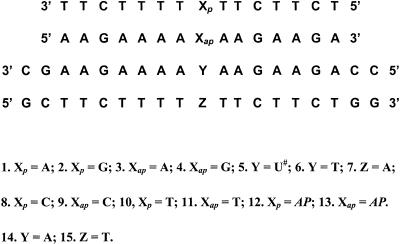Abstract
A necessary feature of the natural base triads for triplex formation is the requirement of a purine (A or G) in the central position, since only these provide sets of two hydrogen bond donors/acceptors in the major groove of the double helix. Pyrimidine bases devoid of this feature have incompatible complementarity and lead to triplexes with lower stability. This paper demonstrates that 5-aminouracil (U#) (I), a pyrimidine nucleobase analogue of T in which 5-methyl is replaced by 5-amino group, with hydrogen bonding sites on both sides, is compatible in the central position of triplex triad X*U#·A, where X = A/G/C/T/2-aminopurine (AP), and * and · represent Hoogsteen and Watson–Crick hydrogen bonding patterns respectively. A novel recognition selectivity based on the orientation (parallel/antiparallel) of the third strand purines A, G or AP with A in the parallel motif (Ap*U#·A), and G/AP in the antiparallel motif (Gap/APap*U#·A) is observed. Similarly for pyrimidines in the third strand, C is accepted only in a parallel mode (Cp*U#·A). Significantly, T is recognised in both parallel and antiparallel modes (Tp/Tap*U#·A), with the antiparallel mode being stable compared to the parallel one. The ‘U#’ triplexes are also more stable than the corresponding control ‘T’ triplexes. The results expand the lexicon of triplex triads with a recognition motif consisting of pyrimidine in the central strand.
INTRODUCTION
The interest in oligonucleotides and their analogues containing chemically modified bases and sugar–phosphate backbone continues unabated, mainly due to their potential applications as antigene/antisense inhibitors in DNA therapeutics (1–4). Among these, the modifications of the naturally occurring nucleobases by substituents or replacing them by designed heterocycles with retention of molecular specificity in complementary recognition offers considerable challenge. With the advent of triple helical DNA, a variety of chemically modified bases or their conjugates incorporated into the third strand have come to the fore as strategies for stabilisation of DNA triplexes (5,6) or for performing sequence-specific cleavage of the double helix (7). DNA triplexes originate from major groove binding of a third strand oligonucleotide that is either pyrimidine (Y) or purine (R) rich, in parallel (p) or antiparallel (ap) orientation respectively (1,8). The specificity in triplex formation is derived from Hoogsteen (HG; *) hydrogen bonding by which T recognises A of A·T Watson–Crick (WC; ·) base pair (T*A·T) and protonated C+ binds to G of G·C base pair (C+*G·C) in the pyrimidine motif. Similarly, in the purine motif, the third strand A binds to A of A·T, while G binds to G of G·C base pair by reverse Hoogsteen mode. A common feature of all these triads is the requirement of a purine (A or G) in the central position, since only these provide two sets of hydrogen bond donors/acceptors in the major groove of the double helix. Pyrimidine bases devoid of this feature are not generally compatible in the middle position and lead to a decreased triplex stability from HG mismatches. Among the eight possible triads with T or C in the middle, only G*T·A and T*C·G are accommodated with reasonable stability within the established motifs (9–12). This limitation of triplex formation has led to an exploration of new triad combinations involving unnatural base components (5,6,13–18) located in the third position of the triad, that are sterically and electronically complementary, to recognise T and C of the base pairs TA and CG. Pyrimidines can also be engineered to endow dual recognition properties for placement as the central bases of triplex triads, realised in the pseudonucleobases, ψU, ψC and ψ–isoC that possess extra hydrogen bonding sites in the major groove of the derived WC type duplex (19,20).
We envisaged that the pyrimidine analogue 5-aminouracil (U#) I, derived from a simple replacement of the hydrophobic 5-methyl group of T by the hydrophilic 5-amino moiety (21,22), would be suitable as the middle base of a triplex triad, since it has the electronic requirements for simultaneous recognition of complementary bases of triad. In preliminary communications (23,24), we have reported the recruitment of U# in the generation of triplex triads R*U#·A (R = A or G) and Y*U#·A (Y = C or T) and demonstrated a specificity in its recognition by third strand A/G/C/T which is dependent on the orientation of the unmodified HG strand. This paper describes detailed biophysical studies on the novel features of U# in its recognition selectivity for third strand nucleobases A/T/C/G and an unnatural nucleobase 2-aminopurine (AP). It is demonstrated that the observed parallel selectivity for A can be switched to antiparallel by either G or AP and triple helix formation is shown to occur with T in both the pyrimidine (parallel) and purine (antiparallel) motifs, but with C only in the pyrimidine motif. Apart from the UV thermal profiles suggesting that triplexes containing U# are more stable than the corresponding controls with T in identical position, additional evidence from circular dichroism (CD) is presented to confirm the formation of specific triple helices from U# in the central strand. With this finding, U# emerges as a novel pyrimidine nucleobase analogue that recognises all the bases A, G, C and T of third strand in DNA triplex, in an orientation-specific manner.
MATERIALS AND METHODS
All chemicals used were of reagent quality or better grade. Base-protected standard nucleoside phosphoramidites and 5′-O-DMT nucleoside-derived control pore glass supports (CPG) were procured from Cruachem, UK. All oligonucleotides were synthesised on 1.3 µmol scale on a Pharmacia GA plus DNA synthesiser using CPG and nucleobase-protected 5′-O-(4,4′-dimethoxytrityl)-2′-deoxyribonucleoside-3′-O-[N,N-(diisopropylamino)-β-cyanoethyl phosphoramidite] monomers, followed by deprotection with aqueous NH3. The protected N5-trifluoroacetyl-5-amino-dU and N2-isobutyryl-2-aminopurine deoxyribonucleoside-3′-O-phosphoramidite monomers were synthesised according to the reported procedures (21) and (25) respectively. All oligonucleotides were purified by reverse-phase FPLC on a C18 column and the purity rechecked on reverse-phase HPLC using the buffer systems: A, 5% CH3CN in 0.1 M triethylammonium acetate (TEAA) and B, 30% CH3CN in 0.1 M TEAA with a gradient of A to B of 1.5%/min at a flow rate of 1.5 ml/min.
The various triplexes containing U# or T in the central strand (Table 1) were individually constituted by taking 1 µM each of the appropriate single strands based on the UV absorbance at 260 nm calculated using the molar extinction coefficients of A/AP = 15.4, C = 7.3, G = 11.7 and T/U# = 8.8 cm2/µmol. These were annealed by heating at 80°C in 100 mM sodium cacodylate buffer containing 20 mM MgCl2 and 1 M NaCl at pH 5.8 or pH 7.1 for 3 min, followed by slow cooling. Similarly, the duplexes 5·7 and 6·7 were constituted by taking equimolar amounts of appropriate single strands. The thermal stability of duplexes and triplexes was measured by following UV absorbance changes at 260 nm in the temperature range 5–80°C with a heating rate of 0.5°C/min, using a Perkin Elmer Lambda 15 UV/VIS spectrophotometer, fitted with a water jacketed cell holder and a Julabo temperature programmer. The triplex disassociation temperatures (tms) were determined from the midpoint of the first transition in the plots of percent hyperchromicity versus temperature, further confirmed by maxima in differential (δA/δT versus temperature) curves, and are accurate to ±0.5°C over the reported values.
Table 1. UV-tm of triplexesa.
| Entry | X*Y·Z | Triplex | tm (°C) |
|---|---|---|---|
| 1 | Gp*T·A | 2*6·7 | 26 (25) |
| 2 | Ap*T·A | 1*6·7 | 24 |
| 3 | Ap*U#·A | 1*5·7 | 28 |
| 4 | Aap*U#·A | 3*5·7 | nd |
| 5 | Gp*U#·A | 2*5·7 | nd |
| 6 | Gap*U#·A | 4*5·7 | 35 (37) |
| 7 | APp*U#·A | 12*5·7 | 26 |
| 8 | APp*T·A | 12*6·7 | 26 |
| 9 | APap*U#·A | 13*5·7 | 38 (39) |
| 10 | Cp*U#·A | 8*5·7 | 26 |
| 11 | Cp*T·A | 8*6·7 | 20 |
| 12 | Cap*U#·A | 9*5·7 | nd |
| 13 | Tp*U#·A | 10*5·7 | 26 |
| 14 | Tp*T·A | 10*6·7 | 19 |
| 15 | Tap*U#·A | 11*5·7 | 36 (37) |
aAll at pH 5.8. Values in parentheses are tm at pH 7.1. tms are accurate to ±0.5°C. No triplex formation was observed for the following control triads with T in the central position Gap*T·A (4*6·7), Aap*T·A (3*6·7), APap*T·A (13*6·7), Cap*T·A (9*6·7) and Tap*T·A (11*6·7). nd, not detected.
CD spectra were recorded on a JASCO J-715 spectropolarimeter attached to a Julabo water circulator for maintaining the temperatures at 10°C for all measurements using thermostated cells. The samples taken in 10 mm cells, were scanned in the range of 320–200 nm with the following instrumental parameters: scan speed 200 nm/min, band width 1.0 nm, sensitivity 10 mdeg, resolution 0.1 nm and response factor 2 s. Each spectrum was collected as an average of five scans. The samples were made in a similar manner to that for UV melting experiments by mixing 1 µM of each appropriate strand. The Job plots for stoichiometry determination were obtained by the continuous variation method (26) using equimolar solutions of third strand and duplex mixed together at increasing third strand/duplex ratios, keeping the total DNA concentration constant.
RESULTS AND DISCUSSION
Design of U# and AP oligonucleotides for triplex studies
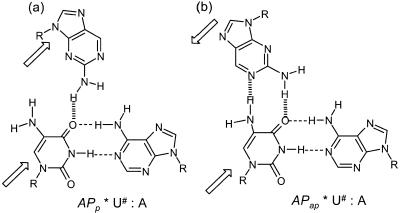
The different possible triads consisting of U# in the central position are shown in Schemes 123. It is seen that accommodation of U# in middle strand of the established pyrimidine and purine motifs is possible only when the HG strand containing A is parallel (Ap*U#·A) (Scheme 1a) and that with G is antiparallel (Gap*U#·A) (Scheme 1d) to the central strand containing U#. The alternative orientations of the third strand (Aap, Gp; Scheme 1b and c) do not provide HG complementation of A and G with U# within the triads due to non-complementarity of donor/acceptor pairs. In contrast to A (6-aminopurine), its isomer AP, which has the amino function participating in hydrogen bonding in a different position, can be accommodated better when the third strand is antiparallel (Scheme 2b) than in a parallel mode (Scheme 2a). Similarly, U# complementation with third strand C is much better in a parallel orientation (Scheme 3a) than in an antiparallel mode (Scheme 3b). In comparison, T in the third strand can be accommodated equally well with U# in both parallel and antiparallel modes (Scheme 3c and d). The oligodeoxyribonucleotide sequences 1–15 were designed to combinatorially generate all the desired triplexes that differ with respect to third strand orientation and a base within a single triad site X*Y·Z [X = A, G, C, T or AP and X strand in parallel (Xp) or antiparallel (Xap) mode]. In each case, the triplex sets corresponding to the oligonucleotides with Y = U# or T provided the test and controls respectively. While designing the sequences, care was taken to avoid the self-complementation and a few Cs were introduced into third strand to break the continuous stretches of T and thereby prevent its slippage on the duplex. This was further ensured by the addition of CG locks at the duplex ends. Such a duplex–triplex system also provided tm in a convenient range for studying all the combinations of triads.
Scheme 1. H-bonding representation of third strand A in parallel (a) and antiparallel (b) motifs and that of G in parallel (c) and antiparallel (d) motifs.
Scheme 2. H-bonding representation of third strand AP in parallel (a) and antiparallel (b) motifs.
Scheme 3. H-bonding representation of third strand C in parallel (a) and antiparallel (b) motifs and that of T in parallel (c) and antiparallel (d) motifs.
Chemical synthesis and characterisation of U# and AP oligonucleotides
We have previously reported the synthesis and base pairing properties of the modified nucleoside 5-amino dU (21), and its chemical incorporation into DNA by solid phase phosphoramidite synthesis, using trifluoroacetyl as a protector for the 5-amino group (22). The coupling efficiency of the modified amidite was similar to that of the phosphoramidites of normal nucleosides. Subsequent to our work, it was reported that during the capping reaction, a small percentage of N5-trifluoroacetyl gets transformed into an N5-acetyl group that remains stable to the final ammonia deblocking step and this led to the use of 2-(4-nitrophenyl)ethoxy as a protector for the 5-NH2 group in U# (27). The product oligonucleotides containing U# in our case, were purified by HPLC and the mass spectral data corresponded to the expected molecular composition of fully deblocked 5-NH2-dU oligonucleotide (molecular mass for C188H211N87O105P18: Mcalc 5923.9, Mobs 5923), thus ruling out the formation of any dU-N5-acetylated oligonucleotides. The U# oligonucleotide 5 was therefore synthesised with the N5-trifluoroacetyl protected monomer, without any changes in the synthesis protocols. The oligonucleotides 12 and 13 having AP in the sequence were synthesised similarly by using 5- (4,4′-dimethoxytrityl)-N2-isobutyryl-2-aminopurine-2′-deoxyribonucleoside phosphoramidite synthon (25) for coupling at the desired position, followed by deprotection and HPLC purification. This product also possessed the molecular weight expected for its molecular composition (12, MWcalc 4464, MWobs 4466; 13, MWcalc 4684, MWobs 4690) as seen by the MALDI-TOF data. The U# and AP oligonucleotides underwent complete digestion with the nucleases snake venom phosphodiesterase and alkaline phosphatase to give the expected base compositions.
Triplex formation with U# in the central strand
The designed oligonucleotides 1–15 were used to combinatorially constitute the various desired triplexes in purine and pyrimidine motifs with the duplex 5·7. The substitution of T by U# in duplexes led to only a slight decrease in tm of 1.5°C/substitution (22): 6·7, 61.0°C; 5·7, 59.5°C in 10 mM sodium cacodylate pH 7.0, 100 mM NaCl. The successful formation of triplexes was characterised by the presence of well-defined biphasic transitions in percent hyperchromicity versus temperature profiles (Figs 1 and 2). The lower temperature transition corresponds to triplex↔duplex melting and the higher temperature one is due to melting of duplex to single strands. The magnitude of percent hyperchromicity changes, in general for triplex melting to duplex (7.5%) was about half of that accompanying the melting of the underlying duplex (15%). The accurate tms (Table 1) were obtained from the corresponding maximum in their first derivative curves. In case of non-triplex formation, the observed hyperchromicity is a sum of that arising from duplex melting (15%) and that due to single strand melting (third strand polypyrimidine <4%). Thus, the absence of triplex formation resulted in a decrease of ∼5% less hyperchromicity changes in comparison to that seen in the presence of triplex. However in the case of third strand polypurines, the relative difference in the hyperchromicity for the presence and absence of triplexes was negligible, since polypurines exhibit considerable hyperchromicity changes even in single-strand melting due to self-stacking. The percent hyperchromicity seen during the thermal melting reflects the extent of base stacking, which differ for polypurine and polypyrimidine sequences. The distinguishable feature for the presence of triplex is the observance of characteristic double sigmoidal melting curves with an additional peak in the first derivative plots, which is well precedented in literature (3). All melting experiments including controls were carried out in the presence of 1 M NaCl since triplexes from U# sequences showed optimum stability under these conditions. The formation of triplexes was also supported by the mixing curves, which showed an inflection point at 0.5 mole fraction, thus establishing a 1:1 stoichiometry of third strand and duplex.
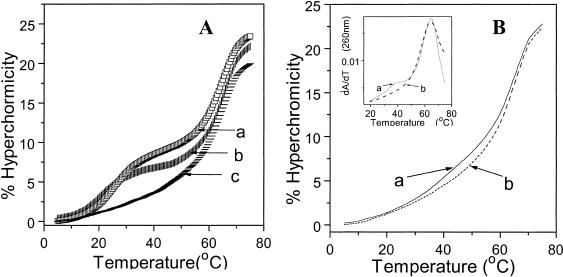
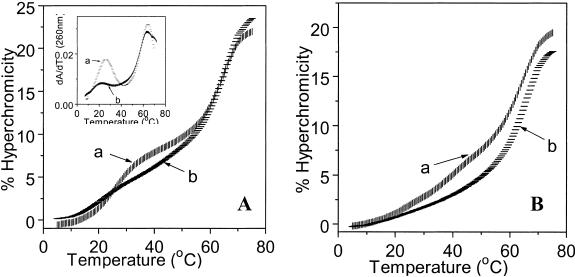
Figure 1.
(A) UV melting profiles in buffer containing 100 mM sodium cacodylate, 20 mM magnesium chloride and 1 M sodium chloride at pH 5.8 for (a) Ap*U#·A (1*5·7); (b) Ap*T·A (1*6·7); (c) Aap*U#·A (3*5·7). (B) (a) APap*U#·A (13*5·7); (b) APap*T·A (13*6·7). Inset: (a) and (b) are first derivatives respectively.
Figure 2.
(A) UV melting profiles of (a) Tp*U#·A (10*5·7); (b) Tp*T·A (10*6·7). Inset: (a) and (b) are first derivatives respectively. (B) (a) Tap*U#·A (11*5·7); (b) Tap*T·A (11*6·7).
Selective recognition of third strand purines (A/G) by central U#
The UV-tm data obtained for different triplexes at pH 5.8 and pH 7.1 are shown in Table 1. At pH 5.8, stable triplex formation is observed for the control triplex with Gp*T·A triad (2*6·7) within the parallel pyrimidine motif (Table 1, entry 1) but not in the antiparallel purine motif (4*6·7). The analogous parallel triplex (1*6·7) with triad Ap*T·A (entry 2) is slightly less stable, in agreement with the literature (3,4). In comparison, the parallel triplex having triad Ap*U#·A (1*5·7) with modified base in central strand (entry 3), has a higher tm than the control triplex Ap*T·A (1*6·7) in the pyrimidine motif; no triplex is detected in the corresponding antiparallel mode (Fig. 1A). In the case of triad Gap*U#·A, triplex formation is seen only in the antiparallel orientation (4*5·7) (entry 6) and not in the parallel form (2*5·7). The Gap*U#·A antiparallel triplex has a higher thermal stability (Δtm = 9°C) than the Gp*T·A triplex (2*6·7) seen in the parallel motif (entry 1). Thus, triplexes with modified base U# in the central strand (1*5·7 and 4*5·7) exhibit not only a higher tm compared to the corresponding control T analogue, but also display a remarkable orientation selectivity in third strand recognition. The tms of the duplex in T and U# triplexes differ only marginally (Δtm = 1–2°C) and are similar to that of the duplex alone. The replacement of T by U# in duplexes alone leads to a small destabilisation of the latter (22) by 1.5°C and in triplexes they are not influenced much by the nature of the third base of the triad. The percent hyperchromicity change for triplex melting in the antiparallel motif is also much less than that in the parallel motif as expected from a poorer base stack in the former as compared to the latter (3). Due to the molecular necessity of N3 protonation in C, triplex formation in the pyrimidine motif is pH dependent with a higher stability at lower pH, which is not the case with purine motif triplexes (3). The pyrimidine motif triplexes containing Ap*U#·A triad and Ap*T·A triad are formed only at pH 5.8 and are not detected at pH 7.1. In contrast, the antiparallel purine motif triplex (4*5·7) devoid of the base C in the third strand is observed in both pH ranges, with a slightly higher stability at pH 7.1 (tm = 37°C) compared to that at pH 5.8 (tm = 35°C).
Recognition of third strand AP by central U# (AP*U#·A)
The hydrogen bonding complementarity (Scheme 2) suggests that if A in the third strand is replaced by AP, specific recognition of U# with two hydrogen bonds occurs only in an antiparallel orientation but with only one hydrogen bond in a parallel mode. The triplexes (12*5·7) and (13*5·7) were therefore constituted to test this possibility and the melting curves shown in Figure 1B are in agreement with the proposed hydrogen bonding scheme. Triplex formation is observed in antiparallel mode with triad APap*U#·A (13*5·7) (Table 1, entry 9) whereas the control APap*T·A (13*6·7) does not show the presence of triplex (Fig. 1B). In the parallel mode, the formation of triplex with either U# (12*5·7, entry 7) or T (12*6·7, entry 8) in the centre also occurs with almost identical stability, but much less than than that seen in antiparallel mode. This indicates no additional hydrogen bonding contribution from U# in parallel mode.
Selective recognition of third strand pyrimidines (C/T) by central U#
The parallel triplex having the triad Cp*U#·A (8*5·7) shows a higher tm compared to the control triplex Cp*T·A (8*6·7, Δtm = 6°C) (Table 1, entry 10,11), while no triplex formation is detected for the antiparallel triplex Cap*U#·A (9*5·7). The triplex stability is better at pH 5.8 than at pH 7.1 as expected for the C*G·C triplexes. In contrast to the selectivity observed for the recognition of C by U# in the parallel mode, triplex formation is observed for third strand T in both, parallel (entry 13) and antiparallel (entry 15) motifs. The parallel triplex with U# in the middle (10*5·7) shows a higher tm (Δtm = 6.5°C) compared to the control T (10*6·7) (Fig. 2A; Table 1, entry 14). The antiparallel triplex with U# (11*5·7) showed a stable triplex with tm of 36°C, while no triplex formation is noticed for the antiparallel control (11*6·7) containing T in the central position instead of U# (Fig. 2B).
These results are in accordance with the hydrogen bonding proposed in Scheme 3. The observance of triplex with T in the middle strand (8*6·7) may be due to one hydrogen bond still possible between O4 of T and 4-NH2 of C. When U# is in the central position, two hydrogen bonds are possible with C in the parallel orientation (Scheme 3a), leading to a higher tm for the Cp*U#·A triplex than that in the antiparallel mode (Scheme 3b), where only one hydrogen bond is possible. It is also noticed that the C2 carbonyl of T is involved in hydrogen bonding with the 5-NH2 of U# in parallel binding (Scheme 3c) while the C4 carbonyl of T hydrogen bonds with U# in antiparallel mode (Scheme 3d).
Circular dichroic spectral studies of U# triplexes
CD spectroscopy is a useful diagnostic tool for the characterisation of conformation in DNA duplexes and triplexes (28). It has been shown that the CD spectra of nucleic acid triplexes differ from the sum of the individual spectra of the constituent single- and double-stranded DNA (29). In the case of a parallel pyrimidine motif with high AT content, the appearance of an intense negative band in the wavelength region 210–220 nm is characteristic of the presence of triple-stranded structures (30). The formation of triplex is accompanied by a reduction in intensity of the positive bands at short and long wavelength region, present in the CD of duplexes. In the difference CD spectrum obtained from the CD spectra of the triplex and the sum of the target duplex and the third strand, the appearance of a negative band at 210 nm region can be used to confirm the triplex formation.
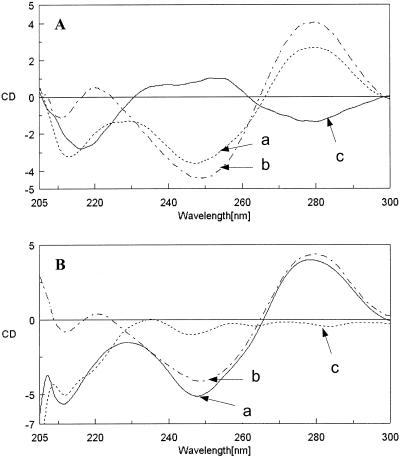
The CD spectra of U# triplexes Ap*U#·A (1*5·7) and Tp*U#·A (10*5·7) along with the sum of CD spectra of duplex (7·5) and the third strands (1 and 10) and the corresponding difference spectra are shown in Figure 3. These triplexes belonging to the parallel pyrimidine motif show an intense negative band at 214 nm for (1*5·7) and at 212 nm for (10*5·7) (Fig. 3, curve a). The general profile of the CD spectra of U#-derived duplexes indicates no major departure from the B-DNA conformation (Fig. 3, curve b). The difference spectra (curve c in Fig. 3) resulted in a large negative band in the region 212–215 nm for triplexes (1*5·7) and (10*5·7). This clearly supports the existence of triplexes seen from their UV-melting curves. This observation which is limited to only the parallel pyrimidine motif, is not seen with U# antiparallel triplexes.
Figure 3.
(A) Circular dichroic spectrum of (a) Ap*U#·A (1*5·7); (b) addition spectrum of duplex U#·A (5·7) and the single strand Ap (1); (c) difference spectrum of (a) – (b). (B) (a) Tp*U#·A (10*5·7); (b) addition spectrum of duplex U#·A (5·7) and the single strand Tp (10); (c) difference spectrum of (a) – (b).
For a standard triplex in the antiparallel motif (e.g. Tap*A·T), the CD spectra consisted of an intense positive band at 217 nm, together with a positive band at 275 nm and a negative band at 250 nm (Fig. 4A). The 217 nm band is characteristic of poly(A)-containing sequences, which are the main constituents of both the middle and third strands (28). Consequently, the difference CD spectrum is devoid of any negative band in 217 nm region and only a slight positive amplitude is seen. In the case of the antiparallel triplex derived from U# in the central strand, i.e. Tap*U#·A (6*3·5), the difference CD spectra is accompanied by a relatively larger positive band at 217 nm (Fig. 4B). Overall, the diagnostic feature of CD at 217 nm that distinguishes standard parallel and antiparallel motifs is also observed with triplexes derived from U#.
Figure 4.
(A) Circular dichroic spectrum of (a) Tap*A·T (11*14·15); (b) addition spectrum of duplex A·T (14·15) and the single strand Tap (11); (c) difference spectrum of (a) – (b). (B) (a) Tap*U#·A (11*5·7); (b) addition spectrum of duplex U#·A (5·7) and the single strand Tap (11); (c) Difference spectrum of (a) – (b).
Novel molecular recognition properties of U# in triplex formation
U# is a pyrimidine analogue in which the hydrogen bonding pattern for WC sites is identical with T or U. The additional H-bonding sites arising from the 5-NH2 function isosteric to 5-CH3 of T in the major groove of duplex are similar to N7 of A and hence the U#·A base pair retains the pseudodyad symmetry of AT. The observed experimental results are in accordance with the proposed hydrogen bonding shown in Schemes 123 and imply a novel molecular recognition of U# of WC duplexes by third strand A, G, C or T, differentiated with respect to HG strand orientation: U# of WC base pair U#·A recognises third strand A and C only in parallel orientation (pyrimidine motif), G recognition occurs in antiparallel orientation (purine motif), while T is recognised both in parallel and antiparallel motifs. The availability of one acceptor (O4 carbonyl) and a donor (5-amino) in U# provides an unambiguous structural basis for orientation specificity in recognition of U#·A duplex by a third base in the major groove. In all possible parallel triplexes, U# in the central position enhances the stability of analogous triplexes derived from T. In contrast, antiparallel triplexes are formed specifically with third strand G, T or AP when U# is present in the centre and do not tolerate T in the identical position, even when one hydrogen bond is possible. The NH2 and CH3 have similar sizes but differ with respect to their polarity [in 5-substituted uracil (31), steric parameter: CH3, 5.63 and NH2, 5.42; hydrophobic parameter: CH3, 0.56 and NH2, –1.23] and consequently, the presence of 5-NH2 group in the major groove of the duplex may effect changes in the local environment. It may be pointed out that hydration sites in the Crick–Hoogsteen groove of a triplex are important determinants for stability in an antiparallel purine motif (32). The replacement of hydrophobic 5-methyl group in T by hydrophilic 5-amino function as in U# may have vital consequences, since the amino group can favourably participate in the hydration network to offer the crucial stability in antiparallel mode. It is interesting to observe that while Gp*T·A forms a stable triplex in parallel mode, the corresponding triplex is not seen in the case of Gp*U#·A. This perhaps suggests that the presence of the 5-methyl group is important in stabilizing the hydrogen bond between the O4 of T and 2-NH2 of G, by providing a local hydrophobic environment. When NH2 is present at 5-position and not able to take part in any productive H-bonding, the induced local hydrophilic environment perhaps weakens the O4-T–2-NH2 G bonding, leading to a destabilised triplex. Recently, we have experimentally demonstrated such shifts in local environment caused by a change of 5-methyl group to 5-NH2 in the major groove of DNA (33). The proposed triad geometries, with non-participation of N7 of purines in the recognition of U# in the central strand are consistent and isomorphous with the established geometries of T*A·T, A*A·T and G*G·C triplexes (34,35). It should be pointed out that the third strand base recognition by U# as shown here also involves nine-membered rings similar to those found in the standard triads. The 5-NH2 is likely to remain largely in free, non-protonated form under the experimental conditions, even at pH 5.8, since its pKa is around 4.2.
An interesting aspect of molecular recognition by unnatural base U# is that it not only pairs predictably with third strand natural bases A, G, C and T in an orientation-dependent manner, but also selectively binds another unnatural base, AP. This is interesting since by shifting the amino group from 6-position in A to 2-position in AP, the orientation selectivity for U# is reversed, while maintaining the electronic complementarity for base pairing. This is a unique case where an engineered molecular recognition leads to a successful HG-type base pairing among two unnatural bases located within the same triad. This also provides a confirmation of the proposed hydrogen bonding schemes and the predictable orientational selectivity in triplex formation for a given triad. The enhancement of triplex stability through engineered hydrogen bonds has recently been seen with 8-amino-dA in the central strand (36–38).
CONCLUSIONS
To our knowledge, the results reported here constitute the first examples where a designed nucleoside base analogue in the central strand such as U#, recognises all four bases A, G, C and T with different selectivities based on parallel and antiparallel orientations of the third strand. It also forms a triad with AP, with reversed selectivity compared to A. The overall results demonstrate the possibilities of generating triplex structures through engineered hydrogen bonds and base triad geometries. The importance and utility of modified bases in the central strand of triplexes was recently shown by use of Ψ-bases to generate triplex base pairing geometries that are isomorphous with the standard triads (39). The experimental realisation of this concept may have a potential outcome: formation of triple-stranded helices at single-strand target sites of unrestricted sequence by addition of two oligonucleotide probes, that contain modified purines and pyrimidines. In this context, the present demonstration that a simple pyrimidine derivative U# in the second strand can selectively tolerate all four natural bases and AP depending on the third strand orientation adds a new repertoire to nucleic acid recognition. Further work involves the study of triplexes with multiple U# substitutions and exploration of other recognition tolerants of U#, that is also emerging as a useful analogue for conjugation of reporter ligands (40,41).
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the Bioorganic Chemistry Department, Centre for Macromolecular Science, Lodz, Poland and Dr V. R. Jadhav (University of Colorado) for providing the mass spectral analysis of the oligonucleotides. K.N.G. thanks the Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore, of which he is an Honorary Senior Faculty. V.S.R. thanks CSIR (New Delhi) for fellowship. NCL Communication Number: 6584.
REFERENCES
- 1.Thoung N.T. and Helene,C. (1993) Angew. Chem. Int. Ed. Eng., 32, 666–690. [Google Scholar]
- 2.Crooke S.T. and Lebleu,B. (1993) Antisense Research and Application. CRC, Boca Raton, Ann Arbor, London.
- 3.Soyfer V.N. and Potman,V.N. (1996) Triple Helical Nucleic Acids. Springer-Verlag, New York, NY.
- 4.Bennett C.F. (1998) In Stein,C.A and Craig,A.M. (eds), Applied Antisense Oligonucleotide Technology. Wiley-Liss Inc., NY, pp. 129–146.
- 5.Ganesh K.N., Kumar,V.A. and Barawkar,D.A. (1996) In Hamilton,A.D. (eds), Supramolecular Control of Structure and Reactivity. John Wiley & Sons, New York, Vol. 3, 2676–2727.
- 6.Luyten I. and Herdewijin,P. (1998) Eur. J. Med. Chem., 33, 515–576. [Google Scholar]
- 7.Knorre D.G., Vlassov,V.V., Zarytova,V.F., Lebedev,A.V. and Federova,O.S. (1993) Design and Targeted Reactions of Oligonucleotide Derivatives. CRC Press, New York.
- 8.Beal P.A. and Dervan,P.B. (1991) Science, 251, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 9.Griffin L.C. and Dervan,P.B. (1989) Science, 245, 967–971. [DOI] [PubMed] [Google Scholar]
- 10.Yoon K., Hobbs,C.A., Koch,J., Sardaro,M., Kutny,R. and Wies,A. (1992) Proc. Natl Acad. Sci. USA., 89, 3840–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang E., Malek,S. and Feigon,J. (1992) Biochemistry, 31, 4838–4846. [DOI] [PubMed] [Google Scholar]
- 12.Dittrich K., Gu,J., Tinder,R., Hogan,M. and Gao,X. (1994) Biochemistry, 33, 4111–4120. [DOI] [PubMed] [Google Scholar]
- 13.Griffin L.C., Kiessling,L.L., Beal,P.A., Gillespie,P. and Dervan,P.B. (1992) J. Am. Chem. Soc., 114, 7976–7982. [Google Scholar]
- 14.Krawczyk S.H., Milligan,J.F., Wadwani,S., Moulds,C., Froehler,B.C. and Matteucci,M.D. (1992) Proc. Natl Acad. Sci. USA., 89, 3761–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshlap K.M., Gillespie,P., Dervan,P.B. and Fiegon,J. (1993) J. Am. Chem. Soc., 115, 7908–7909. [Google Scholar]
- 16.Huang C.-Y., Cushman,C.D. and Miller,P.S. (1993) J. Org. Chem., 58, 5048–5049. [Google Scholar]
- 17.Stilz H.U. and Dervan,P.B. (1993) Biochemistry, 32, 2177–2185. [DOI] [PubMed] [Google Scholar]
- 18.Durland R.H., Rao,T.S., Bodepudi,V., Seth,D.M., Jayaraman,K. and Revanker,G.R. (1995) Nucleic Acids Res., 23, 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trapane T.L., Christopherson,M.S., Roby,C.D., Ts’o,P.O.P. and Wang,D. (1994) J. Am. Chem. Soc., 116, 8412–8413. [Google Scholar]
- 20.Bandaru R., Hashimoto,H. and Switzer,C. (1995) J. Org. Chem., 60, 786–787. [Google Scholar]
- 21.Barawkar D.A., Krishna Kumar,R. and Ganesh,K.N. (1992) Tetrahedron, 48, 8505–8514. [Google Scholar]
- 22.Barawkar D.A. and Ganesh,K.N. (1993) BioMed. Chem. Lett., 3, 347–352. [Google Scholar]
- 23.Rana V.S., Barawkar,D.A. and Ganesh,K.N. (1996) J. Org. Chem., 61, 3578–3579. [DOI] [PubMed] [Google Scholar]
- 24.Rana V.S and Ganesh,K.N. (1999) Org. Lett., 1, 631–634. [DOI] [PubMed] [Google Scholar]
- 25.McLaughlin L.W., Leong,T., Benseler,F. and Piel,N. (1988) Nucleic Acids Res. 16, 5631–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Job P. (1928) Anal. Chim. Acta, 9, 113–120. [Google Scholar]
- 27.Ferrer E., Neubauer,G., Mann,M. and Eritja,R. (1997) JCS Perkin Trans.1, 14, 2051–2057. [Google Scholar]
- 28.Roger A. and Norden,B. (1997) Circular Dichroism and Linear Dichroism. Oxford University Press, NY.
- 29.Manzini G., Xodo,L.E. and Gasparotto,D. (1990) J. Mol. Biol., 213, 833–843. [DOI] [PubMed] [Google Scholar]
- 30.Pilch D.S., Levenson,C. and Shafer,R. (1990) Proc. Natl Acad. Sci. USA., 87, 1942–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansch C., Albert,L., Unger,S.H., Kim,K.H., Nikaitini,D. and Lien,E.J. (1973) J. Med. Chem., 16, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 32.Radhakrishnan I. and Patel,D.J. (1994) Biochemistry, 33, 11405–11415. [DOI] [PubMed] [Google Scholar]
- 33.Jadhav V.R., Barawkar,D.A. and Ganesh,K.N. (1999) J. Phys. Chem. B., 103, 7384–7386. [Google Scholar]
- 34.Best G.C. and Dervan,P.B. (1995) J. Am. Chem. Soc., 117, 1187–1193. [Google Scholar]
- 35.Greenberg W.A. and Dervan,P.B. (1995) J. Am. Chem. Soc., 117, 5016–5022. [Google Scholar]
- 36.Kumar R.K., Gunjal,A.D. and Ganesh,K.N. (1994) Biochem. Biophys. Res. Commun., 204, 788–793. [DOI] [PubMed] [Google Scholar]
- 37.Kawai K., Saito,A. and Sugiyama,H. (1998) Tetrahedron Lett., 39, 5221–5224. [Google Scholar]
- 38.Garcia R.G., Ferrer,E., Macias,M.J., Eritja,R. and Orozo,M. (1999) Nucleic Acids Res., 27, 1991–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapane T.L. and Ts’o,P.O.P. (1994) J. Am. Chem. Soc., 116, 10437–10438. [Google Scholar]
- 40.Barawkar D.A. and Ganesh,K.N. (1995) Nucleic Acids Res., 23, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jadhav V.R., Natu,A.A. and Ganesh,K.N. (1997) Nucleosides Nucleotides, 16, 107–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.