Abstract
Recent technological advances have enabled 3D tissue culture models for fast and affordable HTS. We are no longer bound to 2D models for anti-cancer agent discovery, and it is clear that 3D tumor models provide more predictive data for translation of preclinical studies. In a previous study, we validated a microplate 3D spheroid-based technology for its compatibility with HTS automation. Small-scale screens using approved drugs have demonstrated that drug responses tend to differ between 2D and 3D cancer cell proliferation models. Here, we applied this 3D technology to the first ever large-scale screening effort completing HTS on over 150K molecules against primary pancreatic cancer cells. It is the first demonstration that a screening campaign of this magnitude using clinically relevant, ex-vivo 3D pancreatic tumor models established directly from biopsy, can be readily achieved in a fashion like traditional drug screen using 2D cell models. We identified four unique series of compounds with sub micromolar and even low nanomolar potency against a panel of patient derived pancreatic organoids. We also applied the 3D technology to test lead efficacy in autologous cancer associated fibroblasts and found a favorable profile for better efficacy in the cancer over wild type primary cells, an important milestone towards better leads. Importantly, the initial leads have been further validated in across multiple institutes with concordant outcomes. The work presented here represents the genesis of new small molecule leads found using 3D models of primary pancreas tumor cells.
Keywords: Pancreatic Cancer, Organoid, HTS, Magnetic Bioprinting, Phenotypic, Organoid, Pancreatic, Cancer, Phenotypic, HTS
Introduction
Due to the difficulty in early diagnosis and lack of effective treatment, pancreatic cancer remains one of the most common causes of cancer-related death, with an overall 5-year survival rate of approximately 10%.1,2,3 . The future may be even more grim as it is currently the 3rd leading cause of cancer related deaths in the USA and will rise to 2nd within the next 10 years.4,5 This is particularly worrisome since most patients with advanced stage disease have very limited therapeutic options, resulting in a high degree of mortality within 9-12 months.6 The situation is not entirely bleak with recent advances towards immune checkpoint inhibitors and CAR-T therapies which, while still early in development, so far have met with very limited success due to tumor heterogeneity, the immunosuppressive capacity of the tumor microenvironment (TME), bioavailability, cytokine release syndrome and neurological toxicity.7
New small molecule approaches directed at physiologically relevant pancreas cancer models and their TME remain a serious unmet need.8 3 dimensional (3D) cancer models have become an essential tool for the evaluation of new chemotherapeutics to fight pancreas cancer, yet automated drug screening assays using these models remains challenging for drug discovery. Advances by our group have enabled the screening of cancer 3D models in a rapid, highly miniaturized, and cost-effective fashion that permits direct compound response profiling to be generated in a phenotypic manner.9 3D ex vivo tumor models better recapitulate the features of the disease such as cell interactions, hypoxia, and phenotypic heterogeneity of the cancer tissue and drug response.10,11,12.
Patient tumor derived organoids models can be generated from minimal primary tissue and can be used as models for 3D high-throughput screening (HTS). To effectively deploy 3D models for HTS there are two essential requirements: to produce the required amount of spheroids, and to yield acceptable assay performance.13 In this study, we are focusing on patient derived pancreatic cancer cells and their cancer associated fibroblasts (CAF). All primary cells used in this exercise were originally established directly from pancreatic cancer patient tissue in the Tuveson lab as previously described.9,14 We also apply our HTS platform to exploit its cost effective and rapid data production capabilities to conduct as series of assays in both 2D and 3D formats using autologous cell models. In addition, we provide evidence that while organoids cultures traditionally rely on artificial extracellular matrices (ECMs) to facilitate aggregation into 3D models, we were able to conduct HTS screening in a completely scaffold-free system. The results and the outcomes of this 3D screening using pancreatic spheroids produced as series of potent leads that proved effective across institutions. These leads where subsequently validated for their potency in organoid models in the Tuveson lab and now provide a path forward for future chemistry optimization and the potential for novel drugs.
Material and Methods
Cells
Primary human pancreatic ductal cells hM1A, hT1, and cancer-associated fibroblasts hM1A-CAF and hT1-CAF (immortalized by SV40), were generated from tissues of pancreatic cancer patients in the laboratory of Dr. Tuveson, M.D.14 These cells were cultured in flasks and expanded as 2 dimensional monolayers in RPMI 1640 (Part# 10-040-CV, Life Technology, Carlsbad, CA) with 10% serum (Part# 97068-085, VWR, Radnor, PA), and 1× Anti-Anti (Part# 15240-062, Life Technology, Carlsbad, CA) at 37°C, 5% CO2 and 95% relative humidity. They were harvested and utilized in this format for the purpose of both 2D and 3D testing, using the same media as described above. The Hepatocells used in the assay were from Corning (Part# 354881, Corning Life Sciences, 1 Becton Cir, Durham, NC 27712) along with the special media from Corning (Part# 354882). These cells were developed to retain the morphology and physiological properties of primary human hepatocytes and overcome the inherent shortcomings such as large inter-individual variability and finite supply. The Hepatocells are provided as single-use cryopreserved immortalized cells that were derived from primary human hepatocytes with a high cell purity (>99% hepatocyte-like cells).
SDDL Compound Library
A subset of the Scripps Drug Discovery Library (SDDL) of about 150,000 compounds were screened at the Scripps Research Institute Molecular Screening Center (SRIMSC) and reformatted into 1536-well source plates for automated robotic screening.15 The portion of the SDDL that was chosen out of >666K compounds was based on general diversity and drug like properties which included the Maybridge 14.5K hit finder library.
2D Cell Viability Assay
A 1536-well 2D cell viability assay was optimized and implemented, which determines the number of viable cells based on the amount of ATP present using commercially available luminescence detection reagent CellTiter-Glo (Part# G7573, Promega, Madison, WI) as previously described.16 Prior to plating, cells were grown to 80% confluence in RPMI 1640 complete growth media. After washing once with PBS, cells were detached by TrypLE (Part# 12604021, Life Technologies, Carlsbad, CA) and centrifuged at 300 × g for 5 min. Cells were suspended and filtered through cell strainer. 200 cells in 5 μL culture media were seeded using a Flying Reagent Dispenser (Aurora) in 1536-well plates (Part# 789173-F, Greiner, Monroe, NC). After incubation of the assay plates overnight (~14 hours), cells were treated with compounds and vehicle (10 nL, 0.2% DMSO) via the GNF/Kalypsis robotic pintool (La Jolla, CA). Cell viability was assessed after 72-hour incubation using Cell-Titer Glo reagent according to manufacturer’s instruction. The ViewLux microplate reader (Perkin Elmer, Waltham, MA) was used to quantitate luminescence signal. IC50 values of 3 pharmacological control compounds (Doxorubicin, Gemcitabine, and SN-38; purchased from Sigma) were determined by fitting the concentration response curve data (CRC) with a four-parameter variable slope method in GraphPad Prism (GraphPad software, San Diego, CA).
3D Culture and 3D Viability Assay
The 3D cell viability assay was initially developed at Scripps in a 384-well format and then further miniaturized into 1536-well format.9 The 3D viability assay uses a detection reagent more adapted to spheroids that features a tailored lysis buffer (Cell Titer-Glo 3D, Part# G9683, Promega, Madison, WI). Both assays were optimized by testing different variables including cell number, aggregation time, incubation time, time of drug addition, and Nanoshuttle amount. Nanoshuttle, a reagent obtained from Nano3D Biosciences, contains gold and iron oxide laden nanoparticles attached to poly-L-lysine which non-specifically attached to the cell membrane of all eukaryotic cells. 17 A detailed stepwise protocol of the final conditions is presented in Table 1 which can be found in the publication by Hou et al using automation as described by Baillargeon et. al..9,17 Cells were grown to 80% confluence in RPMI 1640 complete growth media and labelled with NanoShuttle-PL (Greiner Bio-One Part #657846) overnight (~16 hrs.) in the T175 flasks. The next day, labelled cells were harvested and filtered through 70 μm cell strainer. For all types of cells, 1250 cells in 5 μL culture media were seeded in 1536-well Greiner Bio-One flat bottom cell-repellent plates (Part #789979, Greiner Bio-One, NC). For co-culture experiments 1250 cells per well were added for both the cancer and CAF cells, simultaneously. After putting the assay plate on top of the magnetic drive for 4 hours, followed by incubation for 24 hours to allow cells to form 3D structures, cells were treated with compounds or vehicle (or 10 nL, 0.2% DMSO).17 Cell viability was assessed after 72-hour incubation using Cell-Titer Glo 3D reagent according to manufacturer’s instruction. Concentration response curves (CRC) and IC50 values of 3 pharmacological control compounds were used as the guide for assay optimization and drug screening. As a point of comparison, we also tested these cells using Corning spheroid plate technology (Part# 3830, Corning Inc., NY).13 The Corning spheroid-based assay follows the same protocol except that (1) cells are not labelled with Nanoshuttle and also do not need a magnetic drive, and (2) a brief centrifugation was performed immediately after seeding cells into the Corning Spheroid plates to facilitate spheroid formation. The formation of 3D structure was always confirmed by Hoechst staining followed by confocal imaging using an IN Cell Analyzer 6000 high content reader which are then used for Z-stack analysis as previously described.9 In the case of Figure 6C, we incorporated the use of Octadecyl Rhodamine B Chloride (R18) to stain the hT1-CAF cells and CellTracker Green to stain the hT1 cells and monitored their 3D structures at 96hrs post seeding at equal numbers of cells. Images were acquired at 10X using the ThermoFisher CellInsight.
Hepatocells cytotoxicity counterscreen assay
A 1536-well cell viability assay was optimized and implemented, which determines the number of viable cells based on the amount of ATP present using commercially available luminescence detection reagent CellTiter-Glo (Part# G7573, Promega, Madison, WI). Prior to plating, hepatocells were thawed in the Corning Hepatocells growth media and centrifuged at 300 × g for 5 min. Cells were suspended and filtered through cell strainer. For the 2D assay, 400 cells in 5 μL culture media were seeded using a Flying Reagent Dispenser (Aurora) in 1536-well plates (Part# 789173-F, Greiner, Monroe, NC) and for the 3D assay, 1000 cells were seeded in the same volume per well. After 48hrs of incubation of the assay plates, cells were treated with compounds and vehicle (10 nL, 0.2% DMSO) via a pintool and the GNF/Kalypsis platform (La Jolla, CA). Cell viability was assessed after 48-hour incubation using Cell-Titer Glo reagent according to manufacturer’s instruction. The ViewLux microplate reader (Perkin Elmer, Waltham, MA) was used to quantitate luminescence signal. IC50 values of 3 pharmacological control compounds (Doxorubicin, Gemcitabine, SN-38, all purchased from Sigma) were determined by fitting the CRC data with a four-parameter variable slope method in GraphPad Prism.
Luciferase Counterscreen assay
A simple biochemical assay was optimized and implemented in 1536-well which determines the artifacts affecting the activity of luciferase and ATP turnover and was used to identify false positives found in the primary assay. Briefly, 5uL of 1XDPBS containing 2μM of ATP was dispensed on each well. The hits were cherry-picked and these compounds and vehicle (10 nL, 0.02% DMSO) were added to the appropriate wells. The amount of ATP present was measured by using CellTiter-Glo diluted 1:4 in 1XPBS. The ViewLux microplate reader (Perkin Elmer, Waltham, MA) was used to quantitate luminescence signal. IC50 values of two pharmacological control compounds, Resveratrol, and an internal compound SR-389176 were determined by fitting the CRC data as described above.
HTS Campaign and Data Processing
A subset of ~150,000 compounds from the Scripps Drug Discovery library (SDDL) was screened in 1536-well plate format in 3D, at 2 μM nominal concentration against the hT1 pancreatic cancer-spheroid model. We ran at a pace of 25-40 plates per day. All data files obtained were uploaded into Scripps’ institutional database for individual plate quality control and hit selection. Assay plates were determined acceptable only if their Z’ was > 0.5.18 Compound activity was normalized on a per-plate basis using the following equation:19
Test Well refers to those wells with cells treated with test compounds. High Control is defined as wells containing medium only (100% inhibition), and Low Control wells contain cells treated with DMSO only (0% inhibition).
High and Low controls were applied for assay quality evaluation in terms of Z’.18 Day to day assay response and stability was assessed using 3 pharmacological control compounds that we required to be within 3-fold of the expected IC50, on an experiment to experiment basis and across all experiments, for each cell model. An interval-based hit cut-off was used to define active compounds for each assay. This cutoff is calculated as the average percent inhibition plus three times their standard deviation, of all the tested compounds except those showing % inhibition higher than the average+3SD of the high controls or % inhibition lower than the average−3SD of the low controls.20,21 A four-way Venn diagram was used to analyze the hit compounds in hM1A, hM1A-CAF, hT1 and hT1-CAF assays. The tool is freely available at http://www.pangloss.com/seidel/Protocols/venn4.cgi. Active hits against each of the assays were chosen to determine their IC50.
Concentration Response Assays
The selected drugs were prepared as 10-point, 3-fold serial dilutions and tested against 4 pancreatic cancer-derived cells (hM1A, hT1, and hT1-CAF) in 2D or 3D format in triplicate starting from 5 μM nominal concentration. For each test compound, % inhibition was plotted against compound concentration. A four-parameter equation describing a sigmoidal dose-response curve was then fitted with adjustable baseline using Assay Explorer software (Symyx Technologies, Santa Clara, CA). The reported CC50/IC50 values were generated from fitted curves by solving for the X-intercept value at the 50% inhibition level of the Y-intercept value. In cases where the highest concentration tested (i.e., 5 μM) did not result in greater than 50% cytotoxicity, the CC50/IC50 was deemed as greater than 5 μM.
Tanimoto Analysis
Since only 150K compounds were tested in the initial HTS and to encompass the entire SDDL of >666K, in silico structure relationships were determined using Tanimoto similarity score search analysis applying an 80% threshold. This was performed against the entire SDDL using SR-963 as the point of comparison. This analysis found ~3,600 analogs that were cherry picked and tested in the 3D hT1 and hT1-CAF assays in triplicate at 2 μM nominal concentration. The active molecules were then prepared as concentration response curves (see Dose Response Assays method section).
Evaluation of the Leads in the 3D Matrigel Assay
To confirm the activity of the leads Scripps delivered a bolus of each to CSHL for testing in the Tuveson lab using their standardized 3D models of pancreas cancer. Notably these models have never been adapted to 2D models and use Matrigel as a key component of the assay. Thus, while not an exact head-to-head comparison of the technologies, it afforded us with an orthogonal path to confirm efficacy and that the 3D nanoshuttle methods used at Scripps emulated the Matrigel formats. The selected drugs were prepared as 8-point, 3-fold serial dilutions and tested against a panel of pancreatic cancer-derived cells (hM1A, hM1A-CAF, hT1, hT1-CAF, hT102, hF2, hM19B, hM1E, hF39) in 3D format in triplicate starting from 2.99 μM nominal concentration. For each test compound, % inhibition was plotted against compound concentration. IC50 values of the analog compounds (SR-963, SR-115, SR-130 SR-907 and SR-114) were determined by fitting the concentration response curve data (CRC) with a four-parameter variable slope method in GraphPad Prism (GraphPad software, San Diego, CA).
Results
Primary screening of pancreatic spheroid models using m3D bioprinting technology
In this study, we used a 3D culture bioprinting technology for testing primary patient derived pancreas cancer cells in a 1536 well microplate formatted in 3D for large scale screening. Using this 3D nanoshuttle technology, 2D cells originally conditioned from established pancreatic 3D organoids were seeded in cell repellent plates that allows the formation of spheroids in the absence of artificial extracellular matrices (ECM). 22,23 We selected the hT1 line that was derived from a surgically resected primary tumors and used these cells to complete the largest scale high throughput screening campaign seen to date to test >150,000 chemically diverse drug-like small molecules. To accomplish this, we employed a simple phenotypic assay to measure the cell viability in response to cytotoxic drug (Figure 1).13
Figure 1: m3D Spheroid Technology.
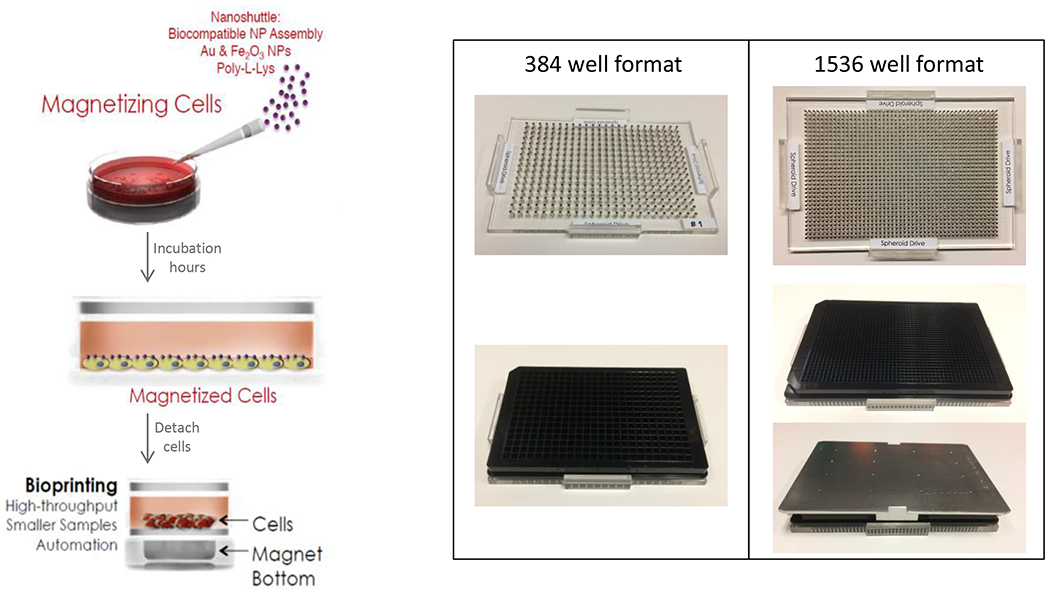
Greiner cell repellent plates readily form spheroids using cells combined with Nanoshuttle. This can be done on the bottom side of the plates using a magnetic driver. Note that the shape of the bottom of the wells is compatible with automated confocal microscopy.
Once the assay was developed in 384 well format, it was then miniaturized to 1,536-well format to perform screening against a large subset of the Scripps Drug Discovery Library (SDDL).1 The primary assay was performed testing each molecule in singlicate at 2 μM (Figure 2). The assay performance was robust across the HTS, yielding an average Z’ of 0.72 ± 0.06 and a signal to background of 188.13 ± 23.07.
Figure 2. Scatter plot analysis of the primary screen.
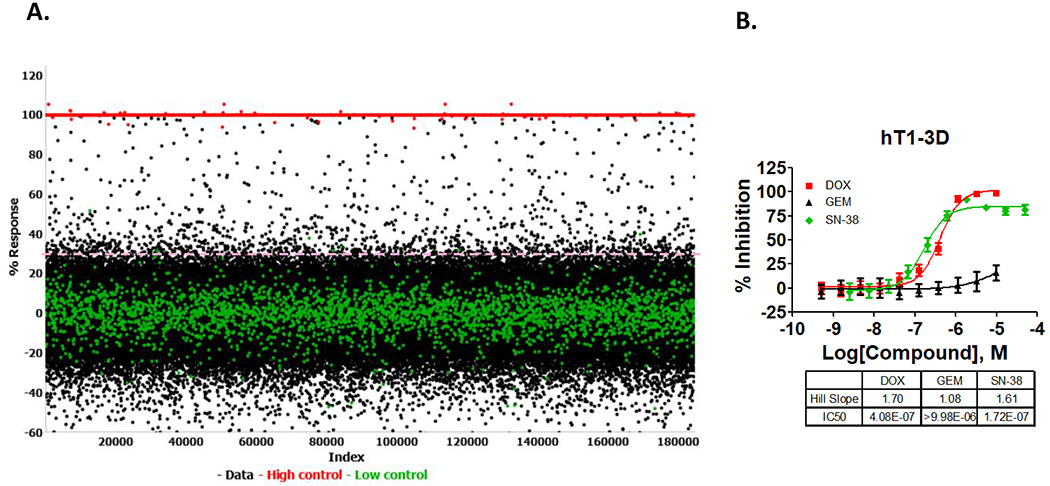
(A) All data from all assay plates, including high and low controls, are displayed. The S:B ratio = 188.13 ± 23.07 and Z’ = 0.72 ± 0.06) between high and low controls wells, as well as broad distribution of hits indicate that the assay is robust. (∆) Represent low control wells containing cells in the presence of DMSO only, (●) represent data wells containing compounds and (■) represent high control wells containing media + DMSO. (B). Concentration response curves for three of the control compounds (Doxorubicin, gemcitabine, and SN-38) versus hT1 in 3D format in 1536 wells. Each curve represents the mean and the standard deviation of 16 replicates. Error bars are included and shown in SD.
Using a primary cutoff calculated using the interval method, 735 compounds were found as primary actives and exhibited a percent inhibition greater than of 34.98%. In addition, CRCs for the control compounds were analyzed (Doxorubicin, gemcitabine, and SN-38) using the same 3D format to verify the sensitivity of the assay on each day for each experiment. Of the 735 primary actives, there were 711 unique hits, from which 703 were available for dose response assays, hence the discrepancy in the number of compounds that progressed to CRC.
CRC Assays
To further evaluate the primary screening hits, 703 compounds were tested in triplicate to obtain IC50s in separate assays testing a panel of pancreas cancer spheroids in both 2D and 3D vs. 4 different cell types (hT1, hT1-CAF, hM1A). The assay statistics Z’ average data for the 3D data are 0.73 ± 0.05, 0.46 ± 0.10, 0.77 ± 0.09 that were obtained for hT1, hT1-CAF and hM1A respectively. The Z’ average for the 2D data is 0.80 ± 0.03, 0.51 ± 0.12, 0.70 ± 0.04 for the same panel of pancreatic cell lines. The counterscreen using the hepatocells was also accessed in 3D and 2D cultures yielding favorable assay statistics of 0.79 ± 0.07 and 0.79 ± 0.03, respectively. In addition, a ninth assay called the luciferase counterscreen was performed at this stage to identify possible luciferase/ATP artifacts. It too was robust with Z’s= 0.97 ± 0.01.
Interestingly, the outcomes of the titration assays displayed reasonable selectivity in terms of compound activity toward the cancer cells over their CAF counterpart and favorable efficacy for some of the compounds to be used as potential leads for follow up assays. From the concentration curve response analysis, initially 3 classes of molecule emerged as the most interesting compounds (Figure 3). This classification was based on the activity of the compounds showing selectivity for hT1, hM1A, or both cancer lines. The pyrazolopyrimidine derivatives show some selectivity for hT1 while the phenylquinolinine derivatives show some selectivity for hM1A. The imidazolidinone derivatives are compounds that are both active and selective for both hT1 and hM1A assays.
Figure 3. SAR Classification of Potential Hits.
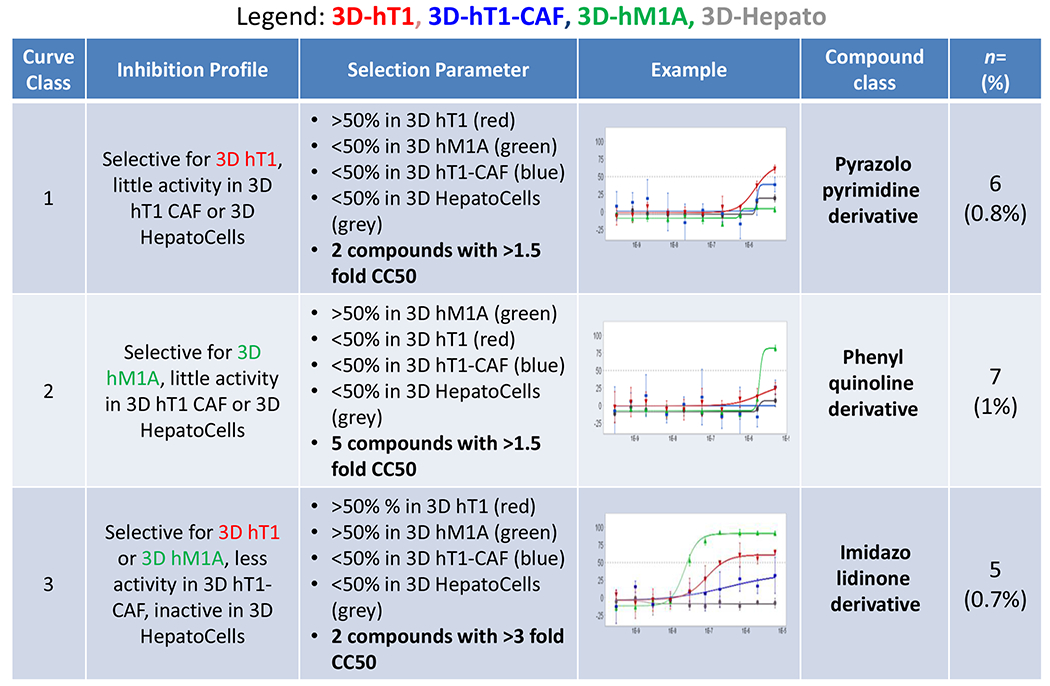
Data analysis was performed to identify compounds that are cancer organoid specific and less active in the CAFs or Hepatocells. Using this method, three compound class were assigned of potential interest for follow up assays.
Another parallel screening effort was done using the Maybridge HIT Finder Library provided by Thermo Fisher against the 3D pancreatic primary and metastatic cell lines. This effort identified yet another attractive scaffold that showed to be highly selective towards the hM1A. This compound was assigned to a fourth classification of pyridinethiol derivatives. Based the compilation of the CRC efforts against all compounds tested, and in collaboration with our chemistry team, we identified a list of 18 compounds that were of further interest that either show selectivity for hT1 primary pancreatic cell line or hM1A. From this set of molecules 4 classes emerged (Figure 4A).
Figure 4. Follow-up assays of the most interesting compounds.
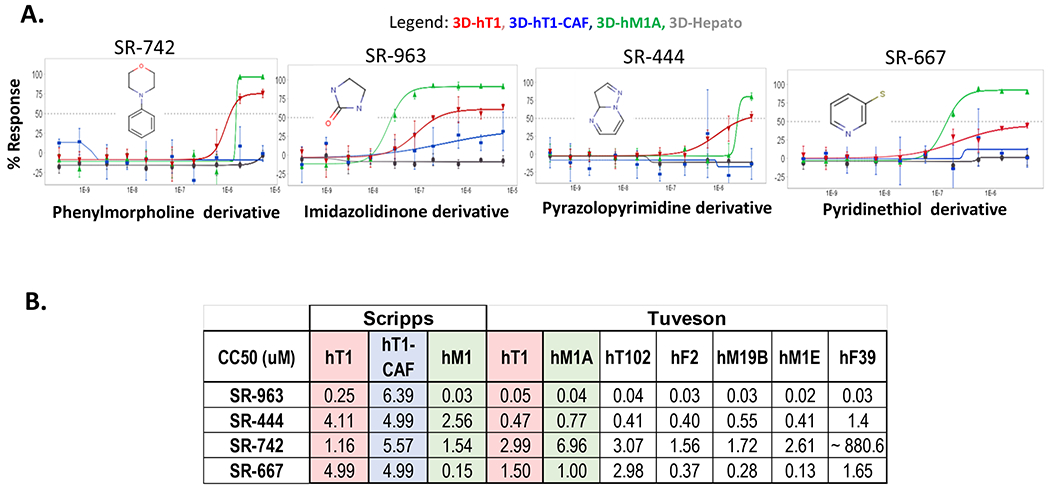
(A). Four chemotypes were determined for the most promising compounds found by 3D HTS with Scripps (>150K) and Maybridge (14.5K) libraries. Data represent 10-point 3-fold serial dilutions starting at 5μM nominal concentration. (n=3 for each concentration tested). Error bars are included and shown in SD. (B). Follow up assays performed for these compounds showed very similar CC50s between the primary and the follow up assays for some of the most promising and selective compounds identified in the HTS campaigns. All data is reported in micromolar concentration.
To improve the efficacy and selectivity of the compounds for these pancreas cancer organoid models, we performed Tanimoto score data analysis across the entire Scripps Drug Discovery Library of >666K compounds to identify compounds with >80% similarity to the most attractive lead (SR-963) for which 3,654 analogs of this compound were identified. These compounds were cherry-picked and tested in triplicate in 3D hT1s assays (Figure 5). The Z’ average obtained for the 3D hT1 is 0.68 ± 0.06, indicating the robustness and consistency of our 3D organoid models. From this, 11 compounds were considered active applying the same primary cutoff of 34.98%. From those, 9 compounds were chosen to be tested in 1536-well plate format as concentration curves responses against the 3D hT1 and hT1-CAF cells as part of our med chem follow up assays. Results from the dose response assays showed four compounds with an CC50/IC50 with at least a 5-fold of difference between hT1 and the CAF CC50/IC50 (Figure 5).
Figure 5. MedChem EC50 data of the analog compounds.
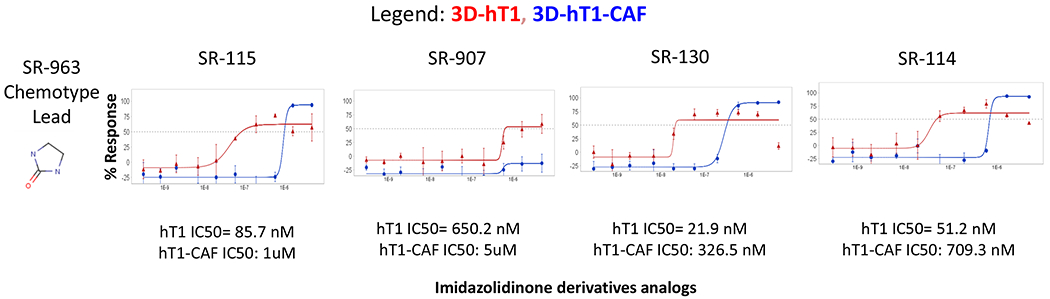
(A). Dose response assays of the analogs of SR-963 chemotype main lead were tested vs. 3D hT1 and hT1-CAF pancreatic cell lines. Data represent 10-point 3-fold serial dilutions starting at 5μM nominal concentration. (n=3 for each concentration tested). Error bars are included and shown in SD.
Low throughput selectivity assays
The four leads (SR-963, SR-742, SR-444 and SR-677) were further tested in the low throughput assays at CSHL in the Tuveson laboratory using their organoid models. The compounds were tested using Matrigel against a panel of 13 pancreatic primary cancer associated organoids. The results presented here (Figure 4B) are for 7 of the cell lines tested in the follow up assays. There is good consistency between the response of these compounds between the results at Scripps using our 3D scaffold free system in 1536-well plate format and the 384-well plate format using Matrigel done at CSHL. SR-963, SR-742 and SR-667 are concordant in CC50 results across the labs and SR-963 was confirmed as a very potent and selective lead (Figure 6A). CSHL also tested the additional four analogs (SR-115, SR-907, SR-130 and SR-114) found active in the dose response med-chem effort at Scripps (Figure 6B). These compounds also provided good reproducibility and potency vs. hT1 organoids at CSHL. The same compounds showed some selectivity differences in the response to the CAFs more so at Scripps than CSHL, and in some cases are even more potent than SR-963, which seems the case for SR-130, that is yielding a CC50 delta between hT1 and CAF of ~ 8-fold at both Scripps and CSHL.
Figure 6. CC50 data of the analogs of SR-963 in the low throughput assays.
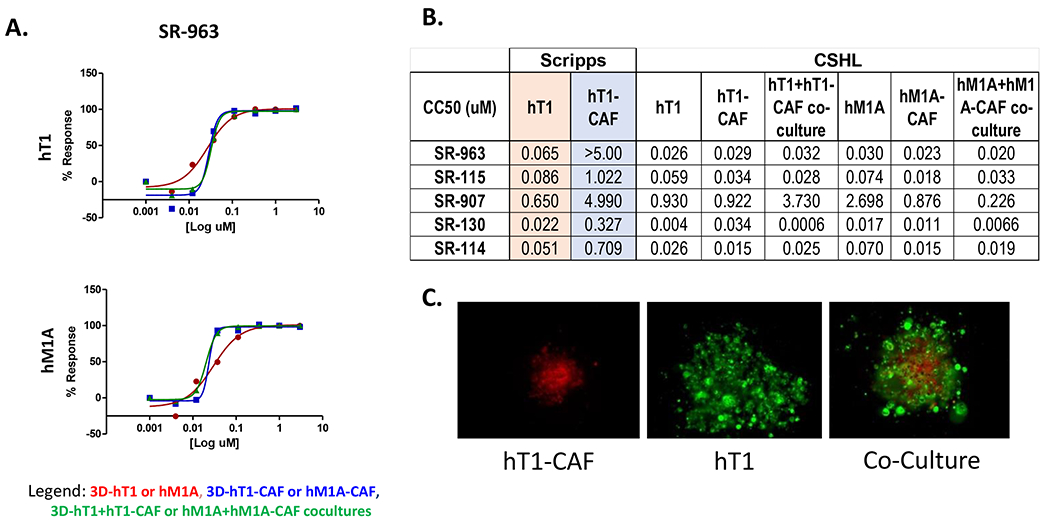
(A). SR-963 compound concentration response curve in the Matrigel 3D scaffold assays using hT1, hM1A, CAF and co-cultures pancreatic organoids. Data represent 8-point 3-fold serial dilutions starting at 2.99 μM nominal concentration. (n=3 for each concentration tested). Error bars are included and shown in SD. (B). Table that summarizes the dose response data results of each of the analogs tested for Scripps and CSHL. (C) Image analysis to demonstrate how spheroids and co-culture spheroids look in the assay.
Finally, these compounds were tested in the co-cultures for hT1 and hM1 cancer cells with their CAFs. The results are generally overlapping with their individual cell response, but SR-130 showed a higher efficacy in the co-cultures compared to the hT1-CAF (Figure 6B).24
Discussion
3D cell models have proven to be a highly effective translational model representing the complexity of solid cancer. The urgency to have a system in place that provides a more accurate representation of what mimics the physiological context of human cancer has been a long-term priority. Creating a three-dimensional (3D) culture system utilizing 3D models of patient-derived spheroids combined with a rapid high throughput screening format that affords large scale screening is an important and essential tool in the drug discovery process. The advancement to rapidly and efficiently screen 3D cell culture models that reflect in vivo tumors will allow us to move faster towards more effective chemotherapeutics.
Primary pancreatic ductal cells hT1 and hM1A and cancer-associated fibroblasts hT1-CAF and hM1A-CAF were generated from tissues of pancreatic cancer patients in the laboratory of Dr. Tuveson at Cold Spring Harbor Laboratory. To generate the number of cells needed for the screening, we used 2D monolayer cells that were conditioned from the established pancreatic 3D organoids. There is evidence that shows that patient derived organoids (PDO) therapeutic profiles correlated with the patient outcomes24,25. These patient derived organoids were also further evaluated to measure the therapeutic sensitivity to different chemotherapeutic agents to reveal the heterogeneity of each of these samples to chemotherapy response. Whether a PDO is sensitive or resistant is dependent of the stage of the disease when isolated and its molecular profiles.24 Largely because the typical diagnosis lags far behind, there are very few curative options for the majority of patients afflicted with pancreas cancer. Hence, we incorporated the validated PDO models used herein to look for new drug leads using our fully automated screening platform outfitted for advanced 1536 well 3D HTS. This system produces physiologically relevant 3D spheroid models that are suitable for high throughput cancer drug discovery.
Previously, we screened 3D pancreatic spheroids9 against small libraries (National Cancer Institute and the Approved Drug Screen). Here, we present the first automated high throughput screening using 3D pancreatic spheroids cell models against a much larger subset of 150,000 compounds of the Scripps Drug Discovery Library. We were able to successfully complete the screening using 3D spheroids and identified promising leads that could be potentially further developed for clinical studies. The results prove our ability to find drug leads indicating the quality and sensitivity of the assay system combined with automation, a powerful combination. Four chemotypes (SR-963, SR-742, SR-667 and SR-444) were identified as leads that were confirmed across institutions in orthogonal 3D models of pancreas cancer, again confirming the robustness of the approach. Importantly, in some cases the leads showed notable efficacy in the cancer cells with reasonable selectivity over there inhibition of CAFs and hepatocells.
Using in-silico analysis afforded us an eye into the full diversity of the Scripps 666K library which found another potent class of molecules alluding to the flexibility and agility screening in 3D. Efforts to expand on the leads and identify more potent molecules across the 3D models was successful and improved our IC50s in some cases to low nanomolar and even picomolar efficacy. This is the case when looking at the co-culture result for SR-130, a truly profound result. Future efforts include target identification using a chemo proteomics approach, further refinement of the lead chemical properties via medchem to improve upon efficacy and potential selectivity, in-vitro and in-vivo DMPK, and testing in pre-clinical animal models of pancreas cancer.
In summary, the work presented here to elucidate new leads targeting cancer of the pancreas represents a successful 3D HTS, on a scale never seen before. Critically, as demonstrated in follow-up procedures, low throughout assays were performed using 3D pancreas derived organoids in Matrigel assays in an alternative lab thus showing both lab to lab reproducibility and a good reproducibility between their independent methods. Exploiting this capability in the future will allow us to focus and move faster towards identification of promising compounds toward a wider array of solid tumor targets.
Acknowledgements
This work was supported by a grant from the National Cancer of Institute of the National Institutes of Health R33CA206949. We thank Lina DeLuca (Scripps, Florida) for her assistance with compound management. Dennis Plenker was supported by the German Research Foundation (DFG, PL 894/1-1).
Footnotes
Declaration of Conflict of Interests
The authors declared no potential conflict of interest with respect to the research, authorship and or publication of this article.
References
- 1.Siegel RL, Miller KD & Jemal A Cancer statistics, 2015. CA: A Cancer Journal for Clinicians 65, 5–29, doi: 10.3322/caac.21254 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Rasheed ZA, M. W, Maitra A in Transworld Research Network (ed Munshi HG, Grippo PJ) Ch. 1, ( Trivandrum (India), 2012). [PubMed] [Google Scholar]
- 3.Jaffee EM, Hruban RH, Canto M & Kern SE Focus on pancreas cancer. Cancer Cell 2, 25–28, doi: 10.1016/S1535-6108(02)00093-4 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD & Jemal A Cancer Statistics, 2017. CA Cancer J Clin 67, 7–30, doi: 10.3322/caac.21387 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rahib L et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74, 2913–2921, doi: 10.1158/0008-5472.CAN-14-0155 (2014). [DOI] [PubMed] [Google Scholar]
- 6.McGuigan A et al. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol 24, 4846–4861, doi: 10.3748/wjg.v24.i43.4846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akce M, Zaidi MY, Waller EK, El-Rayes BF & Lesinski GB The Potential of CAR T Cell Therapy in Pancreatic Cancer. Front Immunol 9, 2166, doi: 10.3389/fimmu.2018.02166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang ZQ, Saluja AK, Dudeja V, Vickers SM & Buchsbaum DJ Molecular targeted approaches for treatment of pancreatic cancer. Curr Pharm Des 17, 2221–2238, doi: 10.2174/138161211796957427 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou S et al. Advanced Development of Primary Pancreatic Organoid Tumor Models for High-Throughput Phenotypic Drug Screening. SLAS Discov 23, 574–584, doi: 10.1177/2472555218766842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powell K Adding depth to cell culture. Science 356, 96–98, doi: 10.1126/science.356.6333.96 (2017). [DOI] [Google Scholar]
- 11.Clevers H Modeling Development and Disease with Organoids. Cell 165, 1586–1597, doi: 10.1016/j.cell.2016.05.082 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Longati P et al. 3D pancreatic carcinoma spheroids induce a matrix-rich, chemoresistant phenotype offering a better model for drug testing. BMC Cancer 13, 95, doi: 10.1186/1471-2407-13-95 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madoux F et al. A 1536-Well 3D Viability Assay to Assess the Cytotoxic Effect of Drugs on Spheroids. SLAS Discov 22, 516–524, doi: 10.1177/2472555216686308 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Boj Sylvia F. et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell 160, 324–338, doi: 10.1016/j.cell.2014.12.021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou S et al. Drug Library Screening for the Identification of Ionophores That Correct the Mistrafficking Disorder Associated with Oxalosis Kidney Disease. SLAS DISCOVERY: Advancing Life Sciences R&D 22, 887–896, doi: 10.1177/2472555217689992 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Pedro-Rosa L et al. Identification of potent inhibitors of the Trypanosoma brucei methionyl-tRNA synthetase via high-throughput orthogonal screening. J Biomol Screen 20, 122–130, doi: 10.1177/1087057114548832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baillargeon P et al. Automating a Magnetic 3D Spheroid Model Technology for High-Throughput Screening. SLAS Technol 24, 420–428, doi: 10.1177/2472630319854337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J-H, Chung TDY & Oldenburg KR A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. Journal of Biomolecular Screening 4, 67–73, doi: 10.1177/108705719900400206 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Smith E et al. Application of Parallel Multiparametric Cell-Based FLIPR Detection Assays for the Identification of Modulators of the Muscarinic Acetylcholine Receptor 4 (M4). Journal of Biomolecular Screening 20, 858–868, doi: 10.1177/1087057115581770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madoux F et al. Discovery of an enzyme and substrate selective inhibitor of ADAM10 using an exosite-binding glycosylated substrate. Scientific Reports 6, 11, doi: 10.1038/s41598-016-0013-4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collia D et al. A Rapid Phenotypic Whole-Cell Screening Approach for the Identification of Small-Molecule Inhibitors That Counter β-Lactamase Resistance in Pseudomonas aeruginosa. SLAS DISCOVERY: Advancing Life Sciences R&D 0, 2472555217728489, doi: 10.1177/2472555217728489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boj SF et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338, doi: 10.1016/j.cell.2014.12.021 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costa EC et al. 3D tumor spheroids: an overview on the tools and techniques used for their analysis. Biotechnology Advances 34, 1427–1441, doi: 10.1016/j.biotechadv.2016.11.002 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Tiriac H et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov 8, 1112–1129, doi: 10.1158/2159-8290.Cd-18-0349 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff RA et al. Dynamic changes during the treatment of pancreatic cancer. Oncotarget 9, 14764–14790, doi: 10.18632/oncotarget.24483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


