Abstract
Read-across is a data gap filling technique utilized to predict the toxicity of a target chemical using data from similar analogues. Recent efforts such as Generalized Read-Across (GenRA) facilitate automated read-across predictions for untested chemicals. GenRA makes predictions of toxicity outcomes based on “neighboring” chemicals characterized by chemical and bioactivity fingerprints. Here we investigated the impact of biological similarities on neighborhood formation and read-across performance in predicting hazard (based on repeat-dose testing outcomes from US EPA ToxRefDB v2.0). We used targeted transcriptomic data on 93 genes for 1060 chemicals in HepaRG™ cells that measure nuclear receptor activation, xenobiotic metabolism, cellular stress, cell cycle progression, and apoptosis. Transcriptomic similarity between chemicals was calculated using binary hit-calls from concentration-response data for each gene. We evaluated GenRA performance in predicting ToxRefDB v2.0 hazard outcomes using the area under the Receiver Operating Characteristic (ROC) curve (AUC) for the baseline approach (chemical fingerprints) versus transcriptomic fingerprints and a combination of both (hybrid). For all endpoints, there were significant but only modest improvements in ROC AUC scores of 0.01 (2.1%) and 0.04 (7.3%) with transcriptomic and hybrid descriptors, respectively. However, for liver-specific toxicity endpoints, ROC AUC scores improved by 10% and 17% for transcriptomic and hybrid descriptors, respectively. Our findings suggest that using hybrid descriptors formed by combining chemical and targeted transcriptomic information can improve in vivo toxicity predictions in the right context.
Keywords: Generalized Read-Across (GenRA), ToxRefDB v2, High throughput transcriptomics (HTTr)
Introduction
The cost and duration of current animal testing approaches only permits a small fraction of the 32,898 chemicals in commerce (US EPA, 2019) to be thoroughly evaluated for human safety. Advances in computing resources, increased access to laboratory automation, and development of new approach methodologies (NAMs) have resulted in the generation of ’big data,’ and the potential for a disruptive change in the field of toxicology. In some jurisdictions and for specific regulatory purposes in particular, there has been a concerted uptake in the application of high-throughput in vitro data and computational models [8,15,46,70]. One example is the US EPA’s Endocrine Disruption Screening Program (EDSP) which permits the use of high throughput assays and computational models to evaluate and screen chemicals. The OECD’s IATA Case studies programme has been a notable activity where Member Countries have submitted case studies in an effort to build capacity and share experiences for how to apply and interpret NAM data for different regulatory purposes. Many of the case studies submitted have been read-across orientated demonstrating how NAM data can be used in conjunction with in vivo data in a weight of evidence approach as well as providing a means to substantiate biological/mechanistic similarity across source analogues ([86]; see also Sakuratani et al. [87] for a summary of the programme and the learnings gained). More recently Health Canada [1] published their scientific approach whereby high throughput data could be coupled with predicted exposure information to establish defined ratios akin to margins of exposure (MoE) for priority setting and risk assessment contexts [53,62,63]. Technologies such as high-throughput and high-content screening methods (HTS/HCS) [29], high-throughput transcriptomics data (HTTr), high-throughput phenotypic profiling (HTPP) for cellular morphology, and high-throughput exposure modelling are broadly referred to as NAMs, providing information about chemical hazards and risks without using intact animals [30]. NAMs also encompass in silico approaches such as (quantitative) structure-activity relationships ((Q)SARs) and read-across [30,55].
There is renewed recognition that in silico approaches can provide practical alternatives to bridge the lack of knowledge about chemical properties and their biological activities [13,65]. In Europe, the REACH regulation calls for the use of non-animal methods to assess chemical toxicity [71,73,74], whereas, in the U.S., the EPA created a NAMs Work Plan to prioritize agency efforts and resources toward activities that will reduce the use of animal testing while continuing to protect human health and the environment [57].
Read-across is a data gap filling technique whereby information from a similar (source) analogue is used to infer the same properties for a substance of interest (target). Read-across has been in broad use for different regulatory purposes for decades [10,38,40,41], but its acceptance has been somewhat thwarted by how to address residual uncertainties [2,3,5,39,45]. The approach itself has always been a subjective expert-driven one, reliant on domain knowledge of the toxicity endpoint of interest and the different considerations that need to be taken on board to rationalize and justify a read-across prediction [10,66]. In recent years, there have been more efforts that seek to better characterize what those uncertainties might be and how to describe and document the considerations underpinning a read-across prediction. While frameworks for the development and acceptance of read-across have been published and extensively discussed (e.g. [5,39,75,76]), there remains a gap in understanding how to apply some of the principles in practice [3,7,37,41,45,47,48]. In the last five years, there has been a concerted move to consider NAMs as a means to enhance read-across. In addition to the work under the OECD IATA Case studies programme, there has been a large programme of work under the auspices of both the EU SEURAT and EU ToxRisk programmes exploring how NAM data can substantiate biological similarity specific case studies [16,49]. However there are many other examples where NAM approaches have been utilized including Brandt et al. [78] who evaluated a WoE approach to assess the persistence characteristics of certain phenolic benzotriazoles. Gautier et al. showcased use of a defined approach as part of a read-across for a resorcinol case study for the skin sensitization endpoint [83]. Gelbke et al. evaluated a category of methacrylates developed to showcase an example assessment prepared to meet the needs of a REACH submission [82]. Grimm et al. [20] used phenotypic and transcriptomic assay data to demonstrate bioactivity similarity in a set of glycol ethers. Pestana et al. [84] showcased how NAM data could reduce uncertainty in the read-across within a category of tetraconazoles to address the information requirements of a 90-day study outcome. Data driven approaches using larger datasets have been also pursued including Sperber et al. [88] who utilized metabolomics, also discussed in Ball et al. [81] as well as exploiting HTS assays from PubChem or ToxCast as investigated in Firman et al. [77].
In our own work, we have also applied a data driven approach to read-across by establishing a baseline in performance using k-nearest neighbors and a similarity weighted activity approach based on chemical fingerprints to make in vivo toxicity predictions [51]. In subsequent analyses, we have been focused on exploring enhancements to read-across through characterizing other similarity considerations, e.g., physicochemical properties as a surrogate for bioavailability [26] and quantifying their relative contribution to improving read-across performance as well as transitioning to predictions of potency [25,27–28]. As noted earlier there have been other works which have explored the concept of biological similarity [16,69]. Here we present a proof of concept study using targeted HTTr data to make repeated dose study-type and liver specific toxicity (see Methods) predictions relative to chemical structural features or a combination of both.
Whilst the use of transcriptomic data is not in of itself novel, indeed the chemical and biological read-across (CBRA) approach, which GenRA was based on, utilized toxicogenomic data for inferring toxicity (Low et al.). With the increasing reliability, reproducibility and scalability of transcriptomic technologies [60]), “connectivity mapping” [33,34] is becoming a powerful approach for inferring the bioactivity of chemicals based on similarity between gene expression profiles. Connectivity mapping has been used to evaluate the mode of action of drugs [31] and to evaluate their safety [54]. More recently, Wang et al. developed “fish connectivity mapping” [59] by linking transcriptomic profiles of known chemicals in ecologically-relevant species with untested chemicals to evaluate their putative mechanisms. Similarly, De Abrew et al. investigated the mode of action for 34 chemicals using transcriptomic profiles produced in multiple cell types against a large reference database [12]. More recently, Harrill et al. used high-throughput transcriptomic (HTTr) data on environmental chemicals to evaluate their putative mechanisms and to estimate potency values using a gene signature-based concentration-response approach based on the concept of connectivity mapping [23]. Despite these advances, the application of transcriptomic data to read-across and especially in the context of traditional read-across remain at an early stage. How these data should be integrated with traditional data streams to make associations with regulatory relevant endpoints for risk assessment, remains an evolving area (see [6,68]).
As a prelude to evaluating the utility of whole transcriptomic data in read-across, we present an analysis using the expression levels measured for a targeted set of 93 transcripts in HepaRG™ cells treated with eight concentrations of 1060 chemicals. HepaRG™ cells express a full repertoire of xenobiotic-metabolizing enzymes, enabling more biological response, particularly from bioactivated or metabolically detoxified chemicals. Using these data, in addition to chemical structure data, repeat-dose toxicity outcomes, and predefined chemical clusters [51], we evaluated predictions of 922 toxicity endpoints using the GenRA approach [27,51] as implemented in genra-py, a new python package [52].
Materials and methods
The workflow followed in this analysis is captured in Fig. 1. Each step in the workflow, namely, data source selection, is described in more detail in the following sections.
Fig. 1.
Workflow for analysis of multiple descriptor types in GenRA.
Chemical libraries
Chemical libraries for this study were the ToxCast Phase I and Phase II libraries; the “phases” here indicate the order of testing by the ToxCast program and are unrelated to metabolism phases. Phase I of ToxCast focused on chemicals, including many pesticides, for which there were extensive in vivo studies for comparison. Phase II included a broader range of chemicals that are common in commerce and the environment. The full list of chemicals is provided as supplemental material, S1(a). Chemical samples were commercially procured, diluted in dimethyl sulfoxide (100% DMSO) to a stock concentration of 20 mM, and plated by Evotec (South San Francisco, CA). Analytical QC for the Phase I chemical inventory was performed using a combination of high-throughput liquid and gas chromatography-mass spectrometry to determine sample purity, parent mass, and sample stability in DMSO over time (https://www.epa.gov/chemical-research/toxcast-chemicals). Similar methods were applied to analyzing the Phase II library in association with the Tox21 project and are publicly available at https://tripod.nih.gov/tox21/samples.
Datasets
Transcriptomic data
The transcriptomic data used in the analysis are described in detail in Franzosa et al. [18] and a brief outline is provided here. These data were generated using the Life Technologies/Expression Analysis (LTEA) assay, which is part of the ToxCast [29,43] HTS data set. The LTEA assay was designed to investigate the role of nuclear receptor activation and key cellular events in hepatotoxicity [50] in a liver-specific in vitro model. In the LTEA assay, HepaRG™ cells were treated with 8 concentrations of 1060 chemicals for 24 h, and the expression levels of genes were measured using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Due to the format of the assay, only a set of 93 transcripts could be concurrently measured and they were targeted to assessing nuclear receptor activation, xenobiotic metabolism, cellular stress, cell cycle progression, and apoptosis, which are key events in different pathways to liver toxicity (the list of genes is provided as Supplemental Material, S2). Concentration-response data for each of the 93 transcripts and cytotoxicity (which was assessed using lactate dehydrogenase (LDH) assay) were analyzed using the ToxCast analysis pipeline package in (R/tcpl) [17]. Each gene was curve-fit twice, once for up-regulation and a second time for down-regulation. After curvefitting, the efficacy, potency (AC50), and hit-call were stored in a MySQL database (invitroDB v3.00) [56]. The hit-call for each chemical and transcript was assigned a binary active (1) or inactive (0) value based on level 5 data [17]. A hit-call was also assigned an “up” or “down” direction depending on whether the efficacy was positive or negative, respectively. The transcriptomic data for each chemical was represented using the hit calls in two ways: firstly, as a vector comprising the binary hit-calls for the 93 genes and secondly, as a vector consisting of the 190 directional activities of the 93 genes (i.e., each transcript was included once for the “up” and once for the “down” direction). The former will be referred to as ‘gene’ (ge) throughout this paper and the latter will be referred to as ‘assay’ (asy).
The 3 LTEA datasets used in the current study are publicly available from the EPA ftp site at: ftp://newftp.epa.gov/COMPTOX/CCTE_Publication_Data/CCED_Publication_Data/Wambaugh/ToxCast_LTEA
These three data sets comprise LTEA_Inucyte_Images.zip (images of each cell culture), LTEA_Level2_20191119.zip (the raw, unnormalized data), and LTEA_Level5_20191119.zip (results of concentration-response curve-fitting). All other data and analysis scripts used are included in [18] and its supplementary information files.
Chemical structure data
Morgan fingerprints (mrgn) [44], Topological Torsion (tptr) fingerprints [42], and ToxPrint (toxp) [67] chemotypes were computed for the 1060 chemicals. These descriptors were represented as binary (bit) vectors where the presence or absence of each structural element was represented as a 1 or 0, respectively. Mrgn and tptr fingerprints were calculated using the freely available python RDKit [35], whereas Tox-Prints [67]; chemotyper.org) were downloaded using the batch search functionality (ChemoTyper format) from the EPA CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard) [64].
Toxicity data
Toxicity data were extracted from ToxRefDB version 2.0 [61], which is accessible as a MySQL Dump file from ftp://newftp.epa.gov/comptox/High_Throughput_Screening_Data/Animal_Tox_Data/current/. ToxRefDB describes the in vivo effects of repeat-dose testing for hundreds of substances observed across various species and target organs. The previous version of ToxRefDB contained only data for positive related effects; however, recent refinements now enable the distinction between untested chemicals and chemicals tested with no effects (negative endpoints) based on adherence to Office of Chemical Safety and Pollution Prevention (OCSPP), National Toxicology Program (NTP) specifications or OECD test guidelines. Chemicals producing significant effects for an endpoint were categorized as positive (1), and those that did not produce significant effects were categorized as negative (0). We found 935 chemicals were assigned with positive or negative toxicity assignments for 252 target organs and effects in 9 guideline repeat dose testing study types in ToxRefDB v2. Endpoints for these target organ and effects are grouped by study type, endpoint category and endpoint type. The 9 study types were namely: chronic toxicity (chr), subchronic toxicity (sub), subacute toxicity (sac), developmental toxicity (dev), multigenerational reproductive toxicity (mgr), reproductive toxicity (rep), developmental neurotoxicity (dnt), acute toxicity (acu), and neurological toxicity (neu). Toxicity studies where a specific guideline was not reported were categorized as “other” (oth). Endpoint categories include cholinesterase, developmental, reproductive, and systemic. For systematic endpoint categories, endpoint types include clinical chemistry, hematological, and in life observation. Overall, there were 922 unique toxicity study targeted effects or endpoints that were used to evaluate the accuracy of GenRA in making in vivo toxicity predictions.
Chemical clustering data
Chemical clusters previously generated in [51] were utilized to assign membership for the total set of 1060 chemicals and chemical controls (i.e. induction positive controls, cytotoxicity controls, and vehicle controls). These clusters were used to explore local domains of chemicals where read-across based on either chemical, biological, or hybrid descriptor performed the best. Nine hundred ninety-four (994) substances were matched by their chemical identifier, CASRN or DTXSID, to clusters [51]. Clusters for the remaining unassigned 71 chemicals were then determined on the basis of their Jaccard pairwise similarity index [32] calculated using Morgan chemical fingerprints. The similarity index helped to identify the most appropriate cluster for each unassigned chemical, i.e., the pairwise similarity between chemicals belonging to clusters and those not belonging defined the most likely cluster assignment. These clusters formed the basis of a ‘local’ performance assessment of the GenRA approach (see SECTION 3.5: Comparing Local Predictive Performance of all Toxicity Outcomes for Morgan, Gene, and Morgan and Gene Hybrid Descriptors).
Summary of GenRA approach
The workflow underpinning the GenRA approach [27] was adapted in this study. Rather than identifying analogues based on structural similarity, transcriptomic data (characterizing mechanistic similarity) or a combination of transcriptomic data and chemical fingerprints were used to identify analogues. In addition, whilst the original GenRA approach used the Jaccard distance, two other distance metrics were explored, the Euclidean distance [11] and the Manhattan distance [4,9]. The similarity weighted activity was calculated using transcriptomic (bio) or chemical (chm) descriptors or a combination of the two (see Fig. 1). A 5-fold grid search cross-validation determined the appropriate number of neighbors (ranging from 1 to 15) and the similarity metric based on the optimal performance, which was assessed using area under the receiver operating characteristic curve (AUC). The impact on the performance of using these different metrics (Jaccard, Euclidean, Manhattan) for each descriptor (bio, chm, chm + bio) combination was evaluated. For each chemical, similarity (Jaccard, Euclidean, Manhattan) was calculated for each of the chemical (chm), biological (bio), and hybrid chemical and biological descriptor (CB) types as listed in Supplemental Material, S3.
Evaluating GenRA performance for different descriptor sets
The prediction accuracy of in vivo toxicity outcomes across all chemicals was evaluated in two ways for each of the three descriptor sets (chm, bio, and CB). 1) A ‘local’ performance evaluation was conducted utilizing predefined clusters of structurally similar chemicals to aggregate the predictions made for all the chemicals in the dataset (as described in section 2.2.4). This was considered a ‘local-validity’ approach whereby the success of read-across for these diverse descriptors was evaluated to determine if chm, bio, or CB predictive performance was specific to certain chemical classes and/or toxicity endpoints. 2) Additionally, an overall, ‘global’ performance evaluation was implemented on the entire data sets to evaluate descriptor performance. For each toxicity endpoint, the AUC measured the prediction accuracy of each descriptor for all chemicals for k-nearest neighbors. Predictive performance for each of the different descriptor types across a neighborhood and a single metric for other toxicity effects were compared in both the global and local analysis. The best similarity metric and the number of nearest neighbors were determined based on the optimal AUC performance. In order to assess the significance of AUC scores, we also calculated empirical p-values based on permutation testing as described in [51]. Lastly, we evaluated the statistical significance of differences between AUC values due to various factors using analysis of variance (ANOVA). One-way ANOVA on AUC scores was used to compare individual differences in performance for the various descriptors, followed by multiple comparisons of mean differences using Tukey’s honest significance difference (HSD) test.
Data analysis and code
Data processing and analysis were conducted in both the R (Version 3.6.1) and Python programming (Version 3.8.2) languages. Specifically, transcriptomics data was retrieved from the EPA Center for Computational Toxicology MySQL invitrodb (Version 3.0) using the tcpl package in R [17] and pre-processed for analysis in python. The RDKit package [35] was used to generate chemical fingerprints or downloaded from the EPA CompTox Chemicals Dashboard, and genra-py [52] was utilized for all toxicity predictions. Jupyter notebooks for the entire analysis workflow are provided on GitHub [https://github.com/i-shah/genra-ltea]. The input data files are provided as Supplemental Material, S1(b–e) from the journal website.
Results
We first conducted a global analysis of all chemicals to determine the optimal choice of GenRA parameters and input descriptor types for classifying all toxicity outcomes. We then conducted the same analysis at a local level using the predefined chemical clusters (described in section 2.2.4) to identify chemistry domains where GenRA performed better (or worse) than on a global level. For the sake of brevity, we refer to all descriptors by their abbreviations (which are listed in the List of Abbreviations).
Predicting toxicity endpoints by optimum metric and nearest neighbors
The classification accuracy of GenRA for each toxicity endpoint depends on the type of descriptor (chm, bio, and CB descriptors), the choice of similarity metric, and the number of nearest neighbors. We used genra-py to systematically explore the relationship between these parameters for every toxicity endpoint. The optimum metric and number of nearest neighbors for each of our descriptors for chronic liver toxicity is shown in Table 1. Although there was no consensus on the optimum number of neighbors across all descriptor types, many of the individual sets of descriptors (biological or chemical structure) and the combined (CB) descriptors performed better with the Jaccard similarity metric. The best performance was observed for MG using the Jaccard metric with ten nearest neighbors. Since the chronic liver toxicity endpoint had the most positive and negative examples among all liver endpoints in our data set, we chose the same GenRA parameters (Jaccard similarity metric with 10 nearest neighbors) that resulted in the best performance outcome for all other toxicity classes.
Table 1.
Appropriate metrics and number of nearest neighbors to assess the performance of various descriptor read-across prediction of chronic liver toxicity. Column 2 represents the descriptors type including: chemical structure (chm), biological (bio), and hybrid (CB). Descriptors names in column 3 include: Biological (B) − Assay (asy) and Gene (ge); Chemical Structure (C) − Morgan (mrgn), Toxprints (toxp), Topological Torsion (tptr), all chemicals combination descriptor (CC), Chemical + Biological Hybrid (CB) − Morgan + Assay (MA), Morgan + Gene (MG), Topological Torsion + Assay (TTA), Topological Torsion + Gene (TTG), Toxprints + Assay (TXA), Toxprints + Gene (TXG), and all chemical and biological descriptors combined (CB). Column 4 denotes the AUC performance values for each descriptor in predicting chronic liver toxicity effect. Column 5–6 denote the appropriate similarity metric and number of neighbors to make accurate prediction for each descriptor. Major descriptors of interest are marked bold.
| Liver Effect | Descriptor Type | Descriptor Name | AUC | Metric | N Neighbors |
|---|---|---|---|---|---|
| Chr_liver | Chm | Tptr | 0.6303 | Euclidean | 9 |
| Chm | Mrgn | 0.64549 | Jaccard | 8 | |
| Chm | Toxp | 0.61379 | Jaccard | 7 | |
| Bio | Ge | 0.648847 | Euclidean | 14 | |
| Bio | Asy | 0.6632 | Euclidean | 11 | |
| CB | mrgn + asy | 0.6883 | Jaccard | 13 | |
| CB | toxp + ge | 0.7044 | Jaccard | 10 | |
| CB | tptr + ge | 0.6818 | Euclidean | 6 | |
| CB | (CB) all | 0.6999 | Jaccard | 14 | |
| Chm | (CC) all | 0.6702 | Jaccard | 10 | |
| CB | mrgn + ge | 0.7049 | Jaccard | 10 | |
| CB | toxp + asy | 0.6992 | Jaccard | 14 | |
| CB | tptr + asy | 0.6721 | Manhattan | 5 |
Comparing global predictive performance of liver toxicity outcomes for different descriptors
The global accuracy of GenRA for predicting liver toxicity outcomes was evaluated using several types of descriptors, and their performance across in vivo outcomes in liver toxicity endpoints, along with statistical significance by permutational testing, shown in Table 2. For all chm, bio, and CB descriptors, the average predictive AUC score for each toxicity endpoint, the number of chemicals with positive and negative study level effects, and the number of chemicals with available toxicity information were calculated. The average AUC scores for all descriptors and study types were close to 0.5, suggesting poor performance overall but there were exceptions. On average, ‘all chemical descriptors combination’ (CC) descriptor outperformed (AUC = 0.61) other descriptors (individual and combinations) by 9%, 15%, and 27% when compared to hybrid descriptors, biological and individual chemical descriptors, respectively. For chronic, developmental, sub-acute, and sub-chronic liver outcomes, the TTG (AUC = 0.7, p < 0.05), MG (AUC = 0.64, p 0.05) and TTG (AUC = 0.74, p < 0.05) hybrid descriptors resulted in the highest respective significant predictive performance scores. The CC descriptors resulted in the best prediction performance (AUC = 0.68, p < 0.05) for multigenerational reproductive liver endpoints. Although the individual topological torsion chemical descriptors (tptr) most accurately predicted reproductive liver endpoints (AUC = 0.78), this result was not statistically significant. Lastly, none of the approaches produced significant AUC scores for developmental neurotoxicological hepatic effects. Despite the low average AUC scores, biological descriptors produced a modest 10% improvement over chemical descriptors. Hybrid descriptors generated an overall 16% increase in predictive performance than individual chemical descriptors and a 6% increase relative to biological descriptors.
Table 2.
Global Prediction Performance (AUC) for All Liver Endpoints Using Various Chemical Structure (C), Biological (B), and Hybrid (CB) Descriptors.
| Liver Toxicity Endpoints | asy (B) | ge (B) | mrgn (C) | toxp (C) | tptr (C) | MA (CB) | MG (CB) | TTA (CB) | TTG (CB) | TXA (CB) | TXG (CB) | CC (C) | CB (CB) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | *0.62 (245+, 134−, 379) | *0.63 (245+, 134−, 379) | *0.47 (236+, 128−, 364) | *0.43 (236+, 128−, 364) | *0.40 (236+, 128−, 364) | *0.69 (236+, 128−, 364) | *0.69 (236+, 128−, 364) | *0.68 (236+, 128−, 364) | *0.70 (236+, 128−, 364) | *0.66 (236+, 128−, 364) | *0.67 (236+, 128−, 364) | *0.63 (236+, 128−, 364) | *0.68 (236+, 128−, 364) |
| Dev | 0.48 (47+, 227−, 274) | 0.51 (47+, 227−, 274) | 0.54 (43+, 219−, 262) | 0.55 (43+, 219−, 262) | 0.56 (43+, 219−, 262) | *0.61 (43+, 219−, 262) | * 0.64 (43+, 219−, 262) | 0.56 (43+, 219−, 262) | 0.56 (43+, 219−, 262) | 0.55 (43+, 219−, 262) | 0.53 (43+, 219−, 262) | 0.58 (43+, 219−, 262) | 0.58 (43+, 219−, 262) |
| Dnt | 0.43 (4+, 63−, 67) | 0.31 (4+, 63−, 67) | 0.42 (4+, 57−, 61) | 0.33 (4+, 57−, 61) | 0.39 (4+, 57−, 61) | 0.48 (4+, 57−, 61) | 0.47 (4+, 57−, 61) | 0.36 (4+, 57−, 61) | 0.38 (4+, 57−, 61) | 0.31 (4+, 57−, 61) | 0.31 (4+, 57−, 61) | 0.54 (4+, 57−, 61) | 0.35 (4+, 57−, 61) |
| Mgr | 0.50 (118+, 90−, 208) | 0.54 (118+, 90−, 208) | *0.52 (115+, 86−, 201) | *0.48 (115+, 86−, 201) | *0.54 (115+, 86−, 201) | *0.63 (115+, 86−, 201) | *0.60 (115+, 86−, 201) | *0.59 (115+, 86−, 201) | *0.59 (115+, 86−, 201) | *0.61 (115+, 86−, 201) | *0.59 (115+, 86−, 201) | * 0.68 (115+, 86−, 201) | *0.62 (115+, 86−, 201) |
| Rep | 0.49 (7+, 47−, 54) | 0.49 (7+, 47−, 54) | *0.47 (7+, 44−, 51) | 0.43 (7+, 44−, 51) | 0.78 (7+, 44−, 51) | 0.40 (7+, 44−, 51) | *0.45 (7+, 44−, 51) | 0.50 (7+, 44−, 51) | 0.51 (7+, 44−, 51) | 0.31 (7+, 44−, 51) | 0.34 (7+, 44−, 51) | 0.48 (7+, 44−, 51) | 0.42 (7+, 44−, 51) |
| Sac | *0.67 (63+, 41, 104) | *0.69 (63+, 41, 104) | 0.49 (58+, 41−, 99) | 0.48 (58+, 41−, 99) | 0.46 (58+, 41−, 99) | *0.74 (58+, 41−, 99) | *0.72 (58+, 41−, 99) | 0.75 (58+, 41−, 99) | * 0.74 (58+, 41−, 99) | *0.69 (58+, 41−, 99) | *0.71 (58+, 41−, 99) | *0.74 (58+, 41−, 99) | *0.74 (58+, 41−, 99) |
| Sub | *0.56 (246+, 86−, 332) | *0.56 (246+, 86−, 332) | *0.48 (237+, 78−, 315) | *0.44 (237+, 78−, 315) | 0.51 (237+, 78−, 315) | *0.57 (237+, 78−, 315) | *0.56 (237+, 78−, 315) | 0.59 (237+, 78−, 315) | 0.56 (237+, 78−, 315) | *0.57 (237+, 78−, 315) | *0.53 (237+, 78−, 315) | 0.56 (237+, 78−, 315) | 0.56 (237+, 78−, 315) |
| Average | 0.537 | 0.530 | 0.483 | 0.449 | 0.520 | 0.588 | 0.590 | 0.576 | 0.575 | 0.529 | 0.525 | 0.609 | 0.563 |
Rows 1–7, all liver endpoint/study types. Row 8, average AUC value for each descriptor type between the various endpoints,
indicates significant p-value (p < 0.05) from permutation testing.
Endpoint AUC (# Chemicals with Positive (+), Negative (−) effects, Total Number of chemicals per descriptor). Liver endpoints include chronic (Chr), developmental (Dev), developmental neurotoxicity (Dnt), multigeneration reproductive (Mgr), reproductive (Rep), subacute (Sac), and sub- chronic (Sub). Descriptors types include: Biological (B) − Assay (asy) and Gene (ge); Chemical Structure (C) − Morgan (mrgn), Toxprints (toxp), Topological Torsion (tptr), all chemicals combination descriptor (CC), Chemical + Biological Hybrid (CB) − Morgan + Assay (MA), Morgan + Gene (MG), Topological Torsion + Assay (TTA), Topological Torsion + Gene (TTG), Toxprints + Assay (TXA), Toxprints + Gene (TXG), and all chemical and biological descriptors combined (CB). Highest significant AUC values for each endpoint are shown in bold font.
Comparing global predictive performance of all toxicity outcomes for different descriptors
GenRA’s global predictive performance for all outcomes in all studies was evaluated with various chemical, biological, and hybrid descriptors. A comparison of the performance for each of these descriptors is shown in Tables 3a and 3b. The columns in both tables denote the individual and hybrid combinations of biological and chemical descriptors, and the rows represent the toxicity endpoints and study types. For Table 3a, each element consists of the AUC (mean and standard deviation (SD)) of each study type for the various descriptors. In addition, we also calculated statistical differences between the AUC scores for descriptors using Tukey’s HSD post hoc test after ANOVA (p < 0.05 are signified with an asterisk). Because average AUC scores were quite low on average, each element within Table 3b includes the number and percentage of instances for all study types where the descriptor AUC greater than 0.70 (which is the same threshold as used in our earlier study [51])). For all study types CC had the greatest average performance (AUC = 0.53, p < 0.05)) compared to all other descriptors (Table 3a). Likewise, the CC descriptor most frequently predicted toxicity for all study/effect types, producing AUC values greater than 0.70 for 8% of the endpoints (Table 3b). Significant differences in AUC score (p < 0.05) were found to be study type specific. in performance were found for chronic, sub-acute, and sub-chronic study types. Individually, biological descriptors had higher mean performance scores than the single chemical descriptors for all study types; however, these difference were not significant.. The CB descriptors exceeded the mean prediction performance for all study types compared with the individual sets of descriptors, resulting in a minimal 6.25% increase in mean performance values overall. These significant differences were also chronic, sub-acute, and sub-chronic study type specific. Of the hybrid descriptors, the tptr + asy class more often accurately predicted toxicity overall for all study types (Table 3b) yet resulted in similar mean AUC scores to the other hybrid descriptors (Table 3a).
Table 3a.
Average Global Prediction Performance (AUC) for all Toxicity Endpoints Using Various Chemical Structure(C), Biological(B), and Hybrid(CB) Descriptors by study type. Rows 1–7, endpoints stratified by study types.
| Study | asy (B) | ge (B) | mrgn (C) | toxp (C) | tptr (C) | MA (CB) | MG (CB) | TTA (CB) | TTG (CB) | TXA (CB) | TXG (CB) | CC(C) | CB (CB) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | |||||||||||||
| Chr | 0.52 | 0.07 (245+, 134−, 379) | 0.51 | 0.07 (245+, 134−, 379) | 0.50 | 0.06 (236+, 128−, 364) | 0.50 | 0.05 (236+, 128−, 364) | 0.51 | 0.06 (236+, 128−, 364) | *0.55 | 0.09 (236+, 128−, 364) | *0.54 | 0.09 (236+, 128−, 364) | *0.55 | 0.09 (236+, 128−, 364) | *0.54 | 0.09 (236+, 128−, 364) | *0.54 | 0.09 (236+, 128−, 364) | *0.54 | 0.09 (236+, 128−, 364) | * 0.57 | 0.11 (236+, 128−, 364) | *0.55 | 0.10 (236+, 128−, 364) |
| Dev | 0.49 | 0.07 (47+, 227−, 274) | 0.49 | 0.07 (47+, 227−, 274) | 0.49 | 0.07 (43+, 219−, 262) | 0.48 | 0.06 (43+, 219−, 262) | 0.49 | 0.06 (43+, 219−, 262) | 0.51 | 0.10 (43+, 219−, 262) | 0.50 | 0.08 (43+, 219−, 262) | 0.51 | 0.08 (43+, 219−, 262) | 0.51 | 0.08 (43+, 219−, 262) | 0.51 | 0.09 (43+, 219−, 262) | 0.50 | 0.09 (43+, 219−, 262) | 0.51 | 0.07 (43+, 219−, 262) | 0.51 | 0.10 (43+, 219−, 262) |
| Dnt | 0.46 | 0.10 (4+, 63−, 67) | 0.46 | 0.11 (4+, 63−, 67) | 0.44 | 0.11 (4+, 57−, 61) | 0.45 | 0.11 (4+, 57−, 61) | 0.46 | 0.09 (4+, 57−, 61) | 0.48 | 0.14 (4+, 57−, 61) | 0.49 | 0.13 (4+, 57−, 61) | 0.49 | 0.12 (4+, 57−, 61) | 0.48 | 0.12 (4+, 57−, 61) | 0.50 | 0.12 (4+, 57−, 61) | 0.49 | 0.12 (4+, 57−, 61) | 0.48 | 0.13 (4+, 57−, 61) | 0.49 | 0.13 (4+, 57−, 61) |
| Mgr | 0.49 | 0.07 (118+, 90−, 208) | 0.49 | 0.08 (118+, 90−, 208) | 0.49 | 0.08 (115+, 86−, 201) | 0.48 | 0.08 (115+, 86−, 201) | 0.50 | 0.08 (115+, 86−, 201) | 0.51 | 0.10 (115+, 86−, 201) | 0.50 | 0.10 (115+, 86−, 201) | 0.51 | 0.10 (115+, 86−, 201) | 0.51 | 0.09 (115+, 86−, 201) | 0.49 | 0.09 (115+, 86−, 201) | 0.49 | 0.09 (115+, 86−, 201) | 0.51 | 0.10 (115+, 86−, 201) | 0.51 | 0.10 (115+, 86−, 201) |
| Rep | 0.44 | 0.12 (7+, 47−, 54) | 0.46 | 0.11 (7+, 47−, 54) | 0.46 | 0.13 (7+, 44−, 51) | 0.43 | 0.13 (7+, 44−, 51) | 0.47 | 0.14 (7+, 44−, 51) | 0.45 | 0.14 (7+, 44−, 51) | 0.45 | 0.14 (7+, 44−, 51) | 0.45 | 0.13 (7+, 44−, 51) | 0.45 | 0.12 (7+, 44−, 51) | 0.44 | 0.14 (7+, 44−, 51) | 0.46 | 0.12 (7+, 44−, 51) | 0.43 | 0.14 (7+, 44−, 51) | 0.45 | 0.13 (7+, 44−, 51) |
| Sac | 0.48 | 0.11 (63+, 41, 104) | 0.49 | 0.11 (63+, 41, 104) | 0.47 | 0.11 (58+, 41−, 99) | 0.46 | 0.10 (58+, 41−, 99) | 0.48 | 0.11 (58+, 41−, 99) | 0.50 | 0.12 (58+, 41−, 99) | 0.51 | 10 (58+, 41−, 99) | * 0.54 | 0.14 (58+, 41−, 99) | *0.53 | 0.13 (58+, 41−, 99) | 0.50 | 0.13 (58+, 41−, 99) | 0.50 | 0.13 (58+, 41−, 99) | *0.54 | 0.15 (58+, 41−, 99) | *0.53 | 0.14 (58+, 41−, 99) |
| Sub | 0.50 | 0.07 (246+, 86−, 332) | 0.50 | 0.06 (246+, 86−, 332) | 0.49 | 0.07 (237+, 78−, 315) | 0.49 | 0.07 (237+, 78−, 315) | 0.48 | 0.06 (237+, 78−, 315) | *0.54 | 0.10 (237+, 78−, 315) | *0.54 | 0.10 (237+, 78−, 315) | *0.54 | 0.09 (237+, 78−, 315) | *0.53 | 0.09 (237+, 78−, 315) | *0.53 | 0.09 (237+, 78−, 315) | *0.53 | 0.09 (237+, 78−, 315) | * 0.56 | 0.11 (237+, 78−, 315) | *0.55 | 0.10 (237+, 78−, 315) |
| All | 0.49 | 0.09 | 0.49 | 0.08 | 0.48 | 0.09 | 0.48 | 0.08 | 0.48 | 0.09 | *0.52 | 0.11 | *0.51 | 0.10 | *0.52 | 0.11 | *0.52 | 0.10 | *0.51 | 0.11 | *0.51 | 0.10 | * 0.53 | 0.12 | *0.52 | 0.11 |
AUC | SD (*) indicates study type specific statistically significant (p < 0.05) differences in mean AUC scores between descriptors and baseline approach (Morgan chemical Structure), (# Chemicals with Positive (+), Negative (−) effects, Total Number of chemicals per descriptor).
Study Types include chronic (Chr), developmental (Dev), developmental neurotoxicity (Dnt), multigeneration reproductive (Mgr), reproductive (Rep), subacute (Sac), and sub- chronic (Sub). Row 8, average AUC | SD value for each descriptor type between the various endpoints. Descriptors types include: Biological (B) − Assay (asy) and Gene (ge); Chemical Structure (C) − Morgan (mrgn), Toxprints (toxp), Topological Torsion (tptr), all chemicals combination descriptor (CC), Chemical + Biological Hybrid (CB) − Morgan + Assay (MA), Morgan + Gene (MG), Topological Torsion + Assay (TTA), Topological Torsion + Gene (TTG), Toxprints + Assay (TXA), Toxprints + Gene (TXG), and all chemical and biological descriptors combined (CB). Highest AUC values for significant study type are marked bold.
Table 3b.
Global Prediction Performance AUC Values Greater than 0.70 for all Toxicity Endpoints Using Various Chemical Structure (C), Biological (B), and Hybrid (CB) Descriptors by study type. Columns denote the Number | Percent for each descriptor type with predictive performance greater than 0.70(70%). Liver endpoints include chronic (Chr), developmental (Dev), developmental neurotoxicity (Dnt), multigeneration reproductive (Mgr), reproductive (Rep), subacute (Sac), and sub- chronic (Sub). Descriptors types include: Biological (B) − Assay (asy) and Gene (ge); Chemical Structure (C) − Morgan (mrgn), Toxprints (toxp), Topological Torsion (tptr), all chemicals combination descriptor (CC), Chemical + Biological Hybrid (CB) − Morgan + Assay (MA), Morgan + Gene (MG), Topological Torsion + Assay (TTA), Topological Torsion + Gene (TTG), Toxprints + Assay (TXA), Toxprints + Gene (TXG), and all chemical and biological descriptors combined (CB). Overall descriptor with highest number and percent AUC values greater than 0.70 is bolded.
| Study Type | asy (B) | ge (B) | mrgn (C) | toxp (C) | tptr (C) | MA (CB) | MG (CB) | TTA (CB) | TTG (CB) | TXA (CB) | TXG (CB) | CC (C) | CB (CB) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | 4 | 2.4% | 2 | 1.2% | 3 | 1.8% | 0 | 0 | 3 | 1.8% | 7 | 4.2% | 6 | 3.6% | 10 | 6.0% | 5 | 3.0% | 10 | 6.0% | 9 | 5.4% | 17|10.2% | 10 | 6.0% |
| Dev | 3 | 2.6% | 2 | 1.7% | 3 | 2.6% | 1 | 0.8% | 2 | 1.7% | 5 | 4.3% | 5 | 4.3% | 3 | 2.6% | 3 | 2.6% | 5 | 4.3% | 5 | 4.3% | 4 | 3.4% | 3 | 2.6% |
| Dnt | 2 | 3.0% | 2 | 3.0% | 3 | 4.6% | 3 | 4.6% | 2 | 3.0% | 4 | 6.1% | 4 | 6.1% | 3 | 4.6% | 2 | 3.0% | 5 | 7.6% | 4 | 6.1% | 3 | 4.6% | 4 | 6.1% |
| Mgr | 1 | 0.8% | 3 | 2.2% | 4 | 3.0% | 4 | 3.0% | 6 | 4.5% | 6 | 4.5% | 6 | 4.5% | 5 | 3.7% | 5 | 3.7% | 4 | 3.0% | 4 | 3.0% | 7 | 5.2% | 6 | 4.5% |
| Rep | 3 | 4.5% | 2 | 3.0% | 6 | 9.0% | 3 | 4.5% | 3 | 4.5% | 5 | 7.5% | 3 | 4.5% | 4 | 6.0% | 3 | 4.5% | 5 | 7.5% | 4 | 6.0% | 3 | 4.5% | 4 | 6.0% |
| Sac | 5 | 3.9% | 6 | 4.7% | 6 | 4.7% | 3 | 2.4% | 4 | 3.2% | 9 | 7.1% | 9 | 7.1% | 16 | 12.6% | 12 | 9.5% | 7 | 5.5% | 9 | 7.1% | 13 |10.2% | 12 | 9.5% |
| Sub | 4 | 2.4% | 2 | 1.2% | 4 | 2.4% | 2 | 1.2% | 3 | 1.8% | 11 | 6.5% | 11 | 6.5% | 11 | 6.5% | 10 | 5.9% | 7 | 4.1% | 6 | 3.6% | 15 | 8.9% | 10 | 5.9% |
| All | 22 | 2.8% | 19 | 2.4% | 29 | 3.6% | 16 | 2.0% | 13 | 2.9% | 47 | 6.3% | 44 | 5.6% | 52 | 6.7% | 41 | 5.3% | 43 | 5.5% | 41 | 5.3% | 62 | 8.0% | 49 | 6.3% |
Comparing global predictive performance of all toxicity outcomes for Morgan, Gene, and Morgan + Gene hybrid descriptors
A summary of GenRA predictions for selected chemical, biological, and hybrid descriptors is given in Table 4. These descriptors, mrgn and ge, were chosen to compare the baseline GenRA approach from [51], which utilizes mrgn fingerprints and bioactivity assay data (821 HTS assays from ToxCast Phase I and II compounds), and the chemical-biological hybrid to make in vivo toxicity predictions. Similar to [51], the hybrid (mg) descriptor class outperformed the others (number and percent of mg > B & C = 376 | 44%, respectively), resulting in a higher average performance value of 0.51 (SD 0.11) and more total cases (46 | 5.9%) in which study types were predicted with AUC > 0.7. In general, the hybrid descriptor generated a 4% increase in the average prediction score for all toxicity endpoints over biological descriptors and 6.25% over chemical descriptors. The mean AUC scores were statistically significant (p < 0.05) for chronic, developmental neurotoxicity (mrgn-mg), sub-acute, and sub-chronic study types. While on average biological descriptors displayed greater performance than chemical descriptors, chemical descriptors more often outperformed biological descriptors (C > B & CB in comparison to B > C & CB) and accurately predicted study type toxicity (27 total >0.70 vs. 18 total >0.70). Chemical descriptors were able to predict study type toxicity more often consistent with the results generated with a comparison of all descriptors and types (Table 3b). Based on Tukey’s HSD post hoc analysis, overall differences mean performance score across all study types was statistically significant (p < 0.05). Likewise, significance was also study type specific (for chr and sub study types).
Table 4.
Prediction Performance (AUC) for All Toxicity Endpoints Using Morgan Chemical Structure(C), Gene Level Biological(B), and Hybrid(CB) Descriptors by study type. Column 1 expresses various toxicity study types and the total number of chemicals with in vivo data available for all descriptors (in parenthesis). Columns 2–3 comprises the average by study type AUC | SD.
| Study | ge (B) | mrgn (C) | MG(CB) | B > =0.70 | C >=0.70 | CB >=0.70 | B > CB & C | C > B & CB | CB > B & C |
|---|---|---|---|---|---|---|---|---|---|
| Chr (364) | *0.51 | 0.07 | 0.50 | 0.07 | *0.54 | 0.09 | 2 | 1.2% | 2 | 1.2% | 7 | 4.2% | 41 | 25% | 40 | 24% | 86 | 51% |
| Dev (43) | 0.49 | 0.07 | 0.49 | 0.08 | 0.50 | 0.09 | 2 | 1.7% | 4 | 3.4% | 3 | 2.6% | 29 | 25% | 45 | 38% | 43 | 37% |
| Dnt (61) | 0.44 | 0.12 | 0.42 | 0.11 | 0.48 | 0.13 | 1 | 1.5% | 2 | 3.0 | 4 | 6.1% | 12 | 18% | 20 | 30% | 34 | 52% |
| Mgr (201) | 0.48 | 0.09 | 0.48 | 0.09 | 0.50 | 0.11 | 3 | 2.2% | 3 | 2.2% | 8 | 6.0% | 39 | 29% | 40 | 30% | 55 | 41% |
| Rep (51) | 0.45 | 0.12 | 0.45 | 0.15 | 0.45 | 0.14 | 1 | 1.5% | 6 | 9.0% | 4 | 6.0% | 23 | 34% | 20 | 30% | 24 | 36% |
| Sac (99) | 0.49 | 0.11 | 0.46 | 0.12 | *0.50 | 0.12 | 6 | 4.7% | 5 | 3.9% | 8 | 6.3% | 41 | 32% | 45 | 35% | 41 | 32% |
| Sub (315) | *0.50 | 0.07 | 0.49 | 0.08 | *0.54 | 0.10 | 3 | 1.8% | 5 | 3.0% | 12 | 7.1% | 36 | 21% | 40 | 24% | 93 | 55% |
| ALL | *0.49 | 0.09 | 0.48 | 0.10 | * 0.51 | 0.11 | 18 | 2.26% | 27 | 3.39% | 46 | 5.9% | 221 | 26% | 250 | 30% | 376 | 44% |
indicates study type specific statistically significant (p < 0.05) differences in mean AUC scores between descriptors and baseline approach (Morgan chemical Structure).
Columns 4–6 comprise the Number | Percent for each descriptor type with predictive performance >0.70(70%). Columns 7–9 contain the frequency | percentage where each descriptor performance value is greater than the others per study/effect type. Descriptors listed include: Biological(B), Gene (ge); Chemical(C) Structure, Morgan (mrgn); Hybrid(CB), Morgan + Gene. Values of major interest are bolded.
Comparing local predictive performance of all toxicity outcomes for Morgan, Gene, and Morgan + Gene hybrid descriptors
For each cluster containing two or more chemicals, the accuracy of GenRA for predicting toxicity outcomes was evaluated utilizing various descriptor types. The best overall performing descriptors for each cluster are available as supplemental material, S1 (f) and S1 (g). A summary of GenRA local predictions for the mrgn, ge, and hybrid descriptor types is given in Table 5 for cluster 67. This cluster comprises many phenyl containing chemicals such as biphenyl, 2-phenylphenol, and ethyl 3 phenylglycidate. Similar to the previous tables, the columns in Table 5 show the study types and the number of endpoints for the cluster within each study class; the average AUC performance scores and SD for each study type (3–4); the number and percentages of study/endpoint types that can accurately (AUC > 0.70) be predicted by each of the three sets of descriptors (6–8); and the number and percentage of cases in which biological descriptors outperformed the others (B > C & CB), chemical descriptors outperformed the others (C > B & CB), and hybrid descriptors outperformed all others (CB > B & C) (columns 9–11). While not practical to discuss the results of utilizing GenRA local analysis to each cluster, we have selected several examples in which each of the descriptor types may outperform the others. Table 5 is an illustrative example, in which the biological (gene-level) descriptor outperforms both chemical structure (mrgn) and hybrid (ge + mrgn). Compared to the mrgn chemical structure descriptors for this cluster, the gene-level biological descriptor more often accurately predicted toxicity with 6 out of 13 endpoints prediction values greater than 0.70 (compared to 3 of 13 for chemical) and a total 23% increase in average AUC value. Of the 13 endpoint predictions, biological descriptors more often outperformed the chemical and hybrid descriptors 7 times as compared to 4 and 3 times. Thus overall, for a chemical in cluster 67, the biological gene-level based descriptors were more accurate than the chemical structure or hybrid descriptors. According to Tukeys HSD however, these results were not significant when broken down by study type and for the overall mean across study types. This is also true when comparing the local performance of all other previously discussed descriptors. Supplemental material S4a and S4b reflect the consistent pattern for cluster 67 in which both biological descriptors (asy and ge) result in higher average predictive values (S4a) and a greater percentage of prediction score above 0.70 (S4b), yet differences in mean remain insignificant.
Table 5.
Local Prediction Performance (AUC) for All Endpoints Using Morgan Chemical Structure (C), Gene Level Biological (B), and Hybrid (CB) Descriptors by study type for Cluster-67. This chemical cluster consisted of several Phenyl containing chemicals such as biphenyl, 2-phenylphenol, and ethyl 3 phenylglycidate, overlapping with 12 chemicals with available toxicity data. This is an example cluster where the bio (gene) descriptor performs the best. Columns 2 includes the study type and the number of endpoints within each study category. Columns 3–5, Average by study type AUC | SD. Columns 6–8, Number | Percent each descriptor type with predictive performance >0.70(70%). Columns 9–11 frequency | percentage where each descriptor performance value is greater than the others. Descriptors listed include: Biological (B), Gene (ge); Chemical (C) Structure, Morgan (mrgn); Hybrid(CB), Morgan/Gene. Values of major interest are bolded.
| Cluster | Study | ge (B) | mrgn (C) | MG (CB) | B>=0.70 | C>=0.70 | CB>=0.70 | B > CB & C | C > B & CB | CB > B & C |
|---|---|---|---|---|---|---|---|---|---|---|
| 67 | Chr(3) | 0.61 | 0.19 | 0.44 | 0.13 | 0.57 | 0.22 | 1 | 33.33% | 0 | 0 | 1 | 33.33% | 1 | 33.33% | 0 | 0 | 2 | 66.67% |
| Dev(3) | 0.6 | 0.3 | 0.62 | 0.38 | 0.36 | 0.23 | 2 | 66.67% | 1 | 33.33% | 0 | 0 | 3 | 100% | 0 | 0 | 0 | 0 | |
| Mgr(2) | 0.75 | 0.35 | 0.48 | 0.26 | 0.24 | 0.22 | 1 | 50% | 0 | 0 | 0 | 0 | 1 | 50% | 1 | 50% | 0 | 0 | |
| Sac(2) | 1 | 0 | 1 | 0 | 0.5 | 0 | 2 | 100% | 2 | 100% | 0 | 0 | 1 | 50% | 1 | 50% | 0 | 0 | |
| Sub(3) | 0.44 | 0.19 | 0.29 | 0.16 | 0.24 | 0.13 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 33.33% | 2 | 66.67% | 0 | 0 | |
| All(13) | 0.65 | 0.27 | 0.54 | 0.30 | 0.38 | 0.21 | 6 | 46.15% | 3 | 23. 08% | 1 | 7.69% | 7 | 53.84% | 4 | 30.77% | 2 | 15.38% |
There are several examples of clusters where chemical structure and/or hybrid descriptors outperformed the biological descriptors, and two examples are provided in Supplemental Material Tables S5 and S6. The summary table in Supplemental Material S5 is an example in which the performance of the chemical structure descriptor class exceeds other descriptor types in predicting toxicity for chemicals in this cluster. This table shows cluster 19 results consisting of several benzene-containing chemicals such as 4-aminoazobenzene, sodium benzoate, benzyl alcohol, and benzyl acetate. In this particular cluster, we found that the chemical structure descriptors outperformed the biological and hybrid descriptors for 8 of the 15 toxicity endpoints and more often accurately predicted (AUC > 0.70) development (dev), developmental neurotoxicity (dnt), and subacute (sac) effects as compared to the other descriptors. The comparison of performance for all descriptors for cluster 19 in Supplemental Material S6 also shows chemical structure descriptors outperforming others. Mrgn and toxp chemical descriptors displayed greater overall performance scores as seen in Table S6a, and toxp chemical descriptors more often accurately predicted study type toxicity (S6b). Similar to cluster-67, Tukey’s HSD determined these differences are not significant.
In cluster 1 the hybrid mrgn + ge (MG) descriptors performs better than the singular descriptors (Supplemental Material S7). This cluster contains a diverse set of chemicals, including folpet, warfarin, fluridone, and salicylamide. For cluster 1, the MG hybrid descriptor generated the best average performance for all toxicity endpoints. MG also more often accurately predicted toxicity outcomes relative to the individual biological and chemical structure descriptors, which both predicted outcomes correctly for 2 of 12 endpoints. Performance scores for other descriptors in cluster 1 are in Supplemental Material Tables S8a and S8b. Overall, the hybrid descriptors, including mrgn + asy (MA), mrgn + ge (MG), and the CA and CB descriptors, performed best in comparison the all other descriptor types (S8a). The MA hybrid also more often accurately predicting toxicity across all study types (S8b); however, difference in mean performance scores were not significant.
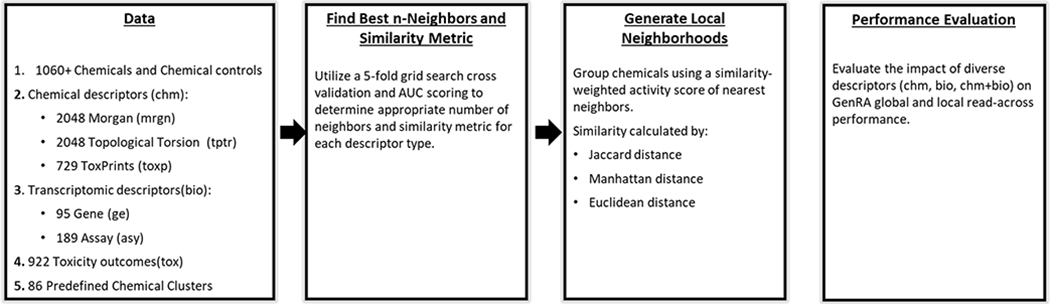
While we identified clusters in which each of the descriptor types outperformed the others, the local GenRA predictive performance using gene descriptors exceeded the chemical (mrgn) and hybrid (mg) descriptors, as seen in Fig. 2. When broken down into cluster and study into organ specific liver targets (Table 6b), differences in performance were not statistically significant. When broken down by study-type and across aggregated study-types significant differences were observed for chr and sub study types, where gene descriptors produced best performance (Table 6a). Additionally, significant differences in performance were observed for rep study-type where mrgn performed the best. Overall, out of the 48 clusters containing more than 2 chemicals with toxicity data, the biological descriptor outperformed the other descriptors’ predictive performance for about 60% (28/48) of the cases. The same was true when stratifying by effect type where the performance of biological descriptors exceeded the others at predicting toxicity endpoints within each cluster (Supplemental Material S9). Overall, of the 13 descriptors evaluated, both individual biological descriptors (asy and ge) outperformed the other descriptor types in 51% of the cases (shown in Supplemental Material S10).
Fig. 2.
Local Prediction Performance (Area Under the ROC Curve) for All Endpoints Using Morgan Chemical Structure (C), Gene Level Biological (B), and Hybrid (CB) Descriptors. The overall performance winners for all clusters. Ge (gene-level) biological descriptor, mg (morgan/gene) hybrid descriptor, mrgn (morgan) structural chemical descriptor.
Table 6.
Local Prediction Performance (AUC) for All Toxicity Endpoints Using Morgan Chemical Structure(C), Gene Level Biological(B), and Hybrid(CB) Descriptors by study type and liver target effects. Column 1 expresses various toxicity study types(6A), or liver effect(6B) and the total number of chemicals with in vivo data available for all descriptors (in parenthesis). Columns 2–3 comprises the average by study type AUC | SD.
| 6A. |
|
|
|
|---|---|---|---|
| Study | ge (B) | mrgn (C) | MG(CB) |
| Chr (276) | *0.58 | 0.33 | 0.39 | 0.33 | 0.44 | 0.34 |
| Dev (273) | 0.40 | 0.38 | 0.38 | 0.37 | 0.40 | 0.34 |
| Dnt (24) | 0.41 | 0.44 | 0.23 | 0.36 | 0.23 | 0.26 |
| Mgr (99) | 0.45 | 0.32 | 0.42 | 0.34 | 0.29 | 0.30 |
| Rep (54) | *0.32 | 0.41 | 0.68 | 0.41 | 0.42 | 0.42 |
| Sac (111) | 0.59 | 0.37 | 0.56 | 0.36 | 0.57 | 0.34 |
| Sub (249) | *0.50 | 0.32 | 0.37 | 0.30 | 0.44 | 0.34 |
| ALL | *0.49 | 0.34 | 0.41 | 0.35 | 0.42 | 0.35 |
| 6B. |
|
|
|
| Target | ge (B) | mrgn (C) | MG(CB) |
|
| |||
| Chr_liver (96) | 0.63 | 0.32 | 0.45 | 0.34 | 0.52 | 0.37 |
| Mgr_liver (75) | 0.46 | 0.36 | 0.46 | 0.36 | 0.33 | 0.33 |
| Sac_liver (33) | 0.58 | 0.43 | 0.72 | 0.34 | 0.62 | 0.33 |
| Sub_liver (78) | 0.52 | 0.35 | 0.37 | 0.28 | 0.45 | 0.34 |
| ALL_liver | 0.55 | 0.35 | 0.47 | 0.34 | 0.46 | 0.35 |
indicates study type specific statistically significant (p < 0.05) differences in mean AUC scores between descriptors and baseline approach (Morgan chemical Structure).
Descriptors listed include: Biological(B), Gene (ge); Chemical(C) Structure, Morgan (mrgn); Hybrid(CB), Morgan + Gene.
Case example of GenRA toxicity predictions using diethyl phthalate
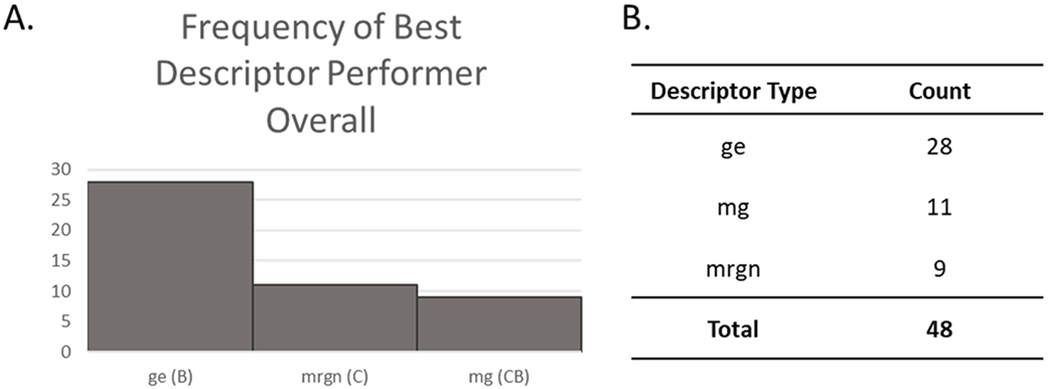
We illustrate the impact of biological similarities on GenRA read-across performance using Diethyl phthalate, a member of cluster-80. The biological and chemical descriptors performed equally well overall. Fig. 3 illustrates the neighborhood for diethyl phthalate based on the application of biological, gene-level descriptors (Fig. 3a), mrgn chemical structure descriptors (Fig. 3b), and hybrid descriptors (Fig. 3c) for toxicity predictions. For each descriptor class, 10 source analogues were identified, comprising cluster-80 chemicals. The membership of the neighborhood varied slightly depending on the descriptor used either in terms of the pairwise similarities themselves or the chemicals.
Fig. 3.
Case Example of GenRA Neighborhood Using Biological, Chemical, and Hybrid Descriptors with the Target Chemical Diethyl phthalate and Cluster 80 Chemicals. The target chemical, anilazine lies in the center of each neighborhood. (A) Numbers in the center represent the pairwise biological (gene-level) similarity scores. (B) Numbers in the center represent the pairwise chemical structure (Morgan) similarity scores. (C) Numbers in the center represent the pairwise hybrid (Morgan H-Gene) similarity scores. (D) Overall summary of cluster-80 untargeted toxicity AUC performance scores for each descriptor.
While the similarity scores for each descriptor neighborhood varied, the chemical descriptor neighborhood more often yielded similarity scores greater than 0.50. The biological descriptors yielded a perfect similarity score (1.0) for diisobutyl phthalate. Overall, this example illustrates that the neighborhoods and the similarity values generated between ‘neighboring’ chemicals depend on the type of descriptors utilized. Similarity scores appear to be comparable to average performance scores for each descriptor in 3d which displayed insignificant differences that varied overall. Likewise, no significant study-type differences were detected. Lack of coverage across sufficient endpoints may account this outcome. There is still merit however in utilizing a combination of biological and chemical descriptors as we saw significant differences in performance with more coverage within study types for both the local and global approaches.
Discussion
The Strategic Plan for NAMs [58] provides an impetus to transition from animal testing to in vitro and in silico approaches. Generation of NAM data has opened the door for a host of computational approaches, including data-driven read-across predictions of hazard and risk [41,55]. Traditionally, read-across relies on chemical similarity to identify candidate source analogues; however, numerous studies demonstrate the utility of characterizing biological similarity with other NAM in vitro data to make read-across predictions of toxicity [19,20,21,22,25,36,51,69]. Here, we used the genra-py package to investigate the role that targeted transcriptomic bioactivity descriptors play in identifying source analogues and predicting hazard in outcomes compared to chemical structure fingerprints and hybrid fingerprints. In all, we systematically evaluated the performance of GenRA using thirteen different types of biological, chemical, and hybrid descriptors to predict 252 apical effects across 10 different guideline study types. We employed two main approaches to summarize read-across performance: 1) a global approach in which we aggregated performance across all chemicals in our data set; and 2) a local approach where performance was aggregated by categories defined by chemical structure clusters. Although average global performance for all descriptors and study types was poor (with AUC = 0.51), we were able to identify specific contexts in which automated read-across performed well with chemical, transcriptomic, or hybrid fingerprints (see Fig. 3b and Supplemental Material, S1(h)).
The global and local performance results for all toxicity endpoints suggest that utilizing a small targeted set of transcriptomic descriptors for read-across predictions can provide only a modest advantage singly or in combination with chemical structure fingerprints. Globally, this is evident in Table 3a and Table 4, where individual biological and hybrid descriptors outperform the individual chemical descriptors on average for all toxicity target studies. However, it is worth noting that combining all chemical descriptors (CC) is advantageous for global read-across analysis, as shown in Tables 3a and 3b. For accurate predictions, CC more often accurately predicted toxicity than all descriptor types (Table 3b) and retained higher average AUC scores amongst the various toxicity study types. One reason for this could be that the much larger number of CC fingerprints (4825 descriptors) provides a more detailed representation of chemicals versus biological or dual hybrid descriptors (See Supplemental Material S3). Increasing the number of individual biological descriptors may be necessary for a fairer comparison. The CC descriptor also outperformed the CB descriptor, which incorporated all individual chm and bio descriptors (a total of 5109 descriptors). Despite an increase in the number of descriptors for CB, there were still fewer biological fingerprints than chemical fingerprints in our analysis. This highlights the importance of expanding the bioactivity data to improve the performance of hybrid read-across predictions or alternative what feature selection procedures might be warranted to target more relevant fingerprints.
It is also noteworthy to consider that while CC had the greatest overall average performance scores for all aggregated study-types and all aggregated liver-specific endpoints, hybrid descriptors more often accurately predicted liver toxicity (Table 2). Since the biological descriptors utilized here were derived from HepaRG cells, which are liver-derived, higher prediction scores for the liver toxicity endpoints (chr_liver, dev_liver, sac_liver, and sub_liver) by biological or hybrid descriptors may be a result of this specificity. Therefore, the biological relevance of the liver-derived targeted transcriptomic descriptors may also explain why the performance for liver toxicity endpoints was higher than CC. The lack of biological relevance of the HepaRG cells to other study types and target organs could also explain why their performance scored lower. A better understanding of cell type-mechanisms’ roles in organ toxicities [79,80] could enable the development of additional targeted assays.
In contrast, the CC combination was not the best performing descriptor for local read-across analysis (See, supplemental material S1 (i) and S9). Local predictions based on biological descriptors frequently outperformed all other descriptors (Supplemental Fig. S9). This was illustrated for phenyl-containing compounds in cluster 67 (Table 5 and Supplemental Table S4(a & b)) where biological descriptors produced the highest overall AUC scores and more accurate predictions (AUC values greater than 0.70). For this example, however, hybrid descriptors performed most poorly. In general, hybrid descriptors elicited increased performance over the individual chemical structure for various toxicity study types; still, the performance of the read-across predictions varied with in study types and depended on specific toxicity effects and the chemical cluster assessed.
This context-dependency of read-across predictions was also reported in Shah et al. [51], where bioactivity fingerprints and hybrid (chemical and bioactivity) outperformed mrgn chemical fingerprints for specific study types. Shah et al. [51] noted that bioactivity might be a more promising predictor due to the association of high dose effects in vivo and the high concentration effects observed in the in vitro activity. The ToxCast HTS assays used in Shah et al. [51] lacked metabolic transformation, whereas this analysis used data from metabolically competent HepaRG™ cells with more xenobiotic responsive metabolizing enzymes [18]. This choice aligns with factors proposed by Thomas et al. [55] for improving the predictive capacity of the ToxCast HTS data and obtaining broader acceptance of predictive approaches [14,55].
Despite the promise of HTS assays for improving read-across predictions, we note [12] two main limitations: the targeted transcriptomic assay only included 93 genes and were only assessed in one cell culture model. While the gene descriptors had the best performance for predicting hepatic outcomes, they did not perform well for other target organ effects. This is likely the result of using transcriptomic data from HepaRG™ cells, which is a liver cell line. Additionally, using a targeted set of 93 genes substantially limits the range of pathway perturbations that can be assessed, resulting in only a partial evaluation of biological similarity. To address these limitations, we are evaluating the utility of HTTr data from multiple cell types for screening thousands of chemicals [23,24]. Using HTTr will expand the scope of biological fingerprints by covering a broader set of biological pathways that can be analyzed with connectivity mapping approaches [12,33,34,59] to automate read-across predictions of hazard classifications for targets beyond the liver.
Conclusion
Generalized Read-Across (GenRA) approach was developed to make automated read-across predictions of toxicity effects, utilizing a similarity weighted average approach [51]. Previously, this approach was shown to be effective in characterizing source analogues using binary and quantitative measures for chemical and bioactivity descriptors, as well as physiochemical property information [25–28,51]. In this present study, we extended the approach to transcriptomic data comprising binary hit-calls (activity calls) from concentration-response data for each gene. Our analysis estimated both global and local performance using diverse individual and combinations of transcriptomic binary hit call measures and chemical structure fingerprints in predicting liver toxicity and overall study-type associated toxicity effects. The global read across performance for all neighborhoods suggests that hybrid combinations of biological and chemical descriptors were effective for several study types (chr, sac, sub). This was also the case for the combination of multiple chemical descriptors. In contrast, for the local read across performance, the individual biological descriptors proved more predictive. Overall, whilst the coverage with this dataset was more limited, in specific cases, the utility of transcriptomic hit-call information in addition to chemical structure information was found to be promising in making in vivo toxicity predictions Next steps will be to extend the scope to a more comprehensive transcriptomic dataset to determine whether the performance significantly improves.
Supplementary Material
Funding
Funding for this work was provided by the US EPA Office of Research and Development.
Abbreviations:
- AC50
Activity concentration at 50% of E-max
- ACU
Acute Toxicity Study
- ANOVA
Analysis of Variance
- ASY
Assay Biological Descriptor
- AUC
Area Under the ROC Curve
- BIO
Biological Descriptor
- CASRN
Chemical Abstract Services Registry Number
- CB
Hybrid Chemical and Biological Descriptor
- CC
Chemical Combination Descriptor of Morgan, Torsion, and Toxprint Descriptors
- CHM
Chemical Descriptor
- CHM
Chronic Toxicity Study
- CHR
Developmental Toxicity Study
- DEV
Developmental Neurotoxicity Study
- DNT
Distributed Structure-Searchable Toxicity
- DSSTOX
Gene-level Biological Descriptor
- GE
High content screening
- HSD
Tukey’s Honest Significance Difference
- HTS
High-throughput screening
- HTPP
High throughput phenotypic profiling
- LOAEL
Lowest observed adverse effect level
- LTEA
Life Technologies/Expression Analysis
- MA
Morgan Chemical and Assay Biological Hybrid Descriptor
- MG
Morgan Chemical and Gene Biological Hybrid Descriptor
- MGR
Multigenerational Toxicity Study
- MRGN
Morgan Chemical Descriptor
- NAM
New approach methodology
- NEU
Neurological Toxicity Study
- NTP
National Toxicology Program
- POD
Point of departure
- OCSPP
Office of Chemical Safety and Pollution Prevention
- OSAR
Quantitative structure-activity relationships
- REP
Reproductive Toxicity Study
- ROC
Receiver Operator Characteristic
- SAC
Sub-Acute Toxicity Study
- SUB
Sub-Chronic Toxicity Study
- TPTR
Torsion Topological Chemical Descriptor
- ToxCast
ToxCast HTS assays
- ToxP
ToxPrint Chemotype Chemical Descriptor
- TTA
Torsion Topological Chemical and Assay Biological Hybrid Descriptor
- TTG
Torsion Topological Chemical and Gene Biological Hybrid Descriptor
- TXA
ToxPrint Chemotype and Assay Biological Hybrid Descriptor
- TXG
ToxPrint Chemotype and Gene Biological Hybrid Descriptor
Footnotes
Disclaimer
The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comtox.2021.100171.
References
- [1].Science approach document-Bioactivity exposure ratio: Application in priority setting and risk assessment. Health Canada, 2021. [Google Scholar]
- [2].Ball N, et al. , The challenge of using read-across within the EU REACH regulatory framework; how much uncertainty is too much? Dipropylene glycol methyl ether acetate, an exemplary case study, Regul. Toxicol. Pharmacol 68 (2014) 212–221. [DOI] [PubMed] [Google Scholar]
- [3].Ball N, et al. , Toward good read-across practice (GRAP) guidance, ALTEX. 33 (2016) 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Black PE, Manhattan Distance. In: Black PE, (Ed.), ed. 11 February 2019, Dictionary of Algorithms and Data Structures, 2019. [Google Scholar]
- [5].Blackburn K, Stuard SB, A framework to facilitate consistent characterization of read across uncertainty, Regul. Toxicol. Pharmacol 68 (2014) 353–362. [DOI] [PubMed] [Google Scholar]
- [6].Bourdon-Lacombe JA, et al. , Technical guide for applications of gene expression profiling in human health risk assessment of environmental chemicals, Regul. Toxicol. Pharmacol 72 (2015) 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chesnut M, et al. , Regulatory acceptance of read-across, ALTEX 35 (2018) 413–419. [DOI] [PubMed] [Google Scholar]
- [8].Collins FS, et al. , Toxicology. Transforming environmental health protection, Science 319 (2008) 906–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Craw S, Manhattan distance, in: Sammut C, Webb GI (Eds.), Encyclopedia of Machine Learning, Springer, US, Boston, MA, 2010, p. 639. [Google Scholar]
- [10].Cronin MT, Computational toxicology is now inseparable from experimental toxicology, Altern. Lab Anim 41 (2013) 1–4. [DOI] [PubMed] [Google Scholar]
- [11].Danielsson P-E, Euclidean distance mapping, Comp. Graphics Image Process 14 (1980) 227–248. [Google Scholar]
- [12].De Abrew KN, et al. , Grouping 34 chemicals based on mode of action using connectivity mapping, Toxicol. Sci 151 (2016) 447–461. [DOI] [PubMed] [Google Scholar]
- [13].Dearden JC, et al. , How not to develop a quantitative structure-activity or structure-property relationship (QSAR/QSPR), SAR QSAR Environ. Res 20 (2009) 241–266. [DOI] [PubMed] [Google Scholar]
- [14].Deisenroth C, et al. , The alginate immobilization of metabolic enzymes platform retrofits an estrogen receptor transactivation assay with metabolic competence, Toxicol. Sci 178 (2020) 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dix DJ, et al. , The ToxCast program for prioritizing toxicity testing of environmental chemicals, Toxicol. Sci 95 (2007) 5–12. [DOI] [PubMed] [Google Scholar]
- [16].Escher SE, et al. , Towards grouping concepts based on new approach methodologies in chemical hazard assessment: the read-across approach of the EU-ToxRisk project, Arch. Toxicol 93 (2019) 3643–3667. [DOI] [PubMed] [Google Scholar]
- [17].Filer DL, et al. , tcpl: the ToxCast pipeline for high-throughput screening data, Bioinformatics 33 (2017) 618–620. [DOI] [PubMed] [Google Scholar]
- [18].Franzosa J, et al. , High-throughput toxicogenomic screening of chemicals in the environment using metabolically competent hepatic cell cultures, NPJ Syst. Biol. Appl (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gadaleta D, et al. , Automated integration of structural, biological and metabolic similarities to improve read-across, ALTEX 37 (2020) 469–481. [DOI] [PubMed] [Google Scholar]
- [20].Grimm FA, et al. , Multi-dimensional in vitro bioactivity profiling for grouping of glycol ethers, Regul. Toxicol. Pharmacol 101 (2019) 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Grimm FA, et al. , A chemical-biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives, Green Chem. 18 (2016) 4407–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Guo Y, et al. , Using a hybrid read-across method to evaluate chemical toxicity based on chemical structure and biological data, Ecotoxicol. Environ. Saf 178 (2019) 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Harrill JA, et al. , High-throughput transcriptomics platform for screening environmental chemicals, Toxicol. Sci (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harrill JS, Imran, Woodrow Setzer R, Haggard Derik, Auerbach Scott, Judson Richard, Thomas Russel S., Considerations for strategic use of high-throughput transcriptomics chemical screening data in regulatory decisions, Curr. Opin. Toxicol 15 (2019) 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Helman G, et al. , Quantitative prediction of repeat dose toxicity values using GenRA, Regul. Toxicol. Pharmacol 109 (2019), 104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Helman G, et al. , Extending the Generalised Read-Across approach (GenRA): a systematic analysis of the impact of physicochemical property information on read-across performance, Comput. Toxicol 8 (2018) 34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Helman G, et al. , Generalized read-across (GenRA): a workflow implemented into the EPA CompTox Chemicals Dashboard, ALTEX 36 (2019) 462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Helman G, Shah I, Padewicz G, Transitioning the generalised read-across approach (GenRA) to quantitative predictions: a case study using acute oral toxicity data, Comput. Toxicol 12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Houck K, et al. , ToxCast: Predicting Toxicity Potential Through Hig Throughput Bioactivity Profiling, 2013.
- [30].ICCVAM A Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States, 2018.
- [31].Iorio F, et al. , Identifying network of drug mode of action by gene expression profiling, J. Comput. Biol 16 (2009) 241–251. [DOI] [PubMed] [Google Scholar]
- [32].Jaccard P, Lois de distribution florale dans la zone alpine, Bull. Soc. Vaudoise Sci. Nat 38 (1902) 69–130. [Google Scholar]
- [33].Lamb J, The Connectivity Map: a new tool for biomedical research, Nat. Rev. Cancer 7 (2007) 54–60. [DOI] [PubMed] [Google Scholar]
- [34].Lamb J, et al. , The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease, Science 313 (2006) 1929–1935. [DOI] [PubMed] [Google Scholar]
- [35].Landrum G, RDKit. 2015.
- [36].Low Y, et al. , Integrative chemical-biological read-across approach for chemical hazard classification, Chem. Res. Toxicol 26 (2013) 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mahony C, et al. , Threshold of toxicological concern (TTC) for botanicals - Concentration data analysis of potentially genotoxic constituents to substantiate and extend the TTC approach to botanicals, Food Chem. Toxicol 138 (2020), 111182. [DOI] [PubMed] [Google Scholar]
- [38].OECD, 2017. Guidance on Grouping of Chemicals, Second Edition. [Google Scholar]
- [39].Padewicz G, et al. , Building scientific confidence in the development and evaluation of read-across, Regul. Toxicol. Pharmacol 72 (2015) 117–133. [DOI] [PubMed] [Google Scholar]
- [40].Padewicz G, et al. , Use of category approaches, read-across and (Q)SAR: general considerations, Regul. Toxicol. Pharmacol 67 (2013) 1–12. [DOI] [PubMed] [Google Scholar]
- [41].Padewicz G, et al. , Navigating through the minefield of read-across tools: a review of in silico tools for grouping, Comput. Toxicol 3 (2018) 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ramaswamy Nilakantan NB, Scott Dixon J, Venkataraghavan R, Topological torsion: a new molecular descriptor for SAR applications. Comparison with other descriptors, J. Chem. Inf. Comput. Sci 27 (1987) 82–85. [Google Scholar]
- [43].Richard AM, et al. , ToxCast chemical landscape: paving the road to 21st century toxicology, Chem. Res. Toxicol 29 (2016) 1225–1251. [DOI] [PubMed] [Google Scholar]
- [44].Rogers D, Hahn M, Extended-connectivity fingerprints, J. Chem. Inf. Model 50 (2010) 742–754. [DOI] [PubMed] [Google Scholar]
- [45].Rovida C, et al. , Internationalization of read-across as a validated new approach method (NAM) for regulatory toxicology, ALTEX 37 (2020) 579–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Schmidt CW, TOX 21: new dimensions of toxicity testing, Environ. Health Perspect 117 (2009) A348–A353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schultz TW, Cronin MTD, Lessons learned from read-across case studies for repeated-dose toxicity, Regul. Toxicol. Pharmacol 88 (2017) 185–191. [DOI] [PubMed] [Google Scholar]
- [48].Schultz TW, et al. , Assessing uncertainty in read-across: Questions to evaluate toxicity predictions based on knowledge gained from case studies, Comput. Toxicol 9 (2019) 1–11. [Google Scholar]
- [49].SEURAT-1, SEURAT-1 – Towards the Replacement of in vivo Repeated Dose Systemic Toxicity Testing News.
- [50].Shah I, et al. , Using nuclear receptor activity to stratify hepatocarcinogens, PLoS ONE 6 (2011), e14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Shah I, et al. , Systematically evaluating read-across prediction and performance using a local validity approach characterized by chemical structure and bioactivity information, Regul. Toxicol. Pharmacol 79 (2016) 12–24. [DOI] [PubMed] [Google Scholar]
- [52].Shah I, et al. , 2021. Generalised Read-Across Prediction using genra-py Bioinformatics. [DOI] [PMC free article] [PubMed]
- [53].Sipes NS, et al. , An intuitive approach for predicting potential human health risk with the Tox21 10k Library, Environ. Sci. Technol 51 (2017) 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Smalley JL, et al. , Application of connectivity mapping in predictive toxicology based on gene-expression similarity, Toxicology 268 (2010) 143–146. [DOI] [PubMed] [Google Scholar]
- [55].Thomas RS, et al. , The next generation blueprint of computational toxicology at the U.S. Environmental Protection Agency, Toxicol. Sci 169 (2019) 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].USEPA, 2018. Toxicology, EPA’s National Center for Computational (2018): ToxCast Database (invitroDB). The United States Environmental Protection Agency’s Center for Computational Toxicology and Exposure. Dataset. [Google Scholar]
- [57].USEPA, 2020a. New approach methods work plan: Reducing use of animals in chemical testing. U.S. Environmental Protection Agency, Washington, DC. EPA 615B2001. [Google Scholar]
- [58].USEPA, 2020b. Strategic Vision for Adopting New Approach Methodologies.
- [59].Wang RL, et al. , Fish connectivity mapping: linking chemical stressors by their mechanisms of action-driven transcriptomic profiles, BMC Genomics 17 (2016) 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wang Z, et al. , RNA-Seq: a revolutionary tool for transcriptomics, Nat. Rev. Genet 10 (2009) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Watford S, et al. , ToxRefDB version 2.0: Improved utility for predictive and retrospective toxicology analyses, Reprod. Toxicol 89 (2019) 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wetmore BA, et al. , Incorporating high-throughput exposure predictions with dosimetry-adjusted in vitro bioactivity to inform chemical toxicity testing, Toxicol. Sci 148 (2015) 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wetmore BA, et al. , Integration of dosimetry, exposure, and high-throughput screening data in chemical toxicity assessment, Toxicol. Sci 125 (2012) 157–174. [DOI] [PubMed] [Google Scholar]
- [64].Williams AJ, et al. , The CompTox Chemistry Dashboard: a community data resource for environmental chemistry, J. Cheminform 9 (2017) 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Worth AP et al. , The Characterisation of (Quantitative) Structure-Activity Relationships: Preliminary Guidance, 2005.
- [66].Wu S, et al. , A framework for using structural, reactivity, metabolic and physicochemical similarity to evaluate the suitability of analogs for SAR-based toxicological assessments, Regul. Toxicol. Pharmacol 56 (2010) 67–81. [DOI] [PubMed] [Google Scholar]
- [67].Yang C, et al. , New publicly available chemical query language, CSRML, to support ehemotype representations for application to data mining and modeling, J. Chem. Inf. Model 55 (2015) 510–528. [DOI] [PubMed] [Google Scholar]
- [68].Yauk CL, et al. , A cross-sector call to improve carcinogenicity risk assessment through use of genomic methodologies, Regul. Toxicol. Pharmacol 110 (2020), 104526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhu H, et al. , Supporting read-across using biological data, ALTEX 33 (2016) 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhu H, et al. , Use of cell viability assay data improves the prediction accuracy of conventional quantitative structure-activity relationship models of animal carcinogenicity, Environ. Health Perspect 116 (2008) 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Benfenati Emilio, et al. , The acceptance of in silico models for REACH: Requirements, barriers, and perspectives, Chem. Cent. J 5 (58) (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].European Commision, Regulation (EC) No 1907/2006 of the European Parliament and of the Council of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/45/EC and repealing Council Regulation (EEC) No 793/93 and Commission Regulation (EC) No 1488/94 as well as Council Directive 76/769/EEC and Commission Directives 91/155/EEC, 93/67/EEC, 93/105/EC and 2000/21/EC. Off J Eur Union, L396/1 of 30.12.2006, Commission of the European Communities; (2006). [Google Scholar]
- [74].Lahl U, Hwxwell KA, REACH–the new European chemicals law, Environ. Sci. Technol 40 (2006) 7115–7121. [DOI] [PubMed] [Google Scholar]
- [75].Schultz TW, et al. , A strategy for structuring and reporting a read-across prediction of toxicity, Regul. Toxicol. Pharmacol 72 (2015) 586–601. [DOI] [PubMed] [Google Scholar]
- [76].ECHA, Read-Across Assessment Framework (RAAF), European Chemicals Agency; (2017). [Google Scholar]
- [77].Firman JW, et al. , Exploring the Potential of ToxCast Data in Supporting Read-Across for Evaluation of Food Chemical Safety, Chem. Res. Toxicol 34 (2021) 300–312. [DOI] [PubMed] [Google Scholar]
- [78].Brandt M, et al. , A weight-of-evidence approach to assess chemicals: case study on the assessment of persistence of 4,6-substituted phenolic benzotriazoles in the environment, Environ Sci Eur 28 (2016) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Gupta R, Xie H, Nanopartides in Daily Life: Applications, Toxicity and Regulations, J Environ Pathol Toxicol Oncol 37 (2018) 209–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Prieto P, et al. , Investigating cell type specific mechanisms contributing to acute oral toxicity, ALTEX 36 (2019) 39–64. [DOI] [PubMed] [Google Scholar]
- [81].Ball N, et al. , Key read across framework components and biology based improvements, Mutat Res 853 (2020) 503172. [DOI] [PubMed] [Google Scholar]
- [82].Gelbke HP, Ellis-Hutchings R, Müllerschön H, Murphy S, Pemberton M, Toxicological assessment of lower alkyl methacrylate esters by a category approach, Regul. Toxicol. Pharmacol 92 (2018) 104–127, 10.1016/j.yrtph.2017.11.013. ISSN 0273-2300. [DOI] [PubMed] [Google Scholar]
- [83].Gautier F, et al. , Read-across can increase confidence in the Next Generation Risk Assessment for skin sensitisation: A case study with resorcinol, Regul. Toxicol. Pharmacol 117 (2020) 104755. [DOI] [PubMed] [Google Scholar]
- [84].Pestana CB, et al. , Incorporating lines of evidence from New Approach Methodologies (NAMs) to reduce uncertainties in a category based read-across: A case study for repeated dose toxicity, Regul. Toxicol. Pharmacol 120 (2021) 104855. [DOI] [PubMed] [Google Scholar]
- [86].Webster F, et al. , Predicting estrogen receptor activation by a group of substituted phenols: An integrated approach to testing and assessment case study, Regul. Toxicol. Pharmacol 106 (2019) 278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sakuratani Y, et al. , Integrated Approaches to Testing and Assessment: OECD Activities on the Development and Use of Adverse Outcome Pathways and Case Studies, Basic Clin. Pharmacol. Toxicol 123 (Suppl. 5) (2018) 20–28. [DOI] [PubMed] [Google Scholar]
- [88].Sperber S, et al. , Metabolomics as read-across tool: An example with 3-amino-propanol and 2-ami noethanol, Regul. Toxicol. Pharmacol 108 (2019) 104442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


