Abstract
Bacterial bloodstream infections (BSIs) resulting in late-onset sepsis affect up to half of extremely preterm infants and have substantial morbidity and mortality. Bacterial species associated with BSIs in neonatal intensive care units (NICUs) commonly colonize the preterm infant gut microbiome. Accordingly, we hypothesized that the gut microbiome is a reservoir of BSI-causing pathogenic strains that increase in abundance before BSI onset. We analyzed 550 previously published fecal metagenomes from 115 hospitalized neonates and found that recent ampicillin, gentamicin, or vancomycin exposure was associated with increased abundance of Enterobacteriaceae and Enterococcaceae in infant guts. We then performed shotgun metagenomic sequencing on 462 longitudinal fecal samples from 19 preterm infants (cases) with BSI and 37 non-BSI controls, along with whole-genome sequencing of the BSI isolates. Infants with BSI caused by Enterobacteriaceae were more likely than infants with BSI caused by other organisms to have had ampicillin, gentamicin, or vancomycin exposure in the 10 days before BSI. Relative to controls, gut microbiomes of cases had increased relative abundance of the BSI-causing species and clustered by Bray-Curtis dissimilarity according to BSI pathogen. We demonstrated that 11 of 19 (58%) of gut microbiomes before BSI, and 15 of 19 (79%) of gut microbiomes at any time, harbored the BSI isolate with fewer than 20 genomic substitutions. Last, BSI strains from the Enterobacteriaceae and Enterococcaceae families were detected in multiple infants, indicating BSI-strain transmission. Our findings support future studies to evaluate BSI risk prediction strategies based on gut microbiome abundance in hospitalized preterm infants.
INTRODUCTION
Preterm birth accounts for 10% of births and 17% of infant mortality in the United States (1, 2). Preterm infants in neonatal intensive care units (NICUs) are particularly vulnerable to infection because of their immature organ systems, breach of cutaneous and mucosal barriers, and considerable antimicrobial exposures (3). The frequency of late-onset sepsis, defined as suspected or confirmed infection after the first 3 days of life (DOL) (3, 4), is inversely proportional to gestational age (GA) at birth, with up to 50% of extremely preterm and very low birth weight (<28 weeks and <1000 g) infants experiencing at least one episode (3, 5). In contrast to early-onset sepsis, the incidence and mortality of late-onset sepsis in preterm infants have not diminished appreciably over the past 30 years (6, 7). It is therefore essential to better understand the pathogen sources and risk factors associated with late-onset sepsis to develop mitigation strategies for this highly consequential complication of preterm birth.
Staphylococcus spp., Enterobacteriaceae (such as Escherichia coli, Klebsiella spp., and Enterobacter spp.), and Enterococcus spp. are the most common causes of bloodstream infection (BSI) in NICUs worldwide (3, 8, 9). Risk factors for late-onset sepsis in general include early GA, low birth weight, presence of an intravenous catheter, formula exposure, total parenteral nutrition, and prolonged duration without oral feeding (10-15). Gram-positive late-onset sepsis (predominately Staphylococcus spp. and Enterococcus spp.) is associated with duration of intravenous catheterization and total parenteral nutrition (13, 14), whereas chorioamnionitis, vaginal birth, prolonged antimicrobial exposure, and necrotizing enterocolitis are associated with Gram-negative, predominately Enterobacteriaceae BSIs (14, 16, 17). Pathogens that cause BSI are commonly found in the NICU environment and in the hospitalized, preterm gut microbiome (18-21). In a subset of preterm infants, the same species or genus can be identified in stool before BSI (15, 22-24). Therefore, a direct gut- or skin-to-bloodstream route of dissemination has been postulated (25). Carl et al. (22) recovered isolates from stool culture that provisionally matched the BSI isolate in 7 of 11 cases of late-onset sepsis. These strains differed by an average of 33.5 nucleotides per 100,000 base pairs [average nucleotide identity (ANI) of 99.7%], a rate that may have overrepresented the nucleotide differences based on sequencing and computational technologies available at the time.
Although these findings are consistent with a gut-to-blood-stream mechanism of BSI pathogenesis, this degree of resolution of strain similarity cannot separate strains that diverged within the past 500 years (26). As such, prior methods that do not avail whole-genome sequencing, especially those relying on 16S ribosomal RNA gene sequencing or multilocus sequence typing, may overestimate strain similarity (26-28). Further, bona fide subspecies that concurrently colonize the same individual on different body sites, such as skin, gut, and mouth, can differ by as few as 30 genomic substitutions, and multiple strains can coexist in the same microenvironment (27, 29, 30). Hence, high genomic precision is essential for accurate pathogen tracking. Recent computational and sequencing methods (26, 31) permit more precise strain tracking to determine whole genome–resolved BSI-gut similarity and identify abundance changes in pathogens within the gut. These methodologic advances are critical to identify the immediate pre-BSI habitat of microbes to focus on appropriate interventions for prevention and diagnosis.
Recent exposure to antibiotics, which are frequently administered in the NICU for suspected late-onset sepsis (32), is associated with an increased abundance of certain pathobionts (commensal organisms that can be opportunistically pathogenic) in the guts of preterm infants (20). Specifically, Enterococcus faecalis and Staphylococcus epidermidis increased in abundance in response to selective pressure of meropenem therapy, and ampicillin, gentamicin, and vancomycin more variably affected overall diversity and specific species abundance (20). Some studies demonstrate diminished gut microbial diversity in neonates before late-onset sepsis versus controls; however, these controls often had more advanced GA, less formula receipt, and less antibiotic exposure, variables known to be associated with lower gut microbiome diversity (15, 24, 33, 34). Because gut, skin, and NICU hospital microbiomes harbor similar species (19, 27), and antibiotics enrich for potential pathogens in the stool of infants residing in NICUs, it is essential to analyze strain relatedness with maximal precision and investigate clinical features distinguishing BSI cases from controls in the NICU to identify risk factors and the proximate source of late-onset sepsis pathogens.
Here, we performed shotgun metagenomic sequencing on 462 stools from 19 preterm infants (cases) and whole-genome sequencing of their BSI isolates and 37 infants without BSI (controls). Our goal was to identify features capable of distinguishing BSI cases and controls without BSI inhabiting the same environments. We used precise BSI strain tracking within the gut microbiome longitudinally to identify sources of BSI pathogens and strain sharing within the NICU. Our results can inform future risk prediction algorithms based on gut microbiome abundance of potential pathogens for hospitalized, preterm infants to anticipate and intervene on at-risk infants.
RESULTS
Abundance of potential pathogens increases in the gut microbiome with recent antibiotics
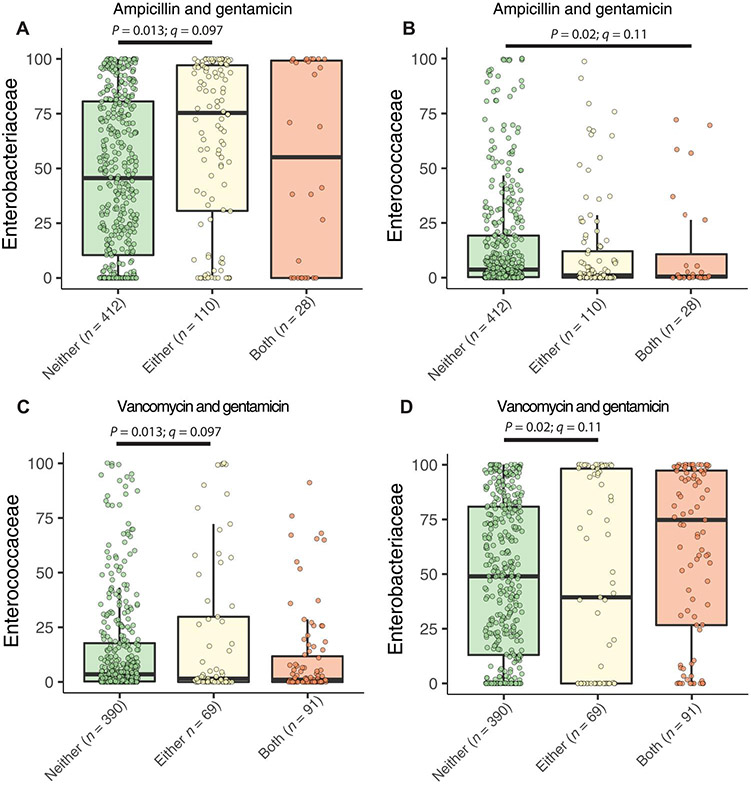
A previous prospective study of 977 infants born at 27 weeks average GA was performed from 2009 to 2013 in three Midwestern NICUs to characterize the development of the preterm infant gut microbiome (20, 22, 35-37). Because antibiotic administration is associated with subsequent BSI, late-onset sepsis, and death (17, 38-40), and infants’ guts harbor potential pathogens (15, 20, 22, 33, 37), we first asked whether pathobionts increased in abundance in the gut after neonates are given common antibiotics. We hypothesized that recent receipt of ampicillin, gentamicin, and vancomycin, the three antibiotics most commonly used in NICUs (20, 40) and often administered in combination, increases the abundance of frequently antibiotic-resistant BSI pathobionts (20). The most common antimicrobials in this cohort immediately after birth were the co-administration of ampicillin and gentamicin, whereas the most common antimicrobials administered for late-onset sepsis rule-out was the co-administration of vancomycin and gentamicin (20) (data file S1). We analyzed 550 previously sequenced stools in the first 100 DOL from 115 hospitalized preterm neonates to determine whether ampicillin, gentamicin, or vancomycin in the 10 days before fecal sample collection were associated with abundance changes in gut microbiome bacterial families using MaAsLin2 (20, 37, 41). We found that only three bacterial families were significantly associated with these antibiotic combinations (data file S2). Ampicillin or gentamicin exposure in the prior 10 days was associated with increased relative abundance of Enterobacteriaceae over receiving neither (Fig. 1A; P = 0.013, q = 0.097), but ampicillin and gentamicin exposure was associated with decreased relative abundance of Enterococcaceae (Fig. 1B; P = 0.02, q = 0.11). Exposure to vancomycin or gentamicin was associated with increased relative abundance of Enterococcaceae (Fig. 1C; P = 0.013, q = 0.097) and decreased Enterobacteriaceae (Fig. 1D; P = 0.02, q = 0.11) relative to receiving neither of these two antibiotics, whereas exposure to vancomycin and gentamicin nonsignificantly increased Enterobacteriaceae relative abundance in the gut microbiome (Fig. 1D). These findings are consistent with at least two nonmutually exclusive hypotheses. Abundance increases of these bacterial families may precede antibiotic delivery secondary to clinical changes prompting antibiotic administration. In addition, these organisms may be resistant to one of these frequently chosen antibiotics (20, 42). Because we found increased relative abundance of potential BSI pathogens in the stools of hospitalized NICU infants agnostic of their BSI status, we designed a case-control cohort study to investigate factors associated with BSI and the gut microbiome in the NICU.
Fig. 1. The abundance of potential pathogens increases in stool of NICU infants after antibiotics.
Family-level changes of bacteria in the gut microbiome were assessed regarding receipt of ampicillin, gentamicin, or vancomycin in the prior 10 days for previously sequenced infants in the cohort irrespective of BSI status. (A) Enterobacteriaceae and (B) Enterococcaceae were the only families found to be changed in the gut after either or both ampicillin and gentamicin. Recent vancomycin or gentamicin increased Enterococcaceae (C) but decreased Enterobacteriaceae (D) abundance, as assessed by MaAsLin2 with DOL as an additional fixed effect and participant as random effect. All P < 0.05 by generalized LME model, q value is Benjamini-Hochberg–corrected for multiple comparisons.
Antibiotic-resistant pathogens cause BSIs in the NICU
Within the overall cohort (36), we identified 19 neonates with BSI who met our enrollment criteria (fig. S1). BSI-causing Gram-positive bacteria were Staphylococcus aureus (n = 6), Streptococcus agalactiae (GBS) (n = 3), and E. faecalis (n = 2). BSI was also caused by Gram-negative Enterobacteriaceae [Serratia marcescens (n = 3), E. coli (n = 2), Klebsiella pneumoniae (n = 2), and Enterobacter cloacae (n = 1)] (Table 1). To control for clinical associations of late-onset sepsis onset and gut microbiome composition, we paired these neonates with control infants without BSI for sex, GA, birth weight, delivery mode, age at onset of BSI, antibiotic days before BSI, and overall antibiotic exposure (data file S1). There were no statistically significant differences for any of these variables between cases and controls (Mann-Whitney, Fisher’s exact, or chi-square test) (Table 1).
Table 1. Demographics and clinical details of case-control cohort.
NA, not applicable; DOL, day of life; NPO: non per os, no oral nutrition.
| Cases | Controls | |
|---|---|---|
| Participants | 19 | 37 |
| Stools | 210 | 252 |
| Number of stools per participant before BSI (25th–75th quartile) | 4 (3–6) | 4 (2–4) |
| Number of stools per participant after BSI (25th–75th quartile) | 6 (3–8) | 3 (3–4) |
| Bacteremia DOL (25th–75th quartile) or matched DOL for controls | 32 (20–44) | 32 (19–46) |
| Antibiotic days before bacteremia or matched control DOL (25th–75th quartile) | 16 (7–25) | 21 (9–28) |
| Overall antibiotic score from birth to BSI or matched control DOL (25th–75th quartile) | 53 (26–112) | 57 (25–85) |
| Gestational age (25th–75th quartile) | 25 (24–27) | 25 (25–26) |
| Birthweight, g (25th–75th quartile) | 740 (620–833) | 810 (676–880) |
| % Female | 68 | 68 |
| % Cesarean | 84 | 84 |
| % First 8 weeks NPO | 26 | 20 |
| % First 8 weeks human milk | 47 | 55 |
| % First 8 weeks donor milk | 6 | 7 |
| % First 8 weeks formula | 21 | 18 |
| Bloodstream infection | 19 | NA |
| Staphylococcus aureus infection | 6 | NA |
| Streptococcus agalactiae | 3 | NA |
| Enterococcus faecalis | 2 | NA |
| Serratia marcescens | 3 | NA |
| Escherichia coli | 2 | NA |
| Klebsiella pneumoniae | 2 | NA |
| Enterobacter cloacae | 1 | NA |
We performed antimicrobial susceptibility testing and whole-genome sequencing on the BSI isolates and used ResFinder to infer antibiotic resistance genes (data file S3). Among S. aureus, two of six (33%) were phenotypically methicillin resistant and harbored mecA, seven of eight (87.5%) of Enterobacteriaceae were ampicillin nonsusceptible, and three of eight (37.5%) were gentamicin nonsusceptible. We identified β-lactam and aminoglycoside antibiotic resistance genes corresponding to the phenotypic antibiotic susceptibility testing for the eight Enterobacteriaceae. Infants whose BSI was caused by Enterobacteriaceae were significantly more likely than infants with BSI caused by other organisms to have had ampicillin, gentamicin, or vancomycin exposure 10 days prior (75% versus 9%; P = 0.0063, Fisher’s exact test). Given our study’s control criteria, infants with BSI had no difference in number of antibiotic days or antibiotic score (43) before bacteremia relative to controls (Table 1 and fig. S2A). However, neonates with BSI had increased antibiotic score after BSI versus before and versus controls owing to antibiotic treatment targeting the BSI organism [fig. S2B and data file S2; linear mixed-effect (LME) model < 0.05]. We found no difference in median antibiotic resistance gene count in the metagenomes of BSI infants before BSI relative to matched controls using ShortBRED (fig. S2C). Specifically, we identified antibiotic resistance genes [in reads per kilobase per million reads (RPKM)] in the metagenomes of cases before BSI and controls during the same matched time span conferring resistance to beta-lactams, aminoglycosides, and vancomycin in decreasing order of prevalence (fig. S3 and data file S3). We could not determine which specific taxa harbor these antibiotic resistance genes (ARGs) from this metagenomic analysis; however, bacteria need not necessarily be resistant to antibiotics to persist because of subinhibitory antibiotic concentrations in the gut (44). We observed a decrease in ARGs in both groups from 2 weeks before BSI to 2 weeks after but no differences between cases and controls in abundance or composition (fig. S4 and data files S2 and S3). Thus, we found antibiotic-resistant organisms causing BSI but no significant differences in the overall gut resistomes of cases before BSI versus controls.
High abundance of bacteremia-causing species in case metagenomes
We hypothesized that BSI-causing species originated in the gut and gained access to the bloodstream or via cutaneous access. Five of 16 (31%) case participants from St. Louis Children’s Hospital (SLCH) had either no intravenous access or only a peripheral intravenous catheter in the 7 days before BSI (data file S3). We determined the relative abundance of the causative species before, during, and after BSI within each case’s gut microbiota. Each of the eight infants with Enterobacteriaceae BSI produced a stool before BSI with >10% relative abundance of the causative species, with 75% producing a stool with >45% relative abundance of the BSI-causing species. After completing the antibiotic course for BSI, both infants with E. coli still had greater than 20% relative abundance of E. coli in their stool (fig. S5). Similarly, the two infants with E. faecalis BSI had several stools with greater than 50% relative abundance of E. faecalis before and after BSI (fig. S6). Conversely, 83% (five of six) of infants with BSI caused by S. aureus had less than 3% relative abundance of S. aureus in all stools before BSI, and only one of six produced a stool dominated by S. aureus, with over 90% relative abundance (fig. S7). Three of five (60%) infants whose stools had minimal S. aureus before BSI had transient increases in fecal S. aureus carriage in the few days after BSI (fig. S7). Only one of three infants with S. agalactiae BSI had substantial fecal carriage before BSI (fig. S8). Thus, we observed at least two separate patterns of gut pathogen abundance before bacteremia; BSI species from Enterobacteriaceae and Enterococcaceae families largely had high gut abundance before bacteremia, whereas BSI from S. aureus and GBS had low or no gut abundance before bacteremia.
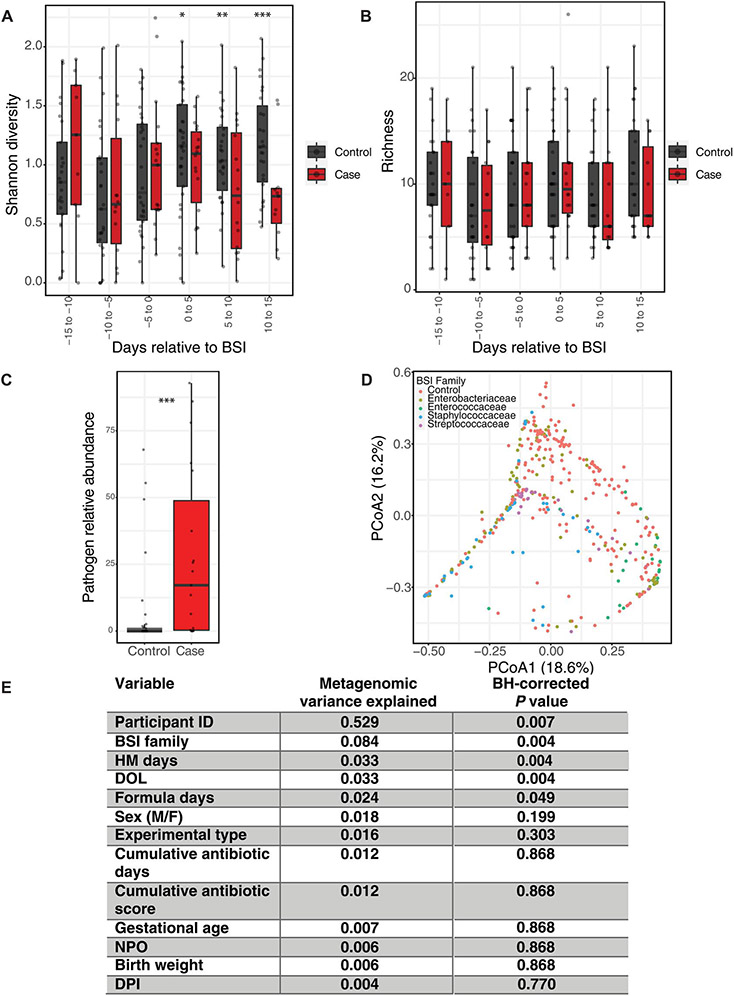
Gut microbiome Shannon diversity was stable 2 weeks before BSI but decreased after BSI during antimicrobial therapy (Fig. 2A and data file S2). We found no difference in Shannon diversity between BSI cases and controls within 5-day windows (bins) before BSI (Fig. 2A). Similarly, there was no difference in the number of species (richness) in the gut microbiome between cases and controls relative to BSI (Fig. 2B). We then compared the mean relative abundance of the BSI-causing species in each case before BSI with the mean relative abundance of the same species in all controls in the same DOL range (Fig. 2C). Despite the low relative abundance before BSI for S. agalactiae and S. aureus, we found significantly increased relative abundance of the causative species in the 2 weeks preceding BSI for all cases versus controls [Fig. 2C; median 17%, interquartile range (IQR) 0.33:48.76 versus median 0%, IQR 0:1.3; P < 0.001, Wilcoxon]. Therefore, the same BSI-causing species is present in stool before BSI onset in a subset of infants, and the relative abundance is higher than in infants not suffering from BSI. We then determined Bray-Curtis dissimilarity between all samples and, using repeated-measures permutational multivariate analysis of variance (PERMANOVA), found that the strongest nonindividual variance was contributed by the BSI-causing organism family (Fig. 2, D and E; 8.4% variance, P < 0.005). This clustering was not attributable to case versus control identity (Fig. 2, D and E) but rather was secondary to differences in the gut microbiome based on the BSI-causing species. Cumulative human milk exposure, DOL, and cumulative formula exposure were the only other metadata significantly contributing to microbiome conformation (Fig. 2E, fig. S9); however, these were not significantly different between cases and controls (Table 1). These data suggest that an organism-specific gut microbial signature might precede BSI.
Fig. 2. Greater abundance of BSI species in case stools before infection onset despite no gross alpha diversity differences.
(A) Shannon diversity and (B) species richness displayed within 5-day windows (bins) relative to BSI. Values are averaged within 5-day windows for infants who produced more than one stool within the same 5-day span. (C) Mean relative abundance of BSI-causing species from each case in the 14 days before BSI versus the same species in controls. Mean value for each case species plotted versus abundance of same BSI-causing species in controls in the same DOL range. (D) PCoA of Bray-Curtis dissimilarity is shown by BSI-causing family. Number in parenthesis is percentage of variance explained. (E) Repeated-measures PERMANOVA results for variance contributed and Benjamini-Hochberg–corrected q value. Boxplots represent 25th to 75th median quartile, with horizontal black bar at the median. (A and B) LME model with participant as a random effect. (C) Wilcoxon signed-rank test. *P < 0.05. HM, human milk; DPI, days post infection.
Gut microbiome features differentiate cases from controls
We next wanted to determine whether any gut microbiome features permitted differentiation between BSI cases caused by these organisms versus others in the same NICUs. We determined gut microbiome enterotypes at the genus level on our 449 samples with sufficient sequencing reads. We identified four enterotypes that minimized Laplace score at the genus level (fig. S10). Enterotypes were variably dominated by key taxa, including Staphylococcus and Enterococcus (m1), Klebsiella and Enterococcus (m2), Veillonella and Klebsiella (m3), and Escherichia and Enterococcus (m4). We found that preinfection gut microbiomes (n = 214) clustered into distinct enterotypes depending on their control status or the organism that caused subsequent BSI (data file S2). Thus, gut microbiome structure before BSI correlates with the causative organism family and control status.
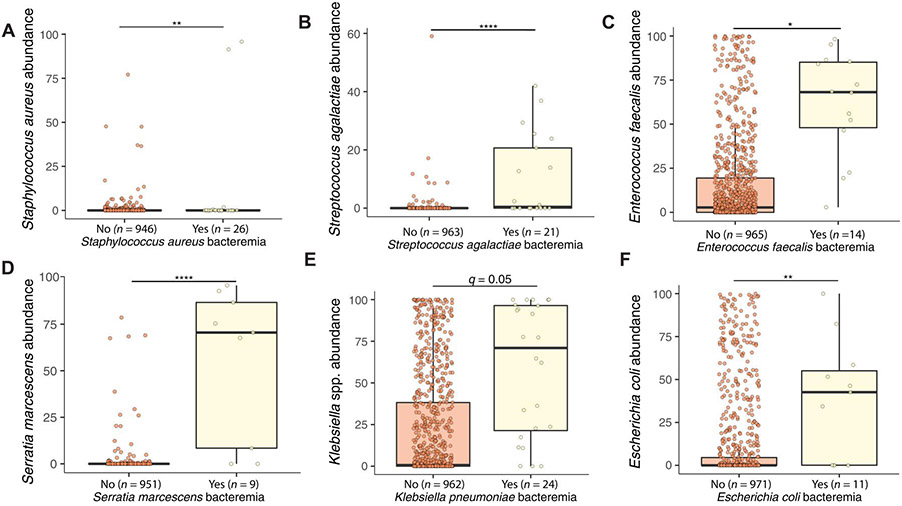
We next postulated that signatures in the gut microbiome might differentiate individuals who experienced BSI from specific organisms from others. We determined metagenomic differences before BSI between cases that later developed BSI caused by specific species versus all others, including previously sequenced samples (20, 37), in the same DOL range. The most significant features in the gut microbiome for individuals with S. aureus BSI were increased S. epidermidis and Cutibacterium avidum abundance (fig. S11, A and B; q < 0.005, coefficient 0.56 and 0.16; data file S2). Despite not being frequently identified in the gut microbiome (fig. S7), we found a small but significant increase in S. aureus in the gut before S. aureus BSI (Fig. 3A; q < 0.01, coefficient 0.10). S. agalactiae was the most significant feature before BSI in the gut microbiomes of cases with GBS BSI (Fig. 3B; q < 0.0001, coefficient 0.2). Veillonella atypica was also increased in the gut microbiome for these same individuals who progressed to GBS BSI (fig. S11C; q < 0.05, coefficient 0.13). E. faecalis was the only significant feature differentiating individuals with E. faecalis BSI from all others in the NICU (Fig. 3C; q < 0.05, coefficient 0.58). S. marcescens abundance was significantly increased in individuals with S. marcescens BSI (Fig. 3D; q < 0.0001, coefficient 0.63). Klebsiella oxytoca (fig. S11D) and Klebsiella michiganensis (fig. S11E) were both reduced in the gut before S. marcescens BSI (q < 0.005, coefficient −0.61 and −0.20). Dermabacter hominis was also slightly increased (fig. S11F; q < 0.05, coefficient 0.03). Klebsiella spp. abundance was the only significant feature differentiating those with K. pneumoniae BSI from all others (Fig. 3E; q < 0.05, coefficient 0.75). Similarly, E. coli abundance was significantly elevated in those with E. coli BSI (Fig. 3F; P < 0.01, coefficient 0.75). Therefore, for every BSI-causing organism, we identified an increase in that species, often the only significant difference, before BSI in affected individuals.
Fig. 3. Gut microbiome signature before infection onset differentiates infants with BSI caused by specific organisms versus controls.
Abundance of the BSI-causing species (A) S. aureus, (B) S. agalactiae, (C) E. faecalis, (D) S. marcescens, (E) Klebsiella spp., and (F) E. coli in the gut microbiome before BSI onset shown for those with BSI from that organism versus from all others. Generalized LME models were used with MaAsLin2 and controlled for DOL and participant with Benjamini-Hochberg–corrected false discovery rate q value. ”Yes” indicates bacteremia from that organism, and “No” indicates absence of bacteremia from the listed organism but could include bacteremia from other organisms. Stools after BSI for participants in the “yes” category were not included in this analysis.
For each of the BSI-causing organisms, we calculated the specificity, sensitivity, and positive and negative predictive values (PPV and NPV, respectively) of BSI on the basis of gut relative abundance thresholds of the causative species. For S. aureus BSI, we found that a gut relative abundance of S. aureus above 0.1% had 6% sensitivity, 98% specificity, 35% PPV, and 86% NPV (P < 0.01, Fisher’s exact test; data file S2). A fecal abundance greater than 0.1% had 29% sensitivity, 99% specificity, 52% PPV, and 97% NPV for GBS BSI (P < 0.0001). E. faecalis abundance of >15% was only 5% sensitive but >99% specific for E. faecalis BSI, with 93% PPV and 72% NPV (P < 0.0001). S. marcescens gut microbiome abundance above 8% was 35% sensitive and >99% specific, with a PPV of 78% and NPV of 99% for S. marcescens BSI (P < 0.0001). For K. pneumoniae BSI, we determined these characteristics at 10, 20, 60, and 90% relative abundance thresholds. The sensitivity ranged from 5 to 13%, and specificity ranged from 98 to >99% with PPV of 42 to 88% and NPV of 62 to 93% (all P < 0.0001, Fisher’s exact test). An E. coli relative abundance above 30% was only 4% sensitive but 99.5% specific for E. coli BSI with 64% PPV and 85% NPV (P < 0.0005). Thus, we have demonstrated that relative abundance thresholds are highly specific for future BSI caused by those same species.
Nearly identical BSI strains present in stool before BSI
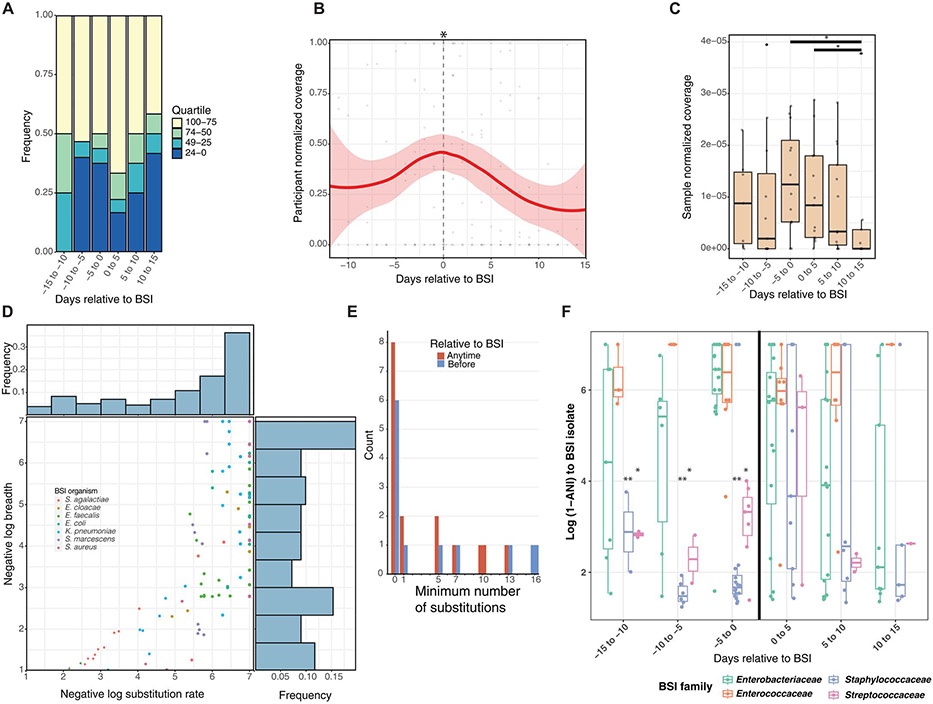
Because of the increased gut relative abundance of the causative species in BSI cases versus controls, and because gut metagenomes clustered according to BSI-causing bacterial family, we investigated whether the BSI species became more abundant in the days preceding BSI. The median species richness within metagenomes of bacteremia cases was 6 to 10 in the 15 days before BSI and did not change before the day of BSI (Fig. 2B). In the 15 days before BSI, in half of cases (4 of 8, 8 of 15, and 8 of 16 within respective 5-day time spans), the future BSI-causing species was in the most abundant quartile within the metagenome (Fig. 4A). At the time of BSI and for the subsequent 5 days, the causative species was in the most abundant quartile in 12 of 18 (67%) of cases. During and after definitive antimicrobial treatment for the BSI organism in the 2 weeks after BSI, the causative species was still in the most abundant quartile in 5 of 12 (42%) of infants. Given these dynamics, we precisely tracked the whole genome of the causative BSI strain in the stool metagenome relative to infection with inStrain (26). We computed the isolate coverage by mapping metagenomic reads and normalized it by read count to allow for cross-cohort comparisons. We found that in the days preceding BSI, the normalized coverage on a scale of 0 to 1 per participant of the BSI isolate within the metagenome increased to an average maximum of 0.48/1 before decreasing once definitive antibiotic treatment was administered (Fig. 4B; LME, P = 0.015). We then determined normalized coverage per metagenomic sample relative to BSI divided by read count for participants who had produced at least one sample with breadth greater than 0.99. In total, 125 of 205 (61%) of metagenomes harbored the cognate BSI isolate at greater than 1× coverage and 0.5 breadth (data file S4). BSI strain coverage was highest in the 5 days before and after BSI, and strain abundance decreased at the end of effective antimicrobial therapy 10 to 15 days after BSI (Fig. 4C; P < 0.05, Wilcoxon). Thus, despite no trends in overall Shannon diversity and richness in case participants before BSI, the coverage of the causative strain within the gut metagenome peaked in the days directly before BSI onset.
Fig. 4. BSI-causing species and strain increases in abundance before BSI.
(A) Rank abundance quartile of the BSI-causing species is shown within the metagenome relative to BSI. (B) Participant-normalized and sample-normalized (C) coverage of the BSI-causing strain increases to a maximum at the day of BSI and declines thereafter. (D) Negative log breadth and substitution rate are plotted by metagenome to same BSI-causing strain with inStrain. Observations below breadth of 0.5 are not displayed. Frequency bar plots represent observations within that bin for negative log substitutions (top) and negative log breadth (right). (E) Minimum number of substitutions if fewer than 20 is plotted for each individual before BSI (blue) or at any time relative to BSI (red). (F) Degree of relatedness as log (1 – population ANI) between BSI strain and metagenome is plotted by family relative to BSI. (B, C, and F) Using LME models with participant as random effect. *P < 0.05.
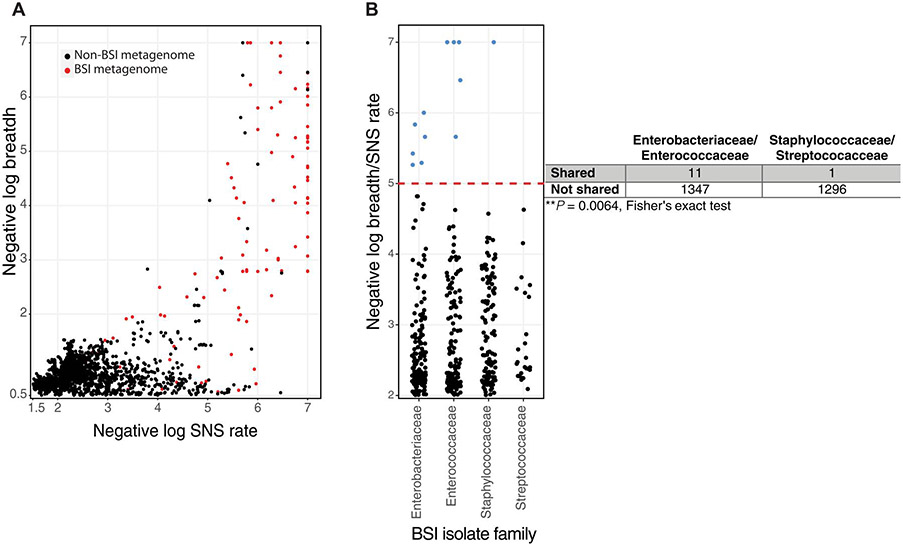
To quantify the genetic relatedness between the BSI isolate and the stool metagenome, we determined the breadth and the number of single-nucleotide substitutions per genome (SNS rate) between each infant’s BSI isolate and their stool metagenome longitudinally using inStrain (26). Figure 4D shows negative log (1 – breadth) and negative log (1 – SNS rate) with a maximum value of 7, where the breadth was 1 and substitutions of 0 in the whole genome. Thus, a y-axis value of 7 signifies that every base pair with sufficient read coverage in the BSI isolate was detected in that cognate metagenome. Among comparisons with breadth >0.5, 45 of 126 (36%) of tested metagenome-BSI isolate pairs had an SNS rate of 0 to 1 (data file S4). Thus, data points in the upper right of Fig. 4D above log breadth of 5 represent those with breadth greater than 0.99999 with 0 to 1 substitutions between their BSI isolate genome and the stool metagenomes over time. We next determined the minimum number of substitutions between the BSI isolate and stool metagenomes before infection or at any time during our sampling interval (Fig. 4E). We found that 11 of 19 (58%) infants produced at least one stool before BSI with fewer than 16 substitutions (Fig. 4D; negative log substitution rate of 5.2 to 7) and at breadth greater than 0.99. This precision confirms that strains present in blood are identical or nearly identical to those in the gut metagenome (26). In addition, the presence of the same strain seemed to correlate with specific bacterial families. Specifically, all Enterobacteriaceae and Enterococcaceae in the stool before BSI had fewer than 16 substitutions at breadth >0.99, whereas only one case with BSI caused by Staphylococcaceae or Streptococcaceae met this threshold (Fig. 4D and E, and data file S4). Although S. aureus or S. agalactiae were not present in the gut before BSI, we found that four infants produced a stool after BSI with these strains harboring fewer than 13 substitutions from the cognate BSI isolate (Fig. 4, D and E). Therefore, the gut may be seeded by the bacteremic event. Thus, we identified most BSI strains in the gut of that same individual before or immediately after BSI.
We then determined the gut and BSI strain similarity by BSI-causing bacterial family using ANI. The genomes of the BSI organisms in our cohort were 2 to 6 Mb, with S. agalactiae being the smallest and K. pneumoniae being the largest (data file S3). Therefore, ANI values of >0.99999 signify fewer than 20 to 50 substitutions between the whole-genome isolate and the metagenome. In the 15 days before infection, individuals with BSI caused by Enterobacteriaceae and Enterococcaceae had significantly higher ANI between the BSI isolate and the stool metagenome than Staphylococcaceae (P < 0.005) and Streptococcaceae (P < 0.01) in a LME model (Fig. 4F and data file S2). Despite effective antimicrobial therapy directed at the BSI pathogen, the same strain with fewer than 20 substitutions at breadth of >0.99 and logANI > 5 (0.99999) persisted in the stool for 6 of 19 (32%) individuals 10 days or more after BSI onset (Fig. 4, E and F, and data file S4). The abundance of the causative strain decreases, but antibiotics do not completely eliminate them, and, often, the causative species still maintains high relative abundance (Fig. 4, A to C). These data demonstrate that, for most cases, the same strain causing BSI is present in the gut microbiome, increases in abundance until BSI onset, and is often not completely cleared during systemic antimicrobial therapy.
BSI strains are shared among infants in the NICU
Environmental surfaces in NICUs are colonized by the same species that are found in gut microbiomes of neonates hospitalized in those NICUs (18, 19, 45). Given this inferred microbial transmission within the NICU environment, we hypothesized that BSI organisms in an infant’s gut could colonize other individuals in the same environment. We mapped metagenomes of sequenced samples from the current cohort and previously sequenced samples from the overall cohort (20, 37) to the BSI isolate genomes using inStrain (26). Most of these mappings (15,554 of 18,356) or 85% resulted in breadth less than 0.5, indicating strain absence in the metagenome. As expected, we found numerous instances of the same or similar BSI strains, with negative log SNS rate > 5 and log breadth > 2 (breadth > 0.99) identified within the same individual over time (Fig. 5A, red dots). We also observed numerous instances of the gut metagenome of unrelated individuals containing a strain causing BSI in an unrelated individual at high breadth with few substitutions (Fig. 5A, black dots). Specifically, we found 19 instances of metagenomes from unrelated children containing reads from the BSI organism at breadth > 0.9 (negative log breadth > 1) and with <50 substitutions (negative log SNS rate > 5). Every one of these 19 comparisons between gut metagenome reads and BSI isolate reads from an unrelated individual shared greater than 0.99999 population ANI establishing identity (26, 46). Among these comparisons, we identified 12 unique participant metagenome-BSI isolate pairs, whereas the other seven observations included the same BSI isolate found at multiple distinct times in the other participant’s metagenomes (data file S5). Only 2 of these 12 events had temporal hospital overlap, whereas the other 10 events were from infants with hospital stays separated by 1 to 36 months. Further, in 5 of 12 of these instances, the BSI case predated the birth of the individual in whom the same strain was later found. These data are consistent with observations in adult hospitals, where identical strains were identified in different individuals over 400 days apart (46). Thus, although a rare occurrence at this stringent level of strain definition, we observed instances of BSI isolates in the gut microbiomes of unrelated individuals, suggesting strain sharing of pathogens in the NICU.
Fig. 5. BSI strains are infrequently shared in the NICU, and sharing differs by causative organism.
(A) InStrain was used to plot BSI isolate reads to metagenomes from the same individual (red) and to all unrelated individuals in the NICU (black). (B) Negative log breadth/substitution rate is plotted by BSI-causing family for each unique non–self-BSI isolate-metagenome pair. Observations above the red dashed line represent negative log breadth/substitution rate above 5 and contain observations with greater than 0.99999 population ANI. Metagenome-BSI mappings below breadth 0.5 were excluded for (A) and (B). Fisher’s exact test for these observations above negative log breadth/substitution rate of 5 compared with all possible nonrelated BSI-metagenome mappings is shown as a two-by-two table.
We hypothesized that because BSI-causing Enterobacteriaceae and Enterococcaceae were more similar to strains within the gut metagenome than were BSI-causing S. aureus and GBS, BSI isolates from these families might be shared more commonly. We plotted the maximum negative log breadth to substitution rate for each unique participant-BSI isolate pair (Fig. 5B). We included observations with negative log breadth/SNS rate > 5 in the numerator with all possible unique participant-BSI isolate mappings in the denominator and restricting to observations with breadth >0.5 to ensure sufficient read-mapping. We found significantly increased likelihoods of BSI isolates from the Enterobacteriaceae and Enterococcaceae identified at high breadth and low SNS rate in the gut metagenome of unrelated individuals versus S. aureus and GBS (P = 0.0064). Because of the overall rarity with which we observed BSI isolates colonizing the guts of other individuals combined with our observations of the same strain increasing in abundance in the gut before BSI, these data support a direct gut-bloodstream or gut-skin-bloodstream translocation route for Enterococcaceae and Enterobacteriaceae that cause BSIs.
DISCUSSION
Late-onset sepsis has devastating consequences, but treating neonates unnecessarily with antibiotics for culture-negative sepsis carries increased aggregate morbidity and mortality, including from future episodes of culture-positive late-onset sepsis and BSI (47). Early identification of at-risk infants can help inform appropriate antibiotic treatment while limiting unnecessary antibiotic use. Other reports have postulated gut-bloodstream overlap in different populations, including in the NICU, with different degrees of genomic precision (22, 31, 46, 48, 49). Here, we precisely track strains at the nucleotide level combined with controls to identify clinical and microbiome correlates of BSI in preterm infants that can be tested in follow-up cohorts and mechanistic models. We found that a gut microbiome signature existed for those individuals who would progress to BSI caused by specific species. We demonstrate that the identical or nearly identical strain causing BSI can be found in the gut microbiome 1 to 2 weeks before late-onset sepsis. In that same interval, we observed increased relative abundance of the cognate species/strain within the gut microbiome without changes in overall richness or diversity.
These findings have several implications for optimizing clinical management to detect and prevent late-onset sepsis in preterm infants. Given the increased abundance of the causative species in cases versus controls and our modeling demonstrating community structure differences between BSI from specific species, gut microbiome profiling could be used to ascribe late-onset sepsis risk to each infant. This finding complements data that show that longitudinal stool volatile profiling could offer late-onset sepsis predictions before onset (50). The possibility of performing rapid sequencing at the start of late-onset sepsis could also help inform antibiotic stewardship and direct more rapid, effective treatment of a causative organism (51).
Our study has several limitations. First, although a direct gut-to-bloodstream translocation is the most likely scenario for Enterobacteriaceae and E. faecalis because of the absence of genomic substitutions between gut metagenome and BSI isolate, we cannot exclude the possibility of short-term cutaneous or tracheal colonization before blood translocation. We note that 5 of 16 case participants from SLCH had no intravenous access or only a peripheral intravenous catheter in the 7 days before BSI. However, on the basis of our identified family-level differences in gut colonization before late-onset sepsis, our data suggest at least two different mechanisms for BSI pathogen origin. This distinction will be especially important in determining the proximate source for coagulase-negative Staphylococcus BSI. These organisms are common gut microbiome and skin colonizers, so distinguishing them as cutaneous contamination or BSI in a blood culture requires clinical adjudication (3, 52). For that reason, we excluded them from this present study. Another important caveat to the family-based differences in pre-bacteremia colonization is the frequency of sampling for certain species. Carl et al. (22) identified a provisionally matched isolate from a single stool, which we did not interrogate here, each from three cases with GBS bacteremia. We did not identify the same strain in pre-BSI stools for GBS, possibly because colonization was transient for these individuals with GBS BSI or because our threshold for same strain declaration (popANI > 0.99999) was stricter. In all three individuals with GBS BSI, popANI between BSI isolate and stool was 0.997 to 0.9999 before BSI, with 13 to 5141 substitutions between BSI isolate and metagenome. Second, we did not include data on maternal prenatal or postnatal antibiotic exposure, which could affect infant microbiome and resistome composition (53). Last, we did not identify a host mechanism of increased susceptibility to BSI between cases and controls, which could be interrogated in clinical studies with serum cytokine or cellular profile measurements and cell culture or animal models.
These data are consistent with other strain-based approaches demonstrating gut colonization before BSI in adults after stem cell transplantation (31) and strain sharing in hospital environments (46). Like those results (31), we did not observe colonization in the gut with S. aureus before BSI apart from one individual. However, in four of six of our cases with S. aureus BSI, the same strain colonized the gut at greater than 20% abundance after BSI. Although the abundance of the causative species and strain decreased during effective antimicrobial treatment, in 30% of individuals, it was still among the most abundant organisms in the gut. For case participants who suffer recurrent BSI from the same species, our data suggest that the gut may be a reservoir for such a relapse (48). These data combined with our observations of transmission events of BSI pathogens are concerning for infection prevention in hospitalized settings. Fortunately, our data demonstrate infrequent sharing of BSI isolates, similar to data recently shown in adults (46). Recent work has also outlined the gut microbiome dynamics in preterm infants (21) showing some intriguing interspecies and interkingdom interactions. Rao et al. (21) found that Klebsiella inhibited Staphylococcus in the gut microbiome, during in vitro growth, and in animal models. We similarly found that enterotype m2 was dominated by Klebsiella and Enterococcus with rare Staphylococcus colonization. Further, we identified an intriguing association that, in individuals with S. marcescens BSI, Klebsiella spp. was essentially absent. A better understanding of how absolute and relative bacterial abundances change in the gut relative to other organisms can help inform risk prediction for late-onset sepsis in the NICU. These data and others provide the framework for longitudinal, selective culturing and/or sequencing to identify neonates at risk of late-onset sepsis (51, 54). The gut microbiome and strain-resolved analyses presented here demonstrate risk factors for BSI caused by specific organisms. Infection prevention interventions can consider these methods to better predict and respond to infections in the NICU.
MATERIALS AND METHODS
Study design
A previously published prospective study was performed from 2009 to 2013 in three Midwestern NICUs to characterize the development of the hospitalized, preterm gut microbiome and its role in necrotizing enterocolitis and other diseases (20, 22, 35-37). Neonates weighing ≤1500 g were enrolled if they were admitted to the NICU and expected to live beyond 1 week of life at SLCH (St. Louis, MO, USA), Children’s Hospital at Oklahoma University Medical Center (Oklahoma City, OK, USA), and Norton (formerly Kosair) Children’s Hospital (Louisville, KY, USA) (55). Stools were collected and, after brief storage at 4°C, stored indefinitely at −80°C until use. Clinical metadata were stored in the REDCap electronic database at Washington University School of Medicine.
Samples and metadata used in this study were previously collected through “the Neonatal Microbiome and Necrotizing Enterocolitis” study funded by National Institutes of Health (NIH) grant UH3AI083265 and approved under Washington University HRPO #201105492 (35-36). This initial study was reviewed by the University of Louisville Institutional Review Board (IRB) (HSPO #11.0136) and University of Oklahoma Health Sciences Center IRB 2472. Each site’s IRB approved the study, and parents were provided informed consent. Secondary analyses as detailed in the current paper were approved under Washington University School of Medicine HRPO #201205152.
Statistical analysis
Analyses were performed using R version 4.0.3, R Studio version 1.4.1103, and GraphPad Prism version 9.3.1. Metadata variables were analyzed with Fisher’s exact (2 × 2 table) or chi-square test (greater than 2 × 2 table) using GraphPad Prism or the R package “stats” version 4.1.1. LME models were performed with the R package “lme4” version 1.1-27.1 with participant ID as the random effect. A post hoc Tukey’s test was performed to identify pairwise differences between groups with the package “emmeans” version 1.7.1.1. MaAsLin2 v1.6.0 was used to determine metadata associations with microbiome features (41). Default parameters were used except that we used arcsine square-root transformation (AST). Participant was always used as a random effect. Significance of principal coordinate analysis (PCoA) of Bray-Curtis dissimilarity was determined using repeated-measures PERMANOVA using the R packages “permute” v0.9-7 and vegan v2.6-2 as previously reported (56). Briefly, the variance and significance of each variable were permuted separately with participant-level blocking due to repeated measures. Two samples for whom feeding data did not exist were excluded from this analysis. Benjamini-Hochberg correction was used to account for multiple comparisons. P < 0.05 and q < 0.25 were considered significant, and all tests were two tailed.
Supplementary Material
Acknowledgments:
We would like to thank S. Fishbein, A. D’Souza, S. Sawhney, R. Thänert, S. Paruthiyil, and members of the Dantas laboratory for suggestions and feedback on the manuscript. We would also like to thank E. Martin, B. Koebbe, J. Hoisington-Lopez, and M.Crosby of the Edison Family Center for Genome Sciences and Systems Biology for computational and sequencing expertise.
Funding:
This research was supported by NIH grants K08 KAI159384 (D.J.S.), R01AI155893 (G.D.), UH3AI083265 (P.I.T.), R01HD092414 (G.D. and P.I.T.), and P30DK052574 (Biobank Core; P.I.T.). D.J.S. was supported by a Thrasher Research Fund Early Career Award, Doris Duke Charitable Foundation Physician Scientist Fellowship grant 2021081, and a Pediatric Infectious Diseases Society St. Jude Children’s Hospital Fellowship in Basic and Translational Research.
Footnotes
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. Shotgun sequencing data, BSI isolate assemblies, and clinical metadata are available in the NCBI repository under BioProject PRJNA884103. Custom code is available at https://zenodo.org/badge/latestdoi/519858068.
Competing interests: P.I.T. is a holder of equity in, a consultant to, and a member of the Scientific Advisory Board of MediBeacon Inc., which is developing a technology to noninvasively measure intestinal permeability in humans. P.I.T. is a coinventor on patents assigned to MediBeacon (U.S. patents 11,285,223 and 11,285,224 titled “Compositions and methods for assessing gut function” and U.S. patent application 2022-0326255, ”Methods of monitoring mucosal healing”), which might earn royalties if the technology is commercialized. P.I.T. receives compensation for his roles as Chair, Scientific Advisory Board of the AGA Center for Microbiome Research and Education, and consultant to Temple University on waterborne enteric infections. He is a member of the Data Safety Monitoring Board of Inmunova, which is developing an immune biologic targeting Shiga toxin–producing E. coli infections, for which he receives no compensation, except for reimbursement of expenses. P.I.T. receives royalties from UpToDate from two sections on intestinal E. coli infections. C.-A.D.B. has consulted for Pattern Bioscience, Cepheid, BioFire, bioMerieux, and Beckman Coulter in the last 5 years, but not presently. G.D. is a cofounder, holder of equity in, a consultant to, and a member of the Scientific Advisory Board of Viosera Therapeutics, which is developing combination antimicrobial therapy against bacterial pathogens. G.D. is a coinventor on a patent assigned to Viosera Therapeutics (“Compositions and methods of use of antibacterial drug combinations,” U.S. Patent Application No. 62/190,588), which might earn royalties if the technology is commercialized. G.D. is a consultant to and a member of the Scientific Advisory Board of Pluton Biosciences, which is developing methods for discovering novel environmental microbes for agricultural and pharmaceutical applications. G.D. has consulted for SNIPR Technologies Ltd. in the last 5 years, but not presently. The authors declare no other competing interests.
REFERENCES AND NOTES
- 1.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE, National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: A systematic analysis and implications. Lancet 379, 2162–2172 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer S, Fleischmann-Struzek C, Machado FR, Reinhart KK, Rowan K, Seymour CW, Watson RS, West TE, Marinho F, Hay SI, Lozano R, Lopez AD, Angus DC, Murray CJL, Naghavi M, Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Y, Speer CP, Late-onset neonatal sepsis: Recent developments. Arch. Dis. Child. Fetal Neonatal Ed 100, F257–F263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shane AL, Stoll BJ, Neonatal sepsis: Progress towards improved outcomes. J. Infect 68, S24–S32 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Boghossian NS, Page GP, Bell EF, Stoll BJ, Murray JC, Cotten CM, Shankaran S, Walsh MC, Laptook AR, Newman NS, Hale EC, McDonald SA, Das A, Higgins RD; Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network, Late-onset sepsis in very low birth weight infants from singleton and multiple-gestation births. J. Pediatr 162, 1120–1124 e1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischmann-Struzek C, Goldfarb DM, Schlattmann P, Schlapbach LJ, Reinhart K, Kissoon N, The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med 6, 223–230 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Hornik CP, Fort P, Clark RH, Watt K, Benjamin DK Jr., Smith PB, Manzoni P, Jacqz-Aigrain E, Kaguelidou F, Cohen-Wolkowiez M, Early and late onset sepsis in very-low-birth-weight infants from a large group of neonatal intensive care units. Early Hum. Dev 88, S69–S74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagstaff JS, Durrant RJ, Newman MG, Eason R, Ward RM, Sherwin CMT, Enioutina DY, Antibiotic treatment of suspected and confirmed neonatal sepsis within 28 days of birth: A retrospective analysis. Front. Pharmacol 10, 1191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reichert F, Piening B, Geffers C, Gastmeier P, Bührer C, Schwab F, Pathogen-specific clustering of nosocomial blood stream infections in very preterm infants. Pediatrics 137, e20152860 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile L-A, Poole WK, Late-onset sepsis in very low birth weight neonates:The experience of the NICHD Neonatal Research Network. Pediatrics 110, 285–291 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Greenberg RG, Kandefer S, Do BT, Smith PB, Stoll BJ, Bell EF, Carlo WA, Laptook AR, Sanchez PJ, Shankaran S, van Meurs KP, Ball MB, Hale EC, Newman NS, Das A, Higgins RD, Cotten CM; Eunice H Kennedy Shriver National Institute of Child and Human Development Neonatal Research Network, Late-onset sepsis in extremely premature infants: 2000-2011. Pediatr. Infect. Dis. J 36, 774–779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlman SE, Saiman L, Larson EL, Risk factors for late-onset health care-associated bloodstream infections in patients in neonatal intensive care units. Am. J. Infect. Control 35, 177–182 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Manouni El Hassani S, Berkhout DJC, Niemarkt HJ, Mann S, de Boode WP, Cossey V, Hulzebos CV, van Kaam AH, Kramer BW, van Lingen RA, van Goudoever JB, Vijlbrief DC, van Weissenbruch MM, Benninga MA, de Boer NKH, de Meij TGJ, Risk factors for late-onset sepsis in preterm infants: A multicenter case-control study. Neonatology 116, 42–51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah J, Jefferies AL, Yoon EW, Lee SK, Shah PS; Canadian Neonatal Network, Risk factors and outcomes of late-onset bacterial sepsis in preterm neonates born at < 32 weeks’ gestation. Am. J. Perinatol 32, 675–682 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Lee C-C, Feng Y, Yeh Y-M, Lien R, Chen C-L, Zhou Y-L, Chiu C-H, Gut dysbiosis, bacterial colonization and translocation, and neonatal sepsis in very-low-birth-weight preterm infants. Front. Microbiol 12, 746111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang S, Yang C, Yang C, Yan W, Shah V, Shah PS, Lee SK, Yang Y, Cao Y; REIN-EPIQ Study Group, Epidemiology and microbiology of late-onset sepsis among preterm infants in China, 2015–2018: A cohort study. Int.J. Infect. Dis 96, 1–9 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Singh N, Patel KM, Leger M-M, Short B, Sprague BM, Kalu N, Campos JM, Risk of resistant infections with Enterobacteriaceae in hospitalized neonates. Pediatr. Infect. Dis.J 21, 1029–1033 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Brooks B, Firek BA, Miller CS, Sharon I, Thomas BC, Baker R, Morowitz MJ, Banfield JF, Microbes in the neonatal intensive care unit resemble those found in the gut of premature infants. Microbiome 2, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks B, Olm MR, Firek BA, Baker R, Geller-McGrath D, Reimer SR, Soenjoyo KR, Yip JS, Dahan D, Thomas BC, Morowitz MJ, Banfield JF, The developing premature infant gut microbiome is a major factor shaping the microbiome of neonatal intensive care unit rooms. Microbiome 6, 112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson MK, Wang B, Ahmadi S, Burnham C-AD, Tarr PI, Warner BB, Dantas G, Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol 1, 16024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao C, Coyte KZ, Bainter W, Geha RS, Martin CR, Rakoff-Nahoum S, Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carl MA, Ndao IM, Springman AC, Manning SD, Johnson JR, Johnston BD, Burnham C-AD, Weinstock ES, Weinstock GM, Wylie TN, Mitreva M, Abubucker S, Zhou Y, Stevens HJ, Hall-Moore C, Julian S, Shaikh N, Warner BB, Tarr PI, Sepsis from the gut: The enteric habitat of bacteria that cause late-onset neonatal bloodstream infections. Clin. Infect. Dis 58, 1211–1218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, Ross MC, Lloyd RE, Doddapaneni H, Metcalf GA, Muzny D, Gibbs RA, Vatanen T, Huttenhower C, Xavier RJ, Rewers M, Hagopian W, Toppari J, Ziegler A-G, She J-X, Akolkar A, Lernmark A, Hyoty H, Vehik K, Krischer JP, Petrosino JF, Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562, 583–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taft DH, Ambalavanan N, Schibler KR, Yu Z, Newburg DS, Deshmukh H, Ward DV, Morrow AL, Center variation in intestinal microbiota prior to late-onset sepsis in preterm infants. PLOS ONE 10, e0130604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Healy DB, Ryan CA, Ross RP, Stanton C, Dempsey EM, Clinical implications of preterm infant gut microbiome development. Nat. Microbiol 7, 22–33 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF, inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol. 39, 727–736 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olm MR, Brown CT, Brooks B, Firek B, Baker R, Burstein D, Soenjoyo K, Thomas BC, Morowitz M, Banfield J, Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 27, 601–612 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olm MR, Crits-Christoph A, Diamond S, Lavy A, Matheus Carnevali PB, Banfield JF, Consistent metagenome-derived metrics verify and delineate bacterial species boundaries. mSystems 5, e00731–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao S, Lieberman TD, Poyet M, Kauffman KM, Gibbons SM, Groussin M, Xavier RJ, Aim DJ, Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe 25, 656–667.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vatanen T, Plichta DR, Somani J, Munch PC, Arthur TD, Hall AB, Rudolf S, Oakeley EJ, Ke X, Young RA, Haiser HJ, Kolde R, Yassour M, Luopajarvi K, Siljander H, Virtanen SM, Ilonen J, Uibo R, Tillmann V, Mokurov S, Dorshakova N, Porter JA, McHardy AC, Lahdesmaki H, Vlamakis H, Huttenhower C, Knip M, Xavier RJ, Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol 4, 470–479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamburini FB, Andermann TM, Tkachenko E, Senchyna F, Banaei N, Bhatt AS, Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat. Med 24, 1809–1814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulman J, Dimand RJ, Lee HC, Duenas GV, Bennett MV, Gould JB, Neonatal intensive care unit antibiotic use. Pediatrics 135, 826–833 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Graspeuntner S, Waschina S, Kunzel S, Twisselmann N, Rausch TK, Cloppenborg-Schmidt K, Zimmermann J, Viemann D, Herting E, Gopel W, Baines JF, Kaleta C, Rupp J, Hartel B, Pagel J, Gut dysbiosis with bacilli dominance and accumulation of fermentation products precedes late-onset sepsis in preterm infants. Clin. Infect. Dis 69, 268–277 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, Shuster J, Sharma R, Hudak ML, Neu J, Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLOS ONE 8, e52876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, Stevens HJ, Bennett WE Jr., Shaikh N, Linneman LA, Hoffmann JA, Hamvas A, Deych E, Shands BA, Shannon WD, Tarr PI, Patterned progression of bacterial populations in the premature infant gut. Proc. Natl. Acad. Sci. U.S.A 111, 12522–12527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warner BB, Deych E, Zhou Y, Hall-Moore C, Weinstock GM, Sodergren E, Shaikh N, Hoffmann JA, Linneman LA, Hamvas A, Khanna G, Rouggly-Nickless LC, Ndao IM, Shands AA, Escobedo M, Sullivan JE, Radmacher PG, Shannon WD, Tarr PI, Gut bacteria dysbiosis and necrotizing enterocolitis in very low birthweight infants: A prospective case-control study. Lancet 387, 1928–1936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, Tarr PI, Warner AB, Dantas G, Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol 4, 2285–2297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotten CM, Taylor S, Stoll B, Goldberg RN, Hansen NI, Sanchez PJ, Ambalavanan N, Benjamin BK Jr.; NICHD Neonatal Research Network, Prolonged duration of initial empirical antibiotic treatment is associated with increased rates of necrotizing enterocolitis and death for extremely low birth weight infants. Pediatrics 123, 58–66 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fajardo C, Alshaikh B, Harabor A, Prolonged use of antibiotics after birth is associated with increased morbidity in preterm infants with negative cultures. J. Matern. Fetal Neonatal Med 32, 4060–4066 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Clark RH, Bloom BT, Spitzer AR, Gerstmann DR, Empiric use of ampicillin and cefotaxime, compared with ampicillin and gentamicin, for neonates at risk for sepsis is associated with an increased risk of neonatal death. Pediatrics 117, 67–74 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Mallick H, Rahnavard A, Mclver LJ, Ma S, Zhang Y, Nguyen LH, Tickle TL, Weingart G, Ren A, Schwager EH, Chatterjee S, Thompson KN, Wilkinson JE, Subramanian A, Lu Y, Waldron L, Paulson JN, Franzosa EA, Bravo HC, Huttenhower C, Multivariable association discovery in population-scale meta-omics studies. PLOS Comput. Biol 17, e1009442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flannery DD, Akinboyo IC, Mukhopadhyay S, Tribble AC, Song L, Chen F, Li Y, Gerber JS, Puopolo KM, Antibiotic susceptibility of Escherichia coli among infants admitted to neonatal intensive care units across the US from 2009 to 2017. JAMA Pediatr. 175, 168–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moehring RAA, Dodds Elizabeth, Developing patient safety outcome measures and measurement tools for antibiotic stewardship programs metric guide (2017). [Google Scholar]
- 44.Zhang L, Huang Y, Zhou Y, Buckley T, Wang HH, Antibiotic administration routes significantly influence the levels of antibiotic resistance in gut microbiota. Antimicrob. Agents Chemother 57, 3659–3666 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brooks B, Olm MR, Firek BA, Baker R, Thomas BC, Morowitz MJ, Banfieid JF, Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat. Commun 8, 1814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siranosian BA, Brooks EF, Andermann T, Rezvani AR, Banaei N, Tang H, Bhatt AS, Rare transmission of commensal and pathogenic bacteria in the gut microbiome of hospitalized adults. Nat Commun. 13, 586 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang S, Yang Z, Shan R, Zhang Y, Yan W, Yang Y, Shah PS, Lee SK, Cao Y; Reduction of Infection in Neonatal Intensive Care Units using the Evidence-based Practice for Improving Quality Study Group, Neonatal outcomes following culture-negative late-onset sepsis among preterm infants. Pediatr. Infect. Dis. J 39, 232–238 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Thanert R, Thanert A, Ou J, Bajinting A, Burnham C-AD, Engelstad HJ, Tecos ME, Ndao HM, Hall-Moore C, Rouggly-Nickless C, Carl MA, Rubin DC, Davidson NO, Tarr PI, Warner BB, Dantas G, Warner BW, Antibiotic-driven intestinal dysbiosis in pediatric short bowel syndrome is associated with persistently altered microbiome functions and gut-derived bloodstream infections. Gut Microbes 13, 1940792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yelin I, Flett KB, Merakou C, Mehrotra P, Stam J, Snesrud E, Hinkle M, Lesho E, McGann P, McAdam AJ, Sandora TJ, Kishony R, Priebe GP, Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat. Med 25, 1728–1732 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deianova N, El Hassani SEM, Niemarkt HJ, Cossey V, van Kaam AH, Jenken F, van Weissenbruch MM, Doedes EM, Baelde K, Menezes R, Benninga MA, de Jonge WJ, de Boer NK, de Meij TG, Fecal volatile organic compound profiles are not influenced by gestational age and mode of delivery: A longitudinal multicenter cohort study. Biosensors (Basel) 10, 50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leggett RM, Alcon-Giner C, Heavens D, Caim S, Brook TC, Kujawska M, Martin S, Peel N, Acford-Palmer H, Hoyles L, Clarke P, Hall LJ, Clark MD, Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat. Microbiol 5, 430–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adeghate JO, Juhasz E, Ivan MA, Pongracz J, Kristof K, Similar strains of coagulase-negative staphylococci found in the gastrointestinal tract and bloodstream of bacteremic neonates. Can. J. Infect. Dis. Med. Microbiol 2020, 3509676 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parnanen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, Virta M, Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun 9, 3891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith A, Saiman L, Zhou J, Della-Latta P, Jia H, Graham PL III, Concordance of gastrointestinal tract colonization and subsequent bloodstream infections with gram-negative bacilli in very low birth weight infants in the neonatal intensive care unit. Pediatr. Infect. Dis. J 29, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warner BB, Tarr PI, Necrotizing enterocolitis and preterm infant gut bacteria. Semin. Fetal Neonatal Med 21, 394–399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beghini F, Mclver LJ, Blanco-Miguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N, Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife 10, e65088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown J, Pirrung M, McCue LA, FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 33, 3137–3139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolger AM, Lohse M, Usadel B, Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmieder R, Edwards R, Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLOS ONE 6, e17288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bushnell B, BBMap: A fast, accurate, splice-aware aligner. Lawrence Berkeley National Laboratory. LBNL Report no. LBNL-7065E; retrieved from https://escholarship.org/uc/item/1h3515gn (2014). [Google Scholar]
- 61.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA, SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM, Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 17, 132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW, CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gurevich A, Saveliev V, Vyahhi N, Tesler G, QUAST: Quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews B, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vazquez-Baeza Y, White RA III; IBDMDB Investigators, Braun H, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C, Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C, High-specificity targeted functional profiling in microbial communities with ShortBRED. PLOS Comput. Biol 11, e1004557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen ALV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire B, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG, CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Sus-ceptibility Testing (Clinical and Laboratory Standards Institute, 2016). [Google Scholar]
- 69.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner B, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh B, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykasenoja S, Olkkola S, Wieczorek H, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM, ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob. Chemother 75, 3491–3500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holmes I, Harris K, Quince C, Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLOS ONE 7, e30126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Souza AW, Boolchandani M, Patel S, Galazzo G, van Hattem JM, Arcilla MS, Melles DC, de Jong MD, Schultsz C; COMBAT Consortium, Dantas G, Penders J, Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers. Genome Med. 13, 79 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.