Abstract
Sequence analysis of Plasmodium falciparum parasites is informative in ensuring sustained success of malaria control programmes. Whole-genome sequencing technologies provide insights into the epidemiology and genome-wide variation of P. falciparum populations and can characterise geographical as well as temporal changes. This is particularly important to monitor the emergence and spread of drug resistant P. falciparum parasites which is threatening malaria control programmes world-wide. Here, we provide a detailed characterisation of genome-wide genetic variation and drug resistance profiles in asymptomatic individuals in South-Western Mali, where malaria transmission is intense and seasonal, and case numbers have recently increased. Samples collected from Ouélessébougou, Mali (2019–2020; n = 87) were sequenced and placed in the context of older Malian (2007–2017; n = 876) and African-wide (n = 711) P. falciparum isolates. Our analysis revealed high multiclonality and low relatedness between isolates, in addition to increased frequencies of molecular markers for sulfadoxine-pyrimethamine and lumefantrine resistance, compared to older Malian isolates. Furthermore, 21 genes under selective pressure were identified, including a transmission-blocking vaccine candidate (pfCelTOS) and an erythrocyte invasion locus (pfdblmsp2). Overall, our work provides the most recent assessment of P. falciparum genetic diversity in Mali, a country with the second highest burden of malaria in West Africa, thereby informing malaria control activities.
Subject terms: Parasite genetics, Parasite genomics, Antimicrobial resistance
Introduction
Malaria was estimated to cause 250 million illnesses worldwide and 619 thousand associated deaths in 2021 alone. Mali is amongst the 9 countries with the highest burden of disease for malaria and its number of malaria cases has increased between 2016 and 20211. Most malaria cases in Mali and in the rest of sub-Saharan Africa are caused by Plasmodium falciparum, the most virulent human malaria parasite. Mali is divided into five ecoclimatic zones2 across which malaria transmission fluctuates. The South-Western zone has the highest P. falciparum incidence rates and transmission is highly seasonal, coinciding with the annual rainy season. However, across different regions in South-Western Mali, the degree of perennial and seasonal transmission varies, as does the timing of the rainy season, causing heterogeneity in transmission seasons and malaria epidemiology2.
In accordance with WHO guidelines, current malaria intervention strategies in Mali include artemisinin combination therapies (ACTs) for uncomplicated P. falciparum malaria, with Artemether-Lumefantrine (AL) as the first-line treatment. Seasonal malaria chemoprevention (SMC) strategies, consisting of sulfadoxine-pyrimethamine with amodiaquine (SPAQ) in children and intermittent preventative treatment for pregnant women (IPTp) with sulfadoxine-pyrimethamine (SP) alone, have been widely implemented in African countries and have been introduced in Mali from 2012 and 2015 onwards, respectively.
In addition to disruption due to the COVID-19 pandemic, increased disease incidence has been linked to the emergence and spread of drug resistant P. falciparum parasites1. Drug-resistant strains against SP, artemisinin derivatives and partner drugs pose a major challenge in the fight against malaria3. Molecular markers of drug resistance are therefore extremely useful in identifying and monitoring drug-resistant P. falciparum parasites and have been described for most antimalarial drugs. Mutations in the multidrug resistance (pfmdr1) gene for example have been associated with various parasite responses to lumefantrine, chloroquine (CQ), amodiaquine, mefloquine and piperaquine. The genetic basis of SP resistance is well documented and involves an accumulation of mutations in the dihydrofolate-reductase (pfdhfr) gene (N51I, C59R, S108N and I164L) and the dihydropteroate-synthase (pfdhps) gene (S436A/F, A437G, K540E, A581G and A613S/T). Infections harbouring the triple dhfr CIRNI mutant (mutations underlined) are common throughout Africa and are pyrimethamine resistant4,5. The combination of this triple dhfr mutant with the double-mutant dhps (A437G and K540E, SGEAA) further increases the risk of SP treatment failure to 50%5,6.
Mutations in the pfkelch13 gene associated with decreased artemisinin susceptibility emerged and spread in South-East Asia and have more recently emerged in several countries in East-Africa7–9. It is expected that this will spread to other parts of Africa and therefore continuous monitoring of pfkelch13 genetic variation is critical. A high prevalence of CQ resistance led to its removal from any treatment guidelines for P. falciparum infections in sub-Saharan Africa. Decades after this, CQ-sensitive P. falciparum parasites have re-emerged in many parts of the world10–13. For that reason, a re-introduction of CQ, in combination with other antimalarials, has been proposed14. Reports from Mali have not observed a substantial decrease in frequency of mutations in the chloroquine resistance transporter (pfcrt) associated with CQ resistance15, although recently a downwards trend was reported in Malian isolates collected in 2016–201716. Continued assessment is therefore important to determine whether CQ could be reintroduced in the region in future.
The majority of malaria infections in endemic areas are asymptomatic17, however, such carriers tend to be underrepresented in genome-wide large-scale genetic analyses, due to both the lack of seeking treatment and technical difficulties with sequencing low density infections. Asymptomatic carriers are the main contributors to the infectious reservoir as they can remain infectious for long periods of time without showing any symptoms, meaning they could unknowingly spread the disease to others while remaining unaware they are infected. In addition to this, their frequency in the population and the characteristics of individuals that are more likely to be asymptomatic, such as a higher risk of mosquito bites, further increase their contribution to the infectious reservoir18,19. Therefore, understanding the genetic characteristics of the asymptomatic reservoir is key to effectively controlling the spread of malaria.
Advances in Next-Generation Sequencing (NGS) technologies have rendered Whole Genome Sequencing (WGS) more accessible and affordable for use in disease management and malaria control. Along with identifying and monitoring molecular markers of drug resistance, investigating genomic variation is useful for understanding transmission dynamics, selective sweeps, and P. falciparum epidemiology. Assessing changes in genomic relatedness within a population, including through using identity-by-descent measures, can provide insights into parasite population demography and transmission intensity over time20. In addition, the identification of genes under selection can offer insights into the selective pressure exhibited by drugs or other unknown agents, which is important for developing effective strategies for prevention, control, and treatment of malaria.
In summary, monitoring of P. falciparum drug resistance is essential to inform drug policies worldwide, particularly in regions of high malaria transmission, and the WHO recommends regular updating and monitoring of antimalarial resistance to support progress made towards malaria control and elimination21. In this report, we provide an in-depth analysis of drug resistance profiles and recent genomic variation, using selection, ancestry and identity-by-descent analysis, in asymptomatic individuals in Ouélessébougou and neighbouring villages, Mali, in 2019 and 2020. Furthermore, we place these in the context of Malian P. falciparum isolates collected between 2007 and 2017, as well as African-wide parasite populations.
Results
Genomic population structure of Malian P. falciparum populations
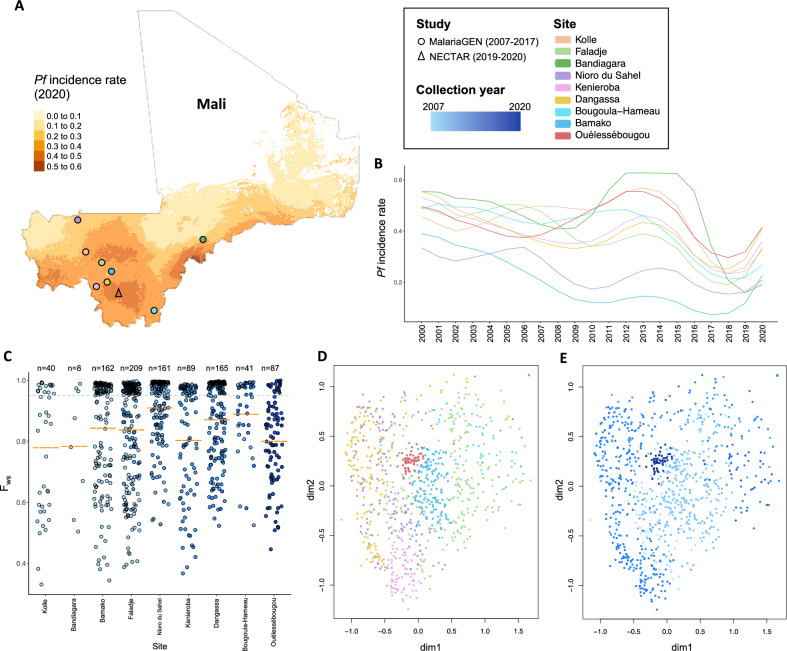
Genome-wide SNP analysis of 962 Malian P. falciparum isolates, collected between 2007 and 2020 in two different studies and originating in 9 locations (Fig. 1A), revealed differences in multiplicity of infection and population structure. Ouélessébougou isolates collected in 2019–2020 originated from asymptomatic P. falciparum infections, while no details on clinical presentation were available for publicly available genomes from the MalariaGEN database. P. falciparum incidence rates varied across sample sites and across two decades (Fig. 1A,B). A total of 863,046 high-quality SNPs were identified. Multiclonality was measured using the Fws metric, or in-breeding coefficient, which is indicative of monoclonality if > 0.95, while a lower Fws metric reflects multiclonality. The mean Fws value for the 2019–2020 Mali samples (n = 87) from Ouélessébougou was 0.80, with only 20% of samples harbouring a single clone. We found a higher multiclonality in the 2019–2020 Ouélessébougou isolates compared to isolates from different sites where collection took place between 2015 and 2017 (Fig. 1C). Using the SNP data, a Uniform Manifold Approximation and Projection (UMAP) statistical analysis to cluster isolates revealed little spatial substructure between populations, although some grouping based on location (Fig. 1D) and collection year (Fig. 1E) could be observed and the Ouélessébougou isolates formed an individual subcluster.
Figure 1.
Population structure of P. falciparum isolates collected in Mali from 2007 to 2020. (A) Map of Mali presenting the number of newly diagnosed Plasmodium falciparum cases per 1,000 population in 2020 (colour scale) and indicating collection sites of the MalariaGEN studies (circle) and the New Drug Combinations for P. falciparum Transmission Reduction (NECTAR) clinical trials (triangle), coloured by village or city name. This map was generated using the tmap R package (version 3.3.3; https://r-tmap.github.io/tmap/) (B) P. falciparum incidence rate for each collection site (by colour) from 2000 to 2020. (C) Complexity of infections estimated by the in-breeding coefficient (Fws metric) per sample, classified per study site, with the colour indicating collection year. Dotted grey trendline is at 0.95, dashed orange marks indicate mean Fws value per group. (D, E) Uniform Manifold Approximation and Projection (UMAP) visualisation of 962 Malian P. falciparum isolates, coloured by site and collection year, respectively.
Genomic population structure of Malian P. falciparum populations in comparison to African-wide populations
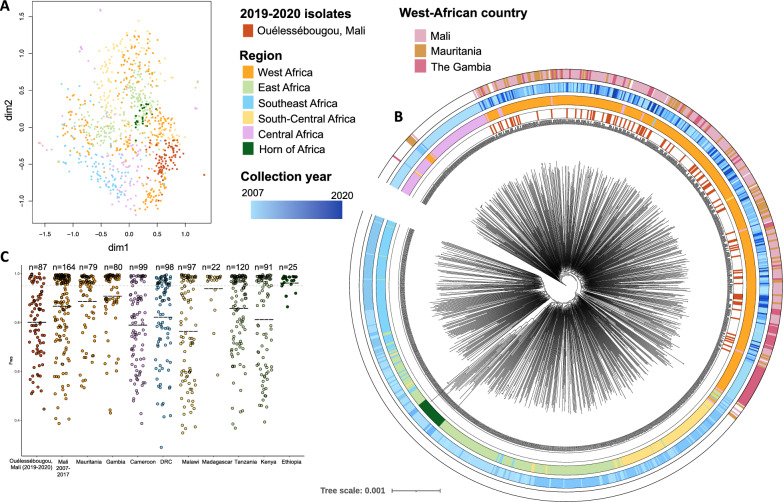
A SNP-based UMAP visualisation and maximum likelihood tree show how the most recently collected Ouélessébougou isolates are related to African-wide populations (n = 875) and revealed similarity with West African and Central African isolates (Fig. 2A,B). No distinguishable clusters could be detected, indicating high relatedness, and suggesting movements of genetic information within these populations, which can occur both through human and vector migration. East African, Southeast African and Central African isolates formed separate clusters, while the Horn of Africa appeared to be a distinct cluster within the East African population. Multiclonality assessment showed that the 2019–2020 Malian isolates were relatively more multiclonal compared to the isolates collected in other West African countries (Fig. 2C).
Figure 2.
Population structure of Malian P. falciparum isolates in the context of African-wide populations. (A) Uniform Manifold Approximation and Projection (UMAP) visualisation of 962 African-wide P. falciparum isolates, including 87 isolates collected in Mali in 2019–2020 and 164 Malian isolates collected in 2007–2020. African regions where the isolates were collected are indicated by colour. (B) Maximum-Likelihood tree of the same dataset, annotated by the 2019–2020 Malian isolates (inner ring), the African region of sample origin (second ring), collection year (third ring) and West-African country of collection (outer ring). (C) Complexity of infections (Fws coefficient) per sample, grouped by country and separating the 2019–2020 Malian isolates from the 2007 to 2017 Malian isolates. Dotted grey trendline is at 0.95, dashed lines indicate mean Fws value per group.
Frequencies of drug resistance molecular markers
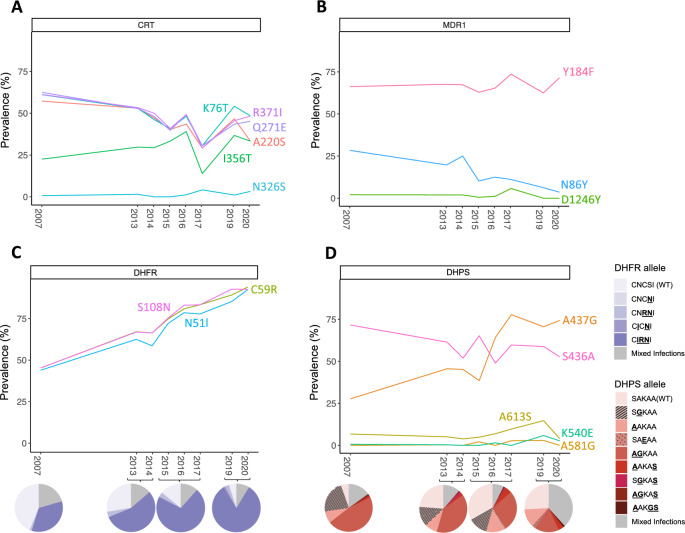
Increased frequencies of molecular markers of sulfadoxine-pyrimethamine and persistence of chloroquine resistance markers were observed in the Ouélessébougou 2019–2020 population (n = 87), compared to Malian isolates from 2007 to 2017 (n = 876) (Table ST1). Polymorphisms causing amino acid changes that confer chloroquine resistance in pfcrt (K76T, A220S, Q271E, N326S, I356T and R371I) persisted at similar frequencies over time (Fig. 3A,B). The N86Y polymorphism on pfmdr1 decreased over time from 28.4% in 2007 to 2.6% in 2020. This 86Y allele has been linked to chloroquine and amodiaquine resistance as well as piperaquine resistance (86Y allele in combination with Y184F), while parasites carrying the N86 allele show lower susceptibility to lumefantrine, piperaquine and mefloquine22,23. Y184F and D1246Y amino acid changes in MDR1 persisted at comparable frequencies. Three non-synonymous SNPs in pfmdr1 that were not previously found in Malian isolates, resulting in amino acid changes S400C, D431Y and K503N, were identified in 2.13% of the 2019 isolates and 2.94% of the 2020 isolates, respectively (Table ST1). The frequency of point mutations in pfdhfr associated with pyrimethamine resistance (N51I, C59R and S108N) approximately doubled since 2007, reaching alarming frequencies of 92.4%, 93.9% and 92.7%, respectively, in 2020. In addition, the CIRNI triple mutant haplotype increased in frequency and made up 82.7% of the parasite population in 2019–2020, while the wild-type haplotype was reduced to 4.9% (Fig. 3C, Table ST2). One mutation in pfdhps conferring sulfadoxine resistance (A437G) increased in frequency from 27.7% in 2007 to 74.3% in 2020, while S436A showed a downwards trend (Fig. 3D). The pfdhps K540E mutation was found in 4 isolates in 2019–2020 and all of these were combined with the CIRNI triple dhfr mutant, leading to a quadruple mutant frequency of 2.47%. A non-synonymous SNP at position 748,145 in pfdhfr (V20I), was newly identified in Malian isolates in 2019, at 2.04% frequency. No known mutations in pfkelch13 associated with artemisinin resistance were identified in any of the Malian isolates. The pfkelch13 mutations R255K, K189N and K189T persisted at similar frequencies to the frequencies observed in 2007 and no new mutations in pfkelch13 were identified (Table ST1).
Figure 3.
Prevalence of single nucleotide polymorphisms known to cause decreased drug susceptibility. Minor allele frequencies (MAFs) are shown for isolates collected from 2007 to 2020 in Mali for genes associated with drug resistance, including (A) pfcrt, (B) pfmdr1, (C) pfdhfr and (D) pfdhps. Pie charts in C and D represent the frequencies at which combinations of pfhdfr and pfdhps mutants were observed in the P. falciparum isolates collected in 2007, 2013–2014, 2015–2017 and 2019–2020.
Regions under selective pressure in Malian isolates
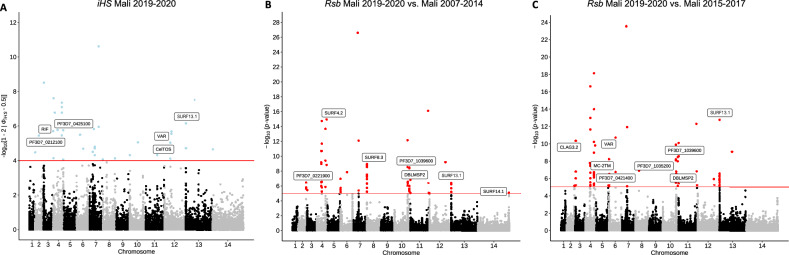
Determination of genomic regions under directional selection by haplotype structure analysis within the Ouélessébougou 2019–2020 isolates and in comparison to the older Malian populations, revealed a number of genes to be under selective pressure. The integrated haplotype score (iHS) metric was used to identify SNPs under selection within the 2019–2020 population (Fig. 4A) and regions of the genome with an elevated number of SNPs under selection (Table ST3). This identified conserved Plasmodium protein coding genes with unknown function (PF3D7_0212100 and PF3D7_0425100), predominantly expressed in ookinetes and ring stages, respectively, as well as the pfCelTOS gene, which encodes a cell-traversal protein for ookinetes and sporozoites and was suggested as an attractive vaccine candidate antigen24,25. The between-population Rsb index was used to identify SNPs under selection when comparing the 2019–2020 population with the older Malian isolates collected in 2007–2014 (n = 414) (Fig. 4B) and 2015–2017 (n = 462) (Fig. 4C). Regions with a high number of SNPs under selection were determined as well (Table ST4). This identified genes associated with erythrocyte invasion (pfdblmsp2, pfmsp3), protein transport (pfMC-2TM), cytoadherence (pfCLAG3.2), and a gene encoding RNA of unknown function (Pf3D7_0421400, RUF6).
Figure 4.
Scan for evidence of recent directional selection. Manhattan plots show analysis of the (A) integrated haplotype score (iHS) for individual SNPs in the 2019–2020 Mali population and Rsb cross-population test for extended haplotypes comparing the 2019–2020 Mali population to the (B) 2007–2014 Mali population and the (C) 2015–2017 Mali population.
Ancestral admixture analysis confirms similar ancestry among Malian isolates
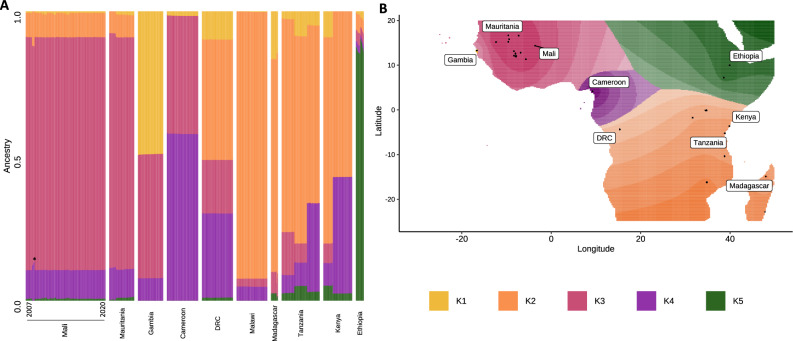
Spatial ancestry estimation of Malian isolates along with African-wide populations found similar ancestral origins among all Malian P. falciparum isolates. The optimum number of ancestral populations was estimated to be 5 (K = 5; K1–K5), based on eigenvalue decay corresponding to K ranging from 1 to 10. The K1 ancestral population was dominant in Gambian samples (49.4%), while the K2 ancestral population appeared to be linked to South-Central and East African populations (Malawi, 92.3%; Madagascar, 74.4%; Tanzania, 69%; Kenya, 61.9%). Malian isolates, along with Mauritanian isolates, seemed to contain mostly the K3 ancestral population (Mali, 80.3%; Mauritania, 80.5%), in addition to smaller portions of K2 ancestry (Mali, 8.5%; Mauritania, 7.7%) and K4 ancestry (Mali, 10%; Mauritania, 10.1%). Very low fractions of K1 and K5 ancestries were present in Malian and Mauritanian isolates, except for isolates from Bandiagara in Mali that appeared to not contain any K5 ancestry (Fig. 5).
Figure 5.
Genome-wide admixture ancestry proportions for P. falciparum populations across the African continent. (A) Ancestries per isolate (columns) for each country, ranked per country by ascending collection year. Asterisk indicates isolates from Bandiagara in Mali. (B) Geographic map of estimated ancestries using K = 5 ancestral populations across the African continent.
Identity-by-descent analysis reveals highly diverse Ouélessébougou population
As a measure of genetic relatedness within populations, identity-by-descent (IBD) analysis revealed that Ouélessébougou isolates exhibit very low fractions of pairwise IBD across the genome (median = 0, range = 0–0.133), while the Malian isolates collected in 2007–2014 and 2015–2017 showed a slightly higher relatedness (median = 0.021 and median = 0.017, respectively) (Fig. S2). The top 5% of IBD positions (classified in 10 kb windows of the genome) in the Ouélessébougou isolates were distributed across 17 regions on 3 chromosomes (chr. 6, 7 and 13) (Fig. S3, Table ST5). Three regions with high IBD on chromosome 6 included the gene encoding histone methyltransferase SET1 and the pfcrt gene on chromosome 7 was also identified as encompassing high IBD.
Discussion
NGS technologies have provided an increasingly feasible method for exploring genome-wide genomic variation and population dynamics of malaria parasites. Here, we have provided a detailed analysis of genome-wide diversity of P. falciparum isolates from asymptomatic gametocyte carriers in 11 villages in Ouélessébougou in 2019 and 2020 and have placed them in the context of previously sequenced Malian isolates, as well as African-wide isolates via genome-wide SNP analysis. We found high multiclonality and low relatedness among isolates and identified genes under selective pressure. We also observed increased frequencies of molecular markers for sulfadoxine-pyrimethamine and lumefantrine resistance, compared to older Malian isolates.
Genomes from Malian P. falciparum isolates collected between 2007 and 2017 have previously been generated16,26. However, a more up-to-date and systematic sampling strategy is required to support efforts toward infection control and elimination. Ouélessébougou is a rural community in the Koulikoro region of South-Western Mali, which is an area with one of the highest P. falciparum incidence rates in the country27. Ideally, in such settings there should be regular monitoring of changes in genomic variation, to determine epidemiological patterns and population dynamics, especially among asymptomatic gametocyte carriers, and to assess the impact of infection control measures. For example, continued surveillance of drug resistance molecular markers is necessary to inform malaria chemotherapy approaches.
Malian isolates show minor clustering using UMAP visualisation, with Ouélessébougou isolates forming a subcluster. On a maximum-likelihood tree, Ouélessébougou and other Malian isolates cluster with West-African populations, as expected. Despite a recent report observing decreasing multiplicity of infection from 2007 to 2017 in Mali16, our findings show a higher multiclonality in 2019–2020 than in isolates from different sites where collection took place between 2015 and 2017. There may be multiple explanations for the highly multiclonal status of the study parasite population, such as the high P. falciparum incidence rates in this region (Fig. 1B), the timing of sample collection at peak transmission season (September-December), and the asymptomatic clinical presentation of the individuals. As asymptomatic P. falciparum gametocyte carriers are less likely to seek treatment, such infections may be prolonged, increasing the likelihood of reinfection and multiclonality. Despite most available sequencing data originating from incident infections, it is important to genetically characterise asymptomatic P. falciparum carriers, as these are the main contributors to the infectious reservoir and carry the infections that escape treatment18.
Malaria treatment strategies world-wide have been altered over time due to the emergence of resistant parasites to former first-line drugs, with the aim of preserving the efficacy of antimalarial drugs and reducing the global burden of malaria. We found that frequencies of molecular markers conferring CQ resistance have persisted in the 2019–2020 Malian isolates at similar frequencies compared to a decade ago, despite its removal from any P. falciparum treatment guidelines since 2006 and a report of a decreasing trend in 2017 in Mali16. This is unlike other areas in the world where a return in CQ sensitivity has been observed11–14,28 and could indicate a low fitness cost associated with maintaining pfcrt resistance polymorphisms in the population or a continued over the counter use of CQ29–31, thereby highlighting the need to investigate and reduce the availability of this drug in Mali and neighbouring countries.
We observed an increase in the N86 variant pfmdr1 from 71.6% in 2007 to 97.4% in 2020. This N86 allele has been linked to lower susceptibility to lumefantrine, piperaquine and mefloquine22,23, while the 86Y allele previously showed chloroquine and amodiaquine resistance, as well as piperaquine resistance (86Y allele in combination with Y184F). As a result, AL was previously found to select for the N86 allele, whereas artesunate-amodiaquine and dihydroartemisinin-piperaquine select for the 86Y allele32–35. N86Y genotyping could therefore be a useful marker to guide rotation of ACTs in a given geographical area36. Thus, the observed increase in the N86 allele may reflect an expanding proportion of isolates with a decreased susceptibility to AL, thereby raising concern for a continued use of AL as the first-line ACT in Mali. A low frequency of the 86Y allele has been reported previously in Mali in 2016, as well as in other African countries where AL is widely used15,34. This finding is in contrast with a previously observed increase in the 86Y allele frequency in children in Ouélessébougou between 2014 and 2016, following 3 years of SMC with SPAQ, which was likely due to amodiaquine selecting for the 86Y allele37. However, it is important to note that, while a decreased susceptibility to AL in isolates with the N86 allele has been observed in isolates from multiple African countries22,32,33, this has not been phenotypically assessed in Malian isolates specifically.
High frequencies of triple pfdhfr CIRNI mutants (82.72%) were observed in the 2019–2020 Ouélessébougou isolates, which is a substantial increase from previous years (34.3%, 55.1% and 70.7% in 2007, 2013–2014 and 2015–2017, respectively). The pfdhps K540E mutant, which was quasi absent in the Malian isolates collected between 2007 and 2017, was found in 5.9% of 2019 isolates and 2.7% of 2020 isolates. In addition, all K540E mutant isolates harboured the triple dhfr mutant. Continued surveillance is therefore needed to monitor levels of SP resistance and the emergence of quadruple and quintuple (in combination with dhps A437G or dhps A581G) mutants. Moreover, online mutation prevalence maps such as wwarn.org SP molecular surveyor will become increasingly useful to assess which drug to use for SMC38. No SNPs at positions in the pfkelch13 gene that have been reported to cause decreased susceptibility to artemisinin derivatives were observed in any Malian isolates, however, further monitoring for pfkelch13 mutants over the next years is needed, as a spread of these alleles from East-Africa or their independent emergence can be expected.
Analysis of haplotype structure in the 2019–2020 Ouélessébougou isolates identified SNPs under selective pressure in the pfCelTOS gene, which is involved in sporozoite gliding motility, cell traversal, and is a transmission-blocking vaccine candidate24,25. Selective pressure has been observed at this genomic location before39, indicating a high degree of diversity and thereby rendering pfCelTOS a less attractive vaccine candidate. Cross-population analysis comparing parasite populations from 2019–2020 to 2007–2014 and 2015–2017 was performed and, despite increasing frequencies of molecular markers of drug resistance over this 13 year period, we did not find any drug resistance associated SNPs under directional selective pressure. We identified SNPs under selective pressure in genes associated with erythrocyte invasion (pfdblmsp2) in both population comparisons and genes associated with protein transport (pfMC-2TM), cytoadherence (pfCLAG3.2), and a gene encoding RNA of unknown function (Pf3D7_0421400, RUF6) in the comparison between the 2019–2020 and 2015–2017 populations. Selective pressure in pfdblmsp2 and members of the clag multigene family has been described before and is likely due to their location as surface proteins and the resulting contact with the host immune system39,40. IBD analysis revealed very low fractions of pairwise IBD across the genome in isolates from 2019–2020 Ouélessébougou, indicating low relatedness between isolates. The pfcrt gene, associated with CQ and piperaquine resistance was found to be in the top 5% of IBD positions, suggesting that this gene is highly conserved among Ouélessébougou isolates. This is in accordance with the observed persistent frequencies of molecular markers for CQ resistance. In addition, the pfSET1 gene, an import histone lysine methyltransferase, was highly conserved as well. Admixture ancestry analysis showed similar ancestries for all Malian isolates, which is in line with a recent report16.
This study had several limitations. Firstly, sampling bias cannot be excluded as we only sequenced parasites from asymptomatic infections and from individuals between the ages of 5 and 50, while publicly available datasets do not specify the clinical presentation or individual’s age. In addition, copy number analysis of plasmepsin1-2, which confers piperaquine resistance, was not performed, due to selective Whole Genome Amplification (sWGA) of parasite DNA prior to sequencing, which prevents adequate analysis of copy number. Lastly, we did not assess phenotypic resistance or any association between genotype and phenotype, however, the molecular markers for drug resistance reported here have been widely proven to predict either in vitro drug resistance levels or patient treatment outcomes4–6,41–43.
After years of progress towards reducing the global burden of malaria, incidence rates and deaths are now on the rise. This rise could be due to disruptions in malaria control programmes during the COVID-19 pandemic, but antimalarial resistance has been linked to this increase as well. In order to progress towards the goal of reducing the global malaria burden by 90% by 203044, we need to generate a comprehensive picture of the genomic variation and the epidemiology of parasite populations. Here, we have provided an updated assessment of genomic diversity of the P. falciparum parasite population in South-Western Mali, a region with very intense malaria transmission. Our results showed that the parasites originating from Mali clustered according to their geographic and temporal origin, with the Ouélessébougou isolates forming a separate subcluster. This suggests a high genetic diversity among Malian isolates. Molecular markers of SP resistance were found to be on the rise, which shows a progression towards failure of this drug combination and necessitates continued monitoring. No decline in CQ resistance over time was observed, opposing the idea of a potential CQ re-introduction in Mali in the near future. Our study contributes valuable data regarding the current epidemiological and drug resistance scenario of malaria in Mali and can aid effective malaria control in Mali. Further applications of sequencing approaches, including new portable technologies and amplicon sequencing assays, in malaria endemic countries are needed to assist disease control and inform treatment guidelines.
Methods
Study sites
In 2019 and 2020 a total of 180 individuals with microscopy detectable P. falciparum gametocytes in the absence of malaria symptoms were recruited into two clinical trials45,46 in Ouélessébougou and 11 villages in Ouélessébougou, Mali (Fig. S1). Ouélessébougou is a commune that includes the town of Ouélessébougou and 44 surrounding villages, which have a total of approximately 50,000 inhabitants. The town is located about 80 km south from Bamako, the capital city of Mali. Malaria transmission in Ouélessébougou is highly seasonal occurring during rainy season from July to November. Publicly available WGS data from Malian isolates originated from an additional 8 locations across Southern Mali, largely consisting of rural villages (Bougoula-Hameau, Dangassa, Faladje, Kenieroba, Kolle, Nioro-du-Sahel, Bandiagara) and one urban area (Bamako). All sites have a subtropical climate with dry and rainy seasons, except Nioro-du-Sahel, which is characterised by a desert climate2,16.
Sample collection and whole genome sequencing
A total of 97 whole blood samples were selected from P. falciparum gametocyte carriers, aged between 5 and 50 years, recruited into two previously published clinical trials in Ouélessébougou45,46. Permission to conduct this study was obtained from the London School of Hygiene and Tropical medicine Research Ethics Committee (reference numbers 17507 and 21905) and the University of Sciences Techniques and Technologies of Bamako Ethical Committee (reference numbers 2019/67/CE/FMPOS and 2020/96/CE/FMPOS/FAPH) and performed in accordance with relevant guidelines and regulations. The trials were registered on ClinicalTrials.gov (NCT04049916 and NCT04609098). Written informed consent was obtained from all subjects and/or their legal guardians prior to sample collection. For minor participants, informed consent for study participation was obtained from their parent and/or legal guardian. Species identification was carried out by microscopy by trained microscopists at the Malaria Research and Training Centre of the University of Bamako (Bamako, Mali). DNA was extracted from 83.3 μL whole blood using a MagNAPure LC automated extractor (Total Nucleic Acid Isolation Kit High Performance; Roche Applied Science, Indianapolis, IN, USA) and amplified using an established selective whole genome amplification (sWGA) primer set and protocol47,48. Whole genome sequencing was performed on an Illumina Novaseq 6000 platform at Eurofins Genomics, Germany, rendering a minimum of 3.75 M paired reads (250 bp reads) per sample.
Data set selection, read mapping, variant detection and quality control
A total of 1701 P. falciparum isolates were included in the analysis (Supplementary File S2), including publicly available whole genome sequences Malaria Genetic Epidemiology Network26 (MalariaGEN) (Pf Community Project, n = 1141; SPOTmalaria project, n = 463) and newly sequenced isolates (n = 97) from asymptomatic P. falciparum infected individuals recruited into two previously published clinical trials in Ouélessébougou, Mali45,46. All raw sequence data was filtered using trimmomatic (version 0.39) and the following parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36. Illumina reads were then mapped to the P. falciparum (Pf3D7; v3) reference genome using bwa-mem software (v0.7.17). SNPs and short insertions and deletions were called using samtools (v1.12) and GATK (v4.1.4.1) software. Mixed call SNPs were assigned genotypes determined by a ratio of coverage in which nucleotide calls were 80% or higher. Samples with more than 40% missingness were not included in any analysis. Of the 97 newly sequenced samples, 9 were removed due to missingness and one was removed after species prediction identified P. malariae (https://github.com/jodyphelan/malaria-profiler), leaving a total of 87 (89.6%) newly sequenced isolates in the final analyses. Of the publicly available datasets, 8 were removed, resulting a total of 1673 samples included in the analyses, including isolates from Cameroon (n = 99), Democratic Republic of Congo (DRC; n = 98), Gambia (n = 80), Kenya (n = 91), Malawi (n = 97), Mali (n = 962), Mauritania (n = 79), Tanzania (n = 120), Madagascar (n = 22) and Ethiopia (n = 25).
Population genetic analyses
Visualisation of sample site geography was performed using the ggmap (version 3.0.0) and tmap (version 3.3.3) R packages. P. falciparum incidence rates from 2000 to 2020 were accessed from the Malaria Atlas Project27. Uniform Manifold Approximation and Projection (UMAP) plots were created using the uwot R package with ‘hamming’ metric and default parameters. A maximum likelihood tree was created by applying iqtree software using genome-wide SNPs, and visualisation was performed in iTOL (version 6)49,50. For population genetic analyses that compare Malian isolates to African-wide populations, a subset of Malian isolates collected between 2007 and 2017 was used (including 10 isolates from each site and each collection year, if available), to obtain comparable number of isolates between populations. Multiclonality was determined by calculating the Fws metric, using an in-house script that utilizes the moimix R package (https://github.com/bahlolab/moimix) to assess within-host diversity in relation to the local population diversity. Only bi-allelic SNPs in coding regions were used for the calculations and Minor Allele frequencies (MAFs) filtering of 0.1% was performed in order to exclusively include robust SNPs. MAFs in drug resistance genes were extracted from the binary matrix and annotated using Bcftools CSQ software, which identifies the mutation as non-synonymous, synonymous, or intergenic as well as the codon and protein shift caused by any non-synonymous mutations51. Only genomic positions with MAFs of at least 2% were retained in the analysis. Data visualisation was performed using the R-based ggplot2 package (R version 4.1.2). Regions of the genome under directional selection were identified using the rehh R package (version 3.2.2), which uses population-based measures of haplotype diversity within (iHS) or between (Rsb) populations52. The R-based Tess3r package (version 1.1.0, using default parameters apart from ‘rep = 25’). was used to calculate admixture based on the spatial modelling of allele sharing using geographical coordinates from sampling sites in addition to genome-wide SNP data53. The optimal number of ancestries was determined across different numbers of sub-populations (K = 1, 2,…, 10). IBD analysis for isolates with Fws > 0.85 was performed to assess connectivity between parasites within populations. This was achieved by estimating the pairwise fraction of shared ancestry between genomic segments, which were inferred to have descended from a recent common ancestor. These IBD fractions were calculated using the hmmIBD software with default parameters, which deploys a hidden Markov model-based approach54. French translation of the manuscript is available in Supplementary File S3 and was assisted by DeepL Translator (https://www.deepl.com/translator).
Supplementary Information
Acknowledgements
We wish to thank the study participants and staff of the community health center in Ouélessébougou and surrounding villages for their cooperation, as well as the local safety monitor and the members of the data and safety and monitoring board for their oversight. We thank the MalariaGEN resource centre for the generation of the published genetic data.
Author contributions
A.D., W.S., C.D., M.J.S., T.B., S.C., A.M. and L.N.V conceived and designed the study; A.M., S.M.N., K.S., A.Y., A.D., M.D., S.O. and A.D. coordinated and conducted recruitment and sample collection; L.N.V. processed samples for sequencing. L.N.V., S.C. and C.D. coordinated the sequencing of samples; L.N.V., E.M., J.P., A.O. and A.S. performed the bioinformatic and statistical analysis, under the supervision of S.C. and T.G.C.; L.N.V. wrote the first draft of the manuscript, and the final version included edits from all authors. The final manuscript was read and approved by all authors.
Funding
This analysis was performed on sequenced genomes from blood samples collected in two clinical trials in Mali, which were funded by the Bill & Melinda Gates Foundation (#INV-002098). LNV is supported by a Biotechnology and Biological Sciences Research Council (BBSRC) LSHTM-tethered PhD award. SC is funded by the Medical Research Council UK (Grant no. MR/M01360X/1). TGC is supported by the Medical Research Council UK (Grant no. MR/M01360X/1, MR/N010469/1, MR/R020973/1, MR/X005895/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets presented in this study can be found in European Nucleotide Archives (ENA). The names and sample accession number(s) can be found in the Supplementary File S1. Raw sequences for the isolates sequenced in this study are available from the ENA website (Project accession PRJEB60381).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Leen N. Vanheer, Email: leen.vanheer@lshtm.ac.uk
Susana Campino, Email: susana.campino@lshtm.ac.uk.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-36002-w.
References
- 1.World Health Organization, World malaria report 2022. Geneva: World Health Organization, 2022. Accessed: Dec. 18, 2022. [Online]. Available: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2022
- 2.Cissoko M, et al. Geo-epidemiology of malaria at the health area level, dire health District, Mali, 2013–2017. Int. J. Environ. Res. Public. Health. 2020;17(11):3982. doi: 10.3390/ijerph17113982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhorda M, Amaratunga C, Dondorp AM. Artemisinin and multidrug-resistant Plasmodium falciparum—A threat for malaria control and elimination. Curr. Opin. Infect. Dis. 2021;34(5):432. doi: 10.1097/QCO.0000000000000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sirawaraporn W, Sathitkul T, Sirawaraporn R, Yuthavong Y, Santi DV. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U.S.A. 1997;94(4):1124–1129. doi: 10.1073/pnas.94.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staedke SG, Sendagire H, Lamola S, Kamya MR, Dorsey G, Rosenthal PJ. Relationship between age, molecular markers, and response to sulphadoxine–pyrimethamine treatment in Kampala, Uganda. Trop. Med. Int. Health. 2004;9(5):624–629. doi: 10.1111/j.1365-3156.2004.01239.x. [DOI] [PubMed] [Google Scholar]
- 6.Kublin JG, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J. Infect. Dis. 2002;185(3):380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 7.Uwimana A, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020;26(10):1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balikagala B, et al. Evidence of artemisinin-resistant malaria in Africa. N. Engl. J. Med. 2021;385(13):1163–1171. doi: 10.1056/NEJMoa2101746. [DOI] [PubMed] [Google Scholar]
- 9.Stokes BH, et al. Plasmodium falciparum K13 mutations in Africa and Asia impact artemisinin resistance and parasite fitness’. Elife. 2021;10:e66277. doi: 10.7554/eLife.66277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frosch AEP, et al. Return of widespread chloroquine-sensitive Plasmodium falciparum to Malawi. J. Infect. Dis. 2014;210(7):1110–1114. doi: 10.1093/infdis/jiu216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asare KK, Africa J, Mbata J, Opoku YK. The emergence of chloroquine-sensitive Plasmodium falciparum is influenced by selected communities in some parts of the Central Region of Ghana. Malar. J. 2021;20(1):447. doi: 10.1186/s12936-021-03985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagnogo O, et al. Towards a re-emergence of chloroquine sensitivity in Côte d’Ivoire? Malar. J. 2018;17(1):413. doi: 10.1186/s12936-018-2551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Njiro BJ, Mutagonda RF, Chamani AT, Mwakyandile T, Sabas D, Bwire GM. Molecular surveillance of chloroquine-resistant Plasmodium falciparum in sub-Saharan African countries after withdrawal of chloroquine for treatment of uncomplicated malaria: A systematic review. J. Infect. Public Health. 2022;15(5):550–557. doi: 10.1016/j.jiph.2022.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 2003;187(12):1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 15.Diakité SAS, et al. A comprehensive analysis of drug resistance molecular markers and Plasmodium falciparum genetic diversity in two malaria endemic sites in Mali. Malar. J. 2019;18(1):361. doi: 10.1186/s12936-019-2986-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulibaly A, et al. Genome-wide SNP analysis of Plasmodium falciparum shows differentiation at drug-resistance-associated loci among malaria transmission settings in southern Mali. Front. Genet. 2022;13:943445. doi: 10.3389/fgene.2022.943445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: Detectability, transmissibility and public health relevance. Nat. Rev. Microbiol. 2014;12(12):833–840. doi: 10.1038/nrmicro3364. [DOI] [PubMed] [Google Scholar]
- 18.Tadesse FG, et al. The relative contribution of symptomatic and asymptomatic plasmodium vivax and Plasmodium falciparum infections to the infectious reservoir in a low-endemic setting in Ethiopia. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2018;66(12):1883–1891. doi: 10.1093/cid/cix1123. [DOI] [PubMed] [Google Scholar]
- 19.Gonçalves BP, et al. Examining the human infectious reservoir for Plasmodium falciparum malaria in areas of differing transmission intensity. Nat. Commun. 2017;8(1):1133. doi: 10.1038/s41467-017-01270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neafsey DE, Taylor AR, MacInnis BL. Advances and opportunities in malaria population genomics. Nat. Rev. Genet. 2021;22(8):502–517. doi: 10.1038/s41576-021-00349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patouillard E, Griffin J, Bhatt S, Ghani A, Cibulskis R. Global investment targets for malaria control and elimination between 2016 and 2030. BMJ Glob. Health. 2017;2(2):e000176. doi: 10.1136/bmjgh-2016-000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malmberg M, et al. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J. Infect. Dis. 2013;207(5):842–847. doi: 10.1093/infdis/jis747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veiga MI, et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016;7(1):11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergmann-Leitner ES, Legler PM, Savranskaya T, Ockenhouse CF, Angov E. Cellular and humoral immune effector mechanisms required for sterile protection against sporozoite challenge induced with the novel malaria vaccine candidate CelTOS. Vaccine. 2011;29(35):5940–5949. doi: 10.1016/j.vaccine.2011.06.053. [DOI] [PubMed] [Google Scholar]
- 25.Bergmann-Leitner ES, et al. Self-adjuvanting bacterial vectors expressing pre-erythrocytic antigens induce sterile protection against malaria. Front. Immunol. 2013;4:176. doi: 10.3389/fimmu.2013.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MalariaGEN, et al. Pf7: An open dataset of Plasmodium falciparum genome variation in 20,000 worldwide samples. Wellcome Open Res. 2023;8:22. doi: 10.12688/wellcomeopenres.18681.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeffer DA, et al. malariaAtlas: An R interface to global malariometric data hosted by the Malaria atlas project. Malar. J. 2018;17(1):352. doi: 10.1186/s12936-018-2500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ocholla H, et al. Whole-genome scans provide evidence of adaptive evolution in Malawian Plasmodium falciparum isolates. J. Infect. Dis. 2014;210(12):1991–2000. doi: 10.1093/infdis/jiu349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frosch AE, Venkatesan M, Laufer MK. Patterns of chloroquine use and resistance in sub-Saharan Africa: A systematic review of household survey and molecular data. Malar. J. 2011;10(1):116. doi: 10.1186/1475-2875-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paloyo A, Reichert A. Biting back at malaria: Assessing health-service providers’ compliance with treatment guidelines. Rev. Dev. Econ. 2017;21(3):591–626. doi: 10.1111/rode.12283. [DOI] [Google Scholar]
- 31.United Nations Office on Drugs and Crime Trafficking in medical products in the Sahel. United Nations. 2023 doi: 10.18356/9789210025409. [DOI] [Google Scholar]
- 32.Otienoburu SD, et al. Selection of Plasmodium falciparum pfcrt and pfmdr1 polymorphisms after treatment with artesunate–amodiaquine fixed dose combination or artemether-lumefantrine in Liberia. Malar. J. 2016;15(1):452. doi: 10.1186/s12936-016-1503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sisowath C, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J. Infect. Dis. 2005;191(6):1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 34.Taylor AR, et al. Artemether-lumefantrine and dihydroartemisinin-piperaquine exert inverse selective pressure on Plasmodium falciparum drug sensitivity-associated haplotypes in Uganda. Open Forum Infect. Dis. 2017 doi: 10.1093/ofid/ofw229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sondo P, et al. Artesunate-amodiaquine and artemether-lumefantrine therapies and selection of Pfcrt and Pfmdr1 alleles in Nanoro, Burkina Faso. PLoS ONE. 2016;11(3):e0151565. doi: 10.1371/journal.pone.0151565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gil JP, Krishna S. pfmdr1 (Plasmodium falciparum multidrug drug resistance gene 1): A pivotal factor in malaria resistance to artemisinin combination therapies. Expert Rev. Anti Infect. Ther. 2017;15(6):527–543. doi: 10.1080/14787210.2017.1313703. [DOI] [PubMed] [Google Scholar]
- 37.Mahamar A, et al. Effect of three years’ seasonal malaria chemoprevention on molecular markers of resistance of Plasmodium falciparum to sulfadoxine-pyrimethamine and amodiaquine in Ouelessebougou, Mali. Malar. J. 2022;21(1):39. doi: 10.1186/s12936-022-04059-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Infectious Diseases Data Observatory (IDDO), ‘SP Molecular Surveyor’, Worldwide Antimalarial Resistance Network, Feb. 02, 2015. https://www.wwarn.org/tracking-resistance/sp-molecular-surveyor (Accessed 10 May, 2023).
- 39.Osborne A, et al. Characterizing the genomic variation and population dynamics of Plasmodium falciparum malaria parasites in and around Lake Victoria, Kenya. Sci. Rep. 2021;11(1):19809. doi: 10.1038/s41598-021-99192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abera D, Kibet CK, Degefa T, Amenga-Etego L, Bargul JL, Golassa L. Genomic analysis reveals independent evolution of Plasmodium falciparum populations in Ethiopia. Malar. J. 2021;20(1):129. doi: 10.1186/s12936-021-03660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J. Infect. Dis. 2001;183(10):1535–1538. doi: 10.1086/320195. [DOI] [PubMed] [Google Scholar]
- 42.Mombo-Ngoma G, et al. High prevalence of dhfr triple mutant and correlation with high rates of sulphadoxine-pyrimethamine treatment failures in vivo in Gabonese children. Malar. J. 2011;10:123. doi: 10.1186/1475-2875-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 1988;85(23):9109–9113. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.World Health Organization, Global Technical Strategy for Malaria 2016–2030. Geneva: World Health Organization, 2015. Accessed: 12 January, 2023. https://apps.who.int/iris/handle/10665/176712
- 45.Stone W, et al. Single low-dose tafenoquine combined with dihydroartemisinin–piperaquine to reduce Plasmodium falciparum transmission in Ouelessebougou, Mali: A phase 2, single-blind, randomised clinical trial’. Lancet Microbe. 2022 doi: 10.1016/S2666-5247(21)00356-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone W, et al. Pyronaridine–artesunate or dihydroartemisinin–piperaquine combined with single low-dose primaquine to prevent Plasmodium falciparum malaria transmission in Ouélessébougou, Mali: A four-arm, single-blind, phase 2/3, randomised trial. Lancet Microbe. 2022;3(1):e41–e51. doi: 10.1016/S2666-5247(21)00192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oyola SO, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar. J. 2016;15(1):597. doi: 10.1186/s12936-016-1641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke EL, Sundararaman SA, Seifert SN, Bushman FD, Hahn BH, Brisson D. swga: A primer design toolkit for selective whole genome amplification. Bioinform. Oxf. Engl. 2017;33(14):2071–2077. doi: 10.1093/bioinformatics/btx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 50.Minh BQ, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danecek P, McCarthy SA. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics. 2017;33(13):2037–2039. doi: 10.1093/bioinformatics/btx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gautier M, Klassmann A, Vitalis R. rehh 2.0: A reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol. Ecol. Resour. 2017;17(1):78–90. doi: 10.1111/1755-0998.12634. [DOI] [PubMed] [Google Scholar]
- 53.Caye K, Deist TM, Martins H, Michel O, François O. TESS3: Fast inference of spatial population structure and genome scans for selection. Mol. Ecol. Resour. 2016;16(2):540–548. doi: 10.1111/1755-0998.12471. [DOI] [PubMed] [Google Scholar]
- 54.Schaffner SF, Taylor AR, Wong W, Wirth DF, Neafsey DE. hmmIBD: Software to infer pairwise identity by descent between haploid genotypes. Malar. J. 2018;17(1):196. doi: 10.1186/s12936-018-2349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in European Nucleotide Archives (ENA). The names and sample accession number(s) can be found in the Supplementary File S1. Raw sequences for the isolates sequenced in this study are available from the ENA website (Project accession PRJEB60381).