) RNA, 6, 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishikawa F., Roy,M., Fauzi,H. and Nishikawa,S. (1999) Nucleic Acids Res., 27, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fersht A.R. (1988) Biochemistry, 27, 1577–1580. [DOI] [PubMed] [Google Scholar]
- 20.Chartrand P., Usman,N. and Cedergren,R. (1997) Biochemistry, 36, 3145–3150. [DOI] [PubMed] [Google Scholar]
- 21.Serra M.J. and Turner,D.H. (1995) Methods Enzymol., 259, 242–261. [DOI] [PubMed] [Google Scholar]
- 22.Xia T., Santa Lucia,J., Burkard,M.E., Kierzek,R., Schroeder,S.J., Jiao,X., Cox,C. and Turner,D.H. (1998) Biochemistry, 37, 14719–14735. [DOI] [PubMed] [Google Scholar]
- 23.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 24.Crothers D.M. (1979) In Schimmel,P.R., Söll,D. and Abelson,J.N. (eds), Transfer RNA: Structure, Properties and Recognition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 163–176.
- 25.Laggerbauer B., Murphy,F.L. and Cech,T.R. (1994) EMBO J., 13, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarrinkar P.P. and Williamson,J.R. (1994) Science, 265, 919–924. [Google Scholar]
- 27.Treiber D.K., Rook,M.T., Zarrinkar,P.P. and Williamson,J.R. (1998) Science, 279, 1943–1945. [DOI] [PubMed] [Google Scholar]
- 28.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt J.A., Puglisi,J.D. and Tinoco,I.Jr (1990) J. Mol. Biol., 214, 455–470. [DOI] [PubMed] [Google Scholar]
- 30.Qiu H., Kaluarachchi,K., Dhu,Z., Hoffman,D.W. and Giedroc,D.P. (1996) Biochemistry, 35, 4176–4186. [DOI] [PubMed] [Google Scholar]
- 31.Puglisi J.D., Wyatt,J.R. and Tinoco,I.Jr (1990) J. Mol. Biol., 214, 437–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gluick T.C., Wills,N.M., Gesteland,R.F. and Draper,D.E. (1997) Biochemistry, 36, 16173–16186. [DOI] [PubMed] [Google Scholar]
- 33.Kumar P.K.R., Taira,K. and Nishikawa,S. (1994) Biochemistry, 33, 583–592. [DOI] [PubMed] [Google Scholar]
- 34.Jeoung Y.H., Kumar,P.K.R., Suh,Y.A., Taira,K. and Nishikawa,S. (1994) Nucleic Acids Res., 22, 3722–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar P.K.R., Jeoung,Y.H. and Nishikawa,S. (2000) In Krupp,G. and Gauer,R.K. (eds), Ribozymes: Biochemistry and Biotechnology. Eaton Publishing, Natick, MA.
- 36.Makino S., Chang,M.-F., Shieh,C.-K., Kamahora,T., Vannier,D. Govindarajan,S. and Lai,M.M.C. (1987) Nature, 329, 343–346. [DOI] [PubMed] [Google Scholar]