Abstract
Introduction
STAT4 is a transcriptional regulator that has been reported to have oncogenic activities in various cancers. In our study, the posttranscriptional regulatory effect of miR-200a-3p on STAT4 and the prognostic significance of miR-200a-3p and STAT4 were evaluated in bladder cancer (BCa).
Material and methods
Proliferation and apoptosis of BCa cell lines were monitored using CCK-8 and Annexin V-FITC assays, respectively. Gene and protein expression levels in BCa tissues and cells were detected using RT-qPCR and western blotting, respectively.
Results
Significant downregulation of miR-200a-3p and upregulation of STAT4 were observed in BCa tissues and cells compared with the corresponding non-tumor adjacent tissues. Both STAT4 and miR-200a-3p were validated as independent prognostic indicators in sixty-nine BCa patients for predicting overall survival and disease-free survival. In vitro experimental analyses revealed that knockdown of STAT4 repressed BCa cell growth and elevated cell apoptosis. Molecular interactive analysis revealed STAT4 as a direct target of miR-200a-3p, which could suppress STAT4 protein expression by posttranscriptional repression. Cotransfection of miR-200a-3p mimics and STAT4 overexpression plasmids into BCa cells demonstrated that the antineoplastic activities of miR-200a-3p in vitro were neutralized by overexpressed STAT4.
Conclusions
The miR-200a-3p/STAT4 signaling cascade plays an important role in the progression of BCa, which provides a new promising target for targeted BCa therapies.
Keywords: bladder cancer, miR-200a-3p, STAT4, prognosis, proliferation, posttranscriptional regulation
Introduction
Bladder cancer (BCa) is the most common urinary tract malignant tumor and the ninth most frequently diagnosed cancer worldwide [1]. In China, it is estimated that 80,500 cases are newly diagnosed every year and 32,900 deaths of BCa patients occurred in 2015, resulting in an approximately 40% mortality rate [2]. Moreover, according to an epidemiological study, approximately one-third of BCa patients are diagnosed with muscle-invasive bladder cancer and have undetected metastasis before hospitalization [3]. Currently, surgical operation combined with chemotherapy is recommended as a first-line treatment for BCa patients [4]. Although considerable progress has been made in clinical practice, there has been no significant improvement in mortality in metastatic BCa patients. Even worse, chemotherapy resistance is frequently reported in the treatment of BCa [5, 6]. Therefore, it is extremely urgent to explore the pathogenesis of BCa, which may provide novel therapeutic strategies for the treatment of BCa.
Signal transducer and activator of transcription 4 (STAT4) is a member of the STAT family and can be activated by interleukin 12 signaling to promote the differentiation of Th1 cells and accelerate the secretion of interferon-γ, thus participating in the regulation of autoimmunity and inflammation [7]. Recent studies have reported that, apart from these roles, STAT4 is deregulated under malignant pathological processes [8–10]. The upregulation of STAT4 has been observed in many types of carcinomas, including ovarian cancer [8], colorectal cancer [9] and gastric cancer [10], and has been correlated with poor prognosis. However, the role of STAT4 and the underlying molecular mechanisms in BCa remain unclear.
MicroRNAs (miRNAs) are a class of small, single-stranded, noncoding RNA molecules that are approximately 22 nucleotides in length and can regulate gene expression or translation by targeting 3′-untranslated regions (3’-UTRs) and are involved in diverse biological functions [11]. Numerous microRNAs (miRNAs) are implicated in the progression of tumorigenesis as tumor suppressors or oncogenes via posttranscriptional repression of target genes [12, 13]. Recent accumulating evidence has demonstrated that miRNAs such as miR-149, miR-1178-3p and miR-558 play crucial regulatory roles in BCa [14–16]. However, the role of miR-200a-3p in BCa is unclear. Downregulation of miR-200a-3p manipulates the expression of multiple genes in favor of malignant transformation in hepatocellular carcinoma (HCC) [17, 18], renal cell carcinoma [19] and gliomas [20].
The present study revealed a significant negative correlation between miR-200a-3p and STAT4 expression in human BCa tissues. We also demonstrated that STAT4 is a direct target of miR-200a-3p and observed that overexpression of miR-200a-3p inhibited proliferation and induced apoptosis in BCa cells by suppressing STAT4 expression.
Material and methods
Sample collection
BCa and adjacent non-tumor specimens were obtained from seventy-two BCa patients at the Fifth People’s Hospital of Dongguan (Dongguan, China) between February 2010 and February 2014. Based on histopathological evaluation, 3 pairs of BCa and adjacent non-tumor tissues were excluded from our study. Therefore, 69 pairs of BCa and adjacent non-tumor tissues were used to evaluate clinicopathological or experiment parameters. The specimens were promptly stored in liquid nitrogen until use for gene or protein detection. Our study was approved by the Ethics Committee of the Fifth People’s Hospital of Dongguan (approval number: FHD2009c0036; Dongguan, China), and signed informed consent forms were obtained from each individual BCa patient.
Cell culture
The human normal bladder epithelial cell line SV-HUC-1 and the BCa cell lines 5637, SW780, T24 and EJ were purchased from ATCC. Cells were cultured as described previously [14].
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The miRNeasy Mini Kit (Qiagen, Inc., Valencia, CA, USA) was used to extract total RNA. The TaqMan RT kit and TaqMan MicroRNA assay (Applied Biosystems) were used to detect miR-200a-3p expression levels, according to the manufacturer’s protocol. The stem-loop sequence of miR-200a-3p was as follows: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACATCGTT-3′. The thermocycling conditions for RT were as follows: 37°C for 5 min, 42°C for 45 min and 85°C for 5 min. The following primers were used for PCR analysis: miR-200a-3p (annealing temperature: 60°C): forward, 5′-GGCTAACACTGTCTGGTAACGATG-3′, and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 (control gene; annealing temperature: 60°C) forward, 5′-CTCGCTTCGGCAGCACA-3′, and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The thermocycling conditions for PCR were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. miR-200a-3p-probe: FAM-CTCGATACGTAGCACGC-MGB; U6-probe: FAM-CCATGCTAATCTTCTCTGTA-MGB.
Moloney murine leukemia virus reverse transcriptase (Invitrogen) was used to synthesize cDNA from 2 µg of total RNA according to the manufacturer’s protocol. Real-time PCR was performed using the Applied Biosystems 7300 System with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific). The relative expression levels of STAT4 were calculated using the 2–ΔΔCq method [21]. The following primers were used: STAT4 (annealing temperature: 58°C): forward, 5′-GCCGGTTGTCAAATCCCTTAC-3′, and reverse, 5′-TGTGACTGCTGTCTTGATTCCCT-3′; GAPDH (control gene; annealing temperature: 60°C) forward, 5′-CAAATTCGTGAAGCGTTCCATA-3′, and reverse, 5′-AGTGCAGGGTCCGAGGTATTC-3′. The thermocycling conditions for PCR were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s, 58°C for 30 s and 72°C for 30 s, and a final extension step at 72°C for 3 min. STAT4-probe: FAM-AGCAGCTACCCGAAGAAGTGCT-MGB; GAPGH-probe:FAM-TGGAGAAGGCTGGGGCTCAT-MGB.
Western blotting
Cells were lysed using RIPA buffer (Beyotime Institute of Biotechnology). Western blotting was performed as described previously [22]. The primary antibodies against STAT4 and β-actin were obtained from Abcam (1 : 1,000 dilution; Cambridge, UK) and Santa Cruz Biotechnology (1 : 2,000 dilution), respectively. The secondary antibody was purchased from Santa Cruz Biotechnology Inc. (cat. no. sc-516102; dilution: 1 : 5,000). Protein bands were visualized by chemiluminescence (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Protein bands were analyzed as described previously [22].
Cell transfection and vector constructs
Short hairpin RNA (shRNA; dsDNA with sense-loop-antisense structure) was designed using an online tool (http://rnaidesigner.thermofisher.com/rnaiexpress/) to knock down STAT4. sh-Control (sh-Con; 5′-TAATTGTCAAATCAGAGTGCT-3′) and sh-STAT4 (5′-GCAATGAAGTCTCCTTATTCT-3′) were synthesized by GenePharma (Shanghai, China). The lentiviral vectors were constructed using the pLV-hU6-hEF1a-EYFP-2A-Puro system to express sh-Con or sh-STAT4, and the manufacturing procedures and transfection of lentivirus vectors were performed as described previously [23]. Moreover, miR-control (miR-Con; 5′-UCUGAUUGGUCUCUGUAUGUAAA-3′) and miR-200a-3p mimics (5′-UAACACUGUCUGGUAACGAUGU-3′) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and transfected into BCa cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocols. STAT4 overexpression plasmids were purchased from GeneCopoeia, Inc. (cat. no. EX-K3038-Lv105; Rockville, MD, USA). An empty vector served as the negative control (vector-Con). Vector-Con and STAT4 overexpression vectors were transfected into BCa cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocols.
Cell counting kit 8 (CCK-8) assay
The CCK-8 kit (Dojindo Laboratories, Japan) was used to measure cell proliferation according to the manufacturer’s instructions.
Cell apoptosis
Apoptotic cells were analyzed using flow cytometry (FACScan) with an Annexin V-FITC kit (BD Biosciences) as described previously [24].
Luciferase reporter assay
STAT4 sequence with wild-type (WT; 5′-ACAACUUUAAGAAACCAGUGUUA-3′) or mutant-type (mut; 5′-ACAACUUUAAGAAACUCACUGGA-3′) 3′-UTR was synthesized by Sangon (Shanghai, China) and inserted into the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega). For the luciferase assay, BCa cells (1 × 105) were seeded into 24-well plates and cotransfected with luciferase reporter vectors (0.5 µg) containing WT or Mut 3′-UTR of STAT4 and miR-Con or miR-200a-3p (100 nM) using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h. Luciferase activity was measured using a dual luciferase reporter assay kit (Beyotime Institute of Biotechnology) according to the manufacturer’s protocol.
Ethics approval
Written informed consent was obtained from all of the participants prior to sample collection. The study was approved by the Ethics Committee of the Fifth People’s Hospital of Dongguan (Dongguan China) according to the Helsinki Declaration.
Statistical analysis
Data are presented as the mean ± standard error (SE). Statistical analysis was calculated using GraphPad Prism version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). Two-group differences were analyzed using Student’s t-test. Intergroup differences were analyzed by one-way analysis of variance followed by a post hoc Tukey test. The differences between the clinical characteristics and STAT4 or miR-200a-3p expression levels in BCa patients were evaluated by Pearson χ2 tests. The Kaplan-Meier method with log-rank test was applied for overall survival (OA) and disease-free survival (DFS) analyses. P < 0.05 represents a statistically significant difference.
Results
miR-200a-3p expression and prognostic significance in BCa patients
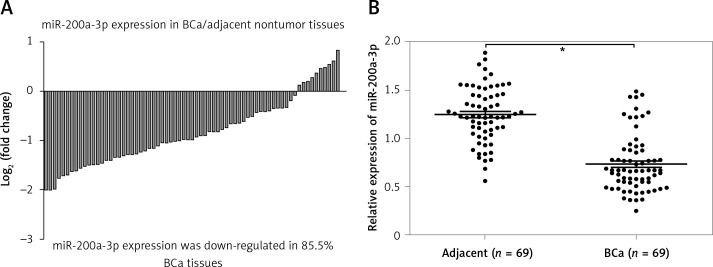
miR-200a-3p expression in BCa tissues and paired normal bladder tissues was measured using RT-qPCR, and the results revealed that miR-200a-3p expression levels were reduced in 85.5% BCa tissues (59/69) compared with paired normal bladder tissues (Figure 1 A). A paired t test showed a significant difference in miR-200a-3p expression in 69 pairs of BCa and adjacent nontumor tissues (Figure 1 B). A χ2 test was performed to investigate the differential expression of miR-200a-3p in various clinicopathological subgroups. Lower expression of miR-200a-3p was significantly correlated with tumor stage (T) (p = 0.007) and histological grade (p = 0.001) in BCa patients (Table I).
Figure 1.
miR-200a-3p was downregulated in BCa tissues. A – The log2 (fold change) of miR-200a-3p expression in 69 pairs of BCa and adjacent nontumor tissues was calculated. B – RT-qPCR was performed to measure the expression of miR-200a-3p in 69 pairs of BCa and adjacent nontumor tissues, and the results demonstrated that miR-200a-3p was significantly lower in BCa tissues compared with adjacent nontumor tissues. *P < 0.05, n = 69 in each group. The lines represent that a statistical analysis was performed between BCa tissues and adjacent non-tumor tissues. Data are presented as the mean ± SE
Table I.
Correlation between clinicopathological variables and expression of miR-200a-3p and STAT4
| Variables | N | miR-200a-3p expression | P-value | STAT4 expression | P-value | ||
|---|---|---|---|---|---|---|---|
| Low (n = 31) | High (n = 38) | Low (n = 34) | High (n = 35) | ||||
| Age [years]: | 0.510 | 0.555 | |||||
| ≤ 55 | 43 | 18 | 25 | 20 | 23 | ||
| > 55 | 26 | 13 | 13 | 14 | 12 | ||
| Gender: | 0.646 | 0.103 | |||||
| Male | 47 | 22 | 25 | 20 | 27 | ||
| Female | 22 | 9 | 13 | 14 | 8 | ||
| Tumor size [cm]: | 0.602 | 0.537 | |||||
| ≤ 3 | 38 | 16 | 22 | 20 | 18 | ||
| > 3 | 31 | 15 | 16 | 14 | 17 | ||
| Tumor stage T: | 0.007 | 0.011 | |||||
| Ta-T1 | 30 | 8 | 22 | 20 | 10 | ||
| T2-T4 | 39 | 23 | 16 | 14 | 25 | ||
| Histological grade: | 0.001 | 0.002 | |||||
| L | 28 | 6 | 22 | 20 | 8 | ||
| H | 41 | 25 | 16 | 14 | 27 | ||
| Lymph node metastasis: | 0.102 | 0.053 | |||||
| No | 45 | 17 | 28 | 26 | 19 | ||
| Yes | 24 | 14 | 10 | 8 | 16 | ||
| Vascular invasion: | 0.061 | 0.005 | |||||
| No | 48 | 18 | 30 | 29 | 19 | ||
| Yes | 21 | 13 | 8 | 5 | 16 |
STAT4 – signal transducer and activator of transcription 4/
Next, to determine the role of miR-200a-3p in BCa prognosis, Kaplan-Meier survival analyses were performed to explore the prognostic significance of 8 demographic and clinical factors. Age (Figure 2 A), gender (Figure 2 B), tumor size (Figure 2 C) and lymph node metastasis (Figure 2 F) were not correlated with OS of BCa patients. Tumor stage (p = 0.011; Figure 2 D), histological grade (p = 0.015; Figure 2 E) and vascular invasion (p = 0.007; Figure 2 G) could function as prognostic factors in BCa patients. We also found that low miR-200a-3p expression levels were correlated with poor OS and DFS in BCa patients (Figures 2 H and I). These findings indicated that miR-200a-3p might be an effective predictor for OS and DFS in BCa patients.
Figure 2.
Prognostic significance of miR-200a-3p in BCa patients. Kaplan-Meier analysis and logrank test indicated that age (A), gender (B), tumor size (C) and lymph node metastasis (F) had no effect on survival prognosis in BCa patients. Tumor stage (D), histological grade (E) are correlated with OS in BCa patients Vascular invasion (G) is correlated with OS in BCa patients. Low miR-200a-3p expression was correlated with poor OS (H) and DFS (I) in BCa patients
STAT4 expression and prognostic significance in BCa patients
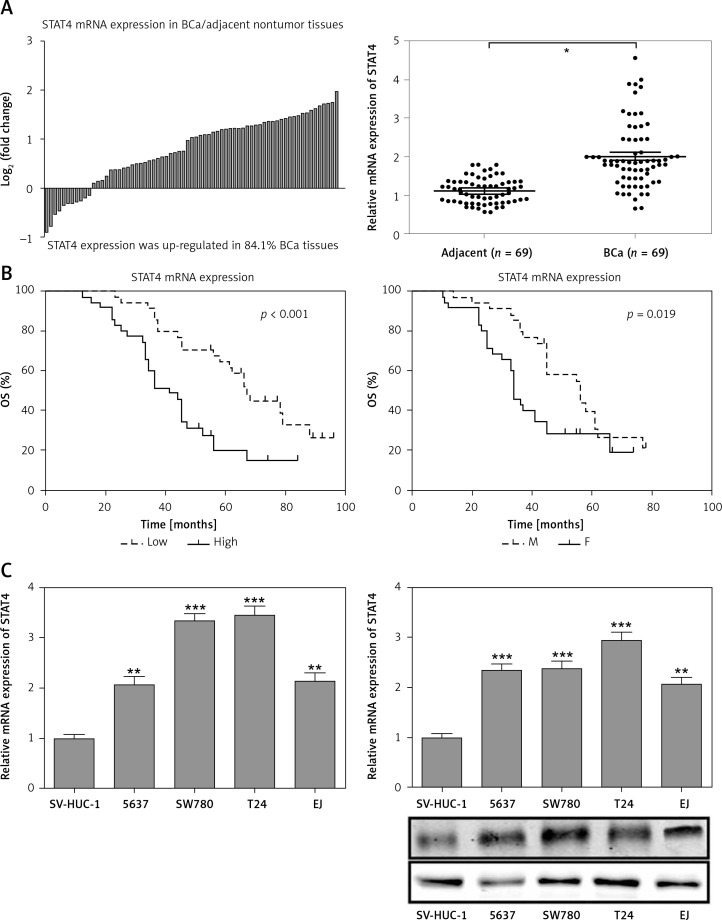
For most of the BCa patients (58/69, 84.1%) there was an increase in STAT4 mRNA expression in BCa tissues compared to paired normal bladder tissues (Figure 3 A). Clinicopathological statistical data showed that high STAT4 expression was correlated with poor tumor stage (p = 0.011), histological grade (p = 0.002) and vascular invasion (p = 0.002) but was not correlated with age, gender, tumor size or lymph node metastasis in BCa patients (Table I). We also found that BCa patients with high STAT4 expression had worse OS and DFS (Figure 3 B). To investigate the role of STAT4 in the tumorigenesis of BCa, we detected the expression level of STAT4 in the normal bladder epithelial cell line SV-HUC-1 and 4 human BCa cell lines, 5637, SW780, T24 and EJ. RT-qPCR and western blotting assays demonstrated that STAT4 mRNA and protein levels were significantly upregulated in BCa cell lines compared with human normal bladder epithelial cells (Figure 3 C). Next, we investigated whether STAT4 could regulate cell proliferation and apoptosis in vitro. We transfected SW780 and T24 cells with shRNA to inhibit the expression of STAT4 and observed that the expression of STAT4 was downregulated by approximately 70% (Figure 3 D). The STAT4 inhibition experiments indicated that STAT4 loss-of-function significantly inhibited proliferation (Figure 3 E) and induced apoptosis (Figure 3 F) in SW780 and T24 cells.
Figure 3.
STAT4 expression and prognostic significance in BCa patients. A – The log2 (fold change) of STAT4 mRNA expression in 69 pairs of BCa and adjacent nontumor tissues was calculated, and the results demonstrated that STAT4 was significantly higher in BCa tissues compared with adjacent nontumor tissues. B – Kaplan-Meier analysis and logrank test were performed to analyze the association between STAT4 expression and OS or DFS. C – The mRNA and protein expression levels of STAT4 in the normal bladder epithelial cell line SV-HUC-1 and 4 human BCa cell lines, 5637, SW780, T24 and EJ, were measured using RT-qPCR and western blotting, respectively (n = 3 in each group) D – An shRNA was designed to silence the expression of STAT4 in SW780 and T24 cells. After transfection of SW780 and T24 cells with sh-Con or sh-STAT4, cell proliferation (E) and apoptosis (F) were measured using CCK-8 assay and Annexin V-FITC double staining, respectively. *P < 0.05, **p < 0.01, ***p < 0.001 compared with the control group. The lines represent that a statistical analysis was performed between BCa tissues and adjacent non-tumor tissues. Data are presented as the mean ± SE
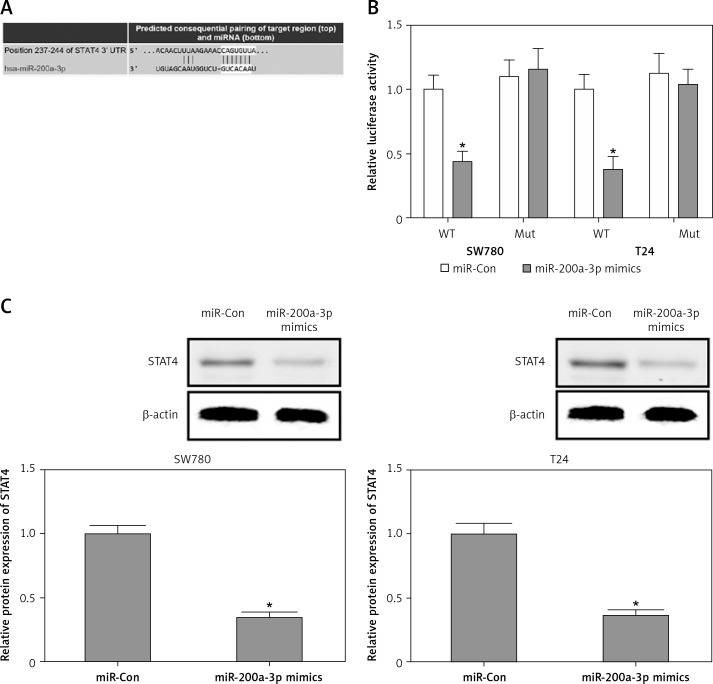
STAT4 is a direct target of miR-200a-3p
To investigate whether miR-200a-3p as a posttranscriptional regulator targeted the inhibition of STAT4, a bioinformatics analysis was performed in TargetScan (www.targetscan.org) to predict the binding sites of miR-200a-3p in the 3′-UTR of STAT4. As shown in Figure 4 A, the conserved complementary sequences between miR-200a-3p and the 3′-UTR of STAT4 were predicted. Subsequently, luciferase reporter assays were performed to validate the ability of miR-200a-3p to target the 3′-UTR of STAT4. The results demonstrated that the luciferase activity was markedly reduced in SW780 and T24 cells co-transfected with miR-200a-3p mimics and the WT 3′-UTR of STAT4, while the luciferase activity caused no obvious change in the cells cotransfected with the mut 3′-UTR of STAT4 (Figure 4 B). In addition, downregulation of STAT4 protein expression was observed in SW780 and T24 cells transfected with miR-200a-3p mimics (Figure 4 C). These findings suggested that miR-200a-3p had the ability to suppress STAT4 protein expression by post-transcriptional repression.
Figure 4.
STAT4 is a direct target of miR-200a-3p. A – The bioinformatics analysis was performed in TargetScan (www.targetscan.org) to predict the binding sites of miR-200a-3p in the 3′-UTR of STAT4. B – A luciferase reporter assay was performed in SW780 and T24 cells cotransfected with miR-200a-3p mimics and a plasmid containing the WT or mut 3′-UTR of STAT4. C – After transfection of miR-200a-3p mimics into SW780 and T24 cells, the protein expression of STAT4 was measured using western blotting. *P < 0.05 compared with the control group
miR-200a-3p-induced growth inhibition and apoptosis were reversed by STAT4 overexpression
The above results suggested that miR-200a-3p and STAT4 might play reciprocal roles in the carcinogenesis of BCa. To further validate this conclusion, miR-200a-3p mimics and STAT4 overexpression plasmids were transfected into SW780 and T24 cells. Western blotting analysis indicated that the protein expression of STAT4 was remarkably increased after cotransfection of SW780 and T24 cells with miR-200a-3p mimics and STAT4 overexpression plasmids (Figure 5 A). The effects of miR-200a-3p on proliferation and apoptosis were evaluated using a CCK-8 assay and Annexin V-FITC/PI double staining. The results indicated that the miR-200a-3p mimics dramatically repressed proliferation and induced apoptosis in SW780 and T24 cells (Figures 5 B and C), while the anti-proliferative and pro-apoptotic effects of miR-200a-3p were neutralized by STAT4 overexpression plasmids in SW780 and T24 cells (Figures 5 B and C).
Figure 5.
The miR-200a-3p-induced growth inhibition and apoptosis were reversed by STAT4 overexpression. After cotransfection of STAT4 overexpression plasmids and miR-200a-3p mimics into SW780 and T24 cells, the protein expression of STAT4 was measured using western blotting (A); cell proliferation (B) and apoptosis (C) were measured using CCK-8 assay and Annexin V-FITC double staining, respectively
*P < 0.05.
Discussion
As immune and inflammatory regulators STAT family members (STAT1-6) have been linked to cancer metastasis, and STAT inhibitors may be potential therapeutic drugs for cancer [25–27]. In tumorigenesis, STAT proteins become constitutively active by phosphorylation, and phosphorylated STATs dimerize, ultimately leading to transcriptional activation of target genes [27–29]. For example, STAT3-mediated increased expression of RhoU enhances breast cancer cell migration [28]. STAT2 cooperates with nuclear factor-κB to drive IL6 expression, which can promote the protumorigenic activation of STAT3 [29]. STAT1-mediated pentraxin3 secretion in fibroblasts promotes cell proliferation in breast cancer [27]. Unlike other STAT proteins (for example, STAT1, STAT2 and STAT3), which appear to be widely expressed in a variety of cancer tissues, STAT4 has recently emerged in a few tumor types, including gastric cancer, ovarian cancer, colorectal cancer and lymphoma [7–9, 30].
In the present study, abnormal STAT4 activation was detected in over 80% of BCa tissues, and STAT4 activation was associated with poor pathological classification and vascular invasion. High STAT4 expression has been shown to correlate with poor OS and DFS in BCa patients. These results indicated that STAT4 seems to promote tumorigenesis and could serve as an independent biomarker for predicting the survival prognosis of BCa patients. The in vitro experiments revealed upregulation of STAT4 expression in BCa cell lines compared with the human normal bladder epithelial cell line. Subsequently, inhibition of STAT4 with shRNA led to growth suppression and an increase in apoptosis in SW780 and T24 cells. These findings provide a good avenue for developing STAT4 inhibitors for the treatment of BCa. Mora Vidal et al. discovered that erlotinib treatment of RT112 cells induced the reduction of STAT4 phosphorylation, while the specific roles and underlying molecular mechanisms of STAT4 in RT112 cell proliferation have not been determined [31].
On the other hand, bioinformatics analysis predicted one conserved binding site of miR-200a-3p in the 3′-UTR of STAT4. Experimental measurements validated STAT4 as a direct target of miR-200a-3p and demonstrated that STAT4 could be posttranscriptionally inhibited by miR-200a-3p mimics. Further studies showed that transfection with miR-200a-3p mimics led to growth inhibition and increase in apoptosis, while cotransfection of miR-200a-3p mimics and STAT4 overexpression plasmids reversed the antiproliferative activity of miR-200a-3p in SW780 and T24 cells. Thus, it could be inferred that miR-200a-3p might serve as a potential treatment target for regulating STAT4 to inhibit the growth of BCa. Similar results were reported by Xiao et al. in gastric cancer [30] where the miR-141-3p/200a-3p family member miR-141-3p, a potential tumor suppressor miRNA, decreased gastric carcinogenesis by targeting STAT4.
The anti-oncogenic function of miR-200a-3p has been validated in several cancer types, such as papillary thyroid carcinoma [32], renal cell carcinoma [19] and HCC [18]. We observed decreased expression of miR-200a-3p in BCa tissues, and high miR-200a-3p expression was correlated with favorable OS and DFS in BCa patients, suggesting that downregulation of miR-200a-3p might be an independent biomarker for predicting poor prognosis of BCa patients. Tak et al. found that the levels of miR-200a-3p were relatively low in cancerous liver tissues and that miR-200a-3p could serve as a potential marker for the diagnosis and prognosis of HCC [33]. Du et al. reported that downregulation of miR-200a-3p in the urine served as a novel noninvasive diagnostic biomarker and was associated with worse recurrence-free survival in BCa patients [34].
In conclusion, as an oncogene, STAT4 might contribute to the progression of BCa, and high STAT4 expression might be independently associated with poor prognosis in BCa patients. We also found that STAT4 is a direct target gene of miR-200a-3p and could be posttranscriptionally inhibited by miR-200a-3p, which had anti-proliferative and pro-apoptotic effects in BCa cells. These results indicated that the miR-200a-3p/STAT4 signaling pathway plays a vital role in the tumorigenesis of BCa. This mechanism provides a valuable and promising therapeutic avenue for the treatment of BCa.
Acknowledgments
This research was supported by the Science and Technology Program of Dongguan Health Bureau of China (Grant. No: 2016105101045).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Antoni S, Ferlay J, Soerjomataram I, Znaor A, Jemal A, Bray F. Bladder cancer incidence and mortality: a global overview and recent trends. Eur Urol 2017; 71: 96-108. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115-32. [DOI] [PubMed] [Google Scholar]
- 3.Alfred Witjes J, Lebret T, Comperat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol 2017; 71: 462-75. [DOI] [PubMed] [Google Scholar]
- 4.Ploeg M, Aben KK, Kiemeney LA. The present and future burden of urinary bladder cancer in the world. World J Urol 2009; 27: 289-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi A, Eto M, Shiota M, et al. Sunitinib enhances antitumor effects against chemotherapy-resistant bladder cancer through suppression of ERK1/2 phosphorylation. Int J Oncol 2012; 40: 1691-6. [DOI] [PubMed] [Google Scholar]
- 6.Miyata Y, Matsuo T, Nakamura Y, et al. Expression of class III beta-tubulin predicts prognosis in patients with cisplatin-resistant bladder cancer receiving paclitaxel-based second-line chemotherapy. Anticancer Res 2018; 38: 1629-35. [DOI] [PubMed] [Google Scholar]
- 7.Lupov IP, Voiles L, Han L, et al. Acquired STAT4 deficiency as a consequence of cancer chemotherapy. Blood 2011; 118: 6097-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao L, Ji G, Le X, et al. An integrated analysis identifies STAT4 as a key regulator of ovarian cancer metastasis. Oncogene 2017; 36: 3384-96. [DOI] [PubMed] [Google Scholar]
- 9.Cheng JM, Yao MR, Zhu Q, et al. Silencing of stat4 gene inhibits cell proliferation and invasion of colorectal cancer cells. J Biol Regul Homeost Agents 2015; 29: 85-92. [PubMed] [Google Scholar]
- 10.Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 induced by helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem 2014; 33: 1003-12. [DOI] [PubMed] [Google Scholar]
- 11.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 2016; 17: 719-32. [DOI] [PubMed] [Google Scholar]
- 12.Kasinski AL, Slack FJ. Epigenetics and genetics. Micro-RNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer 2011; 11: 849-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10: 704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer 2018; 17: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 2017; 18: 1646-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res 2018; 37: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi T, Hua Q, Ma Z, Lv Q. Downregulation of miR-200a-3p induced by hepatitis B Virus X (HBx) Protein promotes cell proliferation and invasion in HBV-infection-associated hepatocarcinoma. Pathol Res Pract 2017; 213: 1464-9. [DOI] [PubMed] [Google Scholar]
- 18.Gong Y, Mao J, Wu D, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 2018; 18: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Jiang F, Song H, Li X, Xian J, Gu X. Micro-RNA-200a-3p suppresses tumor proliferation and induces apoptosis by targeting SPAG9 in renal cell carcinoma. Biochem Biophys Res Commun 2016; 470: 620-6. [DOI] [PubMed] [Google Scholar]
- 20.Berthois Y, Delfino C, Metellus P, et al. Differential expression of miR200a-3p and miR21 in grade II-III and grade IV gliomas: evidence that miR200a-3p is regulated by O(6)-methylguanine methyltransferase and promotes temozolomide responsiveness. Cancer Biol Ther 2014; 15: 938-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402-8. [DOI] [PubMed] [Google Scholar]
- 22.Gong J, Wang ZX, Liu ZY. miRNA1271 inhibits cell proliferation in neuroglioma by targeting fibronectin 1. Mol Med Rep 2017; 16: 143-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q, Liu Y, Xie H, et al. Lentivirus-mediated shRNA targeting MUTYH inhibits malignant phenotypes of bladder cancer SW780 cells. Onco Targets Ther 2018; 11: 6101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin T, Liu MM, Jin RT, Kong J, Wang SH, Sun WB. miR-152-3p modulates hepatic carcinogenesis by targeting cyclin-dependent kinase 8. Pathol Res Pract 2019; 215: 152406. [DOI] [PubMed] [Google Scholar]
- 25.Heppler LN, Frank DA. Targeting oncogenic transcription factors: therapeutic implications of endogenous stat inhibitors. Trends Cancer 2017; 3: 816-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trivedi S, Starz-Gaiano M. Drosophila Jak/STAT signaling: regulation and relevance in human cancer and metastasis. Int J Mol Sci 2018; 19: pii: E4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zellmer VR, Schnepp PM, Fracci SL, Tan X, Howe EN, Zhang S. Tumor-induced stromal STAT1 accelerates breast cancer via deregulating tissue homeostasis. Mol Cancer Res 2017; 15: 585-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monteleone E, Orecchia V, Corrieri P, et al. SP1 and STAT3 functionally synergize to induce the RhoU small GTPase and a subclass of non-canonical WNT responsive genes correlating with poor prognosis in breast cancer. Cancers 2019; 11: pii: E101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nan J, Wang Y, Yang J, Stark GR. IRF9 and unphosphorylated STAT2 cooperate with NF-kappaB to drive IL6 expression. Proc Natl Acad Sci USA 2018; 115: 3906-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao J, Zhong JH, Wei SH, et al. MiR-141-3p suppresses gastric cancer induced transition of normal fibroblast and BMSC to cancer-associated fibroblasts via targeting STAT4. Exp Mol Pathol 2019; 107: 85-94. [DOI] [PubMed] [Google Scholar]
- 31.Mora Vidal R, Regufe da Mota S, Hayden A, et al. Epidermal growth factor receptor family inhibition identifies P38 mitogen-activated protein kinase as a potential therapeutic target in bladder cancer. Urology 2018; 112: 225.e221-225.e227. [DOI] [PubMed] [Google Scholar]
- 32.Wu DM, Wang S, Wen X, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell Death Dis 2018; 9: 947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tak H, Kang H, Ji E, Hong Y, Kim W, Lee EK. Potential use of TIA-1, MFF, microRNA-200a-3p, and microRNA-27 as a novel marker for hepatocellular carcinoma. Biochem Biophys Res Commun 2018; 497: 1117-22. [DOI] [PubMed] [Google Scholar]
- 34.Du L, Jiang X, Duan W, et al. Cell-free microRNA expression signatures in urine serve as novel noninvasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Oncotarget 2017; 8: 40832-42. [DOI] [PMC free article] [PubMed] [Google Scholar]