Abstract
RNA polymerase II from the fission yeast Schizosaccharomyces pombe consists of 12 species of subunits, Rpb1–Rpb12. We expressed these subunits, except Rpb4, simultaneously in cultured insect cells with baculovirus expression vectors. For the isolation of subunit complexes formed in the virus-infected cells, a glutathione S-transferase (GST) sequence was fused to the rpb3 cDNA to produce GST–Rpb3 fusion protein and a decahistidine-tag sequence was inserted into the rpb1 cDNA to produce Rpb1H protein. After successive affinity chromatography on glutathione and Ni2+ columns, complexes consisting of the seven subunits, Rpb1H, Rpb2, GST–Rpb3, Rpb5, Rpb7, Rpb8 and Rpb11, were identified. Omission of the GST–Rpb3 expression resulted in reduced assembly of the Rpb11 into the complex. Direct interaction between Rpb3 and the other six subunits was detected by pairwise coexpression experiments. Coexpression of various combinations of a few subunits revealed that Rpb11 enhances Rpb3–Rpb8 interaction and consequently Rpb8 enhances Rpb1–Rpb3 interaction to some extent. We propose a mechanism in which the assembly of RNA poly-merase II is stabilized through multiple subunit–subunit contacts.
INTRODUCTION
RNA polymerase II (Pol II) purified from the fission yeast Schizosaccharomyces pombe consists of 12 species of subunits designated as Rpb1–Rpb12. We have cloned all the 12 genes, rpb1–rpb12, coding for these subunits (1–7). Among them the largest and the second largest subunits, Rpb1 and Rpb2, have sequence similarities with the β′ and the β subunits of prokaryotic RNA polymerase, respectively. Rpb3 and Rpb11 show limited similarities with the N-terminal domain of α subunit. The four subunits, Rpb1, Rpb2, Rpb3 and Rpb11, are therefore supposed to form an enzyme core which corresponds to the core enzyme, α2ββ′, of the prokaryotic RNA polymerase. In the presence of 6 M ures, the purified S.pombe Pol II dissociates forming an Rpb2–Rpb3–Rpb11 complex (8), which is equivalent to the assembly intermediate, α2β, of prokaryotic RNA polymerases. Furthermore, the two large subunits (Rpb1 and Rpb2) interact with DNA in S.pombe (8), human (9) and Drosophila melanogaster (10,11), as is the case with β and β′ subunits. We have mapped the regions which form the active center of RNA polymerization within these two subunits by photo-crosslinking of nascent RNA 3′ end to the enzyme and peptide mapping of the crosslinked subunits (12).
The function and structure of other eukaryotic Pol II subunits have been studied mainly using the enzyme from Saccharomyces cerevisiae. RPB4, which is essential in the stress response (13), is dissociable as a complex with the RPB7 from Pol II. The Rpb4–Rpb7 complex can stimulate the initiation of transcription at certain promoters (14). Electron crystallography showed a more closed cleft structure for S.cerevisiae Pol II with RPB4 and RPB7 than the enzyme without these two subunits, suggesting that RPB4 and/or RPB7 play a role in the conformational transition of the enzyme (15). RPB5 plays a role in transcriptional activation at some promoters (16) and is considered to be located near the template DNA in the initiation complex (9). A direct interaction has been reported between the human RPB5 and hepatitis B virus X protein (17,18). The yeast RPB6, one of the common subunits of Pol I, II and III, is required for the RNA synthesis activity at least in Pol I (19). RPB9 plays roles in transcription elongation through arrest sites (20), as well as in selection of accurate start sites (21). A high resolution NMR structure was solved for the RPB8 (22).
Except for the fragmentary knowledge of the functions of each subunit, the structure–function organization within Pol II complexes still remains unclear. To elucidate the functional organization, we have analyzed the subunit–subunit interactions within the S.pombe Pol II by chemical crosslinking and Far-western blotting. The results indicate that all small subunits interact with Rpb1 and/or Rpb2; there are also six combinations of pairwise interactions between small subunits: Rpb3–Rpb5, Rpb3–Rpb10, Rpb3–Rpb11, Rpb5–Rpb6, Rpb6–Rpb7 and Rpb6–Rpb8 (23). We also mapped the Rpb5-contact site on the Rpb1 and the Rpb3-contact site on the Rpb2 using a yeast two-hybrid system (24). Reconstitutions of the Rpb3–Rpb11 heterodimer in Arabidopsis thaliana and S.cerevisiae (25,26) and the Rpb4–Rpb7 heterodimer in A.thaliana, S.cerevisiae (27), human (28) and S.pombe (7) have been reported. Moreover, all possible combinations of pairwise interactions between human subunits expressed in insect cells have been analyzed (29). However, these subunit–subunit interactions were limited in a sense that they only showed interactions between two subunits, except in the case with Rpb5, which stimulates the Rpb3–Rpb11 heterodimer formation (30). Here we present the formation of multi-protein complexes of Pol II subunits in insect cells expressing the recombinant subunit proteins. Our results indicate that multiple contacts among the subunits are involved in the formation of stable Pol II.
MATERIALS AND METHODS
Construction of an S.pombe strain carrying the GST–rpb3 gene
A plasmid pUC-GSTrpb3-ura4 was constructed by replacing the decahistidine (His10)-tag sequence between the NheI and ApaI sites in pUC-rpb3H-ura4 (8) with a DNA fragment of glutathione S-transferase (GST) sequence which was amplified by PCR using plasmid pGEX-4X-1 (Pharmacia) as a template and 5′- and 3′-primers carrying an NheI and ApaI sequence, respectively. The inserted fragment encodes amino acid residues 2–219 of the GST protein. Gene replacement in the S.pombe strain JY741 was carried out as described (8).
Purification of GST–Pol II
The S.pombe strain carrying the GST–rpb3 gene was cultured in YE medium, containing 50 µg/ml each of adenine and uracil at 30°C, to 5 × 107 cells/ml. After harvest by centrifugation, the cells were frozen in liquid nitrogen. The frozen cells were disrupted with Cryopress (Microtech Nichion), and suspended in double the cell volume of 1.5× buffer A [50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 20% glycerol, 0.1 M (NH4)2SO4, 1 mM dithiothreitol and 0.5 mM phenylmethylsulfonyl fluoride (PMSF)] containing a proteinase inhibitor mixture (100 µg/ml benzamidine, 1 µg/ml leupeptin, 1 µg/ml aprotinin, 1 µg/ml pepstatin, 1 µg/ml Nα-p-tosyl-l-lysine chlomethylketone, 1 µg/ml N-tosyl-l-phenylalanine chlomethylketone, 0.5 µg/ml chymostatin and 0.25 µg/ml antipain). After sonication, the lysate was centrifuged at 30 000 g for 30 min at 4°C. The supernatant was diluted up to 10 times the original cell volume with the buffer A containing the proteinase inhibitor mixture, and then polyethyleneimine (pH 7.9) was added to 0.1% (v/v). The mixture was incubated on ice for 30 min and centrifuged at 30 000 g for 20 min at 4°C. The precipitant was extracted with double the original cell volume of buffer B [50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 20% glycerol, 0.2 M (NH4)2SO4 and 0.5 mM PMSF]. After centrifugation at 30 000 g for 20 min at 4°C, the supernatant was loaded onto a glutathione (GSH)–Sepharose 4B (Pharmacia) column equilibrated with buffer B. The column was washed with 20× bed volume of buffer C [50 mM Tris–HCl, pH 8.0, 20% glycerol, 0.2 M (NH4)2SO4 and 0.5 mM PMSF] and eluted with buffer G [20 mM GSH, 50 mM Tris–HCl, pH 8.0, 20% glycerol, 0.2 M (NH4)2SO4 and 0.5 mM PMSF].
Baculovirus expression system
Sf9 insect cells were cultured in Grace medium supplemented with 9% fetal bovine serum at 28°C. Bac-to-Bac™ baculovirus expression systems (Gibco BRL) were used to express the Pol II subunit proteins. Plasmid pFastBac1 with a polyhedrin promoter or pFastBac™DUAL with a polyhedrin and a p10 promoter was used for construction of the donor plasmids. The viruses and the donor plasmids used in this study are listed in Table 1. pFastBac-gus was used to make a control virus expressing β-glucuronidase. Each cDNA encoding one of the S.pombe Pol II subunits was inserted under the polyhedrin or the p10 promoter. The recombinant viruses constructed carry one or two subunit cDNAs and in the mixed infection, viruses were selected so as to minimize the number of coinfected virus species. The rpb1 cDNA was modified as to include a sequence coding for a His10-tag at the immediate upstream of the C-terminal domain (CTD) (8) (Rpb1 with His10-tag is designated as Rpb1H), and as a control another artificial rpb1 allele, rpb1ΔC, which lacks both the His10-tag and the CTD sequences, was constructed. The rpb3 cDNA used carries a sequence coding for GST fused to the N-terminus of Rpb3, and expresses an identical protein to that produced in the S.pombe GST–rpb3 strain described above. The recombinant viruses were prepared and amplified according to the manufacturer’s instructions.
Table 1. Recombinant baculoviruses and donor plasmids.
| Virus | Plasmid | Promoter | |
|---|---|---|---|
| Polyhedrin | p10 | ||
| Bv1H |
pFB-rpb1H |
rpb1H10 |
|
| Bv1ΔC |
pFB-rpb1ΔC |
rpb1ΔC |
|
| Bv2 |
pFB-rpb2 |
rpb2 |
|
| BvG3 |
pFB-GSTrpb3 |
GST–rpb3 |
|
| Bv5 |
pFBD-rpb5 |
rpb5 |
– |
| Bv6 |
pFBD-rpb6 |
– |
rpb6 |
| Bv7 |
pFBD-rpb7 |
rpb7 |
– |
| Bv8 |
pFBD-rpb8 |
– |
rpb8 |
| Bv9 |
pFBD-rpb9 |
rpb9 |
– |
| Bv10 |
pFBD-rpb10 |
– |
rpb10 |
| Bv11 |
pFBD-rpb11 |
– |
rpb11 |
| Bv12 |
pFBD-rpb12 |
rpb12 |
– |
| Bv2/10 |
pFBD-rpb2/10 |
rpb2 |
rpb10 |
| BvG3/11 |
pFBD-GSTrpb3/11 |
GST–rpb3 |
rpb11 |
| Bv5/6 |
pFBD-rpb5/6 |
rpb5 |
rpb6 |
| Bv7/9 |
pFBD-rpb7/9 |
rpb7 |
rpb9 |
| Bv12/8 |
pFBD-rpb12/8 |
rpb12 |
rpb8 |
| BvG/11 | pFBD-GST/rpb11 | GST | rpb11 |
cDNAs inserted under each promoter are listed.
GSH-affinity isolation of subunit complexes
Sf9 suspension cultures were infected with various combinations of the expression viruses at a multiplicity of infection of three for each virus and at 66 h post-infection, the cells were harvested and frozen in liquid nitrogen. Complexes containing the GST–Rpb3 protein were purified by the same method as that employed for GST–Pol II purification from the S.pombe strain described above. Western blotting was carried out as described (23) and chemiluminescence was quantitated with a lumino-image analyzer LAS-1000 (Fuji). Anti-Rpb1 antiserum was raised against the N-terminal fragment of the Rpb1 whereas other antisera were against each full length subunit.
Ni2+-affinity chromatography
Eluted fractions from the GSH–Sepharose column, or extracted fractions from the polyethyleneimine precipitation were loaded onto a Ni2+-charged nitrilotriacetic acid–agarose (Qiagen) column equilibrated with buffer D [50 mM Tris–HCl, pH 8.0, 20% glycerol, 0.2 M (NH4)2SO4, 10 mM 2-mercaptoethanol and 0.5 mM PMSF]. The column was washed with 20× bed volume of buffer I-20 (buffer D containing 20 mM imidazole) and eluted with buffer I-100 (buffer D containing 100 mM imidazole).
RESULTS
GST fusion to the N-terminus of Rpb3 does not interfere with the subunit assembly
We have reported the gene cloning of 11 subunits of S.pombe Pol II, excluding Rpb4 [the rpb4 has also been isolated (7)]. In order to analyze the assembly mechanism of these 11 subunits, we attempted to isolate subunit complexes from the extracts of insect cells coinfected with various combinations of the recombinant baculovirus vectors, each carrying one or two of the subunit cDNAs. Because the α-subunit, the prokaryotic counterpart of Rpb3, plays a central role in the assembly of prokaryotic RNA polymerase (31), we used GST–Rpb3 fusion protein, in which GST is fused to the N-terminus of Rpb3, as an anchor subunit for the affinity pull-down assays of S.pombe Pol II subunit complexes.
To confirm that the addition of GST to Rpb3 does not interfere with the Pol II assembly, we constructed an S.pombe haploid strain in which the rpb3 gene on the chromosome was replaced by the sequence encoding the GST–Rpb3 fusion protein. Pol II could be isolated from cell extract of this strain through GSH-affinity chromatography. The purified Pol II fraction contained all 12 subunits (Fig. 1), indicating that the addition of GST did not interfere with Pol II assembly. Furthermore, the GST–Pol II sustains essential functions for cell growth because the strain with the GST–rpb3 gene in place of wild-type rpb3 is viable.
Figure 1.
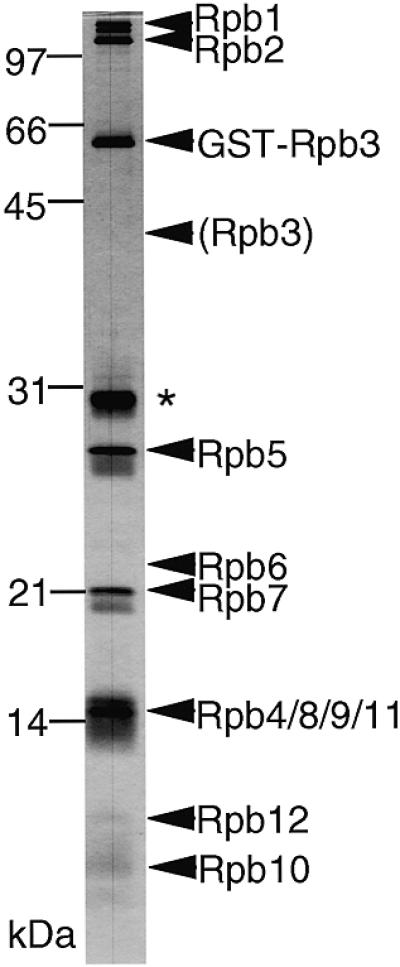
Isolation of Pol II from S.pombe carrying the GST–rpb3 fusion gene. A DNA sequence encoding GST was inserted into the 5′-terminus of the chromosomal rpb3 gene of S.pombe in order to produce the GST–Rpb3 fusion protein. The Pol II fraction purified from this strain by GSH-affinity chromatography was subjected to SDS–PAGE and the gel was silver stained. Positions of the molecular weight markers are indicated on the left and the subunit species are indicated on the right. Two bands of Rpb1 correspond to the intact Rpb1 and the truncated one without CTD, respectively. The migration position expected for the intact Rpb3 is indicated by a parenthesis. Rpb6 cannot be stained by silver. The overlapping bands of the four subunits, Rpb4, Rpb8, Rpb9 and Rpb11, were confirmed by western blotting (data not shown). The asterisk indicates a degraded fragment of GST–Rpb3 as identified by peptide sequencing (data not shown).
For isolation of subunits associated with Rpb1, a His10-tag sequence was inserted in the rpb1 cDNA at the immediate upstream of CTD. The recombinant Rpb1 product was designated as Rpb1H. The insertion of His10-tag into Rpb1 does not interfere with the subunit assembly as we have shown previously (8).
Expression of the subunit proteins using the baculovirus vectors
The 18 species of the recombinant baculovirus and the donor plasmids used for recombinant virus construction are listed in Table 1. Twelve viruses, Bv1H to Bv12, each carrying one of the 11 subunit cDNAs under the control of the polyhedrin or the p10 promoter were constructed. A recombinant virus Bv1ΔC carrying the rpb1 cDNA without both the CTD and the His10-tag was also constructed. The other six viruses, Bv2/10 to Bv12/8, have a pair of cDNAs, and each cDNA was inserted so as to express independently under the control of the polyhedrin or the p10 promoter. As a control for expression of Rpb11 and the GST in place of the GST–Rpb3 fusion protein, the virus BvG/11 was constructed.
To minimize the number of virus species for coinfection, the viruses with two cDNAs were preferentially used. The expression of each subunit protein was detected by western blotting of whole cell lysates using antisera against each subunit. A cell lysate prepared from the cells infected with a baculovirus which expresses a non-Pol II protein, β-glucuronidase, exhibited no immunostained signal by any one of the anti-subunit antisera used (Fig. 2A, lane 1), indicating that the antisera cross-react with none of the cellular or viral proteins.
Figure 2.
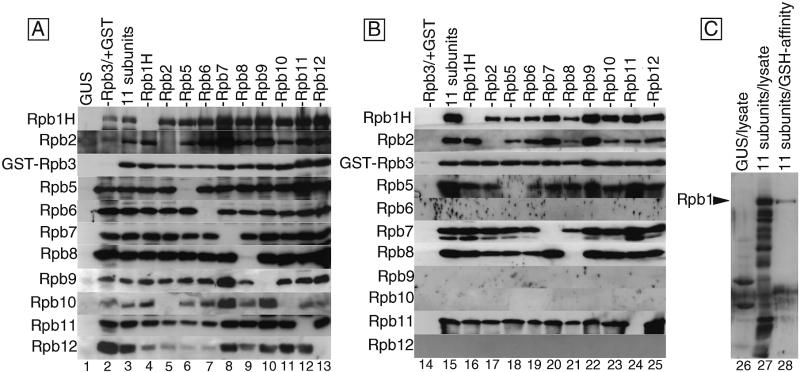
Coexpression of various combinations of the subunits and formation of subunit complexes. (A) Coexpression of Pol II subunits. The complete set of 11 Pol II subunits or various combinations of 10 subunits were expressed by mixed infection of the recombinant viruses. Lysates of the infected cells were subjected to western blotting and detected with antisera against each subunit. Portions of the filters corresponding to the migration positions of each subunit in the SDS–PAGE are arrayed, and the detected subunits are indicated on the left. The proteins expressed in each lane are: GUS, β-glucuronidase; –Rpb3/+GST, GST plus 10 subunits except Rpb3; 11 subunits, 11 subunits including GST–Rpb3; –Rpb1H to –Rpb12, 10 subunits (including the GST–Rpb3) except for the indicated subunit. (B) Isolation of subunit complexes. The cell extracts were subjected to GSH-affinity chromatography. The eluted fractions were analyzed by western blotting as in (A). Lanes correspond to those in (A) with the same designations. (C) Preferential assembly of the intact Rpb1H. Western blotting with anti-Rpb1 antiserum was carried out: GUS/lysate, the lysate of cells expressing GUS; 11 subunits/lysate, the lysate of cells expressing the 11 subunits; 11 subunits/GSH-affinity, the subunit complex fraction isolated by GSH-affinity chromatography.
Simultaneous expression and assembly of 11 Pol II subunits
To express the 11 subunits simultaneously in the same cells, an Sf9 culture was coinfected with six species of recombinant virus, Bv1H, Bv2/10, BvG3/11, Bv5/6, Bv7/9 and Bv12/8. Western blotting of the infected cell lysate indicated that all 11 subunits were successfully expressed albeit at different levels (Fig. 2A, lane 3) with the expression of Rpb1 and Rpb2 being the lowest as judged by silver staining (data not shown). The coexpression of GST, in place of GST–Rpb3, with the other 10 subunits was also successful (Fig. 2A, lane 2), in which the expression of GST was confirmed by silver staining of the electrophoresis gel after GSH-affinity chromatography (data not shown).
To examine the assembly state, the cell extracts were subjected to GSH-affinity column chromatography and the subunit proteins bound to GST–Rpb3 were eluted with GSH-buffer and identified by western blotting (Fig. 2B). No subunit protein was detected in the eluted fraction of the control extract with GST expression in place of GST–Rpb3 (Fig. 2B, lane 14). In contrast, at least six subunits, Rpb1H, Rpb2, Rpb5, Rpb7, Rpb8 and Rpb11, were clearly detected in addition to the GST–Rpb3 in the GSH-eluted fraction from the cells in which all 11 subunits were coexpressed (Fig. 2B, lane 15), indicating that these seven subunits formed assemblies in the virus-infected cells. Though the coinfected cell lysate contained various species of degradation products of Rpb1H, only the intact Rpb1H protein was detected in the subunit assemblies co-eluted with GST–Rpb3 (Fig. 2C), indicating that the assemblies detected in this assay represent specific complexes, and not artificial aggregates.
The four subunits, Rpb6, Rpb9, Rpb10 and Rpb12, were not immunodetected in the eluted fraction, although they were present in the column-loaded extract (data not shown). This implies that: (i) these four subunits were not assembled into the complex; (ii) these subunits were assembled but at low efficiencies below the detection level; or (iii) these subunits were dissociated during the isolation of subunit assemblies.
Characterization of the sub-assemblies
In order to elucidate the assembly mode, we have examined a total of 10 combinations of subunit, each lacking one other than GST–Rpb3. In all the combinations tested, all 10 subunits were successfully expressed (Fig. 2A, lanes 4–13), implying that none of the present subunits depend on other subunits for expression. After GSH-affinity purification, the six subunits among the seven subunits, Rpb1H, Rpb2, GST–Rpb3, Rpb5, Rpb7, Rpb8 and Rpb11, were always detected in the assembly fractions, except for the omitted subunit (Fig. 2B, lanes 16–25). The results indicate that the omission of any one of the six subunits does not interfere with the assembly of the other sub-units to GST–Rpb3. Thus, two possible mechanisms can be suggested: (i) each of the six subunits binds to Rpb3 independently; and/or (ii) the seven subunits form a network involving multiple contacts.
Since Rpb1H contains a His10-tag, we further characterized the sub-assemblies eluted from the GSH-column by successive Ni2+-affinity chromatography. The sub-assemblies were retained on the Ni2+–agarose column and all the seven subunits were recovered in the imidazole eluted fraction (Fig. 3A, lane 4). When the Rpb1ΔC lacking both the His10-tag and the CTD was coexpressed with the other 10 subunits, assemblies containing the same set of seven subunits were obtained by GSH-affinity chromatography (lane 1), but none of the subunit proteins bound to the Ni2+ resin (lane 3). These observations indicate that the sub-assemblies containing both GST–Rpb3 and the Rpb1H also contain Rpb2, Rpb5, Rpb7, Rpb8 and Rpb11. However, after the Ni2+-affinity chromatography, the recovery of the other six subunits was lower than that of Rpb1H. In particular, the reduction of Rpb2 was more than that of the other five subunits. It is feasible, therefore, that Rpb2 is associated with only a small portion of the subunit complexes. The recovery of both Rpb8 and Rpb5 in the Ni2+-affinity fraction was higher than others, consistent with the tight binding of Rpb1–Rpb8 as previously observed (8,23).
Figure 3.
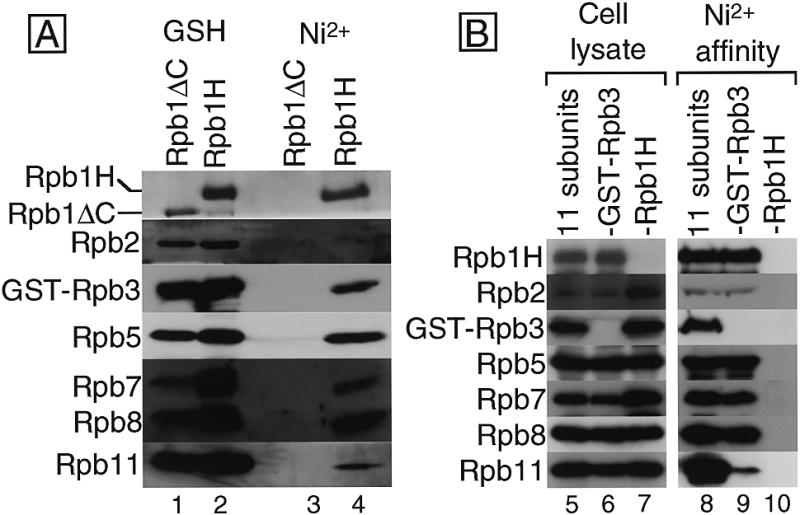
(A) Ni2+-affinity chromatography of the GSH-affinity fractions. Two sets of the 11 subunits, each including Rpb1H or Rpb1ΔC, were coexpressed in two cell cultures and the cell extracts were subjected to GSH-affinity chromatography. The eluted fractions were further purified by Ni2+-affinity chromatography. Each eluted fraction was analyzed by western blotting with antisera against each subunit. Portions of the filters corresponding to the migration positions of each subunit in the SDS–PAGE are arrayed, and the detected subunits are indicated on the left. The affinity columns and the expressed subunits used are indicated on the top: GSH, GSH–Sepharose column; Ni2+, Ni2+–agarose column; Rpb1H, 11 subunits including GST–Rpb3 and Rpb1H; Rpb1ΔC, 11 subunits including GST–Rpb3 and Rpb1ΔC, which lacks both the His10-tag and the CTD. (B) Complex formation without Rpb3. Eleven subunits, or 10 subunits without GST–Rpb3 or Rpb1H were coexpressed and the cell extracts were subjected to Ni2+-affinity chromatography. Cell lysates (cell lysate) and imidazole-eluted fractions from Ni2+–agarose (Ni2+-affinity) were analyzed by western blotting and the portions of the filter are arrayed: 11 subunits, 11 subunits including GST–Rpb3 and Rpb1H; –Rpb3, 10 subunits excluding GST–Rpb3; –Rpb1H, 10 subunits excluding Rpb1H.
Sub-assembly formation in the absence of Rpb3
To detect interactions among subunits other than Rpb3, we expressed 10 subunits excluding GST–Rpb3 and isolated, by Ni2+-affinity chromatography, subunit complexes containing Rpb1H. When the 11 subunits were coexpressed (Fig. 3B, lane 5), the same set of seven subunits as isolated by the GSH-affinity method were isolated by Ni2+-affinity purification (lane 8). In agreement with the result of GSH-affinity assay, the level of Rpb2 recovery was lower than those of the other six. When Rpb1H expression was omitted (Fig. 3B, lane 7), no subunit was recovered (lane 10), indicating that this system can be used for the isolation of subunit assemblies. In the absence of GST–Rpb3 expression (Fig. 3B, lane 6), four subunits, Rpb2, Rpb5, Rpb7 and Rpb8, were recovered in the Ni2+-affinity fractions at the same level as those in the presence of Rpb3, but the Rpb11 recovery was much reduced (Fig. 3B, lane 9). This indicates that there are Rpb3-independent direct and/or indirect interactions among the five subunits, Rpb1, Rpb2, Rpb5, Rpb7 and Rpb8, and that Rpb3 is indispensable for the Rpb1–Rpb11 connection.
Interactions among the four conserved subunits
Next we analyzed interactions among the four subunits, Rpb1H, Rpb2, GST–Rpb3 and Rpb11, which correspond to the prokaryotic β′, β, α and α subunit, respectively. When the four subunits were coexpressed, all the four subunits were copurified by GSH-affinity chromatography (Fig. 4A and B, lanes 2 and 7), and when the three subunits, Rpb1H, Rpb2 and GST–Rpb3, were coexpressed in the absence of Rpb11, the GSH-affinity fractions contained all the three subunits (Fig. 4A and B, lanes 3 and 8). Furthermore, binary complexes, Rpb1H–(GST–Rpb3) and Rpb2–(GST–Rpb3), were recovered when the respective subunit pairs were coexpressed (Fig. 4A and B, lanes 4, 5, 9 and 10). This indicates that both Rpb1 and Rpb2 is able to bind to Rpb3 independently.
Figure 4.
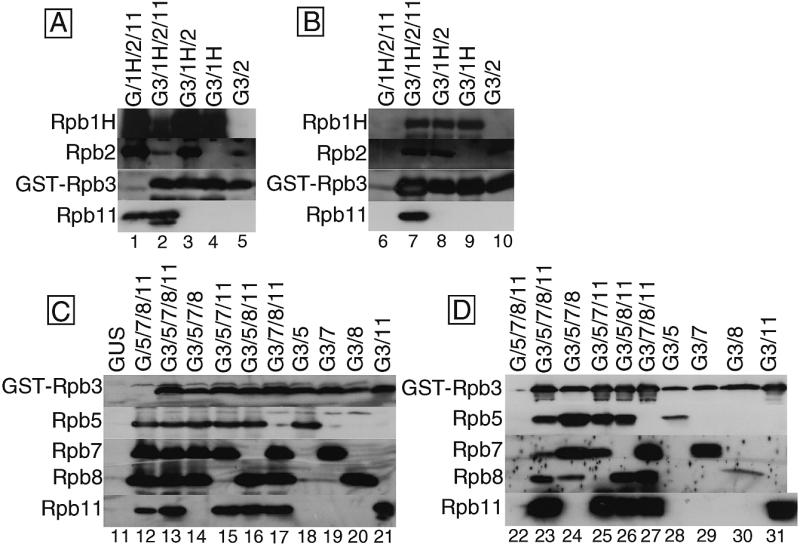
Interaction among the subunits. (A) Coexpression of the four conserved subunits. Expression of the four subunits, Rpb1H, Rpb2, GST–Rpb3 and Rpb11, were detected by western blotting of the lysate with antisera against each subunit. The coexpressed proteins are indicated on the top: G, GST; G3, GST–Rpb3; 1H, Rpb1H; 2, Rpb2; 11, Rpb11, and the detected subunits are indicated on the left. As a control, GST was expressed in place of GST–Rpb3 (lane 1). Portions of the filters corresponding to the migration positions of each subunit in the SDS–PAGE are arrayed. (B) Analysis of the assemblies of four subunits. The extracts of the coinfected cells in various combinations were subjected to GSH-affinity chromatography and the eluted fractions were analyzed by western blotting as in (A). (C) Coexpression of the five small subunits. Expression of the five subunits, GST–Rpb3, Rpb5, Rpb7, Rpb8 and Rpb11, in the coinfected cells was detected by western blotting with antisera against each subunit. The proteins coexpressed are indicated on the top: GUS, β-glucuronidase as a control for antisera cross-reaction; G, GST; G3, GST–Rpb3; 5, Rpb5; 7, Rpb7; 8, Rpb8; 11, Rpb11, and the detected subunits are indicated on the left. Portions of the filters are arrayed as in (A). (D) Analysis of the assemblies of five small subunits. The extracts of the coinfected cells were subjected to GSH-affinity chromatography and the eluted fractions were analyzed by western blotting as in (C).
Interactions among the five small subunits
We also analyzed interactions among the five small subunits, GST–Rpb3, Rpb5, Rpb7, Rpb8 and Rpb11, which were included in the assemblies isolated from the cells expressing all 11 subunits. When these five subunits were coexpressed, all of them were recovered in the assembly fraction after GSH-affinity chromatography (Fig. 4C and D, lanes 13 and 23). In the control experiment expressing the GST in place of GST–Rpb3, no subunit protein was recovered in the column bound fraction (Fig. 4C and D, lanes 12 and 22).
When one of the four subunits other than GST–Rpb3 was omitted, all the expressed subunits were recovered together in the assembly fractions (Fig. 4C and D, lanes 14–17 and 24–27). In the absence of Rpb11 expression, however, the level of Rpb8 recovery in the assembly fraction was significantly reduced (Fig. 4C and D, lane 24), though the expression level of Rpb8 was as high as those in other combinations of coexpression (Fig. 4C and D, lane 14). We also carried out pairwise expression of each small subunit with GST–Rpb3 (Fig. 4C and D, lanes 18–21). All the four subunits, Rpb5, Rpb7, Rpb8 and Rpb11, formed binary complexes with GST–Rpb3 (Fig. 4C and D, lanes 28–31). However, the level of (GST–Rpb3)–Rpb8 complex formation was less than other binary complexes (Fig. 4C and D, lane 30). These observations indicate that the binding of Rpb8 to the Rpb3 is enhanced by association of Rpb11.
The recovery of Rpb5 in the assembly fraction was also reduced in the pairwise expression with GST–Rpb3 (Fig. 4D, lane 28). Nevertheless, omission of any single subunit from the coexpression of five small subunits had virtually no effect on the high level recovery of Rpb5 in the assembly fractions (Fig. 4D, lanes 24–26). This implies that the Rpb3–Rpb5 interaction can be supported by any one or two of the other three subunits. In the coexpression experiments of two or four sub-units, Rpb7 bound strongly to GST–Rpb3 (Fig. 4D, lanes 24, 25, 27 and 29), but Rpb7 binding was reduced in the coexpression of five subunits (Fig. 4D, lane 23). Coexistence of three other subunits restricted Rpb7 access to Rpb3.
Effect of Rpb8 on the Rpb1–Rpb3 interaction
Rpb8 binds strongly to Rpb1 (Fig. 3A) (8,24). Rpb8 also binds tightly to Rpb3 with the support of Rpb11 (Fig. 4D). To understand the relationship between these observations, we have examined the effect of Rpb8 on the Rpb1–Rpb3 interaction. Rpb1H and GST–Rpb3 were coexpressed in the presence or the absence of Rpb8 (Fig. 5, lanes 1 and 2). Rpb8 expression did not affect the expression levels of Rpb1H and GST–Rpb3, with the difference <3% as measured by the intensity of chemiluminescence in western blotting (data not shown). After GSH-affinity chromatography, Rpb1H from the cells coexpressing Rpb8 was recovered more than that without Rpb8 expression (Fig. 5, lanes 3 and 4). The Rpb1H/GST–Rpb3 ratio between the assembly fractions with or without Rpb8 was reproducibly 1.3:1.0 in two independent experiments as measured by chemiluminescence (data not shown). This indicates the significant, albeit at low level, enhancement of the Rpb1–Rpb3 interaction by Rpb8.
Figure 5.
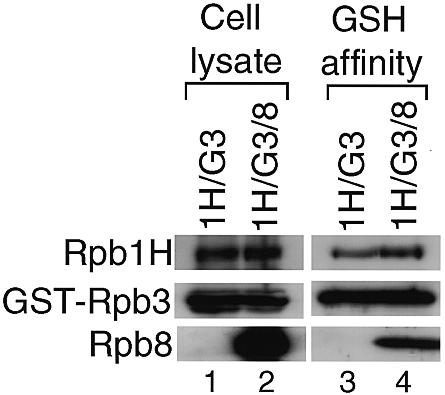
Enhancement of the Rpb1–Rpb3 interaction by Rpb8. Rpb1H and GST–Rpb3 were coexpressed with and without Rpb8. The expression was detected by western blotting with antisera against each subunit (cell lysate). The extracts of the coinfected cells were subjected to GSH-affinity chromatography and the eluted fractions were analyzed by western blotting (GSH affinity). The proteins coexpressed are indicated on the top: 1H, Rpb1H; G3, GST–Rpb3; 8, Rpb8, and the detected subunits are indicated on the left. Portions of the filters corresponding to the migration positions of each subunit in the SDS–PAGE are arrayed.
DISCUSSION
The formation of a multi-subunit sub-complex of S.pombe Pol II has been analyzed through the coexpression of 11 species of Pol II subunit in insect cells. Since the fusion of GST to the Rpb3 subunit does not prevent the assembly and the essential functions of Pol II in S.pombe (Fig. 1), we expressed the GST–Rpb3 fusion subunit for the isolation of subunit assemblies by GSH-affinity chromatography.
Using six different recombinant baculoviruses, all the 11 subunits were successfully expressed in the same cells. In the cells simultaneously expressing the 11 subunits, the six subunits, Rpb1H, Rpb2, Rpb5, Rpb7, Rpb8 and Rpb11, formed complexes with GST–Rpb3 (Fig. 2). The coexpression of all the 11 subunits in the same cells was supported, because each of the unassembled subunits, Rpb6, Rpb9, Rpb10 and Rpb12, was expressed from the same viruses with the assembled subunits, Rpb5, Rpb7, Rpb2 and Rpb8, respectively. The assembly in vitro in cell extracts is unlikely, because Acker et al. (29) observed that independently expressed human Pol II subunits did not form complexes after mixing the cell lysates.
Interactions between the S.pombe and the insect Pol II subunits may be possible because some Pol II subunits are highly conserved among organisms (1–7). However, western blot analysis indicated that the major contaminants in the assembly fractions were degradation products of the S.pombe small subunit, especially degraded GST–Rpb3 which can bind to GSH–Sepharose (data not shown). The possibility of hybrid Pol II formation may be excluded because: (i) the expression levels of S.pombe subunit proteins were higher than the endogenous Pol II [the level of even the least expressed subunit, Rpb1H, was 30–50-fold higher than that of the cellular Rpb1 as revealed by western blotting using an anti-CTD antibody (data not shown)] and therefore the recombinant subunits must dominate in the assembly; (ii) the host cell gene expression is almost completely shut off at the late stage of infection when the subunit proteins are expressed from the polyhedrin and the p10 promoters; (iii) dissociation and reassociation of the cellular Pol II are improbable because the assembled Pol II enzyme is metabolically stable; (iv) the association of pre-existing unassembled cellular subunits must also be rare because the pool of unassembled subunits, if any, is small and must degrade rapidly after the virus infection.
None of the antisera used showed detectable cross-reaction with the cellular Pol II subunits in cell lysates (Figs 2A and 4C), but the possibility remains that cross-reactive signals are detected at high protein concentrations after the chromatography. However, this possibility could be ruled out, because the omission of each S.pombe subunit resulted in disappearance of the corresponding signal in the assembly fractions (Figs 2 and 4). Thus, all the immunostained signals in the assembly fractions are attributable to the expressed S.pombe subunits.
Pairwise expression experiments indicated that each of the six assembled subunits binds directly to Rpb3 (Fig. 4). The other four subunits, Rpb6, Rpb9, Rpb10 and Rpb12, were expressed at high levels and extracted in the soluble fractions (data not shown) but failed to bind to GST–Rpb3. This group of subunits might not interact directly with Rpb3, but be assembled into Pol II by binding to a subunit(s) other than Rpb3. In fact, we detected the interaction of the three subunits, Rpb6, Rpb10 and Rpb12, with both of the two large subunits (23). In the chemical crosslinking experiments, we also observed the Rpb3–Rpb10 interaction, but the major contact site of Rpb10 might be located on Rpb2. If the assembly of these four subunits depends on Rpb1–Rpb2 complex formation, the level of those subunits in the assemblies might be lower than other subunits which bind directly to Rpb3, because only a small portion of the complexes contained the two large subunits.
In this study Rpb4 was not included because the rpb4 cDNA was not available. We postulate that the absence of Rpb4 does not affect the assembly of other subunits, because a considerable portion of Pol II molecules in S.cerevisiae does not contain RPB4 (32). However, another possibility is that Rpb4 plays a role in the integration of four subunits, because recently we found that most of the S.pombe Pol II molecules contain Rpb4 (7). The failure of detection of the four subunits might also be due to: (i) fortuitous interaction with some cellular or viral components preventing the subunits from assembling; or (ii) some cellular components which are essential for the Pol II assembly are lost after the virus infection.
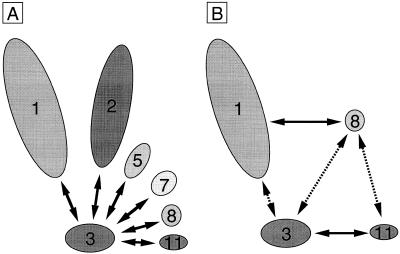
In this study we detected the Rpb3–Rpb7 and Rpb3–Rpb8 interactions, which had not been detected by chemical crosslinking or Far-western blotting (23). The method employed in this work has advantages over previously reported in vitro methods, because: (i) the interaction of native proteins can be assayed in vivo; and (ii) the detection by chemical crosslinking depends on the nature of crosslinkers or the distance between specific amino acid side chains. The direct interaction of Rpb3 with as many as six subunits suggests that Rpb3 is one of the central subunits in the assembly, and thus it provides multiple contact surfaces for other subunits (Fig. 6A), although multiple contacts among other subunits also play a role in the assembly (Fig. 3B). This is reminiscent of the prokaryotic α subunit, the counterpart of Rpb3 and Rpb11, which provides contact surfaces for α dimerization and ββ′ binding (33–36).
Figure 6.
Multiple contacts including Rpb3. (A) Rpb3 interacts with six subunits. Rpb3 provides contact surfaces for the six subunits, Rpb1, Rpb2, Rpb5, Rpb7, Rpb8 and Rpb11, and plays one of the central roles in the subunit assembly. Interactions not including Rpb3, which may also be of importance for the assembly, are not indicated here. (B) Involvement of multiple contacts for stable complex formation. The formation of the Rpb3–Rpb11 heterodimer facilitates the binding of Rpb8 to Rpb3, and thereby Rpb8 promotes Rpb1 binding to Rpb3 to some extent. Rpb3 is essential for Rpb11 to be incorporated to the complex efficiently. Solid lines indicate relatively strong bindings whereas dashed lines indicate interactions which are enhanced by multiple contacts.
Since all the four small subunits, Rpb5, Rpb7, Rpb8 and Rpb11, were found to make direct contacts with both Rpb3 (this study) and one or both of the two large subunits, Rpb1 and Rpb2 (23), the multiple contacts among Pol II subunits must be important for the assembly of Pol II. In agreement with this idea, the assembly state of Pol II subunits is fortified concomitantly with the increase in the number of subunits for coexpression. For instance, the weak Rpb3–Rpb8 interaction was reinforced by Rpb11, and the Rpb1–Rpb3 interaction was done by Rpb8 though at a low level (Fig. 6B). In addition, Rpb3 is essential for the efficient binding of Rpb11 to the complex, even though the Rpb1–Rpb8–Rpb11 interaction is possible (Fig. 3B). A promotion of assembly of the largest subunit by Rpb8 agrees with the in vivo observation that overexpression of C160, the largest Pol III subunit in S.cerevisiae, recruits the heterologously expressed S.pombe Rpb8 into the Pol III (37). In accordance with the stabilization of subunit complexes by multiple subunit–subunit contacts, Rpb8 is efficiently assembled even in the absence of Rpb11 or Rpb3, if all other subunits are coexpressed (Figs 2 and 3B). The Rpb3–Rpb5 interaction was also intensified in the presence of the three subunits, Rpb7, Rpb8 and Rpb11 (Fig. 4C and D), suggesting the involvement of multiple contacts of Rpb3 and Rpb5 with other subunits for the stabilization of complex.
One exception to this rule is that the Rpb3–Rpb7 interaction was slightly decreased when the other three small subunits were coexpressed (Fig. 4A and B). One explanation is that Rpb7 binding is restricted in the presence of a subset of subunits, but in the complete Pol II, this negative effect is neutralized or balanced by a full set of multiple contacts. The Rpb4-independent interaction of Rpb7 with Pol II has been reported (38) and here we propose that Rpb3 is the target for Rpb7 binding.
So far we failed to detect the enzymatic activity of RNA synthesis for the sub-assemblies formed in the insect cells (data not shown). Rpb1 and Rpb2 together form the active center of the enzyme, but only a small fraction of the sub-assemblies isolated in this study contained both of them, probably owing to the low expression levels. At present we reserve the conclusion whether the sub-assemblies are active in RNA synthesis or not. To reconstitute an active Pol II, the expression levels of the two large subunits must be improved.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ramesh Wigneshweraraj for critical reading of the manuscript. We also thank N. Fujita, H. Mitsuzawa and H. Sakurai for advice and discussions, and H. Suzuki for technical assistance. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, and CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation (JST).
REFERENCES
- 1.Azuma Y., Yamagishi,M., Ueshima,R. and Ishihama,A. (1991) Nucleic Acids Res., 19, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawagishi M., Yamagishi,M. and Ishihama,A. (1993) Nucleic Acids Res., 21, 469–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azuma Y., Yamagishi,M. and Ishihama,A. (1993) Nucleic Acids Res., 21, 3749–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyao T., Yasui,K., Sakurai,H., Yamagishi,M. and Ishihama,A. (1996) Gene Cell, 1, 843–854. [DOI] [PubMed] [Google Scholar]
- 5.Sakurai H. and Ishihama,A. (1997) Gene, 196, 165–174. [DOI] [PubMed] [Google Scholar]
- 6.Sakurai H., Kimura,M. and Ishihama,A. (1998) Gene, 221, 11–16. [DOI] [PubMed] [Google Scholar]
- 7.Sakurai H., Mitsuzawa,H., Kimura,M. and Ishihama,A. (1999) Mol. Cell. Biol., 19, 7511–7518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M., Ishiguro,A. and Ishihama,A. (1997) J. Biol. Chem., 272, 25851–25855. [DOI] [PubMed] [Google Scholar]
- 9.Kim T.-K., Lagrange,T., Wang,Y.-H., Griffith,J.D., Reinberg,D. and Ebright,R.H. (1997) Proc. Natl Acad. Sci. USA, 94, 12268–12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kontermann R.E., Kobor,M. and Bautz,E.K.F. (1993) Protein Sci., 2, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontermann R.E. and Bautz,E.K.F. (1994) FEBS Lett., 344, 166–170. [DOI] [PubMed] [Google Scholar]
- 12.Wlassoff W.A., Kimura,M. and Ishihama,A. (1999) J. Biol. Chem., 274, 5104–5113. [DOI] [PubMed] [Google Scholar]
- 13.Woychik N.A. and Young,R.A. (1989) Mol. Cell. Biol., 9, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards A.M., Kane,C.M., Young,R.A. and Kornberg,R.D. (1991) J. Biol. Chem., 266, 71–75. [PubMed] [Google Scholar]
- 15.Jensen G.J., Meredith,G., Bushnell,D.A. and Kornberg,R.D. (1998) EMBO J., 17, 2353–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyao T. and Woychik,N.A. (1998) Proc. Natl Acad. Sci. USA, 95, 15281–15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheong J.-H., Yi,M.-K., Lin,Y. and Murakami,S. (1995) EMBO J., 14, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y., Nomura,T., Cheong,J.-H., Dorjsuren,D., Iida,K. and Murakami,S. (1997) J. Biol. Chem., 272, 7132–7139. [DOI] [PubMed] [Google Scholar]
- 19.Lanzendörfer M., Smid,A., Klinger,C., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (1997) Genes Dev., 11, 1037–1047. [DOI] [PubMed] [Google Scholar]
- 20.Awrey D.E., Weilbaecher,R.G., Hemming,S.A., Orlicky,S.M., Kane,C.M. and Edwards,C.M. (1997) J. Biol. Chem., 272, 14747–14754. [DOI] [PubMed] [Google Scholar]
- 21.Hull M.W., McKune,K. and Woychik,N.A. (1995) Genes Dev., 9, 481–490. [DOI] [PubMed] [Google Scholar]
- 22.Krapp S., Kelly,G., Reischl,J., Weinzierl,R.O.J. and Matthews,S. (1998) Nature Struct. Biol., 5, 110–114. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro A., Kimura,M., Yasui,K., Iwata,A., Ueda,S. and Ishihama,A. (1998) J. Mol. Biol., 279, 703–712. [DOI] [PubMed] [Google Scholar]
- 24.Miyao T., Honda,A., Qu,Z. and Ishihama,A. (1998) Mol. Gen. Genet., 259, 123–129. [DOI] [PubMed] [Google Scholar]
- 25.Ulmasov T., Larkin,R.M. and Guilfoyle,T.J. (1996) J. Biol. Chem., 271, 5085–5094. [DOI] [PubMed] [Google Scholar]
- 26.Larkin R.M. and Guilfoyle,T.J. (1997) J. Biol. Chem., 272, 12824–12830. [DOI] [PubMed] [Google Scholar]
- 27.Larkin R.M. and Guilfoyle,T.J. (1998) J. Biol. Chem., 273, 5631–5637. [DOI] [PubMed] [Google Scholar]
- 28.Khazak V., Estojak,J., Cho,H., Majors,J., Sonoda,G., Testa,J.R. and Golemis,E.A. (1998) Mol. Cell. Biol., 18, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Acker J., Graaff,M., Cheynel,I., Khazak,V., Kedinger,C. and Vigneron,M. (1997) J. Biol. Chem., 272, 16815–16821. [DOI] [PubMed] [Google Scholar]
- 30.Yasui K., Ishiguro,A. and Ishihama,A. (1998) Biochemistry, 37, 5542–5548. [DOI] [PubMed] [Google Scholar]
- 31.Ishihama A. (1981) Adv. Biophys., 14, 1–35. [PubMed] [Google Scholar]
- 32.Choder M. and Young,R.A. (1993) Mol. Cell. Biol., 13, 6984–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura M., Fujita,N. and Ishihama,A. (1994) J. Mol. Biol., 242, 107–115. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M. and Ishihama,A. (1995) J. Mol. Biol., 248, 756–767. [DOI] [PubMed] [Google Scholar]
- 35.Kimura M. and Ishihama,A. (1995) J. Mol. Biol., 254, 342–349. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. and Ishihama,A. (1996) Gene Cell, 1, 517–528. [DOI] [PubMed] [Google Scholar]
- 37.Voutsina A., Riva,M., Carles,C. and Alexandraki,D. (1999) Nucleic Acids Res., 27, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheffer A., Varon,M. and Choder,M. (1999) Mol. Cell. Biol., 19, 2672–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]