Abstract
Efficient delivery of antibacterial agents directly to sites of tissue injury faces challenges such as poor drug stability and fast degradation by biological mechanisms. Biocompatible nanocarrier systems can help sustain and control the delivery of antibacterial compounds while reducing the chances of antibacterial resistance or accumulation in unwanted tissues. In this study, we report the application of tailored polyionic nanoparticles via ionic interactions between negatively charged heparin and positively charge chitosan for efficient encapsulation of polyhexamethylene biguanide (PHMB) antibiotic. Negative zeta potential was required to encapsulate the positively charged PHMB. We demonstrate that the ratio of heparin to chitosan can be employed to create tuned surface charge and maximize the bonding of the drug of choice as well as appropriate particle distribution and uniform morphology. Different formulations were evaluated in terms of size, polydispersity, surface charge and morphology. Out of all these formulations, the best, negatively charged formulation at four-parts heparin to one-part chitosan, was successfully encapsulated with PHMB and showed a sustained and controlled release in vitro for around 10 days. Reduced toxic responses (around 48% reduction) were observed from PHMB-loaded nanoparticles in contact with human dermal fibroblasts as compared to the soluble form of PHMB. Finally, in terms of antibacterial properties, the particles resulted in growth inhibition as well as the direct killing of both Gram-positive (Enterococcus faecalis) and Gram-negative (Escherichia coli) bacterial strains. The minimum inhibitory concentrations required to inhibit bacterial growth were determined by microplate dilution and LIVE/DEAD bacterial evaluation.
Keywords: Antibiotic delivery, Polyionic particles, Polysaccharide nanocarriers, Release kinetics, Wound bacteria
1. Introduction
Polymeric nanoparticles offer numerous applications including controlled release of different drugs or active biomolecules as well as theranostic nanomedicine 1-2. These particles present outstanding advantages, such as size/charge tunability 3, sensitivity to different stimuli (e.g. thermal, pH, light, etc.) 4, excellent ability to bind with and/or absorb biomolecules and drugs, as well as the fact that they can pass through the vasculature to deliver timed payloads or to function as targeted theranostic systems 5. These advantages of nanoparticles are useful to overcome issues associated with drugs/active molecules, such as poor solubility, high toxicity, accumulation in a non-targeted tissue, as well as short half-life resulting in limited bioavailability 5. Additionally, nanoparticles can allow the encapsulated active drug/material to be entrapped and protected from degradation, allowing the payload to safely reach pathological sites and increase therapeutic outcomes with significantly lower dosing required 6.
A variety of natural and synthetic polymers have been used to prepare nanoparticles for controlled-release applications, among which polysaccharides are prominent owing to their excellent biocompatibility, stability and ability to interact with a wide variety of biomolecules. These properties are mainly due to the presence of the hydroxyl, carboxyl and amino functional groups in polysaccharide backbones 1. Heparin is a sulfated polysaccharide 7-8 with a strong anionic structure, owing to the presence of sulfated and carboxylated groups, which provides binding sites for various biomolecules 9. Heparin and its derivates have been widely studied for controlled release of growth factors, such as VEGF 10 and FGF 11, along with cationic drugs. Despite the numerous advantages offered by heparin-based controlled-release systems, its application is often limited by short biological half-life because of rapid release and clearance of some encapsulated agents 10. Heparin’s anionic nature in biological solutions is advantageous for binding to positively charged molecules, but this often results in poor controlled release behavior 9. In order to overcome these drawbacks, as well as to be able to tailor the surface charge of the obtained nanoparticles, researchers have exploited the negative surface charge of heparin in conjugation with cationic polysaccharides, such as chitosan, to form nanoparticles via ionic interactions 9. As a natural linear polysaccharide, chitosan owes its cationic nature in dilute acidic conditions to the availability of structural free amine groups allowing it to interact with anions 12. When conjugated together in a controlled-release platform, heparin and chitosan can synergize to improve encapsulation efficiency, release and bioavailability of different drugs. This conjugate can also be easily shaped into nanoparticles via conventional techniques, such as polyelectrolyte complex formation methods 13. The concept of polyelectrolyte complex formation between oppositely charged polymers has been known and used for decades in different applications, including the development of spherical carriers for drug delivery.
Nanoparticles are of particular interest as antibacterial drug delivery systems, for the treatment of complications, such as osteomyelitis14 or wound infections15. Such systems represent novel strategies of targeted drug delivery to alleviate the increasing antimicrobial resistance crisis, which limits the ability of clinicians to effectively treat bacterial infections, and thus, calling for novel, more efficient delivery strategies 16. Accordingly, the large specific surface area of nanoparticles allows for enhanced interaction with target tissues. Therefore, applying the treatment directly to tissues, rather than delivery via bodily fluids, allows for the same antimicrobial effect at a lower administrated dose 17. PHMB is a broad-spectrum antibiotic with a wide range of activity against bacteria, yeast, fungi and protozoa, with a low toxicity for mammalian cells. Further, due to the net positive charge caused by the biguanide repeating units in biological conditions, PHMB could be a candidate for affinity-based nano-encapsulation using a heparin-based nanocarrier to create an antibacterial controlled release system 18.
The goal of this study was to employ a polyionic complexation approach using chitosan and heparin to create nanoparticles to encapsulate and release PHMB, and subsequently demonstrate their antibacterial activity. As such, we show that polyelectrolyte formation is tunable to provide an efficient antibacterial platform with sustained and controlled release of our antibiotic of choice. More specifically, we demonstrated the ability to tailor the surface charge of our nanoparticles to match the requirements of PHMB, but this in theory could be tuned for any charged molecule. Further, this system successfully demonstrated a controlled and sustained release profile of encapsulated PHMB for around 10 days, while successfully limiting drug toxicity to healthy mammalian cells. Moreover, the HP-CS NPs demonstrated significant antibacterial activity against both the Gram-positive Enterococcus faecalis and Gram-negative Escherichia coli bacteria, which are important pathogens of wound environments.
2. Materials and Methods
2.1. Materials
Highly purified chitosan (90 kDa, 90% deacetylation) was purchased from Mycodev Group (Fredericton, NB, Canada). Heparin sodium salt (17-19 kDa, from porcine intestinal mucosa), sodium bicarbonate and DMSO were purchased from Sigma Aldrich (St. Louis, MO, USA). PHMB (Mw = 185.275 g/mol) was purchased from BOC Sciences (Shirley, NY, USA). Sodium acetate was purchased from VWR (Radnor, PA, USA). Glacial acetic acid was a product of EMD Millipore (Darmstadt, Germany). For all our cell studies, we obtained FBS from Gibco Laboratories (Montgomery County, Maryland, USA). 0.25% trypsin with EDTA was a product of Corning (Corning, NY, USA). High glucose DMEM, Penicillin-Streptomycin and DMSO were all from Sigma Aldrich. PrestoBlue cell viability reagent was purchased from ThermoFisher Scientific (Waltham, MA, USA) to evaluate cytotoxicity responses. For the bacterial experiments, Mueller Hinton broth and agar were both obtained from BD Diagnostic Systems (Sparks, MD, USA). Ampicillin sodium salt was purchased from Sigma. We used the LIVE/DEAD Baclight™ Bacterial Viability kit from Invitrogen (Carlsbad, CA, USA). All polymers, reagents, salts and solvents were used without any further purification. Aqueous solutions were prepared with ultrapure water (18.2 MΩ.cm).
2.2. Preparation of the polyionic heparin-chitosan nanoparticles
The fabrication procedures for heparin-chitosan nanoparticles were adopted from Boddohi et al. 19. Briefly, the polysaccharide solutions were prepared as 0.9 mg/ml of chitosan and 0.95 mg/ml of heparin in 0.1 M acetate buffer at pH 5.0. After complete dissolution, both solutions were filtered using 0.2 μm syringe filters (Whatman, UK) to ensure the complete removal of any dust or undissolved particles. Next, under vigorous stirring of heparin solution (20 ml) at 800 rpm, the chitosan solution was added at a determined ratio using a syringe with an 18G needle. Four different ratios (v/v) of heparin to chitosan were tested (2/1, 3/1, 4/1, 5/1 and 6/1) to determine the best formulation in terms of stability, morphology and polydispersity. The mixture was then allowed to stir for another 3 hours at the same speed at room temperature and was then put aside to settle overnight. The next day, the suspension was decanted, re-dispersed in ultrapure water and centrifuged at 4500 ×g for 20 minutes (Avanti J-26 XPI, Beckman, Brea, CA) 20. This washing step was repeated three times each time by taking the supernatant off, adding more ultrapure water, vortexing briefly and then re-centrifuging. For the plain NPs, the resulting pure suspension of particles was frozen at this step and freeze-dried for 48 hours (FreeZone, Labconco, MO, USA). After freeze-drying the particles formed a white fine powder.
The diameter and surface charge of these nanoparticles were next measured by a zeta and DLS instrument (Zetasizer Nano S90, Malvern, UK). In order to perform DLS and zeta potential measurements, 5 mg of the particles were re-dispersed in 5 ml of ultrapure water and sonicated in an ultrasonic water bath (VWR, PA, USA) for 5 minutes with a power of 590 W right before sizing measurements. To measure the zeta potential, folded capillary zeta cells with electrodes were utilized (Malvern). Standard latex particles (zeta potential transfer standard, Malvern) were used before all measurements to ensure proper calibration of the zetasizer instrument and to confirm accuracy of results.
To encapsulate the NPs with PHMB, NPs were first re-dispersed in a total of 15 ml ultrapure water directly after washing to avoid an extra unnecessary freeze-drying step. A saturated solution of PHMB at a concentration of 10 mg/ml was then prepared in ultrapure water, which was then added to a stirring NP suspension (500 rpm) at a ratio of 15 to 10, NPs to PHMB solution. Stirring was continued for 5 h to maximize binding of PHMB to NPs. The unbound PHMB was then removed by two consecutive washing and centrifugation steps. The supernatant after each washing step was saved to measure the amount of unbound PHMB. Subsequently, to determine the efficiency of drug encapsulation, the unbound PHMB was deducted from the original amount of PHMB to give the amount of PHMB bound to the particles. The ratio of the bound PHMB and the original PHMB content introduced to the mixture was reported as the “encapsulation efficiency”. The particles were then re-dispersed in ultrapure water, freeze-dried for 48 h, and kept in a desiccator at RT to avoid moisture absorbance until further use.
2.3. Morphological assessment of the NPs using TEM
The size and morphology of NPs were confirmed using TEM (JEOL 1230, Tokyo, Japan) with an accelerating voltage of 120 kV. To prepare TEM samples a droplet of nanoparticle suspension at pH = 5 was placed on a carbon-coated copper 200 mesh (Ted Pella, CA, USA) and allowed to dry completely in a vacuum chamber before being imaged.
2.4. PHMB release kinetics
To perform the release studies, 500 mg of PHMB-loaded NPs were re-suspended in 5 ml of PBS (Sigma Aldrich) and transferred into a dialysis membrane (molecular weight cut-off = 12-14 kDa, Spectrum Labs, MA, USA). The dialysis membrane was then sealed and placed in the center of the closed release chamber containing 100 ml of PBS while stirring at 100 rpm to satisfy sink conditions at 37 °C throughout the release experiments. At pre-determined time intervals, a 300 μl volume was collected from the release medium and replaced with fresh PBS. The withdrawn release samples were kept frozen to be measured.
A UV-spectrophotometry method was used to determine the release kinetics of PHMB from the polyionic HP-CS NPs. At the end of the release experiments, all samples were defrosted at RT, vortexed to ensure homogenous mixing, and the concentration of the PHMB in each sample was measured using a UV spectrometer plate-reader (Infinite M200 microplate reader, Tecan, Männedorf, Switzerland). The UV absorbance data was then compared with the prepared PHMB calibration curve to report the concentration of PHMB in each sample.
Furthermore, to investigate the physical mechanism through which PHMB is being released from the NPs, mathematical models of zero order, first order, Korsmeyer-Peppas, Higuchi and Hixson-Crowell kinetic models were applied respectively on the release data, in Equations (1) to (5) at a temperature of 37°C a pH of 7.4 ± 0.2 21.
| (Eq.1) |
where is the amount of drug present in the media at time , is the initial amount of the drug present in the media and is the constant for zero-order release model 22.
| (Eq.2) |
here, represents the amount of drug present in the solution at time and is the release constant for the first-order model.
| (Eq.3) |
where is defined as the fractional release of drug, which is the ratio of the amount of drug released at time and is the total amount of encapsulated drug and is the kinetic constant. value, which determines the release mechanism is the called the release exponent 22.
| (Eq.4) |
where is the dissolution constant for the Higuchi model.
| (Eq.5) |
here, is the initial amount of the drug loaded inside the particles, and is the remaining amount of drug inside the NPs at each timepoint. is the kinetic constant that considers the surface-volume ratio 23.
2.5. FTIR Spectroscopy
FTIR (Thermo Scientific Nicolet 6700 FTIR spectrometer) with DRIFTs sampling technique was performed to further examine the physio-chemical properties of the NPs. FTIR helps to analyze the peak variations on both heparin and chitosan after forming the polyelectrolyte complexes as compared to their plain states. To prepare FTIR specimens, all samples, including the lyophilized nanoparticles, were dispersed as a monolayer and placed directly under the FTIR probe. Each spectrum was obtained by 256 scans at 4 cm−1 resolution at RT.
2.6. In vitro cytotoxicity studies
HDFs used for our cytotoxicity studies were harvested from deidentified human neonatal foreskin acquired from Akron General Hospital (Akron, Ohio) approved by the IRB, as previously described 24. These cells were grown in DMEM supplemented with 10% FBS along with 10% penicillin-streptomycin (5000 U/ml, Gibco, MA) in a 5% CO2 incubator at 37 °C. PrestoBlue assay (Invitrogen) was used to assess viability of cells after 24 hours of being in contact with soluble PHMB vs. PHMB-loaded NPs of different concentrations. Briefly, HDFs were seeded in a 96-well plate at a density of 105 cells/cm2 in normal media and allowed to attach and spread overnight. After aspirating off the media, different concentrations of soluble PHMB (plain) were introduced to the cells. The selected concentrations were 400, 200, 100, 50, 20, 10, 5 and 2 μg/ml of PHMB dissolved in media. A non-treated cell group (NT, regular media only) was also considered along with a negative control of 20% DMSO treated cells. Based on the encapsulation efficiency calculations, the same concentrations of encapsulated and soluble PHMB were used as treatments. Both plates were incubated for 24 h before evaluating the viability results with PrestoBlue according to the manufacturer’s protocol. Media-only blank values were subtracted from all other values and the results were then normalized based on the no-treatment values and reported as viability (%) ratio to non-treated control group 25. To better understand the morphological differences between each treatment group as well as to visualize the proliferation trends, a light microscope (Olympus CKX41 inverted microscope system, Tokyo, Japan) was used to obtain images of all treatments at a magnification of 20X. The wells treated with NPs were washed with PBS three times immediately before imaging to remove the NPs and make it possible to view the attached cells.
2.7. Bacterial cultures
To prepare the bacterial inocula, a single bacterial colony was introduced to 3 ml of TSB for E. coli K-12 (ATCC 25404) and BHI broth for E. faecalis (ATCC 47077) and incubated overnight at 37°C at 100 rpm. E. coli was cultured on tryptic soy agar (TSA) plates at 37°C and E. faecalis was similarly grown on BHI agar 26.
2.8. MIC of the PHMB-loaded NPs
The inhibition of bacterial growth by NPs was examined using a microplate serial dilution assay to obtain the MIC 27-29. Briefly, overnight bacterial cultures of E. coli and E. faecalis were prepared in MHB. The concentration of these inocula was measured by serial dilution on MHA plates. The number of colonies were then counted, and the number of bacteria was then calculated as 1.32×109 CFU/ml for E. coli and 1.04×109 CFU/ml for E. faecalis. For all other treatments, the same number of bacteria was used in the stock culture. Knowing the encapsulation ratio of PHMB to NPs, different concentrations of PHMB-loaded NPs were uniformly dispersed in 4.5 ml of MHB to yield a final starting concentration of 200 μg/ml of PHMB. This stock solution was then serially diluted down to 2 pg/ml. All samples were incubated at 37°C while shaking at 80 rpm to allow the encapsulated PHMB to release out of the NPs. The next day, all samples were inoculated with 500 μl of the E. coli or E. faecalis stock cultures and allowed to incubate for another 24 hours to let the drug act on the bacteria. To calculate the MIC, each inoculated sample of PHMB NPs was then plated on the surface of MHA plates and allowed to sit for 48-72 h at room temperature to let the colonies form. A group of unloaded NPs (plain NPs) was also included to serve as a vehicle control to distinguish between the effects of plain NPs and PHMB on growth inhibition. A group treated with ampicillin sodium salt (with the starting concentration of 10 μg/ml for E. coli and 500 μg/ml for E. faecalis) with serial dilutions was considered as the positive control (AMP). Based on this assay, the MIC is considered as the concentration of sample on the plate with no bacterial growth, while visible growth is seen on the next immediate dilution 30. The reason behind performing the cell counts instead of the conventional UV-spectrophotometry technique was to prevent misinterpretation of the MIC. This is because of the cloudiness of the solutions containing the solid NPs, even before inoculation, which would make it inaccurate to count on the UV-based methods 22.
2.9. LIVE/DEAD bacterial viability assays
To further evaluate the effect of NPs on E. coli and E. faecalis viability, a LIVE/DEAD Baclight™ L7012 bacterial viability assay kit (Invitrogen, Carlsbad, CA) was used according to the manufacturer’s protocol 31. The LIVE/DEAD stained cells were imaged immediately using an Olympus IX81 inverted fluorescent microscope (Center Valley, PA, USA) using a GFP filter to capture the green-fluorescence and CY3 for the red-fluorescence. Multiple images were captured of each prepared sample and overlayed to show both the live and dead population in a single image. For quantification, the images were analyzed in separate channels using color thresholding method on 8-bit images using the particle size analyzer plugin on the ImageJ software (National Institutes of Health, Bethesda, MD, USA) 32-33. This method helped us remove the unspecific background staining and to count the live and dead cells showing up in green and red, respectively. Next, the ratio of the areas covered by either the live or dead populations to the total area covered by cells was calculated for each treatment and reported as a percentage to the total.
2.10. Data analysis
All values are reported as mean ± standard deviation (SD). All statistical analyses were performed using JMP pro 12 (SAS Institute, Cary, NC) software. Either one-way ANOVA with Tukey’s post hoc analysis or Student’s t-tests were performed to detect significant differences between groups with an α level of 0.05.
3. Results
3.1. Preparation and characterization of polyionic HP-CS nanoparticles
Five different formulations of HP-CS NPs were prepared via polyelectrolyte complexation to form polyionic NPs, with heparin as the polymer to which chitosan is added. The reason behind adding the chitosan to the heparin solution (as opposed to adding heparin to chitosan solution) was to obtain a final negative surface charge on the NPs, considering the anionic nature of heparin molecules to facilitate the encapsulation of positively-charged PHMB, via ionic interactions. Different formulations were prepared by increasing the heparin to chitosan ratio from 2/1, 3/1, 4/1, 5/1 to 6/1, to reach the best formulation in terms of size, zeta potential, morphology and PDI.
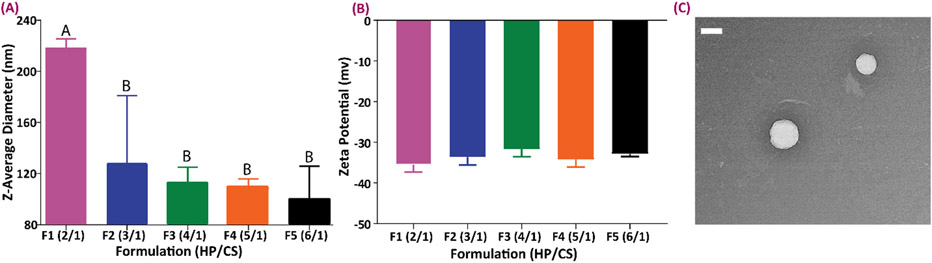
Figure 1 shows the results of the size and surface charge measurements in all formulations. As demonstrated, increasing the amount of heparin in the formulation results in the production of smaller particles. Accordingly, the largest particles belonged to the group with a 2/1 ratio. However, zeta potential measurements demonstrated no significant effects caused by varying the heparin to chitosan ratio in terms of the resulting measured surface electrical charge of the particles (p>0.05), but all formulations resulted in negative zeta potentials. By comparing all the formulations studied, the F3 formulation (4 to 1 heparin to chitosan ratio), showed a monomodal distribution with the PDI of 0.20 ± 0.01, confirming the narrow size spread of the particles. In all other formulations the PDI values were higher than 0.4 and thus considered unfavorable. This formulation was therefore selected as the best formulation to use for PHMB loading and all subsequent experimentation. Fig. 1C shows a TEM micrograph of representative nanoparticles made with this formulation before drug loading. The uniform, non-agglomerated and spherical morphology of these particles validated our choice of the formulation 34.
Figure 1.
Size and zeta potential characterization of the NPs with different formulations. (A) Size variations in particles with different HP to CS ratio. (B) Zeta potential characterization of different nanoparticle formulations. All the DLS and zeta potential measurements were done using 10 acquisitions of six sample, a laser intensity of 850 nm and the fixed angle of 90° was used at RT. Data all n = 6, mean ± SD. Different letters indicate statistically significant differences between groups as determined by one-way ANOVA with Tukey’s post hoc (p < 0.05). (C) Representative TEM image of NPs from the F3 formulation (scale bar = 200 nm).
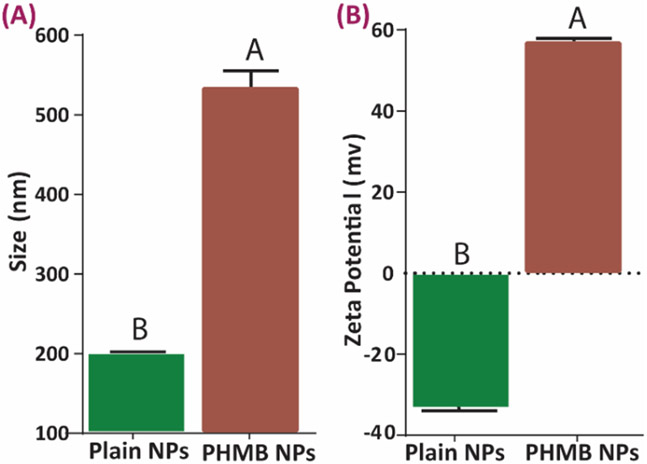
Achieving a negative surface charge is critical to enhancing the encapsulation of cationic molecules, such as PHMB. The results of the size and zeta potential comparison of the NPs before and after PHMB loading are shown in Figure 2. The notable increase in the size of the nanoparticles (Fig. 2A) for PHMB NPs confirms encapsulation. Also, good particle distribution with no agglomeration was observed when PHMB was added to the nanoparticles (PDI after loading = 0.14 ± 0.05). Surface charge further confirms loading (Fig. 2B) as the negative zeta potential value of the plain NPs flipped to a positive value after loading with PHMB indicating a strong positive surfaces charge on the loaded particles. In addition, PHMB encapsulation efficiency was calculated as 75.0 ± 5.2%.
Figure 2.
Size and zeta potential comparison of the PHMB-loaded and plain nanoparticles. (A) size comparison in the formulation with HP/CS ratio of 4/1 before and after loading, (B) zeta potential variations before and after drug loading. Data all n = 3, mean ± SD Different letters indicate statistically significant differences between groups as determined by pairwise t-tests (p < 0.05).
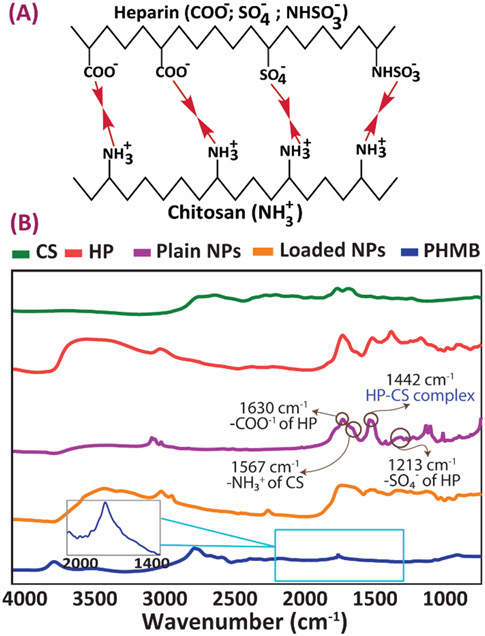
FTIR spectra confirm the successful formation of polyelectrolyte complexes between heparin and chitosan molecules, as well as PHMB incorporation (Fig. 3). Plain HP-CS NPs gave a sharp FTIR peak at 1630 cm−1 attributed to the anionic carboxyl groups (-COO−) of heparin (Fig. 3B). The -SO4− groups of heparin were also observed at 1213 cm−1. The characteristic peak present at 1560 cm−1 was caused by the cationic amine groups (-NH3+) of chitosan 34. Apart from the polyelectrolyte complex peaks, all other spectral peaks were also present plain heparin and chitosan constituents. Importantly, the peak observed at 1442 cm−1 confirms complexation via electrolyte interactions between the ionic groups of heparin and chitosan, which resulted in the formation of a carboxyl-amine salt as shown in Fig 3A 35. PHMB demonstrates a sharp characteristic peak at 1660 cm−1 (via the imine groups), which is also present in the corresponding region of PHMB-loaded NPs spectrum, but has partially overlapped with a chitosan and heparin characteristic peak 18, 36-37.
Figure 3.
FTIR characterization of the NPs before and after drug loading (A) Expected chemical bonds forming between heparin and chitosan as a result of the formation of the polyelectrolyte complex. (B) Corresponding FTIR spectra of chitosan, heparin and PHMB, as well as the NPs before and after loading.
3.2. Release kinetics of PHMB from the polyionic HP-CS nanoparticles
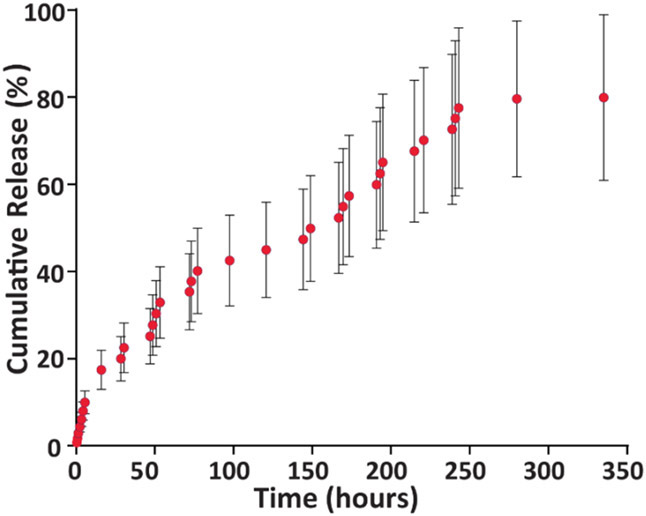
To evaluate the release kinetics of PHMB from HP-CS NPs, a 14-day release study was conducted. The cumulative release profile of 500 mg of particles containing 150 mg of PHMB at 350 h is presented in Figure 4. When bound to the nanoparticle network PHMB shows a sustained release profile with a uniform trend up to a minimum period of 10 days. Afterward, the release reached an equilibrium and remained constant up to the study endpoint.
Figure 4.
PHMB release profile from the HP/CS NPs over two weeks into PBS (n = 3, mean ± SD). Amount of PHMB per sample used for release studies equaled 150 mg.
The results of the kinetic modeling of drug delivery from our system is shown in Table 1 for each of the mathematical models. In regards to the Korsmeyer-Peppas model, only the portion of the release curve with values of less than 0.6 were used to calculate the exponent 21, 38. Based on these results, among all the tested models, the highest coefficient of determination () belongs to the Higuchi model, which indicates that the release of PHMB from the matrix is mostly based on a Fickian diffusion mechanism and is dependent on the square root of time 21, 39.
Table 1.
Kinetic release variables derived from mathematical models to describe PHMB release from the nanoparticles
| Model | Constant | Value |
|---|---|---|
| Zero Order | K 0 | 0.27 |
| R 2 | 0.92 | |
| First Order | K 1 | 0.002 |
| R 2 | 0.98 | |
| Korsmeyer-Peppas | n | 0.7 |
| K | 1.99 | |
| R 2 | 0.97 | |
| Higuchi | K H | 5.02 |
| R 2 | 0.99 | |
| Hixon-Crowell | K s | 0.007 |
| R 2 | 0.97 |
3.3. Effects of soluble versus encapsulated PHMB on cytocompatibility
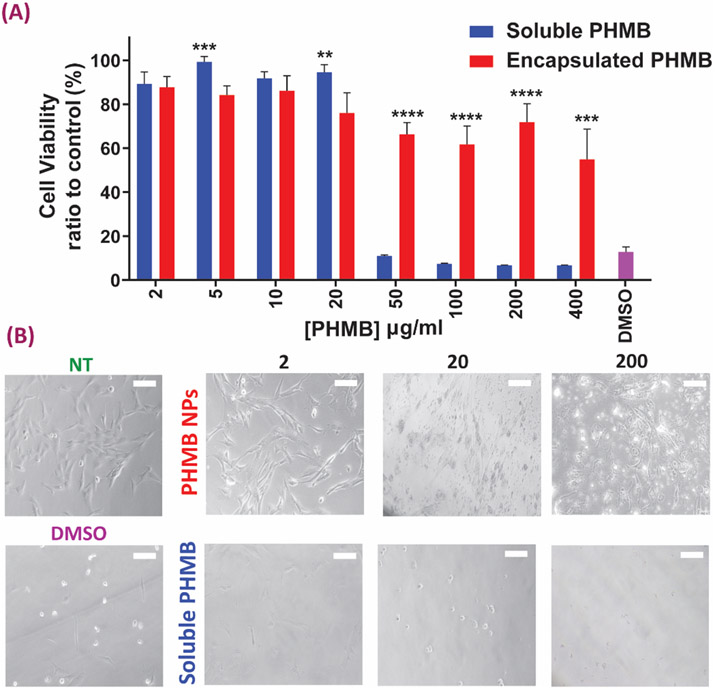
To evaluate the effect of PHMB concentration on HDF cell viability, we compared soluble PHMB to PHMB released from NPs of different concentrations. The main objective of this experiment was to study potential cell toxicity responses to PHMB-loaded NPs and investigate whether or not the encapsulated NPs are capable of lowering the cytotoxic responses as compared to the soluble form of PHMB. Figure 5A shows 8 different PHMB concentrations as well as their equivalent loaded NP concentrations (400, 200, 100, 50, 20, 10, 5 and 2 μg/ml) compared to a no-treatment (media only) control and a known cytotoxic control of 20% DMSO in a 24-hour cytotoxicity assay via PrestoBlue. Representative light microscopy images of some treatments in both NP and soluble PHMB groups (2, 20 and 200 μg/ml) are shown in Figure 5B, to help better visualize morphological differences as well as potential proliferation trends in both groups as compared to controls.
Figure 5.
Cytotoxicity evaluation of the NPs with different concentrations plain vs. loaded (A) Results of cell viability after soluble and encapsulated PHMB treatment of human dermal fibroblasts (HDFs). Data all n = 6, mean ± S.D. Asterisks denote significance as determined by pairwise t-tests with ** p = 0.01, *** p < 0.001 and **** p <0.0001 . (B) Morphological changes of HDF cells after soluble/encapsulated PHMB exposure for 24 h (scale bars = 50 μm). NT indicates the no-treatment media only control group.
Results showed that for all PHMB doses higher than 20 μg/ml, the encapsulated group resulted in lower toxicity than the soluble PHMB group. Importantly, differences between the two groups increased with the concentration of PHMB in the media. In terms of cell spreading and morphology, trends similar to cytotoxicity assay are seen in the microscopy images as well, where lower cell numbers were observed in the group treated with soluble PHMB at all doses. Notably, the bright dots in the PHMB NPs group are the HP-CS nanoparticles still remaining in the well even after three washes. Importantly, presence of dead cells or cells with irregular morphologies are lower in the PHMP-NP group as compared to corresponding free PHMB treatments. Also, proliferation is visibly improved in the encapsulated groups, especially at the lower concentrations.
3.4. Antibacterial performance of the polyionic HP-CS nanoparticles
To obtain MIC values for the HP-CS NPs, stock cultures of E. coli and E. faecalis with 1.32×109 and 1.04×109 CFU/ml were used, respectively. In Table 2, the ‘+’ sign indicates that growth was observed in the corresponding concentration, while the ‘−’ sign indicates no growth. Also, as our positive control we used ampicillin with the MIC value equal to 10−7 μg/ml for E. coli and 50 μg/ml for E. faecalis. From the results presented in Table 2, PHMB-loaded HP-CS NPs showed stronger inhibitory activity against E. coli K12 resulting in an MIC of 0.2 μg/ml after 24 hours, which is lower than the concentration of the PHMB-loaded NPs required to inhibit the proliferation of the E. faecalis strain (MIC = 2 μg/ml). As expected, the particles with no PHMB (plain NPs) showed significant bacterial growth agar surface at all dilutions indicating no inhibitory effects. This supports the fact that the PHMB loaded inside the NPs is responsible for providing the antibacterial effect.
Table 2.
Microplate dilution results of different samples against E. coli and E. faecalis
| Sample | Concentration (μg/ml) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 200 | 20 | 2 | 2×10−1 | 2×10−2 | 2×10−3 | 2×10−4 | 2×10−5 | 2×10−6 | |
| Plain NPs against E. coli | + | + | + | + | + | + | + | + | + |
| Plain NPs against E. faecalis | + | + | + | + | + | + | + | + | + |
| PHMB- NPs against E. coli | − | − | − | − | + | + | + | + | + |
| PHMB- NPs against E. faecalis | − | − | − | + | + | + | + | + | + |
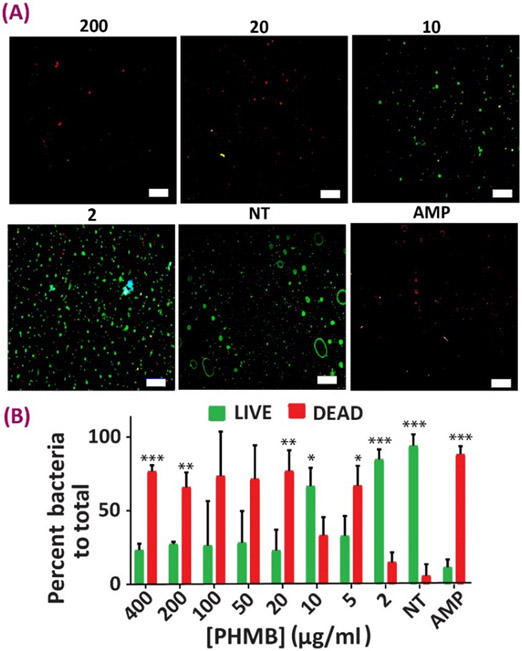
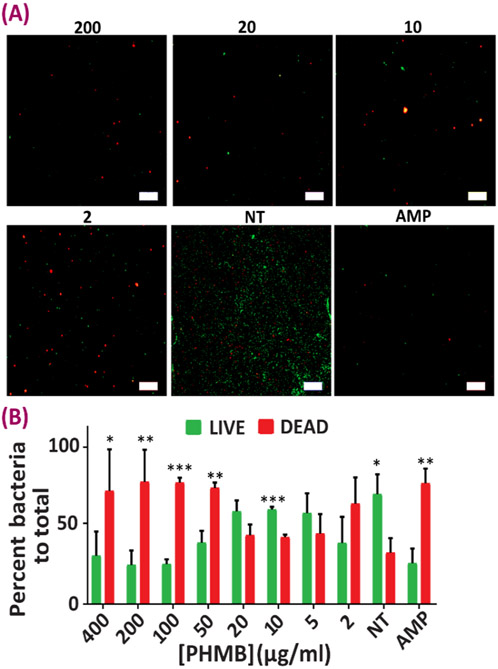
Subsequently, the interaction between the bacteria and the antibacterial NPs was studied via a LIVE/DEAD experiment (Figs. 6, 7). This assay helps to further study whether there is an effect beyond bacterial growth inhibition associated with the HP-CS NPs, such that these antibacterial carriers are also capable of destroying the selected flora. Experiments were carried out both in the presence and absence of NPs (NT) and also against ampicillin sodium (AMP), as a known antibacterial agent to determine the cell viability of and Gram-positive and Gram-negative cultures. In both E. coli and E. faecalis groups, the MIC obtained from the previous method is confirmed as there were lower numbers of live cells as compared to dead ones in those NP concentrations. LIVE/DEAD images were quantified using image analysis to show the percentage of the area covered by green (live) vs. red (dead) in each treatment group (Figs. 6B, 7B). Substantial bactericidal activity was witnessed after 24 h of bacterial incubation with the HP-CS NPs incapsulated with PHMB as compared to the NT group.
Figure 6.
Antibacterial evaluation of the NPs against E.coli. (A) Representative images from Live-Dead analysis of E.coli after being in contact with different nanoparticle concentrations for 24 hours (live cells are stained green and dead cells are showing up as red). (B) Quantification analysis results of Live-Dead analysis of E.coli after being in contact with different nanoparticle concentrations for 24 hours. Data all n = 4, mean ± SD. Asterisks denote significance as determined by pairwise t-tests with * showing p < 0.05, ** p < 0.01 and *** p < 0.001.
Figure 7.
Antibacterial evaluation of the NPs against E. faecalis. (A) Representative images from Live-Dead analysis of E. faecalis after being in contact with different nanoparticle concentrations for 24 hours (live cells are stained green and dead cells are showing up as red). (B) Quantification analysis results of Live-Dead analysis of E. faecalis after being in contact with different nanoparticle concentrations for 24 hours. Data all n = 4, mean ± SD. Asterisks denote significance as determined by pairwise t-tests with * showing p < 0.05, ** p < 0.01 and *** p < 0.001.
4. Discussion
In the past decade significant effort has focused on developing natural nanomaterials for delivering antibacterial agents 40-42. These nanocarriers are beneficial due to their inherent biocompatibility, as well as their promising abilities to encapsulate and protect the loaded antibacterial drugs from destructive mechanisms, such as oxidation, hydrolysis and enzymatic degradation, followed by delivering them in controlled doses to a targeted area 43. These nanoparticles have been used as anti-infective platforms in several applications. To highlight a few of these reports, Saidy khan et al. 44 have developed a nanoantibiotic system based on aragonite NPs loaded with vancomycin antibiotic for the treatment of osteomyelitis with the ability to sustain the antibiotic release for 5 days showing antibacterial properties against methicillin-resistant Staphylococcus aureus . As in our study, Chen et al. 45 used a polyelectrolyte complex wound dressing based on chitosan and alginate incorporated with gelatin microspheres to control the release of a tetracycline hydrochloride antibiotic. This antibiotic wound treatment was able to inhibit the growth of E. coli and S. aureus. Based on these applications and importance, we report the development of a tunable natural antibacterial delivery platform using HP-CS NPs, which notably reduces cytotoxic effects on mammalian cells.
In this study, heparin-chitosan nanocarriers were prepared by polyelectrolyte complexation, which occurs due to the electrostatic interactions between the positively charged amine groups of chitosan with the abundant sulfated and carboxylated groups present in the heparin backbone. However, not all environmental conditions are supportive of polyelectrolyte complexation to form spherical nanoparticles using heparin and chitosan 34. Specifically, Lin et al. demonstrated that a pH range of 4.5-6.5 is favorable for forming stable nanoparticles, whereas above this range large aggregates are formed instead. They attributed this phenomenon to the deprotonization of chitosan molecules at pH 7.0 causing nanoparticle instability. Therefore, in our study, we selected a mixing pH equal to 5.0. Additional work by Boddohi et al. 19, 46 has investigated the importance of heparin to chitosan ratio in nanoparticle formation and stability. This ratio importantly determines the proportion of the electrical charge (both positive and negative) that is ultimately mixed together in solution during complexation. They demonstrated that a cation to anion mixing proportion of 0.5 results in the formation of fine and stable particles with a mass yield of 38%. Moving above this ratio and closer to a ratio of 1, decreased the mass yield and resulted in flocculation as well as wider size distribution (higher PDI) of particles. Accordingly, over the range of heparin to chitosan ratios assessed in our study, a ratio of 4/1 showed the best size distribution, stability and morphology among all other formulations (Fig. 1). The formation of fine particles at this specific ratio of heparin to chitosan could be attributed to the favorable proportions of positive and negative charges within the polyelectrolyte complex, which resulted in the formation of nanoparticles rather than aggregates 34. A possible explanation for this could be that when chitosan is added to the mixing solution the excess volume of heparin molecules prevents charges within the larger chitosan molecules to uniformly match the negative groups of heparin. Therefore, this creates inhomogeneity within the network, where chitosan is ionically bound to more heparin chains forming anionic, more water-soluble regions in the outer parts of the particles 46. Consequently, there are areas close to the NP cores that remain more positive in charge due to proportionally more heparin per volume 47. This provides support for why the particle sizes increase with increasing heparin content (Fig. 1A). Gumustas et al. 23, have reported that particles with zeta potential values of are not electrostatically stable and require surfactants or other agents to create more repulsive forces between the NPs to prevent agglomeration. Four our particles, both before and after PHMB encapsulation, we obtained a surface charge () of 44 mV, further confirming their monodispersity and stability. In terms of the effects of charge density on the antibacterial properties, Murata et al. 48 have shown that high density cationic surfaces are even more efficient surfaces to kill bacterial cells. In addition, in terms of size, the application of large particles, such as microspheres, as antibacterial carriers are very common 49-50. The large change in size of the nanoparticles after being loaded with PHMB, (Fig. 2A), could also be attributed to the diffusion of the PHMB molecules inside the hydrogel polyelectrolyte substrate, which has the ability to expand and trap PHMB within the network. Moreover, some of the PHMB content can also attach to the negatively-charged outer surface of the nanoparticles, which is likely partially responsible for the size variations observed 51-52. FTIR analysis of the NPs also confirmed these ionic interactions between heparin and chitosan chains (Fig. 3). Furthermore, similar circumstances occur when PHMB molecules are introduced during heparin-chitosan complexation. The abundant density of negative charges due to heparin allows PHMB to easily bind to the anionic chains within the NPs, while also diffusing into their gel-like cores to become encapsulated (Fig. 3). In addition, given the polyionic nature of HP-CS NPs, the surface charge conversion is due to the electrostatic interaction between the negative sulfated and carboxylated groups in the structure of nanoparticles and the cationic biguanides of PHMB [23]. This process, in turn, reverts the electrical surface charge of the particles to a highly positive value, while enlarging the overall particle size due to the drug diffusion into the NP cores (Fig. 2). Apart from the disposition of the PHMB molecules inside the nanoparticle network, the significant increase in particle size (p < 0.01) could be explained by the absorption and bonding of biguanides to the sulfated and carboxylic residues of heparin that are oriented outwards of the NP spherical structure [40].
As intended, the encapsulated PHMB can then be released gradually into a biologically relevant environment. Accordingly, from our controlled release experiment, we demonstrated the sustained release of antibacterial PHMB molecules over a period of more than 10 days (Fig. 4). It is interesting to note that there was steady delivery over the time course with no dramatic burst release at the first 24 hours of the experiment. Furthermore, only 80% of the total drug content was released even at the equilibrium state, which could be due to the strong ionic interactions between the negative NPs and the cationic PHMB molecules, which traps the remaining drug [40]. Also, as shown in the release study graph, the particles are still ionically bound to approximateley 15% of the initially loaded drug, which indicates their great stability even after the 2-week study period.
Apart from our choice of PHMB as the encapsulated agent, these optimized and tailorable NPs are capable of encapsulating a large variety of charged biomolecules and drugs. For instance, Liu et al. 53 have investigated the application of these NPs to entrap bovine serum albumin (BSA) as a model protein and showed enhanced BSA entrapment efficiency as a result of increasing heparin content. In a separate study, HP-CS nanoparticles were used as a vehicle for successful localized lentivirus delivery of regenerative factors, namely sonic hedgehog (Shh) and vascular endothelial growth factor (VEGF) 54. Also, of particular relevance to our controlled release approach, Volpato et al. 20 demonstrated the conjugation of FGF-2 with polyelectrolyte complexed heparin-based nanoparticles and were able to preserve FGF-2 mitogenic activity with controlled release of over 30 days. In addition, Tan et al. 10 showed HP-CS NPs embedded in a decellularized bovine jugular vein scaffolds to control the release of VEGF for 30 days to improve tissue regeneration and angiogenesis followed by subcutaneous implantation in mice. Finally, in an antibacterial application, Lin et al. tried the application of HP-CS NPs, in a pH-responsive manner to protect their antibacterial bismuth subnitrate drug from gastric digestion and elimination 34.
Mathematical modeling of drug release data helps to recognize possible governing physical mechanisms that dictate the kinetics of drug release from a polymeric matrix. Swelling, diffusion and erosion are the main mechanisms that are known to control this process 21. For this study we used several conventional models, namely the zero-order kinetic model, first-order, Korsmeyer-Peppas, Higuchi and Hixon-Crowell models 21, 23 to try to describe the kinetic behavior of our drug delivery system. Such models can also be advantageous in predicting and further optimizing the matrix design parameters to achieve a desired release profile and rate 3, 30. Starting with the Korsmeyer-Peppas model, we obtained a release exponent value of 0.7 and an anomalous release profile (Table 1), which indicated a release mechanism controlled by both swelling and diffusion mechanisms 15, 45. To further investigate whether diffusion or swelling was the governing mechanism of PHMB release out of the NPs, the Higuchi model was next applied. This model turned out to best model of those fit for our data (R2 = 0.99) and worked for the entire release period, indicating that diffusion has a greater effect than swelling on release kinetics 44. This finding makes sense as PHMB is a highly water-soluble drug and the Higuchi model is often an appropriate model to describe systems containing hydrophilic drugs 14, 21. It is worth mentioning that although the Higuchi model seems to be a good fit to our drug delivery system, further and more detailed mathematical analysis are required to draw more definitive conclusions 55. It is also worth pointing out that our choice of a dialysis-based drug diffusion model could also be affected by the dialysis membrane itself, especially when the drug shows low affinity to the polymeric substrate 51. Accordingly, Lapitsky 51 has noted that data obtained from release studies performed via dialysis-based methods is only valid when the time of drug release from the nanocarrier is much longer compared to the time that it takes for the drug to pass through the dialysis membrane pores. Otherwise, the kinetics of drug release will be most affected by the dialysis membrane diffusion barrier rather than of the polymeric carrier. In case of our release measurement system, since there is a significant difference between the size of the drug molecules (around 185 Da) and the pore size of the dialysis membrane (12-14 kDa) our release kinetics should have not been mainly driven by the dialysis membrane resistance. However, considering the difficulty and inaccuracies associated with other release measurement methods, such as direct NP dispersion in solvent, dialysis method seems to still be the most reliable and generally agreed upon method for studying drug diffusion 51-52, 56-57.
A successful antibacterial strategy should only be toxic to target bacterial cells and not the healthy cells of the surrounding tissue. Accordingly, in our results human fibroblasts in contact with PHMB doses disturbed directly into the media higher than 50 μg/ml showed significant toxicity responses. However, at most of the evaluated concentrations, the toxicity response was lowered in the group to which PHMB was delivered via the NP carriers. Overall, even though minimal toxicity in the NP delivery groups were observed for 50 μg/ml and above, this dose seems safe to use for the future studies, provided that the antibacterial studies also prove efficiency at this concentration.
Considering the problem of antibiotic resistance of bacteria and its implications to healthcare, NPs present a potent strategy to confront this problem and an alternative to oral or intravenous antibiotics by delivering stronger topical doses of antibiotic to the target areas 58. Among antibacterial agents, PHMB has shown promising broad-spectrum results in controlling infection caused by both Gram-positive and Gram-negative flora. Interestingly, the structure of PHMB is very similar to antimicrobial peptides that are produced by immune cells, such as neutrophils, which gives PHMB the ability to enter the bacterial cell walls/membranes to help disintegrate them, with little to no adverse effects on healthy mammalian cells 59-60. However, the controlled release approach for this potent antibacterial polymer has not been well-studied. In a recent study, Ahani et al. 61 have developed nano-liposomes based on phosphatidylcholine, cholesterol and stearylamine as PHMB carriers showing a significant reduction in cell toxicity on primary human skin fibroblasts as compared to the soluble form of PHMB, which is in line with the findings of the our study (Fig. 5). As observed in Figure 5, HDFs have maintained more than 90% viability at drug concentrations lower than 20 μg/ml in the soluble PHMB group, whereas there was a decrease in viability in higher concentration treatments. This indicates the toxicity of the soluble form of PHMB in concentrations greater than 20 μg/ml in a 24-hour timepoint 56-57. Similarly, we observed a decreasing trend in cell viability at concentrations of NPs higher than 20 μg/ml. These observed viability responses could also be partially due to the fact that the HDFs are adherent cells with their proliferation and growth dependent on cell anchorage and attachment 62. However, the presence of the NPs covering the culture area prevents the attachment to the surfaces and thus leading to decreased cell viability 63. Ahani et al. have also reported effective antibacterial activity of similar liposomes against Staphylococcus aureus and Escherichia coli, matching the results obtained in our study (Figs. 6, 7, Table 2). However, they have not looked at the release profile of PHMB from their nanocarrier systems and have not statistically confirmed the antibacterial effects of PHMB against E. coli or S.s aureus.
Regarding the differences between the Gram-positive and Gram-negative strains, the thicker, multilayer, proteoglycan cell walls of the Gram-positive flora, makes it harder for the antibacterial agent to penetrate through the membrane 64. This confirms the fact that larger MIC values are seen in case of E. faecalis as compared to that of E.coli, although the drug delivery system demonstrated effective antibacterial action in both cases, as confirmed by our LIVE/DEAD experiments (Figs 6, 7, Table 2). The results of these LIVE/DEAD studies also approved that the antibacterial effects provided by the PHMB-loaded NPs is beyond growth inhibition only, and instead it is inducing bacterial destruction possibly by causing membrane disintegration to both Gram-positive and Gram-negative species 27, 65. We acknowledge the differences in the values derived by the microplate dilution and the LIVE/DEAD assay. These differences might be attributed to some non-specific staining related to the remaining culture media, errors in image quantification and experimental/imaging conditions that might have been slightly different between samples. However, the LIVE/DEAD assay still supports the effective antibacterial properties of our PHMB-loaded nanoparticles, and these findings are in agreement with other studies 66-67.
5. Conclusions
In this study we report the synthesis and characterization of polyionic heparin-chitosan nanoparticles as a valuable platform for controlled release of the antibacterial PHMB. Multiple formulations with different heparin to chitosan proportions were used and characterized in terms of their size, zeta potential and morphology before reaching the optimum formulation of 4 parts heparin to one-part chitosan. This ratio created a negative surface charge to improve the efficiency of positively charged PHMB loading. Therefore, it is hypothesized that the resulting surface charge can also be tailored to any desired value based on the application/drug of choice by altering the starting ratio at which the charges are mixed. The selected NP formulation showed a sustained and controlled release of PHMB for over 10 days. As compared to the soluble plain PHMB, the NPs show significantly better biocompatibility of human dermal fibroblasts. Moreover, these antibacterial NPs were capable of effectively destroying Gram-positive and Gram-negative bacterial strains that can be problematic in human infections. In total, the novel antibacterial HP-CS NPs provide a means for increasing drug viability and stability while reducing the chances of drug accumulation into non-targeted tissues taking advantage of their sustained release properties. We are aiming to extend the application of this antibacterial platform to use in a multifunctional wound dressing in the future.
ACKNOWLEDGMENT
The time and efforts of Edmund Burke in bacterial experiments is also appreciated. We would also like to thank Dr. Yakov Lapitsky form the University of Toledo for his valuable input on interpretation of our results.
Funding Sources
We would like to acknowledge partial financial support for this study from the NIH for SA and NDL (R41 DK105704-01A1). The opinions expressed in this article are the author’s own and do not reflect the view of the NIH or the NIDDK.
ABBREVIATIONS
- AMP
Ampicillin
- BHI
Brain Heart Infusion
- CFU
Colony-Forming Unit
- CS
Chitosan
- DLS
Dynamic Light Scattering
- DMEM
Dulbecco’s Modified Eagle’s Medium
- DMSO
Dimethyl Sulfoxide
- DRIFT
Diffuse Reflectance Infrared Fourier Transform Spectroscopy
- E. coli
Escherichia coli
- E. faecalis
Enterococcus faecalis
- EDTA
Ethylenediaminetetraacetic Acid
- FBS
Fetal Bovine Serum
- FGF
Fibroblast Growth Factor
- FTIR
Fourier Transform Infrared Spectroscopy
- HDF
Human Dermal Fibroblasts
- HP
Heparin
- IRB
Institutional Review Board
- MHA
Muller Hinton Agar
- MHB
Mueller Hinton Broth
- MIC
Minimum Inhibitory Concentration
- NIDDK
Diabetes and Digestive and Kidney Diseases
- NIH
National Institute of Health
- NP
Nanoparticle
- NT
Non-treated Cell Group
- PBS
Phosphate Buffered Saline
- PDI
Polydispersity Index
- PHMB
Polyhexamethylene Biguanide
- TEM
Transmission Electron Microscopy
- TSA
Tryptic Soy Agar
- TSB
Tryptic Soy Broth
- VEGF
Vascular Endothelial Growth Factor
REFERENCES
- (1).Swierczewska M; Han HS; Kim K; Park JH; Lee S Polysaccharide-based nanoparticles for theranostic nanomedicine. Adv Drug Deliv Rev 2016, 99 (Pt A), 70–84, DOI: 10.1016/j.addr.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Tan C; Xie JH; Zhang XM; Cai JB; Xia SQ Polysaccharide-based nanoparticles by chitosan and gum arabic polyelectrolyte complexation as carriers for curcumin. Food Hydrocolloids 2016, 57, 236–245, DOI: 10.1016/j.foodhyd.2016.01.021. [DOI] [Google Scholar]
- (3).Pandey R; Khuller GK Nanoparticle-Based Oral Drug Delivery System for an Injectable Antibiotic – Streptomycin. Chemotherapy 2007, 53 (6), 437–441, DOI: 10.1159/000110009. [DOI] [PubMed] [Google Scholar]
- (4).Karimi M; Ghasemi A; Sahandi Zangabad P; Rahighi R; Moosavi Basri SM; Mirshekari H; Amiri M; Shafaei Pishabad Z; Aslani A; Bozorgomid M; Ghosh D; Beyzavi A; Vaseghi A; Aref AR; Haghani L; Bahrami S; Hamblin MR Smart micro/nanoparticles in stimulus-responsive drug/gene delivery systems. Chem Soc Rev 2016, 45 (5), 1457–501, DOI: 10.1039/c5cs00798d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sahoo SK; Misra R; Parveen S Nanoparticles: a boon to drug delivery, therapeutics, diagnostics and imaging. In Nanomedicine in Cancer; Pan Stanford: 2017; pp 73–124. [DOI] [PubMed] [Google Scholar]
- (6).Kumari A; Yadav SK; Yadav SC Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf B Biointerfaces 2010, 75 (1), 1–18, DOI: 10.1016/j.colsurfb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- (7).Kweon DK; Song SB; Park YY Preparation of water-soluble chitosan/heparin complex and its application as wound healing accelerator. Biomaterials 2003, 24 (9), 1595–601, DOI: 10.1016/S0142-9612(02)00566-5. [DOI] [PubMed] [Google Scholar]
- (8).Betz G; Nowbakht P; Imboden R; Imanidis G Heparin penetration into and permeation through human skin from aqueous and liposomal formulations in vitro. International Journal of Pharmaceutics 2001, 228 (1-2), 147–159, DOI: Doi 10.1016/S0378-5173(01)00832-8. [DOI] [PubMed] [Google Scholar]
- (9).Liang Y; Kiick KL Heparin-functionalized polymeric biomaterials in tissue engineering and drug delivery applications. Acta biomaterialia 2014, 10 (4), 1588–600, DOI: 10.1016/j.actbio.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tan Q; Tang H; Hu J; Hu Y; Zhou X; Tao Y; Wu Z Controlled release of chitosan/heparin nanoparticle-delivered VEGF enhances regeneration of decellularized tissue-engineered scaffolds. Int J Nanomedicine 2011, 6, 929–42, DOI: 10.2147/IJN.S18753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Liu LS; Ng CK; Thompson AY; Poser JW; Spiro RC Hyaluronate-heparin conjugate gels for the delivery of basic fibroblast growth factor (FGF-2). J Biomed Mater Res 2002, 62 (1), 128–35, DOI: 10.1002/jbm.10238. [DOI] [PubMed] [Google Scholar]
- (12).Ahsan SM; Thomas M; Reddy KK; Sooraparaju SG; Asthana A; Bhatnagar I Chitosan as biomaterial in drug delivery and tissue engineering. Int J Biol Macromol 2018, 110, 97–109, DOI: 10.1016/j.ijbiomac.2017.08.140. [DOI] [PubMed] [Google Scholar]
- (13).Liu ZH; Jiao YP; Wang YF; Zhou CR; Zhang ZY Polysaccharides-based nanoparticles as drug delivery systems. Advanced drug delivery reviews 2008, 60 (15), 1650–1662, DOI: 10.1016/j.addr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- (14).Dorati R; DeTrizio A; Modena T; Conti B; Benazzo F; Gastaldi G; Genta I Biodegradable scaffolds for bone regeneration combined with drug-delivery systems in osteomyelitis therapy. Pharmaceuticals 2017, 10 (4), 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ranjbar-Mohammadi M; Rabbani S; Bahrami SH; Joghataei M; Moayer F Antibacterial performance and in vivo diabetic wound healing of curcumin loaded gum tragacanth/poly (ε-caprolactone) electrospun nanofibers. Materials Science and Engineering: C 2016, 69, 1183–1191. [DOI] [PubMed] [Google Scholar]
- (16).Bessa LJ; Fazii P; Di Giulio M; Cellini L Bacterial isolates from infected wounds and their antibiotic susceptibility pattern: some remarks about wound infection. Int Wound J 2015, 12 (1), 47–52, DOI: 10.1111/iwj.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Seil JT; Webster TJ Antimicrobial applications of nanotechnology: methods and literature. Int J Nanomedicine 2012, 7, 2767–81, DOI: 10.2147/IJN.S24805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Ashraf S; Akhtar N; Ghauri MA; Rajoka MI; Khalid ZM; Hussain I Polyhexamethylene biguanide functionalized cationic silver nanoparticles for enhanced antimicrobial activity. Nanoscale research letters 2012, 7 (1), 267, DOI: 10.1186/1556-276X-7-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Boddohi S; Almodovar J; Zhang H; Johnson PA; Kipper MJ Layer-by-layer assembly of polysaccharide-based nanostructured surfaces containing polyelectrolyte complex nanoparticles. Colloids Surf B Biointerfaces 2010, 77 (1), 60–8, DOI: 10.1016/j.colsurfb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- (20).Zomer Volpato F; Almodovar J; Erickson K; Popat KC; Migliaresi C; Kipper MJ Preservation of FGF-2 bioactivity using heparin-based nanoparticles, and their delivery from electrospun chitosan fibers. Acta biomaterialia 2012, 8 (4), 1551–9, DOI: 10.1016/j.actbio.2011.12.023. [DOI] [PubMed] [Google Scholar]
- (21).Korsmeyer RW; Lustig SR; Peppas NA Solute and penetrant diffusion in swellable polymers. I. Mathematical modeling. Journal of Polymer Science Part B: Polymer Physics 1986, 24 (2), 395–408. [Google Scholar]
- (22).Gunalan S; Sivaraj R; Rajendran V Green synthesized ZnO nanoparticles against bacterial and fungal pathogens. Progress in Natural Science: Materials International 2012, 22 (6), 693–700, DOI: 10.1016/j.pnsc.2012.11.015. [DOI] [Google Scholar]
- (23).Gumustas M; Sengel-Turk CT; Gumustas A; Ozkan SA; Uslu B Effect of Polymer-Based Nanoparticles on the Assay of Antimicrobial Drug Delivery Systems. In Multifunctional Systems for Combined Delivery, Biosensing and Diagnostics; Elsevier: 2017; pp 67–108. [Google Scholar]
- (24).Akula S; Brosch IK; Leipzig ND Fluorinated Methacrylamide Chitosan Hydrogels Enhance Cellular Wound Healing Processes. Annals of biomedical engineering 2017, 45 (11), 2693–2702, DOI: 10.1007/s10439-017-1893-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Lawrence PG; Patil PS; Leipzig ND; Lapitsky Y Ionically Cross-Linked Polymer Networks for the Multiple-Month Release of Small Molecules. ACS applied materials & interfaces 2016, 8 (7), 4323–35, DOI: 10.1021/acsami.5b10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Retamozo B; Shabahang S; Johnson N; Aprecio RM; Torabinejad M Minimum contact time and concentration of sodium hypochlorite required to eliminate Enterococcus faecalis. J Endod 2010, 36 (3), 520–3, DOI: 10.1016/j.joen.2009.12.005. [DOI] [PubMed] [Google Scholar]
- (27).Gopinath V; Priyadarshini S; Loke MF; Arunkumar J; Marsili E; MubarakAli D; Velusamy P; Vadivelu J Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arabian journal of chemistry 2017, 10 (8), 1107–1117, DOI: 10.1016/j.arabjc.2015.11.011. [DOI] [Google Scholar]
- (28).Nicas TI; Wu CY; Hobbs JN Jr.; Preston DA; Allen NE Characterization of vancomycin resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob Agents Chemother 1989, 33 (7), 1121–4, DOI: 10.1128/AAC.33.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Elisha IL; Botha FS; McGaw LJ; Eloff JN The antibacterial activity of extracts of nine plant species with good activity against Escherichia coli against five other bacteria and cytotoxicity of extracts. BMC Complementary and Alternative Medicine 2017, 17 (1), 133, DOI: 10.1186/s12906-017-1645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kavanagh A; Ramu S; Gong Y; Cooper MA; Blaskovich MAT Effects of Microplate Type and Broth Additives on Microdilution MIC Susceptibility Assays. Antimicrobial agents and chemotherapy 2019, 63 (1), e01760–18, DOI: 10.1128/aac.01760-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Choi O; Deng KK; Kim NJ; Ross L Jr.; Surampalli RY; Hu Z The inhibitory effects of silver nanoparticles, silver ions, and silver chloride colloids on microbial growth. Water Res 2008, 42 (12), 3066–74, DOI: 10.1016/j.watres.2008.02.021. [DOI] [PubMed] [Google Scholar]
- (32).Musken M; Di Fiore S; Romling U; Haussler S A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nature protocols 2010, 5 (8), 1460–9, DOI: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- (33).Shah KM; Stern MM; Stern AR; Pathak JL; Bravenboer N; Bakker AD Osteocyte isolation and culture methods. Bonekey Rep 2016, 5, 838, DOI: 10.1038/bonekey.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Lin YH; Chang CH; Wu YS; Hsu YM; Chiou SF; Chen YJ Development of pH-responsive chitosan/heparin nanoparticles for stomach-specific anti-Helicobacter pylori therapy. Biomaterials 2009, 30 (19), 3332–42, DOI: 10.1016/j.biomaterials.2009.02.036. [DOI] [PubMed] [Google Scholar]
- (35).Shahbazi MA; Hamidi M; Mohammadi-Samani S Preparation, optimization, and in-vitro/in-vivo/ex-vivo characterization of chitosan-heparin nanoparticles: drug-induced gelation. The Journal of pharmacy and pharmacology 2013, 65 (8), 1118–33, DOI: 10.1111/jphp.12076. [DOI] [PubMed] [Google Scholar]
- (36).Napavichayanun S; Amornsudthiwat P; Pienpinijtham P; Aramwit P Interaction and effectiveness of antimicrobials along with healing-promoting agents in a novel biocellulose wound dressing. Materials science & engineering. C, Materials for biological applications 2015, 55, 95–104, DOI: 10.1016/j.msec.2015.05.026. [DOI] [PubMed] [Google Scholar]
- (37).Shyichuk A; Ziolkowska D; Mroczyska K Quantitation of Polyhexamethylene Biguanide Biocide on Cotton Fabric Surface. Cellulose Chemistry and Technology 2015, 49 (3-4), 387–391. [Google Scholar]
- (38).Craig DQ The mechanisms of drug release from solid dispersions in water-soluble polymers. International journal of pharmaceutics 2002, 231 (2), 131–144. [DOI] [PubMed] [Google Scholar]
- (39).Fu Y; Kao WJ Drug release kinetics and transport mechanisms of non-degradable and degradable polymeric delivery systems. Expert opinion on drug delivery 2010, 7 (4), 429–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Santos C; Albuquerque A; Sampaio F; Keyson D Nanomaterials with antimicrobial properties: applications in health sciences. Microbial pathogens and strategies for combating them: science, technology and education. Volume 2013, 4, 2. [Google Scholar]
- (41).Veerapandian M; Yun K Functionalization of biomolecules on nanoparticles: specialized for antibacterial applications. Appl Microbiol Biotechnol 2011, 90 (5), 1655–67, DOI: 10.1007/s00253-011-3291-6. [DOI] [PubMed] [Google Scholar]
- (42).Ravishankar Rai V; Jamuna Bai A Nanoparticles and their potential application as antimicrobials. A Méndez-Vilas A, editor. Mysore: Formatex; 2011. [Google Scholar]
- (43).Nimesh S; Manchanda R; Kumar R; Saxena A; Chaudhary P; Yadav V; Mozumdar S; Chandra R Preparation, characterization and in vitro drug release studies of novel polymeric nanoparticles. Int J Pharm 2006, 323 (1-2), 146–52, DOI: 10.1016/j.ijpharm.2006.05.065. [DOI] [PubMed] [Google Scholar]
- (44).Saidykhan L; Abu Bakar MZB; Rukayadi Y; Kura AU; Latifah SY Development of nanoantibiotic delivery system using cockle shell-derived aragonite nanoparticles for treatment of osteomyelitis. International journal of nanomedicine 2016, 11, 661–673, DOI: 10.2147/IJN.S95885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chen H; Xing X; Tan H; Jia Y; Zhou T; Chen Y; Ling Z; Hu X Covalently antibacterial alginate-chitosan hydrogel dressing integrated gelatin microspheres containing tetracycline hydrochloride for wound healing. Materials Science and Engineering: C 2017, 70, 287–295, DOI: 10.1016/j.msec.2016.08.086. [DOI] [PubMed] [Google Scholar]
- (46).Boddohi S; Moore N; Johnson PA; Kipper MJ Polysaccharide-based polyelectrolyte complex nanoparticles from chitosan, heparin, and hyaluronan. Biomacromolecules 2009, 10 (6), 1402–9, DOI: 10.1021/bm801513e. [DOI] [PubMed] [Google Scholar]
- (47).Ordikhani F; Dehghani M; Simchi A Antibiotic-loaded chitosan–laponite films for local drug delivery by titanium implants: cell proliferation and drug release studies. Journal of Materials Science: Materials in Medicine 2015, 26 (12), 269. [DOI] [PubMed] [Google Scholar]
- (48).Murata H; Koepsel RR; Matyjaszewski K; Russell AJ Permanent, non-leaching antibacterial surfaces—2: How high density cationic surfaces kill bacterial cells. Biomaterials 2007, 28 (32), 4870–4879. [DOI] [PubMed] [Google Scholar]
- (49).Chifiriuc CM; Grumezescu AM; Saviuc C; Croitoru C; Mihaiescu DE; Lazar V Improved antibacterial activity of cephalosporins loaded in magnetic chitosan microspheres. International journal of pharmaceutics 2012, 436 (1-2), 201–205. [DOI] [PubMed] [Google Scholar]
- (50).Tang Y; Xu J; Liu W; Xu L; Li H Preparation of monodispersed core-shell microspheres with surface antibacterial property employing N-(4-vinylbenzyl)-N, N-diethylamine hydrochloride as surfmer. International Journal of Polymeric Materials and Polymeric Biomaterials 2016, 65 (3), 143–150. [Google Scholar]
- (51).Hu F-Q; Jiang S-P; Du Y-Z; Yuan H; Ye Y-Q; Zeng S Preparation and characterization of stearic acid nanostructured lipid carriers by solvent diffusion method in an aqueous system. Colloids and surfaces. B, Biointerfaces 2005, 45 (3), 167–173, DOI: 10.1016/j.colsurfb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- (52).Venier-Julienne MC; Benoît JP Preparation, purification and morphology of polymeric nanoparticles as drug carriers. Pharmaceutica Acta Helvetiae 1996, 71 (2), 121–128, DOI: 10.1016/0031-6865(95)00059-3. [DOI] [PubMed] [Google Scholar]
- (53).Liu Z; Jiao Y; Liu F; Zhang Z Heparin/chitosan nanoparticle carriers prepared by polyelectrolyte complexation. Journal of biomedical materials research. Part A 2007, 83 (3), 806–12, DOI: 10.1002/jbm.a.31407. [DOI] [PubMed] [Google Scholar]
- (54).Thomas AM; Gomez AJ; Palma JL; Yap WT; Shea LD Heparin-chitosan nanoparticle functionalization of porous poly(ethylene glycol) hydrogels for localized lentivirus delivery of angiogenic factors. Biomaterials 2014, 35 (30), 8687–93, DOI: 10.1016/j.biomaterials.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Wei G; Kotoura Y; Oka M; Yamamuro T; Wada R; Hyon S; Ikada Y A bioabsorbable delivery system for antibiotic treatment of osteomyelitis. The use of lactic acid oligomer as a carrier. The Journal of bone and joint surgery. British volume 1991, 73 (2), 246–252. [DOI] [PubMed] [Google Scholar]
- (56).Müller G; Koburger T; Kramer A Interaction of polyhexamethylene biguanide hydrochloride (PHMB) with phosphatidylcholine containing o/w emulsion and consequences for microbicidal efficacy and cytotoxicity. Chemico-biological interactions 2013, 201 (1-3), 58–64. [DOI] [PubMed] [Google Scholar]
- (57).Yabes JM; White BK; Murray CK; Sanchez CJ; Mende K; Beckius ML; Zera WC; Wenke JC; Akers KS In Vitro activity of Manuka Honey and polyhexamethylene biguanide on filamentous fungi and toxicity to human cell lines. Medical mycology 2016, 55 (3), 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Hajipour MJ; Fromm KM; Ashkarran AA; Jimenez de Aberasturi D; de Larramendi IR; Rojo T; Serpooshan V; Parak WJ; Mahmoudi M Antibacterial properties of nanoparticles. Trends Biotechnol 2012, 30 (10), 499–511, DOI: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- (59).Butcher M PHMB: an effective antimicrobial in wound bioburden management. British journal of nursing (Mark Allen Publishing) 2012, 21 (12), S16, S18–21, DOI: 10.12968/bjon.2012.21.Sup12.S16. [DOI] [PubMed] [Google Scholar]
- (60).Chindera K; Mahato M; Sharma AK; Horsley H; Kloc-Muniak K; Kamaruzzaman NF; Kumar S; McFarlane A; Stach J; Bentin T; Good L The antimicrobial polymer PHMB enters cells and selectively condenses bacterial chromosomes. Sci Rep 2016, 6, 23121, DOI: 10.1038/srep23121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ahani E; Montazer M; Toliyat T; Mahmoudi Rad M; Harifi T Preparation of nano cationic liposome as carrier membrane for polyhexamethylene biguanide chloride through various methods utilizing higher antibacterial activities with low cell toxicity. Journal of microencapsulation 2017, 34 (2), 121–131, DOI: 10.1080/02652048.2017.1296500. [DOI] [PubMed] [Google Scholar]
- (62).Singh V; Erady C; Balasubramanian N Cell-matrix adhesion controls Golgi organization and function through Arf1 activation in anchorage-dependent cells. Journal of cell science 2018, 131 (16), DOI: 10.1242/jcs.215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Place LW; Sekyi M; Kipper MJ Aggrecan-mimetic, glycosaminoglycan-containing nanoparticles for growth factor stabilization and delivery. Biomacromolecules 2014, 15 (2), 680–9, DOI: 10.1021/bm401736c. [DOI] [PubMed] [Google Scholar]
- (64).Jung WK; Koo HC; Kim KW; Shin S; Kim SH; Park YH Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Applied and environmental microbiology 2008, 74 (7), 2171–8, DOI: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Damper PD; Epstein W Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob Agents Chemother 1981, 20 (6), 803–8, DOI: 10.1128/AAC.20.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Veiga AS; Sinthuvanich C; Gaspar D; Franquelim HG; Castanho MA; Schneider JP Arginine-rich self-assembling peptides as potent antibacterial gels. Biomaterials 2012, 33 (35), 8907–16, DOI: 10.1016/j.biomaterials.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Li P; Poon YF; Li W; Zhu HY; Yeap SH; Cao Y; Qi X; Zhou C; Lamrani M; Beuerman RW; Kang ET; Mu Y; Li CM; Chang MW; Leong SS; Chan-Park MB A polycationic antimicrobial and biocompatible hydrogel with microbe membrane suctioning ability. Nature materials 2011, 10 (2), 149–56, DOI: 10.1038/nmat2915. [DOI] [PubMed] [Google Scholar]