Abstract
5-Hydroxy tryptamine receptor 1E (5-HTR1E) is reported to activate cAMP and ERK pathways via its ligands and binding partners, but the detailed mechanism underlying the serotonin induced 5-HTR1E signaling is still not known. In the present study, we determined the cellular regulators of ERK and cAMP signaling pathways in response to serotonin induced 5-HTR1E activation in 5-HTR1E overexpressing HEK293 cells. We found that Pertussis Toxin (PTX) treatment completely reversed the effect of serotonin-5-HTR1E mediated signaling on cAMP and ERK pathways, confirming the involvement of a Gαi-linked cascade. We also observed that Gβγ and Gq were not associated with 5-HTR1E activation, while blocking PKA inhibited ERK signaling only, and had no effect on cAMP. Additionally, serotonin-stimulated ERK1/2 phosphorylation was similar in 5-HTR1E overexpressing, β-arrestin-deficient HEK293 cells and is solely dependent on G protein signaling. siRNA mediated gene knockout studies in SH-SY5Y cells revealed that the inhibition of 5-HTR1E reduced the expression of cMyc, Cyclin D1, Cyclin E and BCL2 genes which are related to cell cycle regulation and survival. MTT assays showed that 5-HTR1E knockdown in SHSY-5Y and U118 cells inhibited cell survival significantly. In addition to the signaling mechanism, we also performed RNA-seq analysis in 5-HTR1E overexpressing HEK293 cells and found that 5-HTR1E can regulate the expression of Receptor activity modifying protein 1 (RAMP1), Nuclear receptor 1 (NR4A1) and other Cyclin genes. These findings indicate that serotonin interaction with 5-HTR1E receptor simultaneously activates cAMP and ERK pathway in HEK293 cells and its expression is important for cell survival.
Keywords: GPCR, G proteins, cell signaling, cell survival
Graphical Abstract

5-HTR1E, a Giα linked serotonin receptor can inhibit cAMP pathway idependent of PKA while increasing ERK phosphorylation via PKA/PI3-k activation which is critical for cell survival
Introduction
Serotonin, a monoamine neurotransmitter has thirteen mammalian receptors identified so far. These serotonin receptors are categorized into seven major classes, 5-HT1 to 5-HT7. All serotonin receptors belong to G protein-coupled receptor (GPCR) family except 5-HT3 which is ligand-gated ion channels (1). The largest serotonin receptor family 5-HT1 consists of five members named as 5-HTR1A, 5-HTR1B, 5-HTR1D, 5-HTR1E and 5-HTR1F (2). Several studies done on these 5-HT1 family receptors have shown that they can contribute to various physiological processes like neuronal regulation (3), neuroprotection (4) and ailments like pain, CNS disorders (5, 6), migraine (7) and cancer (8, 9). Despite the enormous work done on 5-HT1 receptors, 5-HTR1E is the least explored serotonin receptor in this family.
Like the other 5-HT1 members, 5-hydroxytryptamine receptor 1-E (5-HTR1E) is also a Gαi-coupled receptor which inhibit cellular cAMP level (1). The 5-HTR1E gene is located on human chromosome 6q14-q15, and does not contain any introns (10). 5-HTR1E is absent in rats and mice but is expressed in guinea pigs, monkeys, and humans (11–14). In terms of function, earlier reports which identified 5-HTR1E as a fifth member of 5-HT1 family were only limited to 5-HTR1E mediated cAMP inhibition. Distribution studies showed the high expression of 5-HTR1E in hippocampus, frontal cortex, and olfactory bulb of guinea pig brain and presumed that it could have a role in memory and learning (14, 15). Since its discovery in 1989, until recently, specific functions of 5-HTR1E largely remain unknown. We recently reported the expression of 5-HTR1E in human hippocampus where it co-localizes with NFα1/CPE. 5-HTR1E can also protect the neurons against oxidative stress via extracellular interaction with NFα1/CPE which is a binding partner of 5-HTR1E (4). In another report, studies in human ovarian cancer SKOV3 cells showed that 5-HTR1E might be able to prevent the stress-promoted progression of ovarian cancer (9). Even after these reports, specific serotonin/5-HTR1E signaling mechanism and downstream functions remain unexplored. In this study, we investigated in detail the signaling mechanism of 5-HTR1E with its natural ligand serotonin (5-HT) in 5-HTR1E overexpressing human embryonic kidney HEK293 cells using various cell and molecular biology assays. Additionally, we also report the effect of 5-HTR1E KO on cell cycle related genes and survival of neuroblastoma SH-SY5Y and glioblastoma U118 cells using as models. To investigate the effect of 5-HTR1E overexpression on gene expression profile, we performed RNA sequencing experiments in 5-HTR1E-HEK293 cells in the presence and absence of serotonin and analyzed the effect of 5HTR1E overexpression and serotonin interaction on many genes related to different pathways.
Materials and method
Cell culture
HEK293 cells stably expressing the human 5-HTR1E were obtained from Dr. Bryan Roth’s laboratory at UNC, North Carolina, USA. These cells carrying human 5-HTR1E cDNA in pcDNA3.0. vector were selected with G418. This cell line was further maintained in DMEM supplemented with 10% serum (FBS) and antibiotics. Human neuroblastoma cell line (SH-SY5Y) was purchased from the ATCC (Manassas, VA, USA) and cultured in Eagle’s minimal essential medium/F12 containing 10% FBS.
5-HTR1E and β-Arrestin Knockdown
SH-SY5Y cells were infected with 40 MOI Ad-5-HTR1E shRNA or control Ad-LacZ (Vector Biolabs) for 72 h in serum free media. U118 cells were transfected with human 50 nM siControl or siHTR1E (sc-42227, Santacruz biotech) using Lipofectamine™ RNAiMAX reagent (Cat. 13778150, Thermo scientific) for 48 h. For β-arrestin knockout, 5-HTR1E stable HEK293 cells were transfected with 50 nM human β-Arrestin1 (sc-29741) and β-Arrestin2 siRNA (sc-29208, Santacruz biotech) for 48 h.
Quantitative real-time PCR (qPCR)
Total RNA was isolated from a 6 well plate using the RNAeasy mini kit (Qiagen). 1μg RNA was reverse transcribed using SensiFAST cDNA Synthesis Kit (cat. no. BIO-65054). Gene specific primers were designed using Primer 3 software and sequence details of these primers are provided in supplementary table 1. PCR reactions were setup in 96 well plates using SYBER green in a Quant Studio (TM) 6 Flex PCR machine (Applied Biosystems, Foster City, CA, USA). qPCR was done with cycling conditions as: 95°C for 10 min, followed by 40 amplification cycles of 95°C for 10 min, 95°C for 15 sec, 60°C for 1 min and 72 °C for 30 s. Threshold values (Ct) were determined by the Quant Studio Real-Time PCR Software and relative mRNA expression was calculated by the comparative ΔΔCt method. Data were normalized to housekeeping gene β-actin, 18S or GAPDH as specified in figure legends.
Western blot
Cells were lysed in Pierce™ IP Lysis Buffer (cat. No. 87787, Thermo scientific, USA) and proteins were quantified using Bradford reagent (BioRad, USA). Total 30 μg proteins were loaded on 4–12% SDS-PAGE gel and transferred onto a nitrocellulose membrane. Blotting membranes were labelled with the protein specific antibodies (details in supple. table 2) and scanned by Odyssey infrared imaging system (Lincoln, NE) and protein bands were visualized by LI-COR Inc software. For the quantification of protein bands Image J software (NIH, Bethesda) was used.
ERK phosphorylation
5-HTR1E stable and HEK293 control cells were seeded in 12 well plate (60–70% confluency) in DMEM media supplemented with 10% FBS and incubated overnight at 37°C in a CO2 incubator. Next day media was changed to serum free medium (SFM) and after 3h. cells were treated with 0 to 1 μM serotonin (5-HT, cat. no. H9523, Sigma-Aldrich) or 5-HTR1E agonist, BRL54443 (cat. no. 1129, Tocris Bioscience) for 4–7 min. For inhibitor experiments, before treatment with 1 μM 5-HT or BRL54443, control and 5-HTR1E expressing HEK293 cells were treated with 200 ng pertussis toxin (Gαi inhibitor, PTX, cat no. 516560, Sigma-Aldrich) for 24 h., 1μm FR900359 (Gq inhibitor, Institute of Pharmaceutical Biology, University of Bonn), 10 μM H-89 (PKA inhibitor, cat. no. 2910, Tocris Bioscience), PI3K inhibitors, 10 μM LY294002 (cat. no. 440202, Sigma-Aldrich) or 0.5 μM Wortmannin (cat. no. W1628, Sigma-Aldrich) for 30 min, 10 μM Gallein (Gβϒ inhibitor, cat. no. 3090, Tocris Bioscience) and 5 μM PD980059 (MEK inhibitor, cat. no. 1213, Tocris Bioscience) for 45 min. Change in pERK was assessed by probing the western blot membranes with pERK1/2 (Thr202/Tyr204) rabbit antibody and tERK1/2 mouse monoclonal antibody simultaneously. Fluorescence labelled anti-rabbit (800 nm) and anti-mouse (680 nm) secondary antibodies (1:10000) were used to visualize the protein bands. pERK was normalized with tERK, and fold change were calculated from three independent experiments after densitometric analysis using image J software, NIH.
cAMP assay
The cAMP assay was performed in 5-HTR1E expressing HEK293 cells. 10,000 cells/well were seeded in a lysine coated 96 well plate in 10% FBS DMEM media. On the next day, cells were incubated with 200 ng PTX (Gi inhibitor) in serum free media for 24 h. or 10 μM H-89 (PKA inhibitor) for 30 min. Before treatment with 1μM 5-HT or BRL54443, these cells were induced with 10 μM forskolin (cAMP activator) in the presence of phosphodiesterase inhibitor (Cat. no. 524718, set I-Calbiochem, Sigma Aldrich, USA) for 15 min. The cAMP assay was performed using cAMP-Glo™ Assay kit (cat. no. V1501, Promega, USA) according to the manufacturer’s protocol and luminescence was recorded on plate reader. All experiments were performed in triplicate and repeated at least three times.
CREB phosphorylation
5-HTR1E stable cells were seeded in 12 well plate (1×105/well) in complete DMEM. On the next day, in SFM media cells were treated with 1μM 5-HT in the presence and absence of 10 μM forskolin for 20 min. Changes in pCREB levels were assessed by western blot using pCREB rabbit antibody against Ser133 and tCREB mouse antibody. Control HEK293 cells were also analyzed for 5-HT mediated CREB phosphorylation.
MTT assay
SHSY-5Y cells were seeded in 35 mm dishes and transduced with 40 MOI AdControl or AdHTR1-shRNA. After 48 h. these cells were seeded in a 96-well plate at a density of 2000 cells/well and treated with or without 1μM 5-HT. For MTT assay in glioma cells, U118 cells transfected with 50 nM 5-HTR1E siRNA (Santa Cruz Biotechnology Inc) or control siRNA for 48 h. and an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed on days 1, 3, 5, and 7. Briefly, 25 μl of MTT reagent (5 mg/ml, Sigma-Aldrich) was added per well and incubated at 37°C in a CO2 incubator for 4h. Supernatant media was removed and 150 μl of DMSO was added to each well. After 5 minutes, the absorbance was measured at 490 nm in a microplate reader (BioTek, Winooski, VT).
LDH assay
5-HTR1E HEK293 cells were seeded in 96 well plate (1×105/well). On the next day, these cells were pretreated with 1μM 5-HT, BRL54443 overnight and then challenged with 300 μM H2O2 for 6h. Cytotoxicity was measured by the amount of LDH released using a CytoTox 96 Non-radioactive cytotoxicity assay kit according to manufacturer’s instruction (Promega, Madison, WI).
RNA-seq analysis
5-HTR1E expressing and control HEK293 cells were seeded in a 6 well plate. On the next morning, cells were serum-starved for 3h. and then treated with 1μM 5-HT for 1h. RNA was extracted from these cells using the RNAeasy mini kit (Qiagen). Poly(A) sequencing libraries were prepared with the Illumina TruSeq stranded mRNA protocol, checking RNA quality with Agilent Technologies 2100 Bioanalyzer. Paired-end, 140-bp reads were sequenced on an Illumina NovaSeq 6000.Paired-end fastq reads underwent quality control checks using FastQC (16), RseQC (17) (18), and MultiQC (19) both before and after trimming reads with cutadapt v3.4 (20) with arguments -q 20 and –minimum-length 25 to perform light quality trimming and retain reads with length > 25 bp following adapter trimming. After confirming all sample libraries passed quality control checks, reads were aligned to the GRCh38 human reference genome using the STAR aligner v2.7.8a (21) with arguments –outFilterType BySJout, --outFilterMultimapNmax 20, --alignSJoverhangMin 8, --alignSJDBoverhangMin 1, --outFilterMismatchNmax 999, --outFilterMismatchNoverReadLmax 0.04, --alignIntronMin 20, --alignIntronMax 1000000, --alignMatesGapMax 1000000 to match ENCODE standard options for long RNA sequencing. Reads were then counted in genes according to the GENCODE release 28 annotation using the featureCounts tool of the subread package v2.0.1 (22) in stranded mode (-s2 argument). Differential expression analysis was conducted on gene counts using DESeq2 v1.34.0 (22). Contrasts were constructed that evaluated differences at the group level, where each group is a combination of the genotype and treatment applied for a given sample (ex: HTR1E-stable_serotonin represents HTR1E-overexpressing cells treated with serotonin). Pairwise comparisons and interaction contrasts were conducted using the model ~group, specifying contrasts using numeric vectors, and using “ashr” log2 fold change shrinkage. Genes were identified as differentially expressed if they had had an adjusted p-value < 0.1. Functional enrichment was performed with the clusterProfiler package v4.2.0. RNA-seq figures were generated using the ggplot2 v3.3.5 package in R v4.1.1.
Results
Gαi linked 5-HTR1E-cAMP pathway is PKA independent
Gαi-linked GPCRs are known to inhibit cAMP levels via protein kinase A (23). Since 5-HTR1E is also a Gαi dependent receptor (24) we investigated the role of PKA in 5-HTR1E mediated cAMP reduction. 5-HTR1E stable cells were pretreated with Gαi inhibitor PTX or PKA inhibitor H-89 which was followed by 1μM 5-HT or BRL54443 treatment, in the presence of forskolin. Results show that both 5-HT and BRL54443 treatment reduced the cAMP levels significantly and this decrease in cAMP was reversed by pretreatment of PTX (p<0.05) (Fig. 1, A1). In H-89 pretreated cells cAMP concentration could not be restored to normal levels and an additional decrease (~20%) was observed in cAMP level as compared to 5-HT (33%) and BRL54443 (43%) treatment alone (p<0.001) (Fig. 1, A2). These results indicate that serotonin-5-HTR1E mediated cAMP pathway is Gαi dependent, but unlike the other classical Gi-protein coupled serotonin receptors (25, 26), 5-HTR1E receptor is not protein kinase A dependent.
Fig: 1. 5-HTR1E-cAMP pathway is Gαi dependent and PKA independent.

5-HTR1E stable cells were incubated with 200 ng PTX (A1) for 24 h. or (A2) 10 μM H-89 (PKA inhibitor) for 30 min. After induction with 10 μM forskolin, cells were treated with 1 μM 5-HT or BRL54443(5-HTR1E agonist) for 20 min. Fold change in cAMP levels were measured by cAMP-Glo™ Assay kit. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test, values are mean ± SD, N=3. Serotonin activates ERK signaling via 5-HTR1E. 5-HTR1E stable cells were treated with up to 1 μM 5-HT or BRL54443 between 4 to 7 min and pERK and pAKT were analyzed by western blotting. Bar graphs showing the Image j quantification of blots as fold change in pERK1/2 (B1–2 and D1–2) and pAKT (C1–2) after normalization with tERK1/2 and tAKT as loading control. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. p values are mean ± SD, N=2. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
Serotonin-5-HTR1E activates ERK pathway
To examine the effect of serotonin on 5-HTR1E mediated ERK and AKT signaling pathways, we treated 5-HTR1E stable HEK293 cells with up to 1μM 5-HT between 4 to 7 min. Fig. 1B shows that 1μM serotonin increased ERK phosphorylation 2.8-fold (p<0.005) (Fig. 1, B1–B2), but no increase was observed in AKT phosphorylation (Fig. 1, C1–C2). To determine if 5-HTR1E agonist BRL54443 (a small molecule like 5-HT, see supple. Fig. S1) could also activate ERK signaling, we treated 5-HTR1E stable cells with up to 1μM BRL54443 and observed that at a lower dose of 10 nM, BRL54443 increased ERK phosphorylation significantly (1.6-fold) and highest increase (~1.9-fold) was achieved at 1μM dose (Fig. 1, D1–D2). While both 5-HT and BRL54443 activated pERK in 5-HTR1E cells, the significant increase with BRL54443 at 10 nM could be attributed to its bias or specificity for 5-HTR1E receptor, as 5-HT (serotonin) showed a significant effect only at a higher dose of 1 μM. This dose dependent effect of serotonin was also checked in control HEK293 cells but no significant increase in ERK1/2 phosphorylation was observed (Supple. fig. S2, A1–A2). Next, we investigated the dose dependent effect of serotonin on SH-SY5Y cells with endogenous 5-HTR1E expression, but the rate and magnitude of 5-HT-induced ERK1/2 activation was considerably less (Supple. Fig. S2, panel B1–B2) in these cells compared to 5-HTR1E stable HEK293 cells. Taken together these results indicate that 1 μM serotonin or agonist (BRL54443) is the optimum dose to activate ERK pathway in 5-HTR1E overexpressing HEK293 cells while AKT pathway remains unaffected by serotonin-5-HTR1E induction in these cells.
Serotonin induced 5-HTR1E/ERK pathway is Gαi dependent
To further explore the involvement of various Gα proteins in serotonin-5-HTR1E induced ERK phosphorylation, we examined the effect of Gαi inhibitor PTX on 5-HTR1E expressing and control HEK293 cells between 4 to 7 min. 1μM 5-HT or BRL54443 induced ERK phosphorylation was completely (above 80%) reversed in cells pretreated with PTX for 24 h. (Fig. 2, A, B) (p<0.05) as compared to the cells not pretreated with PTX (Fig. 2, C1–C2). These data shows that serotonin-5HTR1E mediated ERK activation is activated via Gαi.
Fig: 2. Serotonin-5-HTR1E mediated ERK phosphorylation is Gαi dependent.
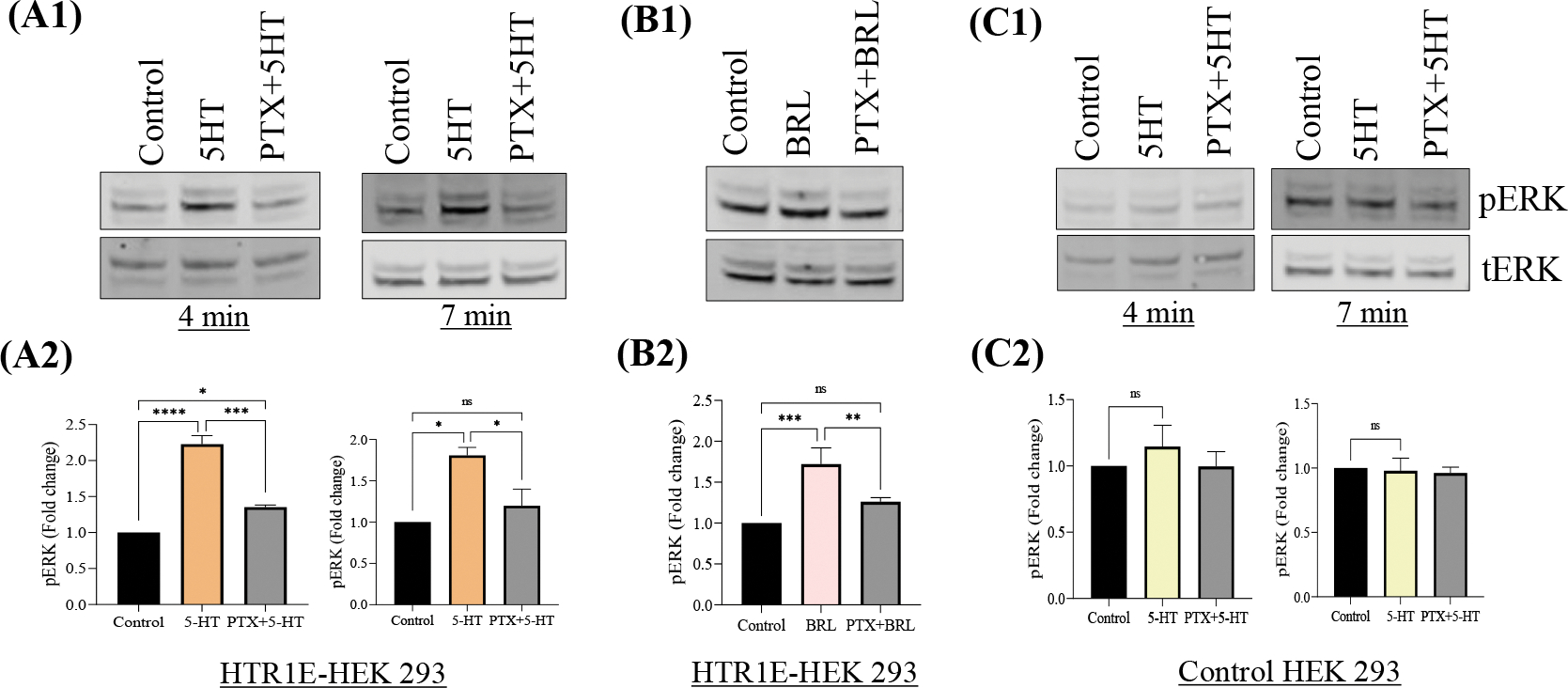
5-HTR1E stable or control HEK293 cells were pretreated with 200 ng PTX (Gi inhibitor) for 24 h. followed by 1μm 5-HT or BRL54443 for 4 to 7 min. Western blot and bar graphs showing the fold change of pERK1/2 in 5-HT (A1–2), BRL54443 (B1–2) and control HEK293 cells (C1–2). Values are mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
Serotonin induced 5-HTR1E/ERK pathway is Gαq independent
To rule out the involvement of Gαq protein in ERK phosphorylation, 5-HTR1E expressing cells were pretreated with 1μM Gαq inhibitor (FR900359) for 30 min. followed by induction with 1μM 5-HT (27). Treatment with Gq inhibitor showed no effect (p=ns) on 5-HT mediated ERK phosphorylation (Fig. 3, A1–A2) which shows that serotonin-5HTR1E mediated ERK activation is Gαq independent.
Fig: 3. 5-HTR1E mediated ERK phosphorylation is independent of Gq or Gβϒ.

5-HTR1E stable cells were pretreated with 1μm FR900359 (Gq inhibitor), 10 μM Gallein (Gβϒ inhibitor) and 5 μM PD98059 (MEK inhibitor) for 30–45 min, followed by 1μm 5-HT or BRL54443 treatment for 4 to 7 min. Western blot and bar graphs showing the effect of Gq inhibitor (A1–2), Gβϒ and MEK inhibitor (B1–2) on pERK level. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. Values mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
Serotonin activated ERK pathway does not involve Gβϒ subunits
In Gαi coupled GPCRs, Gβγ subunits play an important role in the downstream ERK signaling cascade (28–30). To determine the contributions of Gβϒ on Gαi-dependent MAP kinase activation, 5-HTR1E expressing and control HEK293 cells were pretreated with Gβϒ inhibitor Gallein (10 μM) for 30 min or PD980059, MEK inhibitor (5 μM, 45 min) prior to stimulation with 1μM 5-HT. As shown in Fig. 3B1–B2, the Gβϒ inhibitor failed to inhibit 5-HT-stimulated ERK1/2 phosphorylation while MEK inhibitor (positive control) totally blocked this 5-HTR1E mediated effect. Thus, our data suggest that Gβγ subunits are not required for the 5-HT-mediated ERK signaling in 5-HTR1E expressing cells.
Serotonin-5-HTR1E stimulated ERK activity is mediated by PKA
Some of the GPCRs are known to exert their physiological actions through activation of Gi/PKA/PI3-K-dependent ERK signaling (29, 30). To explore the detailed mechanism behind the serotonin induced 5-HTR1E- mediated ERK pathway, we used specific inhibitor against PKA. 5-HTR1E stable HEK293 cells were pretreated with the PKA inhibitor 10μM H-89 before treatment with 5-HT. Results show that H-89 blocked the 5-HT (Fig. 4, A1–A2) or BRL54443 (Fig. 4, B1–B2) stimulated ERK phosphorylation (approx. 80%, p<0.001) in 5-HTR1E overexpressing cells as compared to control HEK293 cells (Fig. 4, C1–C2). These results indicate that 5-HTR1E mediated ERK activation is PKA dependent.
Fig: 4. 5-HTR1E mediated ERK phosphorylation is PKA dependent.

5-HTR1E stable cells were pretreated with 10 μM H-89 (PKA inhibitor for 30 min followed by 1μm 5-HT or BRL54443 treatment for 4 to 7 min. Western blot and bar graphs showing the fold change in 5-HT (A1–2) and BRL54443 (B1–2) induced pERK1/2. (C1–2) Control HEK293 cells. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. Values are mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
PI3-K activation is required for serotonin-5-HTR1E mediated ERK phosphorylation
We then investigated the involvement of PI3K in serotonin induced 5-HTR1E-ERK pathway by using various inhibitors against PI3-K. 5-HTR1E stable HEK293 cells were pretreated with the PI3K inhibitors, LY294002 and wortmannin, prior to stimulation with 5-HT or BRL54443. Both inhibitors showed a limited (~20%, but significant, p<0.05) inhibitory effect against 5-HT (Fig. 5, A1–A2) but when this suppressive effect of PI3-K inhibitors was tested on BRL54443 induced pERK, the inhibition was more potent and 30–40% (p<0.01) decrease was noted in LY294002 or wortmannin pretreated cells (Fig. 5, B1–B2). Since these two 5-HTR1E agonists (5-HT and BRL54443) produce different quantities of a common effect on same receptor active state, this shows their different potency ratios are cell and system independent and BRL54443 is more bias than 5-HT as stated by Kenakin et al., (31). These results indicate that 5-HTR1E mediated ERK activation involves PI3K activation. Similar experiments were also performed in control HEK293 cells (Supple. Fig. S2, C1–C2).
Fig: 5. PI3K mediates serotonin-5-HTR1E induced pERK.

5-HTR1E expressing cells were incubated with 30 μM LY294002 or 0.5 μM wortmannin (PI3K/Akt inhibitor) for 30 min, followed by treatment with 1 μM 5-HT or BRL54443, and effect on pERK was assessed by western blot. Bar graph showing the effect of PI3-K inhibitors on 5-HT (A1–2) and BRL54443 (B1–2) induced pERK. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. Values are mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
Serotonin-5-HTR1E activated ERK pathway is β-arrestin independent
For several GPCRs, β-arrestin 1 and 2 proteins are required for ERK activation (32–34) and previously we also showed that NF-α1/CPE activated 5-HTR1E can increase ERK phosphorylation via β-arrestin recruitment (4). Here we investigated if serotonin-5-HTR1E mediated ERK phosphorylation is also β-arrestin dependent. β-arrestin 1 (Fig. 6, A1 and A2) and β-arrestin 2 (Fig. 6, A1 and A3) were knocked down in 5-HTR1E stable HEK293 cells using siRNA (40–50% reduction, p<0.05) and these cells were then treated with 1μM 5-HT for ERK activation. Serotonin induced ERK 1/2 phosphorylation in 5-HTR1E expressing cells was not affected by β-arrestin 1 and 2 knock down (Fig. 6, A1 and A4). These results indicate that β-arrestin 1 and 2 proteins do not play any role in 5-HTR1E mediated ERK phosphorylation.
Fig: 6. Serotonin-5-HTR1E induced ERK activation is β-arrestin independent.

5-HTR1E expressing HEK293 cells were transfected with 50 μM control or β-arrestin 1 and 2 siRNA. After 48 h. cells were treated with 1 μM 5-HT for 7 min. and changes in β-arrestin 1 and 2 and pERK expression (A1) were analyzed. Bar graphs showing the fold change of expression, β-arrestin 1 (A2) β-arrestin 2 (A3) and pERK1/2 (A4). One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. Values are mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001, p=ns.
5-HTR1E inhibit forskolin stimulated CREB phosphorylation
CREB is a downstream effector of both cAMP and ERK signaling pathways (35, 36) and its phosphorylation is related to various physiological processes via serotonin receptors (37). To further explore the effect of serotonin on 5-HTR1E mediated CREB phosphorylation, we treated 5-HTR1E stable or control HEK293 cells with 1μM 5-HT in the presence and absence of forskolin. After 20 min. of treatment, a significant decrease (31%, p<0.05) was observed in forskolin stimulated CREB phosphorylation (Fig. 7, A1–A2). These data show that serotonin activated 5-HTR1E is a negative regulator of CREB phosphorylation.
Fig: 7. 5-HTR1E inhibit FSK stimulated CREB phosphorylation.

5-HTR1E expressing cells were incubated with 10 μM FSK followed by treatment with 1μm 5-HT for 30 min. pCREB levels were analyzed by western blotting. (A1–2) Bar graphs showing the fold change in pCREB after normalization with tCREB as an internal control. *ATF band which also cross-reacts with this antibody. Lactate dehydrogenase (LDH) cytotoxicity assay. (B) 5-HTR1E expressing HEK 293 cells were treated with 1 μM 5-HT, BRL54443 followed by 300 μM H2O2 treatment for 6h. One-way ANOVA analysis followed by Tukey’s post hoc multiple comparison test. values are mean ± SD, N=3. *p<0.05, **p <0.005, ***p<0.0001, ****p<0.0001.
Effect of serotonin-5-HTR1E signaling on H2O2-induced cytotoxicity
Previously, we have shown that pretreatment with NF-α1/CPE can protect against H2O2-induced cytotoxicity in 5-HTR1E expressing cells (4). Here we sought to determine if such an effect can also occur with serotonin pretreatment. 5-HTR1E stable cells were treated with 1μM 5-HT or BRL54443 for 24 h followed by 300 μM H2O2 treatment for 6 h. Based on the released LDH levels in the media, we observed that 5-HT or BRL54443 pre-treated cells had similar level of cytotoxicity as untreated cells, when challenged with H2O2 (Fig. 7B). Thus, the serotonin-5-HTR1E interaction does not protect HEK293 cells against H2O2-induced oxidative stress.
5-HTR1E regulates the expression of pERK, pAKT and cell cycle genes in SHSY-5Y cells
To determine the effect of 5-HTR1E KO on ERK and AKT phosphorylation in SHSY-5Y cells which have substantial endogenous 5-HTR1E, we used Ad-5HTR1E shRNA to knockdown 5-HTR1E (~60% decrease, Fig. 8, A1–A2). A very little but significant (25%, p<0.001) decrease was observed in both pERK (Fig. 8, B1–B2) and pAkt (Fig. 8, C1–C2) levels in 5-HTR1E KO cells as compared to control. We further investigated the effect of 5-HTR1E KO on early response genes cfos, cJun, cell cycle related genes cMyc, Cyclin D1, Cyclin E and pro survival BCL2. Fig. 8, D1 shows that there was a marked increase in mRNA expression of both cFos (3-fold) and cJun (2-fold) after 5-HTR1E KO while cMyc, Cyclin D1, Cyclin E and BCL2 genes were reduced significantly (p<0.5 to 0.001) (Fig. 8, D2). We also examined the effect of 5-HTR1E KO on the protein levels of these genes and observed that the protein expression was consistent with mRNA data except for BCL2 protein which did not show any reduction (80% decrease in mRNA) after 5-HTR1E knockdown (Fig. 8, E1–E3).
Fig: 8. Role of 5-HTR1E in cell survival.

SH-SY5Y cells were transduced with 40 moi Ad-HTR1E or Ad-Control-shRNA for 72 h. Western blot and bar graphs showing the fold change in the expression levels of HTR1E (A1–2), pERK (B1–2) and pAKT (C1–2). Statistical analysis was performed using student’s t-test, *p<0.0001 in sh-Control vs sh-HTR1E. Data represent mean ± SD from 3 independent experiments. (D1–2) show the fold change in mRNA levels of 5-HTR1E, cMyc, Cyclin D1, Cyclin E, Bcl2, cFos, cJun gene, while representative western blot and bar graph in (E1–3) show the reduced levels of these proteins in SH-SY5Y cells treated with AdHTR1E-shRNA relative to cells treated with AdControl-shRNA. mRNA and protein data were normalized to internal control 18s or β-actin and are presented as mean ± SD, N=3, Effect of 5-HTR1E knock down on SH-SY5Y and U118 cell survival. SH-SY5Y cells (F) were transduced with 40 moi shHTR1E or shControl virus while U118 cells (G1–2) were transfected with 50 nM siHTR1E or siControl. Effect of 5-HTR1E knock down on cell survival was checked but MTT assay over the period of 7 days. *p<0.01, **p <0.001, ***p<0.0001, ****p<0.0001.
5-HTR1E mediates cell survival
Since 5-HTR1E knock-down caused a reduction in cell cycle related genes (Fig 8, D–E), we examined the role of 5-HTR1E in cell survival. The effect of 5-HTR1E knock-down on survival of SHSY-5Y neuroblastoma and U118 glioblastoma cells which express large amounts of 5-HTR1E was investigated by MTT assay. SHSY-5Y treated with Ad-5-HTR1E shRNA reduced the expression of 5-HTR1E significantly (P<0.01) and MTT analysis showed more than 2-fold decrease (p<0.001) in survival in 5-HTR1E knockout cells from day 5 to day 7 (Fig. 8F). In U118 cells treated with siRNA, 5-HTR1E expression was reduced ~65% (Fig. 8, G1). MTT analysis over a 7-day period showed a significant decrease (~3-fold inhibition on day 7) in U118 cell survival after 5-HTR1E knock-down, compared to control cells (Fig. 8, G2). These data indicate that 5-HTR1E plays a key role in promoting survival of SHSY-5Y and U118 cells.
Profiling of 5-HTR1E-regulated gene expression by RNA-seq analysis
To gain further insight into how 5-HTR1E overexpression (Fig. 9A and supple. Fig. S3, A, B) with or without serotonin can induce changes in gene expression profile of HEK293 cells, RNA-seq analysis was performed. 5-HTR1E overexpressing and control HEK293 cells were treated with or without 1μM 5-HT and mRNA was subjected to RNA-seq analysis. 125 genes were found to be up-regulated in 5-HTR1E overexpressing samples at a statistically significant level (1.5 log2FC, FDR<0.1) while 76 genes were down-regulated (Fig. 9B). Modeling the interaction term between overexpression status and 5-HT treatment revealed a total of 229 genes that had a 5-HTR1E-specific response to 5-HT. For example, genes that were unresponsive to 5-HT only when 5-HTR1E was overexpressed tended to be related to innate immune response, chromatin assembly, and clathrin binding. Top genes affected by serotonin interaction in 5-HTR1E overexpressing cells included Acid-sensing ion channel 3 (ASIC3), RNA Component of Signal Recognition Particle 7SL 2 and 3 (RN7SL2 and RN7SL3) lecithin-cholesterol acyltransferase (LCAT), Reticulocalbin-3 (RCN3), Anti-Mullerian Hormone (AMH), Kinesin Family Member C2 (KIFC2,) Tropomyosin 2 (TPM2) which are shown in supple. fig. S4–S5.
Fig: 9. RNA-seq analysis in 5-HTR1E overexpressing HEK293 cells.
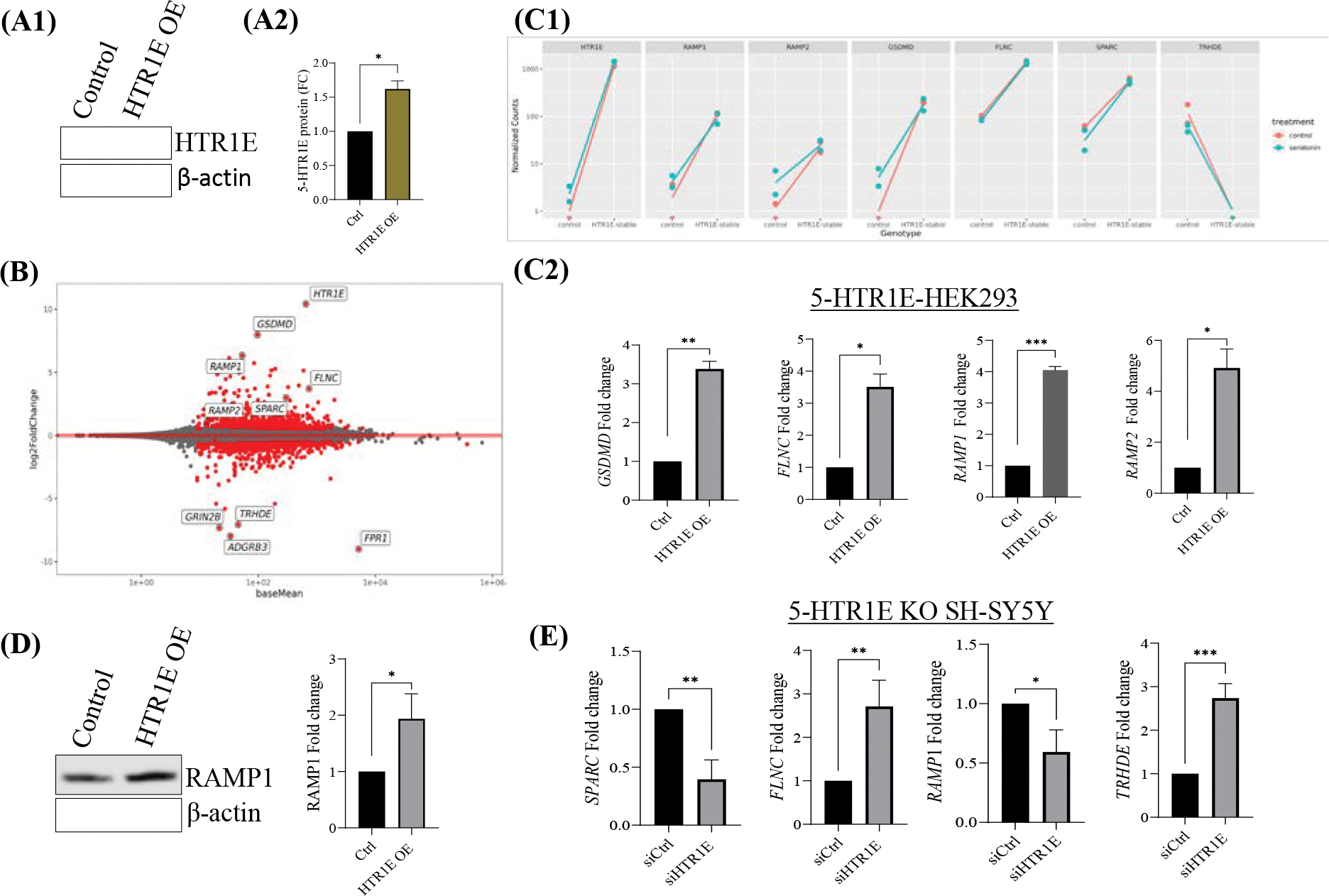
(A) 5-HTR1E overexpression in HEK293 cells. (B) MA plot of RNA-seq analysis showing up and downregulated genes in 5-HTR1E overexpressing in HEK293 cells. Effect of 5-HTR1E overexpression on target genes in (C1) RNA-seq and (C2) qRT-PCR (D) RAMP1 protein expression in 5-HTR1E overexpressing HEK293 cells. (E) Effect of 5-HTR1E knock down on target gene expression in SH-SY5Y cells. Statistical analysis was performed using student’s t-test, mRNA and protein data were normalized to internal control 18s and β-actin. Results are presented as mean ± SD, N=2, *p<0.01, **p <0.001, ***p<0.0001, ****p<0.0001.
RNA-seq library preparation, by necessity, uses a single PCR cycle count for all transcripts but this may not be optimal especially for low-expression genes. Therefore, some of the differentially expressing genes were analyzed by qRT-PCR in 5-HTR1E overexpressing HEK293 and 5-HTR1E KO SHSY5Y cells. Gasdermin D (GSDMD), the second-most upregulated gene after 5-HTR1E itself was increased 7.96 log2FC, in RNA-seq (Fig. 9B) and more than 3-fold in q-PCR (Fig. 9C) analysis when 5-HTR1E overexpressing cells were compared to control. Pore-forming protein Gasdermin D, a member of inflammasome complexes is involved in immune response (38) and the functional enrichment analysis for the interaction term shows evidence of modifying the antimicrobial response (39).
Filamin C (FLNC) which is an actin-binding protein involved in cardiomyopathy (40) was increased by 3.7 log2FC. Secreted protein acidic and rich in cysteine (SPARC) which is also known as Osteonectin binds to calcium in the bone and initiates mineralization and crystal formation (41). SPARC expression was 2.99 logFC up in 5-HTR1E overexpressing cells. Thyrotropin Releasing Hormone Degrading Enzyme (TRHDE) which is a pyroglutamyl- peptidase II enzyme was downregulated −7.063 logFC. 5-HTR1E mediated effect on the expression of these genes is shown in Fig. 9, C–E. Additionally, Receptor activity-modifying proteins, nuclear receptor and cyclins were further studied as described below.
RAMP1 expression is regulated by 5-HTR1E
Receptor activity-modifying proteins (RAMPs) which were originally identified for modifying the activity of Calcitonin receptors, are now known to interact with many GPCRs (42). Our RNA-seq results showed a 6.3-fold (log2FC) (Fig. 9B and C) increase in RAMP1 gene expression in 5-HTR1E overexpressing cells as compared to control HEK293. We examined the effect of 5-HTR1E overexpression on RAMP1 by qPCR and western blot and found a ~ 4-fold mRNA (Fig. 9, C2) and 2-fold increase in RAMP1 protein (Fig. 9, D1–2) in 5-HTR1E expressing cells, as compared to the control HEK293. We also checked the RAMP1 mRNA expression in 5-HTR1E KO SH-SY5Y cells and observed that RAMP1 gene expression was significantly (p<0.05) decreased after 5-HTR1E KO (Fig. 9E). In a reverse experiment, we inhibited RAMP1 expression in SH-SY5Y cells using siRNA and determined if it has any effect on 5-HTR1E. We did not observe any significant change in 5-HTR1E expression (Supple. Fig. S6, A1–2) which suggests that 5-HTR1E is upstream of RAMP1 and can regulate its expression at both gene and protein level.
Serotonin can induce nuclear receptors, Cyclins and Cyclin Dependent kinases via 5-HTR1E
Based on the RNA-seq results, we also tested various genes including Cyclin P and Cyclin L by qRT-PCR. 5-HTR1E overexpressing HEK293 cells were treated with 5-HT or BRL54443 for 90 minutes and expression of NR4A1 (or NURR77), Cyclin P, Cyclin L, CDK10 and CDK11b were examined. In HEK293 cells NR4A1 and CCNP (CNTD2) gene expression was increased almost 2-fold (Fig. 10A) in both 5-HT or BRL54443 treated groups while Cyclin L2 and CDK11b were significantly increased only in BRL54443 groups (Supple. Fig. S6, B1–B2). When SH-SY5Y cells with endogenous 5-HTR1E expression (for mRNA comparison see supple. Fig. S3, C) were treated with 5-HT or BRL54443, expression of NR4A1 mRNA increased up to 3-fold while Cyclin L2 and CDK11b genes were almost 2-fold higher as compared to untreated SH-SY5Y cells (Fig. 10B). These data show that 5-HTR1E can induce the expression of NR4A1, CDK11b, Cyclin P and Cyclin L genes which are involved in various physiological processes (43, 44).
Fig: 10.

(A) mRNA expression of NR4A1 and Cyclin P after 5-HT and BRL54443 treatment in 5-HTR1E overexpressing HEK 293 cells. (B) mRNA expression of NR4A1, Cyclin L2 and CDK11b after 5-HT and BRL54443 treatment. Statistical analysis was performed using student’s t-test, mRNA data were normalized to internal control 18s. Results are presented as mean ± SD, N=2, *p<0.01, **p <0.001, ***p<0.0001, ****p<0.0001. Pathway analysis using ‘GO’ biological process in 5-HTR1E overexpressing HEK 293 cells (C1-C2) without 5-HT (D1-D2) in the presence of 5-HT (serotonin).
Pathway analysis
We also analyzed the pathways changed in response to 5-HTR1E overexpression and serotonin treatment using functional enrichment focusing on the Biological Process (BP) ontology. The pathways enriched in up-regulated genes were related to extracellular matrix and structure organization which also have been reported previously (9) as well as pathways related to axon regulation, connective tissue development and renal absorption (Fig. 10, C1). Pathways enriched in genes down-regulated by 5-HTR1E overexpression were related to embryonic organ morphogenesis and development, pattern specific process and transport across blood brain barrier (Fig. 10, C2). Upon serotonin treatment in 5-HTR1E expressing cells, pathways related to wound healing, glycoprotein metabolism, axon and mesenchymal development were most significantly upregulated (Fig. 10, D1) while down-regulated pathways were mostly related to RNA splicing, RNA metabolism and ribosome biogenesis (Fig. 10, D2).
Discussion
Previously our laboratory reported that 5-HTR1E, a member of the serotonin receptor family, has a robust effect in promoting neuronal cell survival during oxidative and neurotoxic stress via extracellular interaction with CPE/NFα1 (4). In the present study, we investigated the molecular mechanisms associated with the activation of cAMP and ERK signaling cascades by the human 5-HTR1E in response to its natural ligand, serotonin. Our data suggest that serotonin-stimulated 5-HTR1E promotes Gαi activation which in turn inhibit cAMP and stimulate ERK1/2 phosphorylation by two independent signaling mechanisms. 5-HTR1E has a classic cAMP inhibition function like other 5-HT1 family members but the mechanism of this serotonin-5-HTR1E mediated cAMP inhibition had not been studied in detail. Here we report that the effect of serotonin-5-HTR1E on cAMP is through Gαi (Fig. 1, A1), but it does not involve PKA (Fig. 1, A2). Moreover, 5-HTR1E mediated cAMP inhibition is exclusive to its serotonin binding pocket as only 5-HT or BRL54443 showed an inhibitory effect on cAMP but not CPE/NFα1 which interacts with extracellular domains of 5-HTR1E via H-bonds and salt bridges (4). To further explore the effect of 5-HTR1E mediated cAMP inhibition we determined the effect of serotonin on CREB phosphorylation which is a downstream effector of the cAMP pathway (45), and observed that serotonin can reduce the level of forskolin stimulated pCREB significantly (p<0.05) (Fig. 7, A1–A2). These results show that Gαi linked 5-HTR1E can inhibit cAMP and pCREB via serotonin activation.
To analyze the other downstream signaling pathways of 5-HTR1E, we examined the dose dependent effect of 5-HT on 5-HTR1E expressing HEK293 cells at different concentrations and found that 1 μM 5-HT or BRL54443 could induce ERK phosphorylation significantly (Fig. 1). Using specific inhibitors, we found that this serotonin induced ERK activation is Gαi, PKA and PI3K dependent and it does not involve Gβϒ and Gq proteins (Fig. 2 to 5). Previously, we have also shown that the physiological effects of 5-HTR1E stimulation in response to CPE/NFα1 resulted from β-arrestin-dependent ERK activation (4). To evaluate any involvement of β-arrestin in serotonin-induced ERK pathway we used siRNA to knockdown β-arrestin 1/2 protein. We observed that the reduction in β-arrestin expression levels had no impact on 5-HT induced ERK pathway (Fig. 6). Also, the late ERK activity (7–10 min) of CPE/NFα1–5-HTR1E was independent of Gαi (4), while it is much faster in the case of serotonin-5-HTR1E (4–7 min). Moreover, using 5-HTR1E-HEK293 and human neurons, we showed that NF-α1/CPE has no effect on cAMP signaling, but activates ERK pathway via β-arrestin to protect cells against cytotoxicity (4). In contrast, both 5-HT and BRL54443 inhibit cAMP and increase pERK via Gαi, but does not protect cells against oxidative stress, suggesting ligand bias of 5-HTR1E to regulate a specific function. (Fig. 7B). These results show that both serotonin and CPE/NFα1 follow two distinct mechanisms for 5-HTR1E mediated ERK activation. In addition to serotonin induced function in 5-HTR1E overexpressing HEK293, we also found some important effect of 5-HTR1E KO in SHSY-5Y cells which express a high amount of 5-HTR1E. Fig. 8 shows that in 5-HTR1E KO SHSY-5Y cells there was a small but significant decrease in ERK and AKT phosphorylation, which was followed by further reduction in cell cycle related genes cMyc, cyclin D1, cyclin E and pro-survival BCL2. These genes are essential for cell cycle (46) and our results indicate that the expression of 5-HTR1E is important for regulation of these genes/proteins and is crucial for cell cycle. Moreover, MTT assay in 5-HTR1E KO SHSY-5Y and U118 cells showed reduced survival/proliferation of these cells in the absence of 5-HTR1E which highlights its physiological role in these processes.
Interestingly, 5-HTR1E knock-down activated ERK via pSRC in ovarian cancer cells, but inhibited cell proliferation (9) while in HEK293 its overexpression activates ERK in a ligand-dependent manner and is important for survival of SHSY-5Y and U118 cells. All these data and report suggest that 5HT1 receptors have unique coupling mechanism(s) in different cell and tissue types. Our study highlights the 5-HTR1E signaling mechanism specific for serotonin induction in human HEK293, SHSY-5Y and U118 cells.
To gain further insight into the genes and pathways regulated by 5-HTR1E with and without serotonin induction we performed RNA-seq analysis in 5-HTR1E expressing HEK293 cells. Various genes related to important biological processes were changed in response to 5-HTR1E expression (Fig. 9). Notably, Gasdermin D, upon proteolytic cleavage by human caspase-4, triggers activation of caspase-1 in a NLRP3-dependent manner which is important for pyroptosis of innate immune cells. (38). Thus, 5-HTR1E may play a role in enhancing pyroptosis indirectly via increasing GSDMD gene expression, but that will require further investigation. Human RAMP family members are single span transmembrane proteins and can modulate functions of several GPCRs (42). In addition to class B GPCRs like calcitonin receptor, RAMP proteins can also modify the activity and trafficking of class A GPCRs like GPR30 (47). Increased expression of RAMP1 and 2 in response to 5-HTR1E overexpression signals that RAMP proteins might be important for regulation of 5-HTR1E functions. Serotonin treatment increased the expression of CDK 11b, a serine/threonine protein kinases and Cyclin L2, Cyclin P genes which control kinase activity of CDKs (44, 48). NR4A1 which takes part in several biological processes such as cell proliferation, apoptosis (43) was also increased after serotonin treatment. 5-HTR1E induced changes in the expression of several important genes and pathways indicate that serotonin-5-HTR1E might regulate various biological functions. Further exploration of this RNA-seq data will provide detailed information about the specific physiological role of 5-HTR1E in regulation of these pathways.
Conclusions
Our results demonstrated that serotonin stimulation of 5-HTR1E can induce signals in HEK-293 cells by at least two separate pathways: one is Gαi dependent and PKA-independent cAMP pathway (Figure 11A), and another is pERK pathway which is Gαi/PKA/PI3-K-dependent and β-arrestin-independent (Figure 11B). We also found that 5-HTR1E can regulate the expression of various genes and pathways and has a very important role in cell survival.
Fig: 11. Schematic representation of human 5-HTR1E signaling.

Serotonin stimulated 5-HTR1E receptor simultaneously activates cAMP and ERK pathways via Gαi. Upon serotonin treatment 5HTR1E recruits Gαi protein to its intracellular domains and [A] inhibits cAMP levels via inhibition of adenyl cyclase which further downregulates forskolin stimulated pCREB. On the other hand, serotonin-5-HTR1E [B] increases ERK phosphorylation via Gαi/PKA/PI3-K which regulates the expression of cyclin and nuclear receptor genes and plays a role in the survival of various cells.
Supplementary Material
Acknowledgements
We would like to thank Dr. Bryan Roth, UNC, North Carolina for providing 5-HTR1E stable cells and Prof. Evi Kostenis and Prof. Gabriele M. König, Molecular, Cellular and Pharmacobiology Section, Institute of Pharmaceutical Biology, University of Bonn, 53115 Bonn, Germany, for Gq inhibitor FR900359.
Funding
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health, USA.
Footnotes
Declarations: The authors declare no conflict of interest.
Consent for publication: All the authors listed in the article give their consent for the publication of this article.
Availability of data and material
RNA-seq data generated in this study can be found using accession no. GSE223836. The other data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- 1.Nichols DE, and Nichols CD (2008) Serotonin receptors. Chem Rev 108, 1614–1641 [DOI] [PubMed] [Google Scholar]
- 2.Lanfumey L, and Hamon M (2004) 5-HT1 receptors. Curr Drug Targets CNS Neurol Disord 3, 1–10 [DOI] [PubMed] [Google Scholar]
- 3.Rojas PS, and Fiedler JL (2016) What Do We Really Know About 5-HT1A Receptor Signaling in Neuronal Cells? Front Cell Neurosci 10, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma VK, Yang X, Kim SK, Mafi A, Saiz-Sanchez D, Villanueva-Anguita P, Xiao L, Inoue A, Goddard WA 3rd, and Loh YP (2021) Novel interaction between neurotrophic factor-alpha1/carboxypeptidase E and serotonin receptor, 5-HTR1E, protects human neurons against oxidative/neuroexcitotoxic stress via beta-arrestin/ERK signaling. Cell Mol Life Sci 79, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haleem DJ (2018) Serotonin-1A receptor dependent modulation of pain and reward for improving therapy of chronic pain. Pharmacol Res 134, 212–219 [DOI] [PubMed] [Google Scholar]
- 6.Newman-Tancredi A, Depoortere RY, Kleven MS, Kolaczkowski M, and Zimmer L (2022) Translating biased agonists from molecules to medications: Serotonin 5-HT1A receptor functional selectivity for CNS disorders. Pharmacol Ther 229, 107937. [DOI] [PubMed] [Google Scholar]
- 7.Vila-Pueyo M (2018) Targeted 5-HT1F Therapies for Migraine. Neurotherapeutics 15, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karmakar S, and Lal G (2021) Role of serotonin receptor signaling in cancer cells and anti-tumor immunity. Theranostics 11, 5296–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin X, Li J, Wang S, Lv J, Luan F, Liu Y, Chen Y, Chen X, Zhao Y, Zhu J, Piao Y, Zhang W, Shi Y, Xiang R, Qu P, and Wang L (2021) Serotonin/HTR1E signaling blocks chronic stress-promoted progression of ovarian cancer. Theranostics 11, 6950–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy FO, Gudermann T, Birnbaumer M, Kaumann AJ, and Birnbaumer L (1992) Molecular cloning of a human gene (S31) encoding a novel serotonin receptor mediating inhibition of adenylyl cyclase. FEBS Lett 296, 201–206 [DOI] [PubMed] [Google Scholar]
- 11.Leonhardt S, Herrick-Davis K, and Titeler M (1989) Detection of a novel serotonin receptor subtype (5-HT1E) in human brain: interaction with a GTP-binding protein. J Neurochem 53, 465–471 [DOI] [PubMed] [Google Scholar]
- 12.Bai F, Yin T, Johnstone EM, Su C, Varga G, Little SP, and Nelson DL (2004) Molecular cloning and pharmacological characterization of the guinea pig 5-HT1E receptor. Eur J Pharmacol 484, 127–139 [DOI] [PubMed] [Google Scholar]
- 13.Bruinvels AT, Landwehrmeyer B, Gustafson EL, Durkin MM, Mengod G, Branchek TA, Hoyer D, and Palacios JM (1994) Localization of 5-HT1B, 5-HT1D alpha, 5-HT1E and 5-HT1F receptor messenger RNA in rodent and primate brain. Neuropharmacology 33, 367–386 [DOI] [PubMed] [Google Scholar]
- 14.Klein MT, and Teitler M (2009) Guinea pig hippocampal 5-HT(1E) receptors: a tool for selective drug development. J Neurochem 109, 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein MT, and Teitler M (2012) Distribution of 5-ht(1E) receptors in the mammalian brain and cerebral vasculature: an immunohistochemical and pharmacological study. Br J Pharmacol 166, 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andrews S (2010) GitHub Repository. FastQC https://github.com/s-andrews/FastQC. [Google Scholar]
- 17.Wang L, Wang S, and Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 [DOI] [PubMed] [Google Scholar]
- 18.Broad I (2019) Picard Toolkit. GitHub Repository, https://broadinstitute.github.io/picard/ [Google Scholar]
- 19.Ewels P, Magnusson M, Lundin S, and Kaller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M (2011) Cutadapt removed adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12 [Google Scholar]
- 21.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao Y, Smyth GK, and Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 [DOI] [PubMed] [Google Scholar]
- 23.Wang T, Li Z, Cvijic ME, Zhang L, and Sum CS (2004) Measurement of cAMP for G(alphas)- and G(alphai) Protein-Coupled Receptors (GPCRs). In Assay Guidance Manual (Markossian S, Grossman A, Brimacombe K, Arkin M, Auld D, Austin C, Baell J, Chung TDY, Coussens NP, Dahlin JL, Devanarayan V, Foley TL, Glicksman M, Gorshkov K, Haas JV, Hall MD, Hoare S, Inglese J, Iversen PW, Kales SC, Lal-Nag M, Li Z, McGee J, McManus O, Riss T, Saradjian P, Sittampalam GS, Tarselli M, Trask OJ Jr., Wang Y, Weidner JR, Wildey MJ, Wilson K, Xia M, and Xu X, eds), Bethesda (MD) [Google Scholar]
- 24.McAllister G, Charlesworth A, Snodin C, Beer MS, Noble AJ, Middlemiss DN, Iversen LL, and Whiting P (1992) Molecular cloning of a serotonin receptor from human brain (5HT1E): a fifth 5HT1-like subtype. Proc Natl Acad Sci U S A 89, 5517–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Chen L, Li G, Chen X, Hu S, Zheng L, Luria V, Lv J, Sun Y, Xu Y, and Yu Y (2020) Sub-Acute Treatment of Curcumin Derivative J147 Ameliorates Depression-Like Behavior Through 5-HT1A-Mediated cAMP Signaling. Front Neurosci 14, 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang GY, Wang RJ, Guo Y, and Liu J (2022) 5-HT1B receptor-AC-PKA signal pathway in the lateral habenula is involved in the regulation of depressive-like behaviors in 6-hydroxydopamine-induced Parkinson’s rats. Neurol Res, 1–11 [DOI] [PubMed] [Google Scholar]
- 27.Schrage R, Schmitz AL, Gaffal E, Annala S, Kehraus S, Wenzel D, Bullesbach KM, Bald T, Inoue A, Shinjo Y, Galandrin S, Shridhar N, Hesse M, Grundmann M, Merten N, Charpentier TH, Martz M, Butcher AJ, Slodczyk T, Armando S, Effern M, Namkung Y, Jenkins L, Horn V, Stossel A, Dargatz H, Tietze D, Imhof D, Gales C, Drewke C, Muller CE, Holzel M, Milligan G, Tobin AB, Gomeza J, Dohlman HG, Sondek J, Harden TK, Bouvier M, Laporte SA, Aoki J, Fleischmann BK, Mohr K, Konig GM, Tuting T, and Kostenis E (2015) The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun 6, 10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra-Mercado GK, Fuentes-Gonzalez AM, Hernandez-Aranda J, Diaz-Coranguez M, Dautzenberg FM, Catt KJ, Hauger RL, and Olivares-Reyes JA (2019) CRF1 Receptor Signaling via the ERK1/2-MAP and Akt Kinase Cascades: Roles of Src, EGF Receptor, and PI3-Kinase Mechanisms. Front Endocrinol (Lausanne) 10, 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenoy SK, Drake MT, Nelson CD, Houtz DA, Xiao KH, Madabushi S, Reiter E, Premont RT, Lichtarge O, and Lefkowitz RJ (2006) beta-arrestin-dependent, G protein-independent ERK1/2 activation by the beta 2 adrenergic receptor. J Biol Chem 281, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 30.Luttrell LM, Hawes BE, van Biesen T, Luttrell DK, Lansing TJ, and Lefkowitz RJ (1996) Role of c-Src tyrosine kinase in G protein-coupled receptor- and Gbetagamma subunit-mediated activation of mitogen-activated protein kinases. J Biol Chem 271, 19443–19450 [DOI] [PubMed] [Google Scholar]
- 31.Kenakin T (2017) Signaling bias in drug discovery. Expert Opin Drug Discov 12, 321–333 [DOI] [PubMed] [Google Scholar]
- 32.Zhou XE, He Y, de Waal PW, Gao X, Kang Y, Van Eps N, Yin Y, Pal K, Goswami D, White TA, Barty A, Latorraca NR, Chapman HN, Hubbell WL, Dror RO, Stevens RC, Cherezov V, Gurevich VV, Griffin PR, Ernst OP, Melcher K, and Xu HE (2017) Identification of Phosphorylation Codes for Arrestin Recruitment by G Protein-Coupled Receptors. Cell 170, 457–469 e413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain R, Watson U, Vasudevan L, and Saini DK (2018) ERK Activation Pathways Downstream of GPCRs. Int Rev Cell Mol Biol 338, 79–109 [DOI] [PubMed] [Google Scholar]
- 34.Smith JS, and Pack TF (2021) Noncanonical interactions of G proteins and beta-arrestins: from competitors to companions. FEBS J 288, 2550–2561 [DOI] [PubMed] [Google Scholar]
- 35.Arany I, Megyesi JK, Reusch JE, and Safirstein RL (2005) CREB mediates ERK-induced survival of mouse renal tubular cells after oxidant stress. Kidney Int 68, 1573–1582 [DOI] [PubMed] [Google Scholar]
- 36.Li YC, Wang FM, Pan Y, Qiang LQ, Cheng G, Zhang WY, and Kong LD (2009) Antidepressant-like effects of curcumin on serotonergic receptor-coupled AC-cAMP pathway in chronic unpredictable mild stress of rats. Prog Neuropsychopharmacol Biol Psychiatry 33, 435–449 [DOI] [PubMed] [Google Scholar]
- 37.Yue Y, Zhong M, An X, Feng Q, Lai Y, Yu M, Zhang X, Liao Z, Chen M, Dong J, Zhong H, and Shang J (2022) Serotonin (5-HT) 2A Receptor Involvement in Melanin Synthesis and Transfer via Activating the PKA/CREB Signaling Pathway. Int J Mol Sci 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, Liu PS, Lill JR, Li H, Wu J, Kummerfeld S, Zhang J, Lee WP, Snipas SJ, Salvesen GS, Morris LX, Fitzgerald L, Zhang Y, Bertram EM, Goodnow CC, and Dixit VM (2015) Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 [DOI] [PubMed] [Google Scholar]
- 39.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, and Lieberman J (2016) Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S, Shi A, Lian H, Hu S, and Nie Y (2022) Filamin C in cardiomyopathy: from physiological roles to DNA variants. Heart Fail Rev 27, 1373–1385 [DOI] [PubMed] [Google Scholar]
- 41.Rosset EM, and Bradshaw AD (2016) SPARC/osteonectin in mineralized tissue. Matrix Biol 52-54, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pioszak AA, and Hay DL (2020) RAMPs as allosteric modulators of the calcitonin and calcitonin-like class B G protein-coupled receptors. Adv Pharmacol 88, 115–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herring JA, Elison WS, and Tessem JS (2019) Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loyer P, and Trembley JH (2020) Roles of CDK/Cyclin complexes in transcription and pre-mRNA splicing: Cyclins L and CDK11 at the cross-roads of cell cycle and regulation of gene expression. Semin Cell Dev Biol 107, 36–45 [DOI] [PubMed] [Google Scholar]
- 45.Sun C, Wang B, and Hao S (2022) Adenosine-A2A Receptor Pathway in Cancer Immunotherapy. Front Immunol 13, 837230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo J, and Park M (2020) Molecular crosstalk between cancer and neurodegenerative diseases. Cell Mol Life Sci 77, 2659–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lenhart PM, Broselid S, Barrick CJ, Leeb-Lundberg LM, and Caron KM (2013) G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. J Mol Endocrinol 51, 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, Zhao W, Kocak M, Kidd VJ, and Lahti JM (2008) Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J Biol Chem 283, 7721–7732 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), National Institutes of Health, USA.


