Abstract
Objectives
Previous studies identified a number of diseases were associated with 2019 coronavirus disease (COVID-19). However, the associations between these diseases related viral infections and COVID-19 remains unknown now.
Methods
In this study, we utilized single nucleotide polymorphisms (SNPs) related to COVID-19 from genome-wide association study (GWAS) and individual-level genotype data from the UK biobank to calculate polygenic risk scores (PRS) of 487,409 subjects for eight COVID-19 clinical phenotypes. Then, multiple logistic regression models were established to assess the correlation between serological measurements (positive/negative) of 25 viruses and the PRS of eight COVID-19 clinical phenotypes. And we performed stratified analyses by age and gender.
Results
In whole population, we identified 12 viruses associated with the PRS of COVID-19 clinical phenotypes, such as VZV seropositivity for Varicella Zoster Virus (Unscreened/Exposed_Negative: β = 0.1361, P = 0.0142; Hospitalized/Unscreened: β = 0.1167, P = 0.0385) and MCV seropositivity for Merkel Cell Polyomavirus (Unscreened/Exposed_Negative: β = −0.0614, P = 0.0478). After age stratification, we identified seven viruses associated with the PRS of eight COVID-19 clinical phenotypes. After gender stratification, we identified five viruses associated with the PRS of eight COVID-19 clinical phenotypes in the women group.
Conclusion
Our study findings suggest that the genetic susceptibility to different COVID-19 clinical phenotypes is associated with the infection status of various common viruses.
Keywords: common viral infections, coronavirus disease 2019, positive serological measurements, genetic susceptibility, genome-wide association study
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic has resulted in over 603 million infections and more than 6.4 million deaths worldwide as of July 31, 2022, casuing a significant disease burden for countries around the world[1]. COVID-19 is caused by the highly contagious severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which presents with a wide range of clinical symptoms, from asymptomatic infection or mild illness to serious illness requiring hospitalization and mechanical ventilation[2]. Previous study has found that clinical variation in COVID-19 severity and symptom presentation may be due to differences in host genetic factors associated with immune response[3]. Therefore, changes in immune responses may be linked to different clinical manifestations of COVID-19. As research on COVID-19 infection has progressed, several single nucleotide polymorphisms (SNPs) and genes associated with different aspects of susceptibility to infection or disease severity have been identified[4], suggesting that genetic factors in the host could influence its susceptibility and severity to the virus[4, 5] (see Figs. 1 and 2 ).
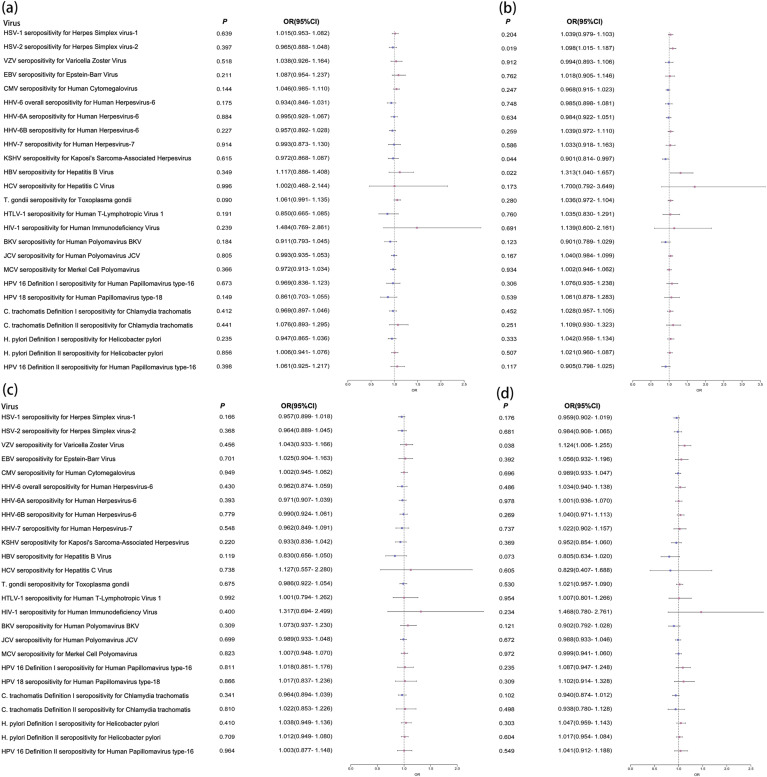
Figure 1.
Forest plots of the results of logistic regression analysis in the whole population. (a) Continuous_Severity_Score; (b) Exposed_Positive/Exposed_ Negative; (c) Hospitalized/Not_Hospitalized; (d) Hospitalized/Unscreened.
Note: Odds ratio (OR) forest plot of the PRS of COVID-19 clinical phenotypes. Use forest plot to visualize logistic regression analysis. The outcome variables were serological measurements of 25 viruses. The instrumental variables were the PRS of COVID-19 clinical phenotypes.
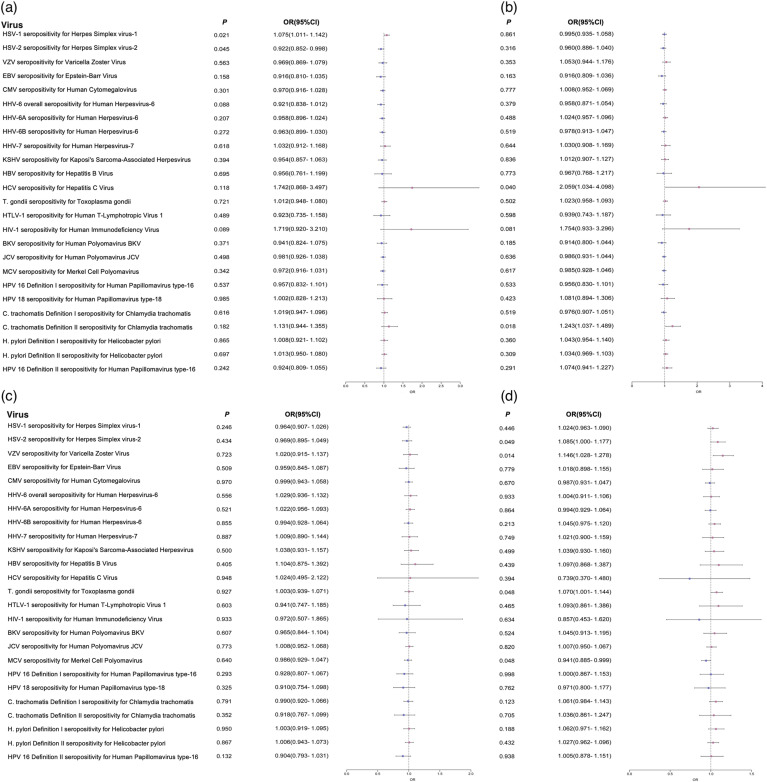
Figure 2.
Forest plots of the results of logistic regression analysis in the whole population. (a) Positive/Negative; (b) Positive/Unscreened; (c) Symptomatic/Paucisymptomatic; (d) Unscreened/Exposed_Negative.
Note: Odds ratio (OR) forest plot of the PRS of COVID-19 clinical phenotypes. Use forest plot to visualize logistic regression analysis. The outcome variables were serological measurements of 25 viruses. The instrumental variables were the PRS of COVID-19 clinical phenotypes.
As COVID-19 prevention measures become normalized, identifying co-infection with one or more respiratory viruses can help us understand the infection status of SARS-CoV-2. During the COVID-19 pandemic, there have been increasing reports of co-infection with other pathogens in COVID-19 patients[6, 7]. A retrospective study found that 242 (94.2%) patients had co-infected with one or more pathogens, the most frequent of which were the Epstein virus Barr virus(EBV), the rhinovirus, and the adenovirus[8]. Herpes simplex virus 1 (HSV-1) and varicella zoster virus (VZV) are DNA viruses of the neurotropic alpha human herpesvirus subfamily (HHV)[9]. The virus remains dormant in the body after recovery from the initial infection and reactivates when the immune system is compromised, causing significant damage to the host organism[10]. Although few case reports of VZV and HSV in patients with COVID-19 have been published, studies have suggested that VZV may be an indicator of potential COVID-19 infection[11]. Previous studies have identified associations between host polymorphisms in genes related to cell entry, cytokine production, and immune response with multiple viruses, and antibody responses have been found to be highly heritable (32%−48%) [12]. The HLA-B*46:01 allele in East Asian patients was associated with infection severity during the 2003 severe acute respiratory syndrome (SARS) outbreak caused by the SARS-CoV-2-related β coronavirus[13].
Researchers have discovered an association between the prevalence of COVID-19 and other viral infections[14]. In addition to age, obesity, hypertension and other common risk factors associated with increased COVID-19 severity[15], a study found that cytomegalovirus (CMV) seropositivity was associated with more than twice the risk of hospitalization due to SARS-CoV-2 infection[14]. Potential CMV infection influences future infection with other viruses and shapes the distribution of adaptive immune cell populations[16, 17]. In SARS-CoV-2 infection, CMV seropositivity results in a severe immunological signature, which is the activation of TEMRA cells[18].
The aim of our study was to investigate which viral infections are associated with the genetic susceptibility to COVID-19 by using polygenic risk scores (PRS) for COVID-19 eight clinical phenotypes. PRS is a score that aggregates genetic variants to predict disease risk. In this work, we used data from the UK Biobank cohort study and established multiple logistic regression models to assess the correlation between serological results of 25 viral infections and the PRS of COVID-19 eight clinical phenotypes.
2. Material and methods
2.1. UK Biobank Cohort
The UK Biobank cohort is a large prospective cohort study that recruited approximately 500,000 participants aged 40−69 between 2006 and 2010 (https://www.ukbiobank.ac.uk/learn-more-about-uk-biobank). Participant characteristics and other health-related indicators were collected through touchscreen questionnaires, short interviews, and a series of body measurements at 22 assessment centers in the United Kingdom[19]. The UK Biobank study was approved by the National Health Service National Research Ethics Service(11/NW/0382), and all participants provided written informed consent to participate[20]. This study was conducted with the permission from the UK Biobank (application number: 46,478).
Multi-batch genotyping was performed using two slightly different arrays, the Applied Biosystems UK BiLEVE Axiom Array from Affymetrix and the Applied Biosystems UK Biobank Axiom Array. For quality control, sex mismatches, departures from Hardy–Weinberg equilibrium, missing genotype rate >0.05 or imputation accuracy score <0.3 were excluded. Samples identified as outliers for heterozygosity and missing rates were removed. Detailed array design, genotyping and quality control procedures can be found in previous studies[19].
2.2. Common Virus Serological Measurements
In this study, we selected serological measurements of 25 common viruses from the UK Biobank (UK Biobank data fields: 23,050–23,071, 23,073–23,075). Detailed information was shown in the Supplemental tables (Table S1). The serological measurements were defined as positive or negative for each virus.
2.3. Genome-wide Association Study (GWAS) Data of COVID-19 Clinical Phenotypes
The GWAS data of COVID-19 were derived from a genetic association study of the expanded phenotypic definition of COVID-19, including eight clinical phenotypes associated with COVID-19 outcomes[5]. Detailed information of eight clinical phenotypes was presented in Supplementary tables (Table S2). Power analysis of case-control discrete traits was performed using the Purcell power calculator. Array-based genotyping and SNP calling were performed by Illumina with the GenotypeStudio platform or Quest/Athena Diagnostics. Genetic principal components were calculated to include in the association studies to control residual population structure and were computed using FlashPCA 2.0.2. Inbred-related participants were removed by using AncestryDNA. Details of the array design, genotyping, and quality control procedures have been described previously[5].
2.4. Polygenic Risk Scores (PRS) Analysis
In this study, we calculated the PRS of eight COVID-19 clinical phenotypes for each subject. SNPs of P < 1.00×10−5 were selected. The PRS of eight COVID-19 clinical phenotype was calculated by PLINK2.0[21], according to the formula[22]:
PRSm represents the PRS value of COVID-19 clinical phenotypes of the mth subject; n denotes the total number of sample size; βi is the effect parameter of risk allele of the ith significant SNP associated with COVID-19 clinical phenotypes, which was obtained from the GWAS of COVID-19 clinical phenotypes; and SNPim denotes the dosage (0, 1, 2) of the risk allele of the ith SNP for the mth individual[22, 23]. The PRS of eight COVID-19 clinical phenotypes were used as instrumental variables to participate in the subsequent statistical analysis.
2.5. Statistical Analysis
We used logistic regression models to evaluate the correlation between common viral infections and genetic predisposition to COVID-19. Serological measurements of common viruses were used as outcome variables, while the calculated PRS of eight COVID-19 clinical phenotypes was used as instrumental variable. Age, sex, Townsend deprivation index(TDI), frequency of alcohol drinking per week, frequency of smoking per day, body mass index (BMI) and 10 principal components of population structure were used as covariates. We also conducted a stratified analysis of age and gender. We set a threshold of P < 0.05 for suggestive significance. All statistical analyses were performed using R software.
3. Results
3.1. Descriptive characteristics of study participants
For C. trachomatis Definition II seropositivity for Chlamydia trachomatis, 3887 subjects were selected, 50.14% of them were woman, mean age was 57.30 years, standard deviation(SD) was 7.91. For H. pylori Definition I seropositivity for Helicobacter pylori, 2394 subjects were selected, 53.88% of them were woman, mean age was 56.81 (SD:7.88) years. For the other viruses, 4800 subjects were selected, 52.92% of them were women, mean age was 56.95 (SD: 7.94) years.
3.2. Viruses Associated with COVID-19 Phenotypes in the Whole Population
We first performed the logistic analysis in the whole population and identified 12 viruses associated with the PRS of COVID-19 clinical phenotypes. For example, VZV seropositivity for Varicella Zoster Virus was associated with Unscreened/Exposed_Negative (β = 0.1361, P = 0.0142), and C. trachomatis Definition II seropositivity for Chlamydia trachomatis was associated with Positive/Unscreened (β = 0.2174, P = 0.0185). The detailed results were shown in Table 1 .
Table 1.
Viral infections associated with COVID-19 clinical phenotypes in whole population.
| Outcome Variable | Instrumental Variable | β | P |
|---|---|---|---|
| Unscreened/Exposed_Negative | VZV seropositivity for Varicella Zoster Virus | 0.1361 | 0.0142 |
| Positive/Unscreened | C. trachomatis Definition II seropositivity for Chlamydia trachomatis | 0.2174 | 0.0185 |
| Exposed_Positive/Exposed_Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.0935 | 0.0192 |
| Positive/Negative | HSV-1 seropositivity for Herpes Simplex virus-1 | 0.0718 | 0.0213 |
| Exposed_Positive/Exposed_Negative | HBV seropositivity for Hepatitis B Virus | 0.2722 | 0.0219 |
| Hospitalized/Unscreened | VZV seropositivity for Varicella Zoster Virus | 0.1167 | 0.0385 |
| Positive/Unscreened | HCV seropositivity for Hepatitis C Virus | 0.7220 | 0.0398 |
| Exposed_Positive/Exposed_Negative | KSHV seropositivity for Kaposi's Sarcoma-Associated Herpesvirus | -0.1044 | 0.0444 |
| Positive/Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | -0.0814 | 0.0446 |
| Unscreened/Exposed_Negative | MCV seropositivity for Merkel Cell Polyomavirus | -0.0614 | 0.0478 |
| Unscreened/Exposed_Negative | T. gondii seropositivity for Toxoplasma gondii | 0.0677 | 0.0481 |
| Unscreened/Exposed_Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.0817 | 0.0493 |
Note: The threshold of significance is P < 0.05.
3.3. Viruses Associated with COVID-19 Phenotype After Age Stratification
In the age < 65 years group, we identified seven viruses associated with PRS of the COVID-19 clinical phenotypes. For example, the association between HSV-2 seropositivity for Herpes Simplex virus-2 and Exposed_Positive/Exposed_Negative (β = 0.1017, P = 0.0177) was significant. Besides, HSV-2 seropositivity for Herpes Simplex virus-2 infection was also associated with Positive/Negative (β = −0.0987, P = 0.0242). In the age > 65 years group, we found 10 viruses associated with PRS of the COVID-19 clinical phenotypes. For example, HSV-2 seropositivity for Herpes Simplex virus-2 was associated with Continuous_Severity_Score (β = 0.3132, P = 0.0084). In addition, HSV-2 seropositivity for Herpes Simplex virus-2 was also associated with Symptomatic/Paucisymptomatic (β = 0.2436, P = 0.0426). The detailed results were shown in Table 2 .
Table 2.
Viral infections associated with COVID-19 clinical phenotypes in the age < 65 years group and the age > 65 years group.
| Group | Outcome Variable | Instrumental Variable | β | P |
|---|---|---|---|---|
| Age < 65 years | Exposed_Positive/Exposed_Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.1017 | 0.0177 |
| Positive/Negative | HHV-6B seropositivity for Human Herpesvirus-6 | -0.0869 | 0.0226 | |
| Positive/Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | -0.0987 | 0.0242 | |
| Positive/Negative | HSV-1 seropositivity for Herpes Simplex virus-1 | 0.0748 | 0.0270 | |
| Exposed_Positive/Exposed_Negative | KSHV seropositivity for Kaposi's Sarcoma-Associated Herpesvirus | -0.1208 | 0.0353 | |
| Unscreened/Exposed_Negative | VZV seropositivity for Varicella Zoster Virus | 0.1206 | 0.0432 | |
| Hospitalized/Not_Hospitalized | HBV seropositivity for Hepatitis B Virus | -0.2565 | 0.0464 | |
| Age > 65 years | Continuous_Severity_Score | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.3132 | 0.0084 |
| Positive/Negative | HHV-6B seropositivity for Human Herpesvirus-6 | 0.2150 | 0.0149 | |
| Hospitalized/Unscreened | KSHV seropositivity for Kaposi's Sarcoma-Associated Herpesvirus | -0.3371 | 0.0171 | |
| Unscreened/Exposed_Negative | BKV seropositivity for Human Polyomavirus BKV | 0.3491 | 0.0208 | |
| Continuous_Severity_Score | HHV-6 overall seropositivity for Human Herpesvirus-6 | -0.2731 | 0.0323 | |
| Exposed_Positive/Exposed_Negative | HPV 18 seropositivity for Human Papillomavirus type-18 | -0.5712 | 0.0343 | |
| Hospitalized/Not_Hospitalized | C. trachomatis Definition I seropositivity for Chlamydia trachomatis | -0.2309 | 0.0367 | |
| Symptomatic/Paucisymptomatic | HPV 16 Definition I seropositivity for Human Papillomavirus type-16 | -0.5834 | 0.0368 | |
| Symptomatic/Paucisymptomatic | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.2436 | 0.0426 | |
| Positive/Unscreened | HHV-6B seropositivity for Human Herpesvirus-6 | 0.1801 | 0.0465 |
Note: The threshold of significance is P < 0.05.
3.4. Viruses Significantly Associated with COVID-19 Phenotype After Gender Stratification
In the women group, we found five viruses associated with the PRS of COVID-19 clinical phenotypes. For example, HPV 16 Definition II seropositivity for Human Papillomavirus type-16 was associated with Exposed_Positive/Exposed_ Negative (β = −0.2766, P = 0.0026), and HPV 18 seropositivity for Human Papillomavirus type-18 was associated with Continuous_Severity_Score (β = −0.3710, P = 0.0054). In the men group, we found 14 viruses associated with the PRS of COVID-19 clinical phenotypes, such as the association between HSV-2 seropositivity for Herpes Simplex virus-2 and Positive/Negative (β = −0.1881, P = 0.0017), and the association between JCV seropositivity for Human Polyomavirus JCV and Exposed_Positive/Exposed_Negative (β = 0.1237, P = 0.0023). The detailed results were shown in Table 3 .
Table 3.
Viral infections associated with COVID-19 clinical phenotypes in the women group and the men group.
| Group | Outcome Variable | Instrumental Variable | β | P |
|---|---|---|---|---|
| Women | Exposed_Positive/Exposed_Negative | HPV 16 Definition II seropositivity for Human Papillomavirus type-16 | -0.2766 | 0.0026 |
| Continuous_Severity_Score | HPV 18 seropositivity for Human Papillomavirus type-18 | -0.3710 | 0.0054 | |
| Hospitalized/Not_Hospitalized | HHV-7 seropositivity for Human Herpesvirus-7 | -0.2380 | 0.0230 | |
| Symptomatic/Paucisymptomatic | HPV 16 Definition II seropositivity for Human Papillomavirus type-16 | -0.2105 | 0.0252 | |
| Hospitalized/Unscreened | BKV seropositivity for Human Polyomavirus BKV | -0.1802 | 0.0356 | |
| Men | Positive/Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | -0.1881 | 0.0017 |
| Exposed_Positive/Exposed_Negative | JCV seropositivity for Human Polyomavirus JCV | 0.1237 | 0.0023 | |
| Positive/Negative | HHV-6 overall seropositivity for Human Herpesvirus-6 | -0.1986 | 0.0048 | |
| Positive/Unscreened | C. trachomatis Definition II seropositivity for Chlamydia trachomatis | 0.3021 | 0.0078 | |
| Unscreened/Exposed_Negative | T. gondii seropositivity for Toxoplasma gondii | 0.1269 | 0.0122 | |
| Positive/Negative | KSHV seropositivity for Kaposi's Sarcoma-Associated Herpesvirus | -0.1936 | 0.0126 | |
| Unscreened/Exposed_Negative | H. pylori Definition I seropositivity for Helicobacter pylori | 0.1633 | 0.0162 | |
| Positive/Unscreened | HPV 16 Definition II seropositivity for Human Papillomavirus type-16 | 0.2164 | 0.0256 | |
| Positive/Negative | HSV-1 seropositivity for Herpes Simplex virus-1 | 0.0989 | 0.0285 | |
| Positive/Negative | HHV-6A seropositivity for Human Herpesvirus-6 | -0.1090 | 0.0308 | |
| Exposed_Positive/Exposed_Negative | H. pylori Definition I seropositivity for Helicobacter pylori | 0.1312 | 0.0333 | |
| Unscreened/Exposed_Negative | VZV seropositivity for Varicella Zoster Virus | 0.1913 | 0.0364 | |
| Hospitalized/Not_Hospitalized | JCV seropositivity for Human Polyomavirus JCV | -0.0876 | 0.0417 | |
| Exposed_Positive/Exposed_Negative | HSV-2 seropositivity for Herpes Simplex virus-2 | 0.1158 | 0.0445 |
Note: The threshold of significance is P < 0.05.
4. Discussion
A previous study found that the severity of COVID-19 infection is influenced by host genetic factors[24]. We are curious if there is a potential correlation between COVID-19-related genetic information and the infection status of other viruses. In this work, we used PRS to represent an individual's genetic susceptibility to COVID-19. Logistic regression models were used to assess the association between multiple common viral infections and the PRS of COVID-19 clinical phenotypes. Our study aimed to detect which viral infections were associated with genetic susceptibility to COVID-19.
Our study builds upon previous research by adding covariates to logistic regression models for correction, in order to explore whether genetic susceptibility to COVID-19 influences the risk of infection by other common viruses. We found that VZV had a significant association with Unscreened/Exposed_Negative in the whole population. Herpes zoster is a viral skin disease in which herpes zoster remains dormant in the dorsal root ganglion of the cutaneous nerve after chickenpox infection[25]. Among reported COVID-19 cases, infected patients exhibited diverse skin manifestations, with varicella-like lesions being one of the major skin manifestations during the COVID-19 outbreak[26]. Cases of herpes zoster infection have been identified in recent symptomatic COVID-19 infections[27]. It is possible that this is SARS-CoV-2 could directly infect lymphocytes and promote apoptosis of lymphocytes, leading to lymphopenia and impaired antiviral response, which may further favor herpes virus recurrence[28].
It has been found that some viruses exhibit a change in incidence with age. For example, the zoster virus demonstrates a steady increase in incidence starting at age 50 years, with the higher incidence in people over 65 years[29]. Therefore, in our study we performed a stratified analysis by controlling for age. After controlling for age variables, we found that the significant associations were not the same between the age < 65 years group and the age > 65 years group. In the age < 65 years group, the most significant association was found between HSV-2 seropositivity for Herpes Simplex virus-2 and Exposed_Positive/Exposed_Negative. However, in the age > 65 years group, the most significant association was found between HSV-2 seropositivity for Herpes Simplex virus-2 and Continuous_Severity_Score. HSV-2 causes ulcerative lesions in adults and primarily affects the genital region through sexual transmission[30]. Following primary infection, Herpes simplex virus enters the latent state in the ganglion and may emerge later, leading to recurrent active infection[31]. Recent studies been found that individuals infected with HSV could affect SARS-CoV-2 IgM/IgG serologic results due to direct binding of IgM antibodies to otherwise detected surface-modified polystyrene particles[32].
The prevalence of common viruses is also gender-dependent. For example, the epidemiology of HSV-2 differs between women and men, with a greater probability of transmission from male-to-female than female-to-male[33]. Therefore, we conducted a gender stratified analysis. After stratifying, we found more virus in the men group may be affected by the genetic susceptibility of COVID-19. For example, 14 significant associations were found in the men group, such as HSV-2 and Positive/Negative, which also consist with previous studies[32]. In the women group, human papillomavirus (HPV)-associated virus infection is associated with genetic susceptibility to COVID-19. In addition, we found human herpesvirus (HHV) is also associated with genetic susceptibility to COVID-19. HHV reactivation was considered a positive polymerase chain reaction result taken at the time of COVID-19 infection[34]. The reactivation of HSV is associated with an increased risk of hospital-acquired pneumonia/ventilator-associated pneumonia (HAP/VAP)[35]. Overall, co-infection with herpes viruses leads to poor clinical outcomes, particularly in critically ill COVID-19 patients[11, 36].
This is a new study that explores which viral infections are associated with genetic susceptibility to COVID-19. Our study finally identified several viral infections that are associated with genetic susceptibility to COVID-19. However, there are some limitations to consider when interpreting these findings. First, our data comes from a UK biobank, which only includes information from people of European descent. Therefore, our conclusions are limited in their applicability to other racial and ethnic populations. Second, our work is only exploratory, and the results can only demonstrate correlation rather than causation. Third, although we controlled for confounding factors, there may still be potential confounding factors that we did not account for. Therefore, the association between viral infections and genetic susceptibility should be interpreted with caution. Finally, more large-scale prospective and biological studies are needed to confirm our results and elucidate the specific mechanisms involved.
In summary, our work identified viral infections that are associated with genetic susceptibility to COVID-19. These findings may help clinicians to prevent and detect the recurrence of other viruses closely related to COVID-19 in a timely manner. Moreover, this association might assist clinicians in identifying patients with a poorer prognosis.
Funding Information
This work was supported by the National Natural Scientific Foundation of China [grant numbers:81922059] and the Natural Science Basic Research Program in Shaanxi Province of China:[grant numbers:2021JCW-08].
Institutional Review Board Statement
This study has been approved by UK Biobank (Application number: 46478) and obtained health-related records of participants.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure
All authors report no biomedical financial interests or potential conflicts of interest.
Author contributions
Na Zhang: Writing – original draft, Conceptualization, Formal analysis. Yujing Chen: Writing – review & editing, Formal analysis. Chun'e Li: Formal analysis. Xiaoyue Qin: Writing – review & editing. Dan He: Methodology. Wenming Wei: Validation. Yijing Zhao: Software. Qingqing Cai: Software. Sirong Shi: Visualization. Xiaoge Chu: Visualization. Yan Wen: Investigation. Yumeng Jia: Investigation. Feng Zhang: Supervision, Project administration.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2023.105170.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.W.H. Organization, Coronavirus disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update, (2022) 1-25.
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velavan T.P., Pallerla S.R., Rüter J., Augustin Y., Kremsner P.G., Krishna S., Meyer C.G. Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine. 2021;72:103629. doi: 10.1016/j.ebiom.2021.103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts G.H.L., Partha R., Rhead B., Knight S.C., Park D.S., Coignet M.V., Zhang M., Berkowitz N., Turrisini D.A., Gaddis M., McCurdy S.R., Pavlovic M., Ruiz L., Sass C., Haug Baltzell A.K., Guturu H., Girshick A.R., Ball C.A., Hong E.L., Rand K.A. Expanded COVID-19 phenotype definitions reveal distinct patterns of genetic association and protective effects. Nat Genet. 2022;54(4):374–381. doi: 10.1038/s41588-022-01042-x. [DOI] [PubMed] [Google Scholar]
- 6.Wu Q., Xing Y., Shi L., Li W., Gao Y., Pan S., Wang Y., Wang W., Xing Q. Coinfection and Other Clinical Characteristics of COVID-19 in Children. Pediatrics. 2020;146(1) doi: 10.1542/peds.2020-0961. [DOI] [PubMed] [Google Scholar]
- 7.Lin D., Liu L., Zhang M., Hu Y., Yang Q., Guo J., Guo Y., Dai Y., Xu Y., Cai Y., Chen X., Zhang Z., Huang K. Co-infections of SARS-CoV-2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. 2020;63(4):606–609. doi: 10.1007/s11427-020-1668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., Zhu F., Zhu B., Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouwendijk W.J., Laing K.J., Verjans G.M., Koelle D.M. T-cell immunity to human alphaherpesviruses. Curr Opin Virol. 2013;3(4):452–460. doi: 10.1016/j.coviro.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell B.M., Bloom D.C., Cohrs R.J., Gilden D.H., Kennedy P.G. Herpes simplex virus-1 and varicella-zoster virus latency in ganglia. J Neurovirol. 2003;9(2):194–204. doi: 10.1080/13550280390194000. [DOI] [PubMed] [Google Scholar]
- 11.Le Balc'h P., Pinceaux K., Pronier C., Seguin P., Tadié J.M., Reizine F. Herpes simplex virus and cytomegalovirus reactivations among severe COVID-19 patients. Crit Care. 2020;24(1):530. doi: 10.1186/s13054-020-03252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenney A.D., Dowdle J.A., Bozzacco L., McMichael T.M., St Gelais C., Panfil A.R., Sun Y., Schlesinger L.S., Anderson M.Z., Green P.L., López C.B., Rosenberg B.R., Wu L., Yount J.S. Human Genetic Determinants of Viral Diseases. Annu Rev Genet. 2017;51:241–263. doi: 10.1146/annurev-genet-120116-023425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin M., Tseng H.K., Trejaut J.A., Lee H.L., Loo J.H., Chu C.C., Chen P.J., Su Y.W., Lim K.H., Tsai Z.U., Lin R.Y., Lin R.S., Huang C.H. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. doi: 10.1186/1471-2350-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alanio C., Verma A., Mathew D., Gouma S., Liang G., Dunn T., Oldridge D.A., Weaver J., Kuri-Cervantes L., Pampena M.B., Betts M.R., Collman R.G., Bushman F.D., Meyer N.J., Hensley S.E., Rader D., Wherry E.J. Cytomegalovirus Latent Infection is Associated with an Increased Risk of COVID-19-Related Hospitalization. J Infect Dis. 2022;226(3):463–473. doi: 10.1093/infdis/jiac020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J.T., Leung K., Bushman M., Kishore N., Niehus R., de Salazar P.M., Cowling B.J., Lipsitch M., Leung G.M. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;26(4):506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cannon M.J., Schmid D.S., Hyde T.B. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20(4):202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 17.Furman D., Jojic V., Sharma S., Shen-Orr S.S., Angel C.J., Onengut-Gumuscu S., Kidd B.A., Maecker H.T., Concannon P., Dekker C.L., Thomas P.G., Davis M.M. Cytomegalovirus infection enhances the immune response to influenza. Sci Transl Med. 2015;7(281):281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patin E., Hasan M., Bergstedt J., Rouilly V., Libri V., Urrutia A., Alanio C., Scepanovic P., Hammer C., Jönsson F., Beitz B., Quach H., Lim Y.W., Hunkapiller J., Zepeda M., Green C., Piasecka B., Leloup C., Rogge L., Huetz F., Peguillet I., Lantz O., Fontes M., Di Santo J.P., Thomas S., Fellay J., Duffy D., Quintana-Murci L., Albert M.L. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol. 2018;19(3):302–314. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 19.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O'Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G., Leslie S., Allen N., Donnelly P., Marchini J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M., Liu B., Matthews P., Ong G., Pell J., Silman A., Young A., Sprosen T., Peakman T., Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3) doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C., PLINK a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng S., Qi X., Ma M., Zhang L., Cheng B., Liang C., Liu L., Li P., Kafle O.P., Wen Y., Zhang F. Assessing the Relationship Between Gut Microbiota and Bone Mineral Density. Front Genet. 2020;11:6. doi: 10.3389/fgene.2020.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudbridge F. Polygenic Epidemiology. Genet Epidemiol. 2016;40(4):268–272. doi: 10.1002/gepi.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu D., Zhao R., Yuan H., Xie Y., Jiang Y., Xu K., Zhang T., Chen X., Suo C. Host Genetic Factors, Comorbidities and the Risk of Severe COVID-19. J Epidemiol Glob Health. 2023:1–13. doi: 10.1007/s44197-023-00106-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dayan R.R., Peleg R. Herpes zoster - typical and atypical presentations. Postgrad Med. 2017;129(6):567–571. doi: 10.1080/00325481.2017.1335574. [DOI] [PubMed] [Google Scholar]
- 26.Wollina U., Karadağ A.S., Rowland-Payne C., Chiriac A., Lotti T. Cutaneous signs in COVID-19 patients: A review. Dermatol Ther. 2020;33(5) doi: 10.1111/dth.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira A., Romão T.T., Macedo Y.S., Pupe C., Nascimento O.J.M. COVID-19 and herpes zoster co-infection presenting with trigeminal neuropathy. Eur J Neurol. 2020;27(9):1748–1750. doi: 10.1111/ene.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai K., Gebremeskel B.G., Acosta C.J. Systematic review of incidence and complications of herpes zoster: towards a global perspective. BMJ Open. 2014;4(6) doi: 10.1136/bmjopen-2014-004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nahmias A.J., Roizman B. Infection with herpes-simplex viruses 1 and 2. II. N Engl J Med. 1973;289(14):719–725. doi: 10.1056/NEJM197310042891404. [DOI] [PubMed] [Google Scholar]
- 31.Fleming D.T., McQuillan G.M., Johnson R.E., Nahmias A.J., Aral S.O., Lee F.K., St Louis M.E. Herpes simplex virus type 2 in the United States, 1976 to 1994. N Engl J Med. 1997;337(16):1105–1111. doi: 10.1056/NEJM199710163371601. [DOI] [PubMed] [Google Scholar]
- 32.Vandervore L., Van Mieghem E., Nowé V., Schouwers S., Steger C., Abrams P., Van Schaeren J., Meskal A., Vandamme T. False positive Herpes Simplex IgM serology in COVID-19 patients correlates with SARS-CoV-2 IgM/IgG seropositivity. Diagn Microbiol Infect Dis. 2022;103(1):115653. doi: 10.1016/j.diagmicrobio.2022.115653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wald A., Langenberg A.G., Link K., Izu A.E., Ashley R., Warren T., Tyring S., Douglas J.M., Jr., Corey L. Effect of condoms on reducing the transmission of herpes simplex virus type 2 from men to women. Jama. 2001;285(24):3100–3106. doi: 10.1001/jama.285.24.3100. [DOI] [PubMed] [Google Scholar]
- 34.Shafiee A., Teymouri Athar M.M., Amini M.J., Hajishah H., Siahvoshi S., Jalali M., Jahanbakhshi B., Mozhgani S.H. Reactivation of herpesviruses during COVID-19: A systematic review and meta-analysis. Rev Med Virol. 2023;33(3):e2437. doi: 10.1002/rmv.2437. [DOI] [PubMed] [Google Scholar]
- 35.Meyer A., Buetti N., Houhou-Fidouh N., Patrier J., Abdel-Nabey M., Jaquet P., Presente S., Girard T., Sayagh F., Ruckly S., Wicky P.H., de Montmollin E., Bouadma L., Sonneville R., Descamps D., Timsit J.F. HSV-1 reactivation is associated with an increased risk of mortality and pneumonia in critically ill COVID-19 patients. Crit Care. 2021;25(1):417. doi: 10.1186/s13054-021-03843-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz J., Yue S., Xue W. Herpes simplex and herpes zoster viruses in COVID-19 patients. Ir J Med Sci. 2022;191(3):1093–1097. doi: 10.1007/s11845-021-02714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.