Abstract
A global mechanism of catabolite repression of the genus Bacillus comprises negative regulation exerted through the binding of the CcpA protein to the catabolite-responsive elements (cres) of the target genes. We searched for cre sequences in the Bacillus subtilis genome using a query sequence, WTGNAANCGNWNNCW (N and W stand for any base and A or T, respectively), picking out 126 putative and known cre sequences. To examine their cre function, we integrated spac promoter (Pspac)-cre-lacZ fusions into the amyE locus. Examination of catabolite repression of β-galactosidase synthesis in the integrants led us to the following conclusions: (i) lower mismatching of cre sequences to the query sequence is required for their function; (ii) although cre sequences are partially palindromic, low mismatching in the same direction as that of transcription of the target genes is more critical for their function than that in the inverse direction; and (iii) yet, a more palindromic nature of cre sequences is desirable for a better function. Furthermore, the alignment of 22 cres that function in vivo implicated a consensus sequence, WWTGNAARCGNWWWCAWW (R stands for G or A). Interestingly, in the case where cre sequences are located in the protein-coding regions of the target genes, their conserved bases are preferentially the third bases of codons where base degeneracy is allowed.
INTRODUCTION
Bacilli as well as low-GC Gram-positive bacteria likely possess a common negative regulatory mechanism of catabolite repression, which is completely different from the positive regulatory one operating in enteric bacteria. This negative regulation of transcription of catabolite-repressive genes, which has been extensively studied in Bacillus subtilis, is exerted through the binding of the CcpA protein (1), which interacts with allosteric effectors, such as P-ser-HPr (2) and P-ser-Crh (3), to their cis-acting catabolite-responsive elements (cres) (4).
The B.subtilis cre was firstly identified in the promoter region of the amyE gene, the consensus sequence of which was deduced by means of site-directed mutagenesis to be TGWAANCGNTNWCA, where N and W stand for any base, and A or T, respectively (5). Another cre was found in the protein-coding region of gntR (6,7). Since then, various cres, including rather classical ones of xylA (8), hutP (9), acsA (10) and ackA (11), have been identified in either the promoter or protein-coding regions of the target genes. The sequences of these cres closely match the consensus sequence described above.
After Hueck et al. (4) had searched and analyzed cre-like sequences among the deposited nucleotide sequences of Gram-positive bacteria, the complete sequence of the B.subtilis genome was reported by Kunst et al. (12). Thus, it was thought to be very interesting to search for cre-like sequences in the B.subtilis genome, and to evaluate and characterize them in more detail. We chose another consensus sequence, WTGNAANCGNWNNCW, as a query sequence for searching for cre-like sequences in the genome, after repeated trials. This sequence is essentially the same as the cre consensus sequence proposed by Weickert and Chambliss (5), but is somewhat degenerate and one base longer. Our in vivo test for the cre function of various cre-like sequences, which had been revealed by our search, led us to find some interesting features of the cre sequence of B.subtilis.
MATERIALS AND METHODS
Bacterial strains and plasmids
The B.subtilis strains constructed in this work were derived from strain GM122 (trpC2 sacB′-′lacZ) (13). Plasmid pCRE-test (Fig. S1, Supplementary Material) was constructed as follows. A region of plasmid pAG58 containing a spac promoter (Pspac) (14) was amplified by PCR using a primer pair designed to produce flanking EcoRI and BamHI sites. In addition, a region of plasmid pMUTIN1 containing a Shine–Dalgarno sequence and the 5′-portion of lacZ (12 codons) (15) was amplified using another primer set designed to produce flanking BamHI and HindIII sites. The resulting PCR products were digested with the respective endonucleases, and then ligated with the EcoRI–HindIII arm of plasmid ptrpBGI (16). The ligated DNA was used for the transformation of Escherichia coli strain JM109 (17) to ampicillin resistance. The correct construction of plasmid pCRE-test was confirmed by sequencing.
cre search of the B.subtilis genome
cre-like sequences in the B.subtilis genome were searched for with an originally developed Perl program on a workstation (Sun SPARC station 20) with the query sequence of WTGNAANCGNWNNCW.
Integration of the Pspac-cre-lacZ fusion into amyE
An appropriate region containing each cre-like sequence (15 bp) and its upstream and downstream flanking sequences (each ∼30 bp long) was amplified by PCR using chromosomal DNA of B.subtilis strain Marburg 168 (trpC2) as a template and a primer pair designed to generate two flanking BamHI sites. The PCR products were digested with BamHI and then ligated with DNA of plasmid pCRE-test, which had been cleaved with the same enzyme. The ligated DNAs were used for the transformation of E.coli strain JM109 to ampicillin resistance. The sequence and orientation of the cloned fragments were determined by sequencing. The constructed plasmids carrying each cre-like sequence in the same direction with respect to the transcription were linearized with PstI or ScaI, and then used for the integration of Pspac-cre-lacZ into the amyE locus of B.subtilis strain GM122 through a double crossover event by selecting chloramphenicol-resistant transformants (Fig. S1).
Examination of catabolite repression of β-galactosidase (β-Gal) synthesis in integrants
The integrants were grown to an optical density at 600 nm (OD600) = 0.6 in S6 medium (18) containing 0.5% Casamino Acids (Difco), which was supplemented with tryptophan (50 µg/ml) and chloramphenicol (5 µg/ml), with or without 10 mM glucose. The cells (OD600 x ml = 3.6) were harvested, and then lysed by lysozyme treatment and brief sonication as described previously (19). The β-Gal activity was spectrophotometrically assayed as described by Atkinson et al. (20).
RESULTS AND DISCUSSION
Search for cre-like sequences in the B.subtilis genome
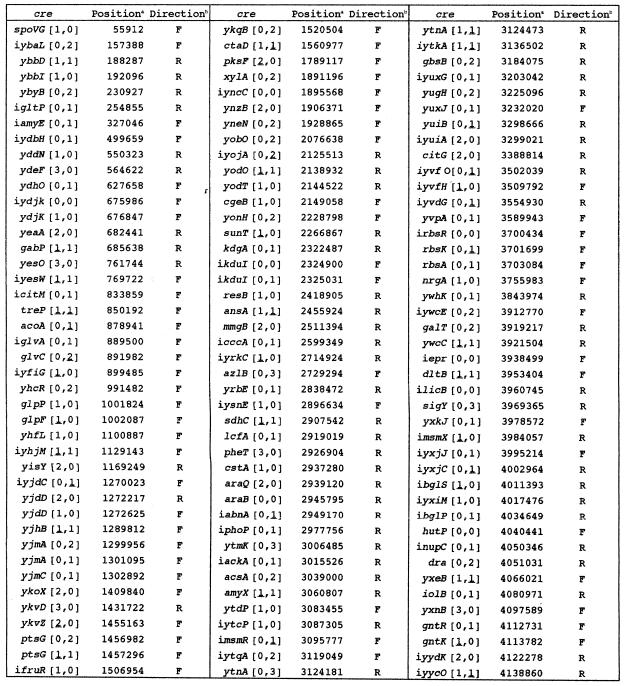
Firstly, we used a well-known cre consensus sequence of 14 bases, TGWAANCGNTNWCA, proposed by Weickert and Chambliss (5) to search for cre-like sequences in the B.subtilis genome. During this search, we picked out 31 cre-like sequences which show no mismatching to this query sequence. However, this number was much lower than we expected, because rough estimation of glucose-repressive protein spots on a two-dimensional gel as well as a search for glucose-repressive genes using hundreds of plasmid pMUTIN-integrants (15) suggested that there might be at least 150 cres in B.subtilis (data not shown). In addition, well-characterized cres such as those located in gntR (6,7), xylA (8) and hutP (9) were not included in these 31 sequences.
We attempted to find a more suitable cre query sequence for a computer search for cre-like sequences in the genome, mainly by degenerating the consensus sequence, TGWAANCGNTNWCA. After repeated trials, we finally chose a 15-base sequence, WTGNAANCGNWNNCW, for a search of cre-like sequences, which is partially palindromic. Our search with this query sequence led us to find 108 cre-like sequences in the genome exhibiting no mismatching to it, which are included in a list of known and putative cres (Table 1).
Table 1. List of known and putative cres.
The cre-like sequences were located in the protein-coding or intergenic regions of the putative target genes. In the latter cases, the names of the genes, the 5′-ends of which are closer to the sequences, are preceded by ‘i’ (Table 1). As described below, orientation of cre with respect to the direction of transcription of its target gene was found to be important for cre to function. The cre-like sequences are partially palindromic, so their mismatch numbers as to the query sequence with regard to the same and inverse directions as that of transcription of the target genes, that is, those on the anti-sense and sense strands, are given as the first and second numbers in brackets, respectively (Table 1). As discussed below, the first and second bases, W and T, of the query sequence were found not to be strictly conserved. So, when the cre-like sequences carry one mismatch at the first and second bases to be G or C and A, respectively, the mismatch numbers are underlined. Among cre-like sequences carrying one mismatch in both directions, those which have at least one underlined mismatch are also listed in Table 1.
Examination of cre function of putative cre sequences
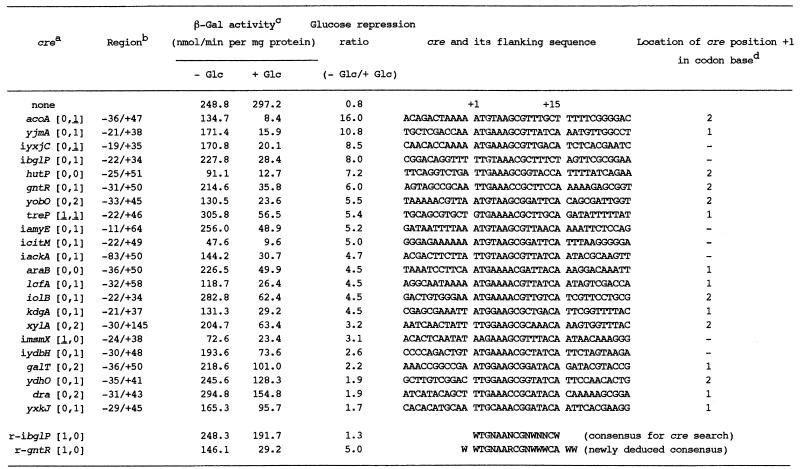
In order to determine whether or not the cre-like sequences function as cre in vivo, we constructed plasmid pCRE-test (Fig. S1). After cloning an appropriate region containing each cre-like sequence into the BamHI site of plasmid pCRE-test, the constructed plasmids were linearized with PstI or ScaI, and then used for Pspac-cre-lacZ integration into the amyE locus through a double crossover event. β-Gal synthesis in the integrant of the Pspac-lacZ fusion without cre was almost constitutive in the presence and absence of glucose in the medium (Table 2). Thus, we were able to test the cre function by examining catabolite repression of β-Gal synthesis in the Pspac-cre-lacZ integrants, which was most likely evoked by the transcription roadblock owing to a complex of CcpA and P-ser-HPr (or another factor) bound to cre (2,7).
Table 2. Catabolite repression of β-Gal synthesis exerted by various cres and their sequence alignment.
aAmong 32 cre candidates examined for their catabolite repressive ability, only functional cres are listed. cre candidates (ybyB [0,2], resB [1,0], ansA [1,1], sdhC [1,1], sunT [1,0], ptsG [1,1], ptsG [0,2], spoVG [1,0], azlB [0,3] and sigY [0,3]) did not function; their catabolite repression ratios were between 0.7 and 1.0.
bcre regions which were cloned into plasmid pCRE-test are indicated; +1 is the first base of each cre-like sequence.
cThe Pspac-cre-lacZ integrants were grown with and without glucose (Glc). The cells were disrupted, and β-Gal was assayed as described in the text. The β-Gal activities (1 and 0.5 nmol/min/mg protein) in cells of strain GM122 grown with and without glucose, respectively, have been subtracted from the corresponding activities in the integrants.
d+1 of each cre in the coding region is the first or second base of codons (1 or 2), which means that not only position +1 but also +4, +7, +10 and +13 are the first or second bases of codons.
Among the 126 cre-like sequences listed in Table 1, 32 were tested for their ability as cres (Table 2). We chose them in a fashion to cover various kinds of mismatches in both directions and to include several known cres. As shown in Table 2, the β-Gal activities fluctuated by 6-fold in various integrants grown without glucose, probably because of the different stabilities of mRNAs carrying each of the 32 cre-like sequences between Pspac and lacZ. However, it is considered that the catabolite repression ratio for each of the cre-like sequences reflects its ability to cause a transcription roadblock. In Table 2, 22 cre-like sequences out of 32 were found to function in vivo (catabolite repression ratio >1), and are listed in the order of their strength from cre-acoA to cre-yxkJ. All of the well known cres tested [cre-ibglP (21), cre-gntR (6,7), cre-hutP (9), cre-iamyE (5), cre-iackA (11) and cre-xylA (8)] were active in our in vivo cre test system, which indicates that this test system is highly reliable.
As shown in Table 2, we tested the repression ability of 28 cre-like sequences which exhibited no mismatching to the consensus sequence, at least in the same or inverse direction as that of transcription of the target genes. All cres classified as [0,0], [0,1] and [0,1] exhibited repression ability, whereas among cres classified as [0,2] and [1,0], some functioned but others did not. But, cres classified as [0,3] and [1,0] did not function. Furthermore, we also tested four cre-like sequences which exhibited one mismatch in both the same and inverse directions as that of transcription of the target genes; a cre classified as [1,1] functioned, but the others classified as [1,1] and [1,1] did not. These results imply that lower mismatching of cre sequences to the query sequence, especially in the same direction as that of transcription of their target genes, is required for their function, and that a more palindromic nature of cre sequences is desirable for a better function. The requirements of lower mismatching of cre sequences to the query sequence and their palindromic nature for their function can be explained by the cre binding strength of CcpA interacting with some effector, which likely depends on their mismatch levels in both directions with respect to that of the transcription of their target genes, because CcpA is supposed to be dimerized in vivo (22).
The results of the above cre tests implied that low mismatching of cre sequences in the same direction as that of the transcription of their target genes is likely more critical for their function than that in the inverse direction. To confirm this, we oppositely placed cre-ibglP and cre-gntR between Pspac and lacZ, and then examined the catabolite repression of β-Gal synthesis in the constructed integrants carrying Pspac-(cre-r-ibglP)-lacZ and Pspac-(cre-r-gntR)-lacZ, respectively (Table 2). Thus, these inversed cres are classified as [1,0] instead of [0,1] for the original cres (Table 2). As shown in Table 2, the inversion of the ibglP- and gntR-cres decreased their catabolite repression ratios from 8.0 to 1.3 and 6.0 to 5.0, respectively. These results as well as those of the above tests suggest that not only the binding strength of CcpA as to cres but also lower mismatching in the same direction as RNA polymerase moves might be determinants for a transcription roadblock to occur.
Although we did not test all the cres listed in Table 1, we could predict from the above results whether or not the cre-like sequences not tested might function in vivo. cres classified as [0,0], [0,1], [0,1] and [1,0] are expected to function in vivo, but those classified as [0,3], [2,0] and [3,0] might not function. However, it is hard to predict whether or not the other cres listed might function. Furthermore, we do not think that the cres listed in Table 1 include all the B.subtilis cres. For example, iynaJ-cre [1,1] for the ynaJ-xynB operon, which is known to function in vivo (3), is not listed in Table 1. Therefore, it is likely that we have to find more cre-like sequences which exhibit high mismatch numbers through careful consideration of their palindromic nature in order to cover all the cres of B.subtilis.
Alignment of cres and their location in target genes
As shown in Table 2, we aligned 22 cre sequences together with the surrounding ones, which were found to function in vivo with our cre test system. From this alignment, we found that the outside bases of these 15-base cre sequences were also conserved. When the first base of cre, which corresponds to the first ‘W’ in the cre query sequence, is assigned as +1, the bases at positions –1, +16 and +17 are W with high probabilities of 21, 18 and 19 bases out of 22, respectively. The importance of the flanking AT-rich sequences of a 14-base cre consensus sequence proposed by Weickert and Chambliss (5) was also pointed out by Zalieckas et al. (23). In addition, preferable bases for the +7, +12, +13 and +15 positions are R (G or A), W, W and A, with high probabilities of 20, 19, 18 and 20 bases out of 22, respectively. Therefore, a consensus sequence for cre and its surrounding region (bases –1 to +17) was deduced to be WWTGNAARCGNWWWCAWW.
Among 22 cres that function in vivo, 15 are located in the protein-coding regions of the target genes, so a very interesting question arose. Where are these 15 cre sequences localized in the three possible protein-coding frames? Thus, we examined the relative localization of the cre sequences in the protein-coding frames of the target genes, finding that in eight and seven genes, positions +1, +4, +7, +10 and +13 of WTGNAANCGNWNNCW are the first and second bases of codons, respectively, but in no case are they their third bases. The bases at these positions are W or N in the cre query sequence, in which relative randomness of base species is allowed, whereas they are the first or second base of codons in the protein-coding frames of the target genes in which relatively strict bases are required. In other words, the other bases of the cre consensus sequence are conserved, so there is a relatively high probability that these positions are the third bases of codons in the protein-coding frames where base degeneracy is allowed. This fact implies the elegant harmony between the establishment of a cre sequence and the evolution of a functional protein encoded by a catabolite-repressive gene.
SUPPLEMENTARY MATERIAL
See Supplementary Material available at NAR Online for a figure showing the in vivo test system.
Acknowledgments
ACKNOWLEDGEMENTS
We thank S. Eguchi, S. Kawahara, K. Okamura, T. Aoki, M. Kou, S. Iijima and K. Kawai for their help in the experiments. This work was supported by a grant, JSPS-RFTF96L00105, from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Henkin T.M., Grundy,F.J., Nicholson,W.L. and Chambliss,G.H. (1991) Mol. Microbiol., 5, 575–584. [DOI] [PubMed] [Google Scholar]
- 2.Fujita Y., Miwa,Y., Galinier,A. and Deutscher,J. (1995) Mol. Microbiol., 17, 953–960. [DOI] [PubMed] [Google Scholar]
- 3.Galinier A., Deutscher,J. and Martin-Verstraete,I. (1999) J. Mol. Biol., 286, 307–314. [DOI] [PubMed] [Google Scholar]
- 4.Hueck C.J., Hillen,W. and Saier,M.H.Jr (1994) Res. Microbio l., 145, 503–518. [DOI] [PubMed] [Google Scholar]
- 5.Weickert M.J. and Chambliss,G.H. (1990) Proc. Natl Acad. Sci. USA, 87, 6238–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miwa Y. and Fujita,Y. (1990) Nucleic Acids Res., 18, 7049–7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miwa Y. and Fujita,Y. (1993) J. Biochem., 113, 665–671. [DOI] [PubMed] [Google Scholar]
- 8.Jacob S., Allmansberger,R., Gärtner,D. and Hillen,W. (1991) Mol. Gen. Genet., 229, 189–196. [DOI] [PubMed] [Google Scholar]
- 9.Wray L.V.Jr, Pettengill,F.K. and Fisher,S.H. (1994) J. Bacteriol., 176, 1894–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy F.J., Waters,D.A., Takova,T.Y. and Henkin,T.M. (1993) Mol. Microbiol., 10, 259–271. [DOI] [PubMed] [Google Scholar]
- 11.Grundy F.J., Waters,D.A., Allen,S.H.G. and Henkin,T.M. (1993) J. Bacteriol., 175, 7348–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunst F., Ogasawara,N., Moszer,I., Albertini,A.M., Alloni,G., Azevedo,V., Bertero,M.G., Bessières,P., Bolotin,A., Borchert,S. et al. (1997) Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 13.Miwa Y., Nagura,K., Eguchi,S., Fukuda,H., Deutscher,J. and Fujita,Y. (1997) Mol. Microbiol., 23, 1203–1213. [DOI] [PubMed] [Google Scholar]
- 14.Jaacks K.J., Healy,J., Losick,R. and Grossman,A.D. (1989) J. Bacteriol., 171, 4121–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vagner V., Dervyn,E. and Ehrlich,S.D. (1998) Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- 16.Shimotsu H. and Henner,D.J. (1986) Gene, 43, 85–94. [DOI] [PubMed] [Google Scholar]
- 17.Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]
- 18.Fujita Y. and Freese,E. (1981) J. Bacteriol., 145, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujita Y. and Fujita,T. (1986) Nucleic Acids Res., 14, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkinson M.R., Wray,L.V.Jr and Fisher,S.H. (1990) J. Bacteriol., 172, 4758–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krüger S. and Hecker,M. (1995) J. Bacteriol., 177, 5590–5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miwa Y., Saikawa M. and Fujita,Y. (1994) Microbiology, 140, 2567–2575. [DOI] [PubMed] [Google Scholar]
- 23.Zalieckas J.L., Wray,L.V.Jr and Fisher,S.H. (1998) J. Bacteriol., 180, 6649–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.