Abstract
The molecular basis for function of the mammalian H19 as a tumor suppressor is poorly understood. Large, conserved open reading frames (ORFs) are absent from both the human and mouse cDNAs, suggesting that it may act as an RNA. Contradicting earlier reports, however, recent studies have shown that the H19 transcript exists in polysomal form and is likely translated. To distinguish between possible functional roles for the gene product, we have characterized the sequence requirements for H19-mediated in vitro suppression of tumor cell clonogenicity and analyzed the sequence of the gene cloned from a range of mammals. A cDNA version of the human gene, lacking the unusually short introns characteristic of imprinted genes, is as effective as a genomic copy in blocking anchorage-independent growth by G401 cells. The first 710 nucleotides of the gene can be deleted with no effect on in vitro activity. Further truncations from either the 5′- or 3′-end, however, cause a loss of suppression of clonogenicity. Using conserved sequences within the H19 gene as PCR primers, genomic DNA fragments were amplified from a range of mammalian species that span the functional domain defined by deletion analysis. Sequences from cat, lynx, elephant, gopher and orangutan complement the previous database of sequences from human, mouse, rat and rabbit. Hypothetical translation of the resulting sequences shows an absence of conserved ORFs of any size. Free energy and covariational analysis of the RNA sequences was used to identify potential helical pairings within the H19 transcript. A set of 16 helices are supported by covariation (i.e. conservation of base pairing potential in the absence of primary sequence conservation). The predicted RNA pairings consist largely of local hairpins but also include several long range interactions that bridge the 5′- and 3′-ends of the functional domain. Given the evolutionary conservation of structure at the RNA level and the absence of conservation at the protein level, we presume that the functional product of the H19 gene is a structured RNA.
INTRODUCTION
The mammalian H19 gene was initially cloned in the course of a subtractive hybridization screen that aimed to identify genes whose expression is coordinately regulated with α-fetoprotein (1). Early studies mapped the human gene to 11p15.5, a chromosomal region with known tumor suppressor activity, and subsequent analysis revealed that H19 expression in several different types of tumors is often altered relative to that in adjacent, non-transformed cells (2–13). Tycko and co-workers (14) directly demonstrated that induced expression of a transfected copy of H19 suppresses cellular proliferation, clonogenicity and tumorigenicity in certain tumor cell lines. Together these results suggest that the H19 gene yields a product that functions under some conditions as a tumor suppressor. Despite extensive analysis of the gene, especially in terms of its parent-of-origin-specific genomic imprinting and its expression patterns (15–18), it remains unclear whether the gene encodes a functional protein or RNA product and, if it does, what its mode of action would be. As outlined below, several alternative, conflicting models have been proposed, yet definitive evidence for each remains lacking.
Initial comparison of the human and mouse H19 sequences by Tilghman and co-workers (1) failed to identify a substantial conserved open reading frame (ORF) expected for a protein-coding mRNA. Fractionation of cytoplasmic extracts treated with cycloheximide, EDTA and/or phenol showed that the abundant transcript forms a non-polysomal, protein-associated complex, further suggesting that it remains non-translated (19). H19 is one of a handful of genes which are genomically imprinted and it lies adjacent to the reciprocally imprinted IGF2 gene (20,21). A large body of evidence suggests that expression of H19 and IGF2 requires a common enhancer lying downstream of H19 and that both genes compete for its utilization (22,23). This model can explain how the simple act of transcribing the H19 gene would yield a phenotype (i.e. reduced transcription of IGF2) without requiring a direct function for the H19 transcript itself.
Several observations, however, suggest a more active role for the transcript. Deletion of the 5′-end of the H19 structural gene blocks imprinting, indicating that expressed sequences are functionally important (the possibility remains that these sequences act at the DNA level, e.g. as internal promoter elements) (24). Transfection with an episome carrying an inducible H19 is able to restore normal cell cycle control to transformed cells (14). In this case, the physical linkage between the H19 and IGF2 loci is clearly broken yet a strong phenotype is observed. Incorporation of IGF2 transcripts into polysomes is inversely correlated with H19 expression, suggesting that H19 may act at both the post-transcriptional as well as transcriptional level to alter IGF2 expression (25).
In principle, a transcript such as H19 lacking a large ORF could function in several different ways to produce a biological phenotype. One possibility is that activity resides in the introns embedded within the primary transcript rather than in the mature mRNA itself. As a precedent, several small nucleolar RNAs (snoRNAs) involved in rRNA processing are known to be derived from the introns of host genes. In the case of the mammalian U22 host gene, the mature transcript for the host gene has no function other than to provide the snoRNAs (its processed transcript is exported to the cytoplasm and rapidly degraded) (26,27). It has been noted that imprinted genes as a group have unusually short introns (those in the human H19 gene range in size from 50 to 120 nucleotides) (28,29). This anomaly might be explained if the introns of these genes served some essential biological function.
Alternatively, the mature transcript may function at the RNA level without being translated. A growing number of non-coding RNAs have recently been identified, many of which appear to function to control cell growth and cell differentiation (30,31). These ‘riboregulators’ include Xist (a nuclear RNA involved in X-chromosome inactivation) (32–34), the yellow crescent (yc) RNA (an inhibitor of PCNA expression) (35), lin-4 (an inhibitor of lin-14) (36,37), enod40 (involved in plant development) (38), gadd7 (involved in cell growth inhibition following DNA damage) (39) and His-1 (mouse gene of unknown function) (40). With the exception of the ycRNA and lin-4, both of which appear to block expression of a target transcript by antisense pairing (37), the means by which these RNAs function remain poorly understood.
A final possibility is that the functional product of the gene is a small protein derived by translation of the mRNA. In vitro analysis of H19 cDNA clones shows that truncated transcripts lacking the first 600 nucleotides can be translated to yield a 26 kDa protein product while translation of full-length transcripts is blocked (24). It is possible that under certain in vivo conditions, translation proceeds despite the presence of the 5′-end (possibly using an internal ribosome entry site defined by the 5′-UTR). While the protein product observed in these studies corresponds to the longest ORF in the human mRNA, this ORF is absent in other mammalian sequences (rat, mouse and rabbit), raising questions about its biological significance.
In the current report, we have attempted to distinguish between the possibilities outlined above using two experimental approaches. Given the observation by Hao et al. (14) that H19 expression in G401 cells blocks soft agar clonogenicity, we have tested the ability of several modified forms of H19 to function similarly. In a separate series of experiments, we have used PCR amplification of genomic DNA from several mammalian species to obtain H19 sequence information. Suppression of clonogenicity by different H19 constructs shows that both the introns and the first 710 nucleotides are non-essential for function. Analysis of an alignment of nine H19 genes makes it possible to deduce evolutionary constraints on the sequence. We conclude that none of the potential ORFs are conserved within the mammalian sequences, strongly arguing against a function as a protein. At the same time, the existence of several RNA helical pairings is supported by covariational analysis (looking for the conservation of base pairing potential in the absence of primary sequence conservation). On the basis of these results, we conclude that the mature RNA transcript is the functional product of the H19 gene and that its function requires the ability to fold into a specific secondary structure.
MATERIALS AND METHODS
H19 deletion constructs
The human H19 genomic DNA cloned into a pMEP-4 vector (Invitrogen) was provided by Dr Benjamin Tycko. The human H19 cDNA (2.3 kb) cloned into a pBluescript vector (Stratagene) was provided by Dr Shirley Tilghman. Digestion of the cDNA clone with KpnI, XhoI or EagI generates 1.6, 1.4 or 2.0 kb fragments of H19 respectively (corresponding to nucleotides 710–2300, 850–2300 or 154–2145). These fragments were ligated into a pMEP-4 vector previously linearized with the corresponding restriction enzyme. Clones were isolated using the Promega Wizard miniprep kit and mapped by restriction digestion to distinguish sense and antisense orientations of the insert.
Cell transfection
G401 cell stocks were obtained from American Type Cell Culture (ATCC# CRL-1441) and maintained in 25 cm2 flasks on Dulbecco’s modified Eagle’s medium (DMEM; Mediatech), supplemented with 10% fetal bovine serum (FBS; Sigma). Cells were cultured in a fully humidified incubator at 37°C supplemented with 5% CO2. Cells were passaged several times with trypsin (Sigma) and maintained at an optimal confluency of 50–70%.
Transfections were carried out in 25 cm2 flasks (Corning, T-25 flasks). G401 cells were passaged with trypsin, washed and counted using a hemacytometer. These were plated at 60% confluency 2 days prior to transfection. Immediately prior to transfection, cells were washed twice with serum free media containing penstrep (SFM-p). A cocktail containing 6–10 mg of supercoiled vector DNA, 15 ml of lipofectin (Gibco BRL) and 600 ml OPTI-MEM (Gibco BRL) was pre-incubated on ice for 30 min and subsequently diluted with 5.4 ml of SFM-p. The entire mixture was then overlaid on adherent G401 cells in T-25 flasks and incubated under culture conditions for 8–12 h. The lipofectin mixture was aspirated and replaced with stock culture media supplemented with 10% FBS and 1% penstrep. Cells were allowed a 2–3 day recovery time between initial transfection and selection with hygromycin B (Sigma, 200 mg/ml). Cells were monitored daily and routinely washed for 7 days after introduction of antibiotic to remove cell debris. After selection, cells were grown at a maintenance concentration of 100 mg/ml hygromycin B. Cells were transferred to new flasks once a stable concentration of 20–30% confluency was established to provide a clean substrate for optimal growth conditions.
Clonogenicity assay
Cells were plated in triplicate in 6 cm diameter Petri dishes incubated inside a 15 cm bacterioplate. A 2× solution of DMEM culture media supplemented with 20% FBS and 1% penstrep was first mixed with an equal volume of a 1.2% solution of low melt agarose cooled to 39°C (Intermountain Scientific Corporation) to make a 0.6% base layer (LMP solution). The base layer mixture was then poured at 4 ml per dish and transferred to 4°C for 10 min to solidify. G401 cells were harvested with trypsin, washed twice and counted with a hemacytometer. Cells were plated at densities of 103 and 104/plate. A total of 4 ml of the 0.32% agarose cell suspension layer, made by mixing equal volumes of the cell suspension in 2× DMEM and a 1.2% LMP solution, was poured over the base layer. Plates were supplemented with a top layer consisting of an equal volume of 2× DMEM and 0.69% LMP every 3–4 days to maintain hydration. Plates were stored in a humidified incubator at 37°C and 5% CO2. Colony formation was assessed 1, 2 and 3 weeks after plating.
Genomic fragment construction
Blood samples were provided by the San Francisco Zoo DNA bank. Genomic DNA was extracted by pre-treatment with proteinase K (200 µg/ml) and 0.5% SDS, followed by extensive phenol/chloroform extraction. Three different conserved regions of the H19 sequence were used to generate primers for PCR: A, 5′-AGG WGA CAT CKT CTC GGG GGG AGC CGA GAC-3′; B, 5′-GAC ATG GTC CGG TGT GAY GGH GAG GAC AGA-3′; C, 5′-CTC CYC ACC AGG GCY NCA NCA GRR GYC CTG G-3′. A–B or B–C primer sets were used to amplify the genomic fragments by PCR using Taq DNA polymerase (Promega). The bands generated using the A–B primer set were ∼1–1.2 kb, while the bands generated using the B–C primer set were ∼0.9–1.1 kb. The bands were extracted from 1% agarose gels using a PEG 8000 precipitation method, followed by phenol/chloroform extraction and ethanol precipitation.
These bands were then cloned into the pGEM-T vector system and screened using the blue/white selection method (GenBank accession nos: cat AF190057; elephant AF190054, gopher AF190055, lynx AF190056, orangutan AF190058). DNA was extracted from these colonies using the Promega Wizard miniprep kit. Insert-containing clones were identified by PCR amplification and automatically sequenced using the Sanger dideoxy method with M13 forward and reverse primers (Genemed Synthesis Incorporated, San Francisco, CA). The location of introns in the newly determined sequences was deduced by alignment to previously determined H19 genomic and cDNA sequences (intron–exon boundary sequences are well conserved in all of the sequences).
Covariational analysis
Intron-deleted sequences were aligned using ClustalW, version 1.6 (41) and automatically analyzed using the X2s program for free energy and covariational analysis (http://tyrant/X2s ) (42). Empirical values optimized previously for RNAs of known secondary structure were used to predict H19 secondary structure. Control random alignments were prepared automatically using X2s as follows. The human H19 sequence was initially permuted at random to yield a new sequence with the same base composition as the original. This random sequence served as the base for a random phylogeny of sequences, generated by adding uncorrelated mutations at each site such that the overall pairwise identity for the new alignment matched that for the true H19 sequences (26% substitution frequency).
RESULTS AND DISCUSSION
Mapping regions required for tumor suppressor activity
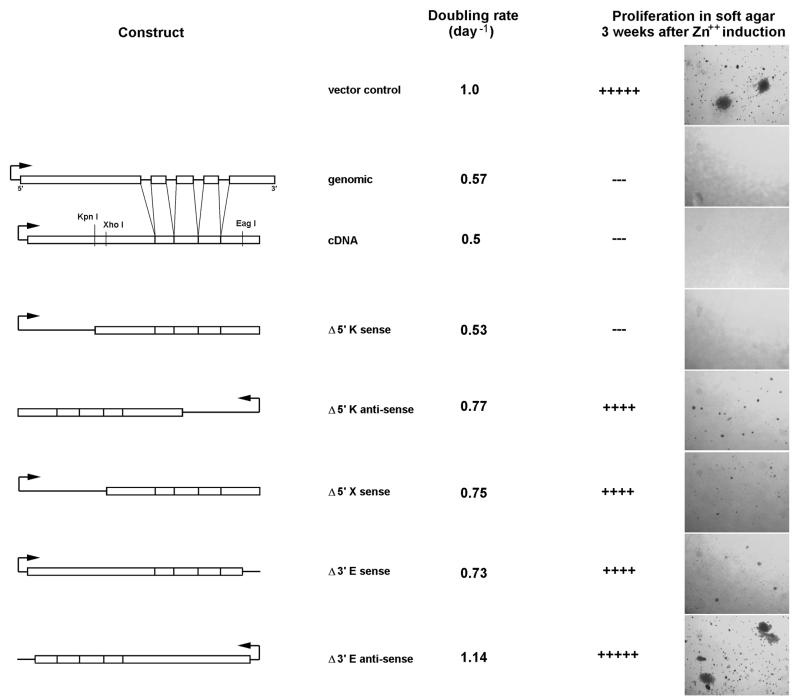
Hao et al. (14) previously demonstrated that transcription of the H19 genomic sequence inhibits the proliferation, clonogenicity and tumorigenicity of the transformed G401 cell line. Using suppression of G401 cell clonogenicity as an in vitro assay for activity, we sought to identify regions of the H19 structural gene which are essential for function. As shown in Figure 1, we are able to reproduce the original results of Hao et al. (i.e. expression of a transfected genomic copy of the H19 gene suppresses anchorage-independent growth of these cells in soft agar) (14). Suppression of clonogenicity is paralleled by a decrease in the rate of cellular proliferation although transfected cells continue to divide and remain generally healthy. In comparison, transfection with vector alone or with the H19 gene in an antisense orientation has no effect.
Figure 1.
Mapping regions of the H19 RNA required for tumor suppressor activity. G401 cells were transfected with the indicated pMEP4-H19 constructs. Proliferation rates were quantified following plating at low density and monitoring growth over the period of 1 week. Effects of H19 on clonogenicity were determined after allowing transfected cells to grow for 3 weeks on soft agar as described in Materials and Methods. Shown are transfection results for vector control, H19 genomic DNA, H19 cDNA and deletion constructs obtained by restriction digestion using KpnI (K), XhoI (X) or EagI (E).
H19 contains a number of unusually short introns, ranging in size from 50 to 120 nucleotides. To test the possibility that these introns are functionally important, we prepared a vector carrying the cDNA version of the gene and tested its effect on G401 cell clonogenicity. Essentially identical results are obtained for the cDNA and genomic versions of H19 (Fig. 1A), suggesting that the intron sequences are dispensable for tumor suppressor activity and arguing against H19 function as a host gene.
Using unique restriction sites present in the cDNA sequence, we generated a series of truncations from the 5′- and 3′-ends of the gene. These constructs were transfected into the G401 cell line and assayed for growth-suppressive effects (Fig. 1B). Deletion of sequences upstream of the KpnI site (position 710) has no effect on the ability to suppress soft agar colony formation. Deletion of an additional 150 nucleotides up to the XhoI site (position 854), however, results in a complete loss of activity, indicating that some essential component is present in the region 710–854. An H19 fragment generated by digestion with EagI lacks 154 nucleotides from the 5′-end and 180 nucleotides from the 3′-end. Cells transfected with the EagI construct show no suppression of clonogenicity or proliferation. Given the prior observation that truncation of the 5′-end in the KpnI construct has no effect in vitro, we presume that the loss of activity in the EagI construct indicates a functional requirement for the last 180 nucleotides. However, this does not rule out the possibility that the lack of growth suppression by the EagI construct may result only from a simultaneous loss of the 154 nucleotide fragment at the 5′-end. Control antisense versions of the KpnI and EagI constructs did not suppress anchorage-independent growth (an XhoI antisense construct was not prepared or tested).
Sequence conservation in H19 RNA
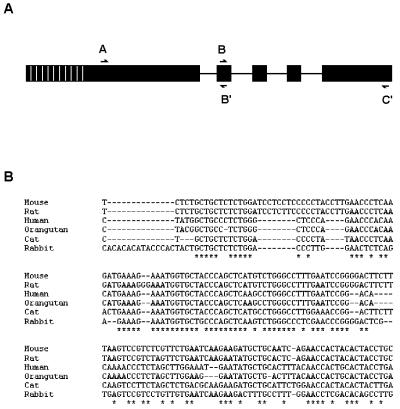
To further understand the sequence requirements for biological function, we used PCR amplification of genomic DNA to clone and sequence the H19 gene from several mammalian species. Three islands of conservation are observed at positions 600–630 (region A), 1540–1570 (region B) and 2460–2480 (region C) within a sequence alignment constructed from the four sequences currently available in GenBank (human, mouse, rat, rabbit; numbering according to the human cDNA). Primers were designed to target these regions and tests with human genomic DNA showed that the A–B and B–C primer pairs could efficiently amplify the H19 gene as two contiguous fragments (corresponding to nucleotides 600–1600 and 1600–2500 respectively) and thus provide essentially the entire gene sequence with the exception of its non-conserved, functionally dispensable 5′-end. Following extensive efforts to optimize PCR conditions, we were able to amplify both fragments from orangutan and cat and the A–B fragment from lynx, elephant and gopher (no conditions or primer sequences tested were capable of amplifying the B–C fragment from these three species). Amplified products were cloned and sequenced as described in Materials and Methods. An analysis of the resulting sequence alignment containing six H19 sequences (four from GenBank and two new) is shown in Figure 2B.
Figure 2.
Sequence conservation in the H19 RNA. (A) Regions marking the primer sites used to generate the PCR products are shown along the H19 gene. The non-conserved 5′-extension in the database H19 sequences is indicated by shading. (B) A representative portion of the ClustalW alignment of H19 cDNAs shows regions of very high and low sequence conservation at the DNA level. Complete conservation is marked by an asterisk.
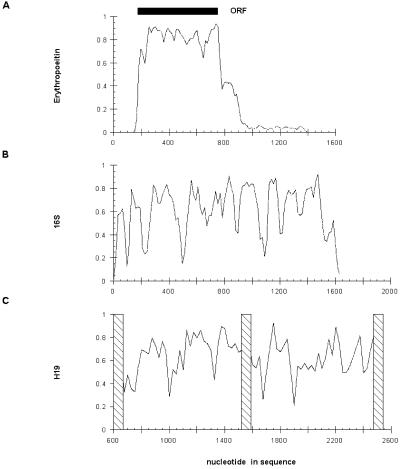
A striking feature of the H19 sequence alignment is the way sequence conservation varies across the gene. In several instances, highly conserved islands of 20–40 nucleotides are flanked by regions of poor conservation (Fig. 3C). A similar overall pattern is observed in an alignment of bacterial 16S ribosomal RNAs (Fig. 3B). In comparison, an alignment of the protein-coding erythropoeitin gene (for which a comparable database of mammalian sequences is available) shows fairly uniform conservation with fluctuations localized to the untranslated regions flanking the ORF (Fig. 3A).
Figure 3.
Average pairwise identity at the nucleotide level. The average pairwise identity of nucleotides in the alignments of erythropoeitin mRNA (A), 16S rRNA (B) and H19 mRNA (C) are calculated as described in Results and plotted as a function position along the sequence.
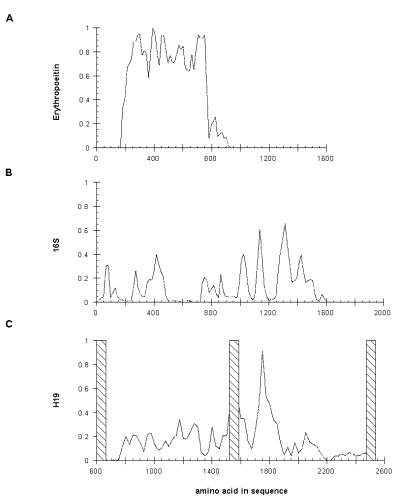
If the functional product of the H19 gene is a protein generated by translation of its mRNA, one would expect to identify an ORF conserved in all mammalian species. With the exception of the human and cat genes, none of the H19 sequences contains an ORF of any substantial length. Start and stop codons appear at regular intervals in all three potential reading frames for every sequence. Conservation of the largest human and cat ORFs is no different from that expected on the basis of the nucleotide similarity throughout the alignment and the positions of start and stop codons for the two ORFs differ widely. A previous analysis of the human and mouse genes noted that several short ORFs overlap in the two sequences although efforts to identify the largest of these using antibodies to synthetic peptides proved unsuccessful (1,19,43). Analysis of the larger database of sequences shows that all ORFs can be discounted on the basis of their poor conservation at the amino acid level. As shown in Figure 4, conservation predicted by hypothetical translation of all reading frames is <40% with the exception of a short region spanning nucleotides 1740–1820. Within this short region of apparent conservation, there are many cases of non-conservative substitutions, including several positions at which glycines and tryptophans are interchanged (evolutionarily, one of the least conservative mutations possible). Similar conservation at the peptide level is observed with 16S rRNA (Fig. 4B).
Figure 4.
Average pairwise identity of ORFs. The average pairwise identity of all predicted ORFs in erythropoeitin (A), 16S rRNA (B) and H19 (C) are plotted as a function of position within the gene.
Identifying secondary structure within the H19 mRNA
Given the absence of evolutionarily conserved ORFs and thus the likelihood that the gene does not yield a functional protein, we re-analyzed the sequences in an attempt to identify RNA secondary structure that would enable the transcript itself to fold and function biologically. This analysis was facilitated using the program X2s to automatically search for all possible helical pairings. The algorithm implemented in X2s assigns secondary structure on the basis of both predicted folding free energy and covariational evidence for Watson–Crick base pairing (42). In previous studies, >80% of all helices in structural RNAs (e.g. SRP RNA, 16S rRNA) were predicted using alignments with as few as five sequences. In the absence of additional experimental information to validate the prediction, we divided the sequence alignment into working and test sets to separately predict the structure and then to test it. The working set consisted of the A–B/B–C fragments from the human, mouse, rat, rabbit, cat and orangutan genes while the test set included the A–B fragment from lynx, elephant and gopher. Covariation observed between proposed paired regions within the test set sequences was taken as experimental support for the proposed pairings.
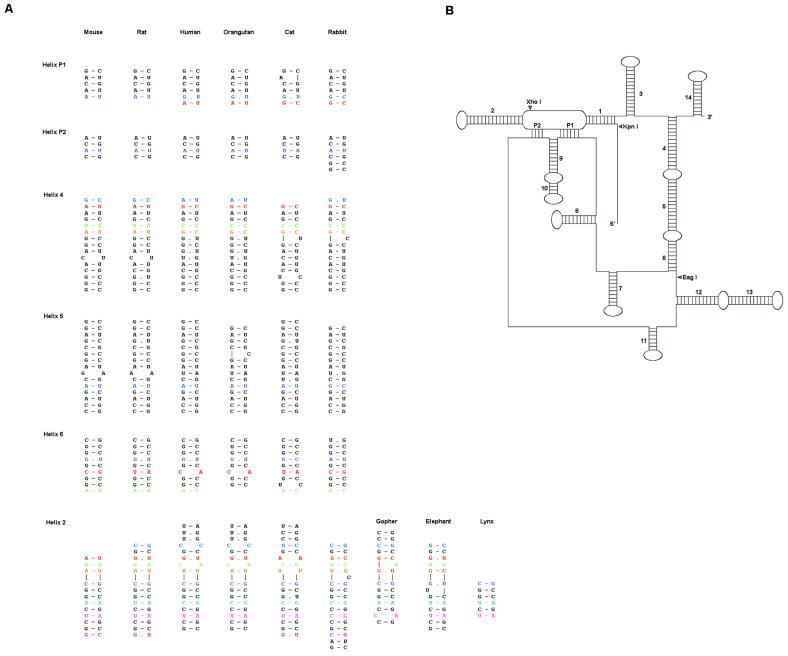
A total of 17 helices were identified in the working set sequences. Of these, 16 exhibit covariation in the test set. The remaining helix is absolutely conserved in the test set but is predicted to form a stable pairing. Most of the helices identified form local hairpins although a few are long range pairings which effectively bring together the 5′- and 3′-ends of the molecule. Figure 5 shows predicted foldings in the working set and test alignments for helices that form long-range pairings.
Figure 5.
Secondary structure of the H19 RNA. (A) A sampling of helical pairings in the H19 alignment identified by X2s. Analogous base pairs are aligned horizontally. Pairs displaying covariation are colored. (B) A schematic diagram of the proposed secondary structure. The location of restriction sites used to generate truncated forms of H19 are marked with arrowheads. The length of unstructured linkers is not to scale.
In searching a long sequence alignment with a limited number of sequences, one might expect to identify by chance pairings that appear to covary with each other. As controls for the prediction algorithm, we attempted to predict secondary structure using (i) the erythropoeitin mRNA alignment (for which there is no previous evidence for secondary structure) and (ii) an artificial, random sequence alignment that maintains the base composition and overall sequence similarity of the H19 alignment. A single helical pairing supported by covariation is predicted for the erythropoeitin alignment and none are predicted for the random alignment, arguing that the algorithm does not substantially over-predict pairings.
The deletion experiments are generally consistent with the proposed secondary structure. The structure predicted for the four full-length H19 sequences contains no additional pairings outside the A–C domain and the predicted structure contains no evolutionarily conserved pairings upstream of the KpnI site. The difference in activity for the KpnI and XhoI sites suggests that a functionally essential element is present within region 710–854. We note that several pairings for which there is extensive covariational support, including both pseudoknots and helix 2, lie within this region and would be deleted or disrupted in the non-functional XhoI truncation. Deletion of 180 nucleotides at the 3′-end in the EagI truncation leads to a loss of activity and is also predicted to disrupt three long-range pairings (helices 4, 5 and 6) that effectively join the ends of the molecule together. Further support for the proposed structure is provided by RNase mapping experiments with in vitro transcribed RNA. In the presence of an oligonucleotide complementary to the 5′-strand of helix 5, the full-length mRNA remains uncleaved by RNase H while the transcript corresponding to the EagI truncation is readily degraded (data not shown).
The role of the proposed secondary structures in facilitating the activity of H19 as a tumor suppressor remains to be determined. Interestingly, we note that absolutely conserved sequences often lie next to helical pairings in both hairpin loops and joining regions, suggesting that the structure serves as a framework for presenting these invariant nucleotides. It is worth noting that because identification of pairings requires covariational support, predicted helices are effectively excluded from absolutely conserved regions. In several cases, pairs of nucleotides appear to covary in many sequences but are disrupted in some. This behavior is typical for RNPs such as the signal recognition particle in which the RNA helices are relatively long and the failure to form a single base pair does not prevent their proper folding. Separating the conserved helices in some regions are sizeable linkers that exhibit poor sequence conservation. Using the clonogenicity assay for H19 function, it should be possible to generate internal truncations in which these regions are systematically deleted and thereby generate a minimal functional transcript for further characterization.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the San Francisco Zoo DNA bank for generously providing blood from elephant, lynx and orangutan, Soule lab for the gopher genomic DNA, and Creekside Veterinary Hospital for providing cat tissue. This work was supported by grants from the NIH (GM52707) and the Packard Foundation to C.W.
DDBJ/EMBL/GenBank accession nos AF190054–AF190058
REFERENCES
- 1.Pachnis V., Brannan,C.I. and Tilghman,S.M. (1988) EMBO J., 7, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg A.P. (1999) Cancer Res., 59 (suppl.), 1743s–1746s. [PubMed] [Google Scholar]
- 3.Dao D., Walsh,C.P., Yuan,L., Gorelov,D., Feng,L., Hensle,T., Nisen,P., Yamashiro,D.J., Bestor,T.H. and Tycko,B. (1999) Hum. Mol. Genet., 8, 1337–1352. [DOI] [PubMed] [Google Scholar]
- 4.Joyce J.A., Lam,W.K., Catchpoole,D.J., Jenks,P., Reik,W., Maher,E.R. and Schofield,P.N. (1997) Hum. Mol. Genet., 6, 1543–1548. [DOI] [PubMed] [Google Scholar]
- 5.Chung W.Y., Yuan,L., Feng,L., Hensle,T. and Tycko,B. (1996) Hum. Mol. Genet., 5, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 6.Dowdy S.F., Fasching,C.L., Araujo,D., Lai,K.M., Livanos,E., Weissman,B.E. and Stanbridge,E.J. (1991) Science, 254, 293–295. [DOI] [PubMed] [Google Scholar]
- 7.Weissman B.E., Saxon,P.J., Pasquale,S.R., Jones,G.R., Geiser,A.G. and Stanbridge,E.J. (1987) Science, 236, 175–180. [DOI] [PubMed] [Google Scholar]
- 8.Reid L., West,A., Gioeli,D., Phillips,K., Araujo,D., Stanbridge,E. and Weissman,B. (1994) Am. J. Hum. Genet., 55 (suppl.), A268. [Google Scholar]
- 9.Cui H., Hedborg,F., He,L., Nordenskjold,A., Sandstedt,B., Pfeifer-Ohlsson,S. and Ohlsson,R. (1997) Cancer Res., 57, 4469–4473. [PubMed] [Google Scholar]
- 10.Biran H., Ariel,I., De Groot,N., Shani,A. and Hochberg,A. (1994) Tumor Biol., 15, 123–134. [DOI] [PubMed] [Google Scholar]
- 11.Verkerk A.J.M.H., Ariel,I., Hochberg,A., Dekker,M.C., Van Gurp,R.J.H.L.M., Gillis,A.J.M., Oosterhuis,J.W. and Looijenga,L.H.J. (1997) Tumor Biol., 18 (suppl. 1), 32. [Google Scholar]
- 12.Ariel I., Ayesh,S., Perlman,E.J., Pizov,G., Tanos,V., Schneider,T., Erdmann,V.A., Podeh,D., Komitowski,D., Quasem,A.S., De Groot,N. and Hochberg,A. (1997) Mol. Pathol., 50, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonomura N., Miki,T., Nishimura,K., Kanno,N., Kojima,Y. and Okuyama,A. (1997) J. Urol., 157, 1977–1979. [PubMed] [Google Scholar]
- 14.Hao Y., Crenshaw,T., Moulton,T., Newcomb,E. and Tycko,B. (1993) Nature, 365, 764–767. [DOI] [PubMed] [Google Scholar]
- 15.Casola S., Pedone,P.V., Cavazzana,A.O., Basso,G., Luksch,R., D’Amore,E.S., Carli,M., Bruni,C.B. and Riccio,A. (1997) Oncogene, 14, 1503–1510. [DOI] [PubMed] [Google Scholar]
- 16.Bartolomei M.S., Zemel,S. and Tilghman,S.M. (1991) Nature, 351, 153–155. [DOI] [PubMed] [Google Scholar]
- 17.Ripoche M.A., Kress,C., Poirier,F. and Dandolo,L. (1996) Int. J. Dev. Biol., 40, 36S. [Google Scholar]
- 18.Ferguson-Smith A.C., Sasaki,H., Cattanach,B.M. and Surani,M.A. (1993) Nature, 362, 751–755. [DOI] [PubMed] [Google Scholar]
- 19.Brannan C.I., Dees,E.C., Ingram,R.S. and Tilghman,S.M. (1990) Mol. Cell. Biol., 10, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zemel S., Bartolomei,M.S. and Tilghman,S.M. (1992) Nature Genet., 2, 61–65. [DOI] [PubMed] [Google Scholar]
- 21.Tilghman S.M., Bartolomei,M.S., Webber,A.L., Brunkow,M.E., Saam,J., Leighton,P.A., Pfeifer,K. and Zemel,S. (1993) Cold Spring Harb. Symp. Quant. Biol., 58, 287–295. [DOI] [PubMed] [Google Scholar]
- 22.Ripoche M.A., Kress,C., Poirier,F. and Dandolo,L. (1997) Genes Dev., 11, 1596–1604. [DOI] [PubMed] [Google Scholar]
- 23.Yoo-Warren H., Pachnis,V., Ingram,R.S. and Tilghman,S.M. (1988) Mol. Cell. Biol., 8, 4707–4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joubel A., Curgy,J.J., Pelczar,H., Begue,A., Lagrou,C., Stehelin,D. and Coll,J. (1996) Cell. Mol. Biol., 42, 1159–1172. [PubMed] [Google Scholar]
- 25.Li Y.-M., Franklin,G., Cui,H.-M., Svensson,K., He,X.-B., Adam,G., Ohlsson,R. and Pfeifer,S. (1998) J. Biol. Chem., 273, 28247–28252. [DOI] [PubMed] [Google Scholar]
- 26.Tycowski K.T., Shu,M.D. and Steitz,J.A. (1994) Science, 266, 1558–1561. [DOI] [PubMed] [Google Scholar]
- 27.Frey M.R., Wu,W., Dunn,J.M. and Matera,A.G. (1997) Histochem. Cell Biol., 108, 365–370. [DOI] [PubMed] [Google Scholar]
- 28.Hurst L.D., McVean,G. and Moore,T. (1996) Nature Genet., 12, 234–237. [DOI] [PubMed] [Google Scholar]
- 29.Haig D. (1996) Bioessays, 18, 351–353. [DOI] [PubMed] [Google Scholar]
- 30.Erdmann V.A., Szymanski,M., Hochberg,A., De Groot,N. and Barciszewski,J. (1999) Nucleic Acids Res., 27, 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Askew D.S. and Xu,F. (1999) Histol. Histopathol., 14, 235–241. [DOI] [PubMed] [Google Scholar]
- 32.Kay G.F. (1998) Mol. Cell Endocrinol., 140, 71–76. [DOI] [PubMed] [Google Scholar]
- 33.Endo Y., Watanabe,T., Mishima,Y., Yoshimura,A., Takagi,N. and Kominami,R. (1999) Mamm. Genome, 10, 606–610. [DOI] [PubMed] [Google Scholar]
- 34.Hong Y.K., Ontiveros,S.D., Chen,C. and Strauss,W.M. (1999) Proc. Natl Acad. Sci. USA, 96, 6829–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeffery W.R. and Swalla,B.J. (1995) Mol. Biol. Cell, 6 (suppl.), 345A.7612968 [Google Scholar]
- 36.Lee R.C., Feinbaum,R.L. and Ambros,V. (1993) Cell, 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 37.Ha I., Wightman,B. and Ruvkun,G. (1996) Genes Dev., 10, 3041–3050. [DOI] [PubMed] [Google Scholar]
- 38.Barciszewski J. and Legocki,A.B. (1997) Acta Biochim. Pol., 44, 795–802. [PubMed] [Google Scholar]
- 39.Hollander M.C., Alamo,I. and Fornace,A.J.Jr (1996) Nucleic Acids Res., 24, 1589–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Askew D.S., Li,J. and Ihle,J.N. (1994) Mol. Cell. Biol., 14, 1743–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higgins D.G., Thompson,J.D. and Gibson,T.J. (1996) Methods Enzymol., 266, 383–402. [DOI] [PubMed] [Google Scholar]
- 42.Juan V. and Wilson,C. (1999) J. Mol. Biol., 289, 935–947. [DOI] [PubMed] [Google Scholar]
- 43.Leibovitch M.P., Nguyen,V.C., Gross,M.S., Solhonne,B., Leibovitch,S.A. and Bernheim,A. (1991) Biochem. Biophys. Res. Commun., 180, 1241–1250. [DOI] [PubMed] [Google Scholar]