Abstract
Auditory neuropathy spectrum disorder (ANSD) represents a variety of sensorineural deafness conditions characterized by abnormal inner hair cells and/or auditory nerve function, but with the preservation of outer hair cell function. ANSD represents up to 15% of individuals with hearing impairments. Through mutation screening, bioinformatic analysis and expression studies, we have previously identified several apoptosis-inducing factor (AIF) mitochondria-associated 1 (AIFM1) variants in ANSD families and in some other sporadic cases. Here, to elucidate the pathogenic mechanisms underlying each AIFM1 variant, we generated AIF-null cells using the clustered regularly interspersed short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system and constructed AIF-wild type (WT) and AIF-mutant (mut) (p.T260A, p.R422W, and p.R451Q) stable transfection cell lines. We then analyzed AIF structure, coenzyme-binding affinity, apoptosis, and other aspects. Results revealed that these variants resulted in impaired dimerization, compromising AIF function. The reduction reaction of AIF variants had proceeded slower than that of AIF-WT. The average levels of AIF dimerization in AIF variant cells were only 34.5%‒49.7% of that of AIF-WT cells, resulting in caspase-independent apoptosis. The average percentage of apoptotic cells in the variants was 12.3%‒17.9%, which was significantly higher than that (6.9%‒7.4%) in controls. However, nicotinamide adenine dinucleotide (NADH) treatment promoted the reduction of apoptosis by rescuing AIF dimerization in AIF variant cells. Our findings show that the impairment of AIF dimerization by AIFM1 variants causes apoptosis contributing to ANSD, and introduce NADH as a potential drug for ANSD treatment. Our results help elucidate the mechanisms of ANSD and may lead to the provision of novel therapies.
Keywords: Auditory neuropathy spectrum disorder, Apoptosis-inducing factor (AIF) mitochondria-associated 1 (AIFM1) variants, Dimerization, Caspase-independent apoptosis, Nicotinamide adenine dinucleotide (NADH) treatment
Abstract
听神经病谱系障碍(ANSD)属于感音神经性耳聋,其特征为内毛细胞和/或听觉神经元的功能异常,但外毛细胞的功能正常。在听力障碍患者中,听神经病谱系障碍的发病率高达15%。我们前期通过突变筛查、生物信息学分析和蛋白表达等检测,在ANSD家系和某些散发病例中发现了凋亡诱导因子1(AIFM1)基因的几种点突变。为阐明AIFM1突变体的致病机制,本文使用CRISPR/Cas9系统构建了凋亡诱导因子(AIF)蛋白敲除的细胞系,及其稳定转染野生型和突变型AIF 蛋白(p.T260A、p.R422W和p.R451Q)的细胞系,并且分析了AIF蛋白结构、AIF与辅酶的亲和力及细胞凋亡等情况。结果显示,上述AIF突变体可导致AIF蛋白二聚体形成障碍,损害AIF蛋白的生理功能。突变型AIF蛋白的还原速率显著低于野生型AIF蛋白。且在AIF突变型细胞系中,AIF蛋白的二聚体含量仅为AIF野生型细胞系的34.5%~49.7%,导致非caspase依赖性细胞凋亡。AIF突变型细胞系中凋亡细胞的平均百分比为12.3%~17.9%,显著高于对照组的6.9%~7.4%。特别是,烟酰胺腺嘌呤二核苷酸(NADH)处理显著提高AIF突变型细胞中的AIF蛋白二聚体含量,从而降低细胞凋亡。结果表明:AIFM1突变引起AIF蛋白二聚体形成障碍,使得细胞凋亡增加,导致ANSD发生;NADH是ANSD的潜在治疗药物。我们的研究结果有助于阐明ANSD的发病机制,并可能提供新的治疗方案。
Keywords: 听神经病谱系障碍, 凋亡诱导因子1(AIFM1)基因突变, 二聚化, 非caspase依赖性细胞凋亡, 烟酰胺腺嘌呤二核苷酸(NADH)处理
1. Introduction
Auditory neuropathy spectrum disorder (ANSD) is a particular kind of hearing disorder characterized by disruption of the transmission of sound information to the auditory nerve and brain, but normal outer hair cells within the cochlea. The prevalence of ANSD ranges from ≤1% to almost 15% of individuals with hearing impairment (Penido and Isaac, 2013). About 42% of ANSD occurrences have a genetic basis (Starr et al., 2000). Several genes associated with ANSD have been discovered including auditory neuropathy, dominant 1 (AUNA1)/diaphanous homolog 3 (DIAPH3) (Schoen et al., 2010; Sánchez-Martínez et al., 2017), protocadherin 9 (PCDH9) (Starr et al., 2004), otoferlin (OTOF) (Varga et al., 2003; Tekin et al., 2005; Romanos et al., 2009; Chiu et al., 2010; Al-Wardy et al., 2016), deafness, autosomal recessive 59 (DFNB59)(Delmaghani et al., 2006), connexin 26 (GJB2) (Cheng et al., 2005; Santarelli et al., 2008), auditory neuropathy, dominant 2 (AUNA2)(Lang-Roth et al., 2017), Na+/K+ ATPase α3(ATP1A3)(Matsuoka et al., 2017; Tranebjærg et al., 2018), mitochondrial 12S ribosomal RNA (rRNA) gene (Wang et al., 2005), anda novel auditory neuropathy, X-chromosomal 1 (AUNX1)/apoptosis-inducing factor (AIF) mitochondria-associated 1 (AIFM1) (Wang et al., 2006; Zong et al., 2015). However, the underlying molecular mechanisms contributing to ANSD remain elusive.
The AIFM1 gene maps to the q25–26 region of the human X chromosome and encodes a flavoprotein, named AIF. The most ubiquitously expressed AIF isoform has 16 exons. AIF locates in the mitochondrial intermembrane space (IMS) and consists of a mitochondrial localization sequence (MLS) in the N-terminal, two nuclear localization sequences (NLSs), a nicotinamide adenine dinucleotide (NADH)/nicotinamide adenine dinucleotide phosphate (NADPH)-binding domain, two flanking flavin adenine dinucleotide (FAD)-binding domains, and a C-terminal domain (Susin et al., 1999). It was initially described as a death-related factor associated with mitochondrial-nuclear translocation (Susin et al., 1999; Yu et al., 2002). Several studies have indicated that AIF undergoes dimerization and a conformational change, resulting in the formation of an air-stable FADH•–NAD+ charge transfer complex (CTC) (Ferreira et al., 2014; Brosey et al., 2016), which is vital to its function. AIF has three functions at the cellular level: first, to participate in the regulation of the caspase-independent programmed cell death pathway (Tang et al., 2019); second, as an important oxidoreductase to maintain mitochondrial structure (Cheung et al., 2006); third, to take part in the biogenesis of mitochondrial respiratory chain complexes (Hangen et al., 2010; Bano and Prehn, 2018; Herrmann and Riemer, 2020). In 2010, the first AIFM1 pathogenic mutation was identified in two male infant patients with mitochondrial encephalomyopathy (p.Arg201del) (Ghezzi et al., 2010). Several nerve-related diseases were reported with AIFM1 mutations, such as an early prenatal ventriculomegaly associated with the p.Gly308Glu variant (Berger et al., 2011), cowchock syndrome associated with p.Glu493Val (Rinaldi et al., 2012), motor nerve-related diseases associated with p.Phe210Ser (Sancho et al., 2017), peripheral nervous-related diseases associated with p.Phe210Leu (Hu et al., 2017), neurodegenerative diseases associated with p.Asp237Gly (Mierzewska et al., 2017), and cerebellar ataxia associated with p.Met340Thr or p.Thr141Ile (Heimer et al., 2018). Given that different AIFM1 mutations may lead to various diseases, investigating its detailed pathogenesis becomes extremely challenging.
We previously reported several novel variants, such as AIFM1 c.778A>G/p.T260A, c.1264C>T/p.R422W, and c.1352G>A/p.R451Q, which were associated with ANSD in five unrelated families and in 93 other sporadic cases (Zong et al., 2015). In this study, we investigated the effects of these variants on AIF dimerization, coenzyme-binding affinity, AIF dimer structure, and apoptosis to provide insight into the pathogenesis of ANSD. We also identified NADH as a potential drug for ANSD therapy.
2. Results
2.1. Construction of stable transfection cell lines and characterization of their growth properties
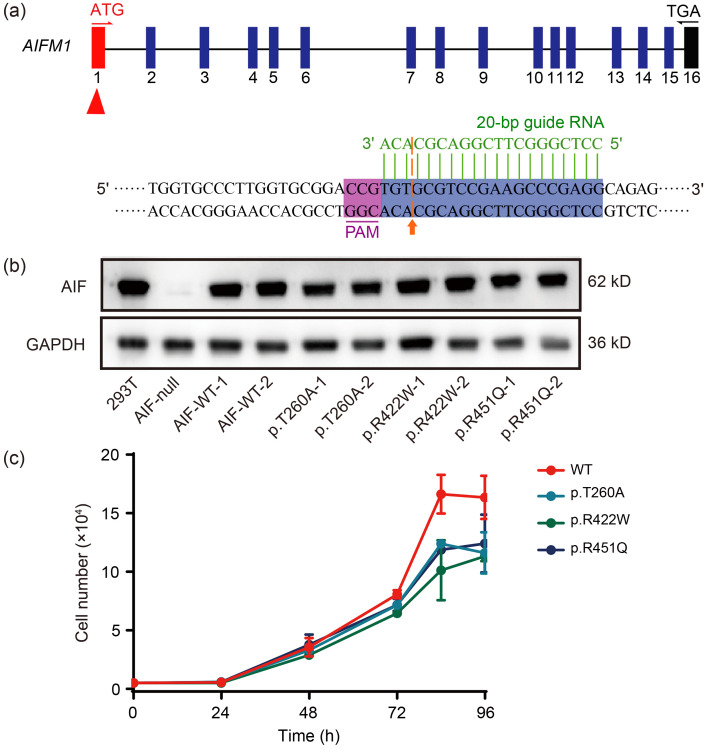
To investigate the effect of the AIFM1 variants on cellular growth properties, we generated AIF-null cells using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system (Yang et al., 2015; Li et al., 2021) (Fig. 1a), and then constructed AIF-wild type (WT) and AIF-mutant (mut) (p.T260A, p.R422W, and p.R451Q) stable transfection cell lines using the lentivirus infection system. These cells were checked for expression of the AIF variants (Fig. 1b). AIFM1 variant sites were present in the p.T260A, p.R422W, and p.R451Q cell lines, but absent from the AIF-WT cell lines (Fig. S1). Based on the cell growth curve (Fig. 1c), the average doubling time was (24.7±1.9) h (P=0.024) in p.T260A cells, (20.3±1.7) h (P=0.04) in p.R422W cells, and (20.6±0.3) h (P=0.0024) in p.R451Q cells, whereas that of AIF-WT cells was (16.0±0.2) h. These results indicated that all these variants had poor physiological status and survivability compared to AIF-WT cells, suggesting that the AIF variants affect cell growth properties.
Fig. 1. Construction of AIF-WT and AIF-mut (p.T260A, p.R422W, and p.R451Q) stable transfection cell lines, and their growth characteristics. (a) Sketch of the guide RNA sequence. The 20-bp sequence indicated with a green line is the guide RNA. The cleavage site is indicated with an orange arrow. PAM is in purple. (b) Western blotting for the presence and levels of AIF in the 293T, AIF-null, AIF-WT, and AIF variant cell lines. The AIF band was absent from AIF-null cells, but appeared upon stable transfection of the variant constructs. GAPDH is shown as a reference. (c) Growth curve of AIF-WT and AIF variant cell lines. Data are represented as mean±SEM (n=3). AIF: apoptosis-inducing factor; AIFM1: apoptosis-inducing factor mitochondria-associated 1; WT: wild type; mut: mutant; PAM: protospacer adjacent motif; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; SEM: standard error of the mean.
2.2. Activated caspase-independent apoptosis in AIF variants
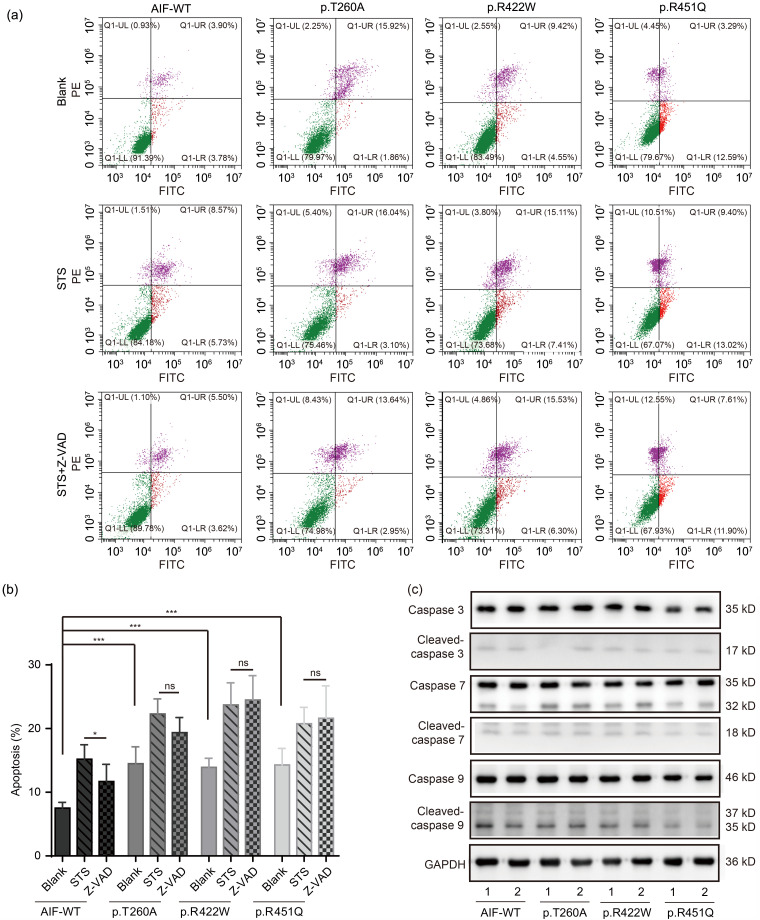
To investigate whether apoptosis was activated in the AIF p.T260A, p.R422W, and p.R451Q variant cell lines, we performed flow cytometry. The results showed that the percentage of apoptotic cells was (14.5±2.6)% (P<0.001) in p.T260A, (13.9±1.4)% (P<0.001) in p.R422W, and (14.3±2.6)% (P<0.001) in p.R451Q variant cells, but only (7.5±0.9)% in the AIF-WT cells (Figs. 2a and 2b). To investigate any patterns of increased apoptosis in the AIF variants, the levels of activated caspases 3, 7, and 9 were measured. Interestingly, activated caspase 3, 7, or 9 level was not appreciably different between AIF-WT and AIF variant cells (Fig. 2c), indicating that increased apoptosis in variant cells had not occurred according to a classical caspase-dependent pathway. To confirm caspase-independent and AIF-mediated apoptosis, cells were treated with an apoptosis inducer staurosporine (STS) and a caspase-dependent apoptosis-specific inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (Z-VAD-FMK). Flow cytometry results showed that when treated with STS only, cell apoptosis was induced in both cell types (Figs. 2a and 2b). However, when treating variants and WT cells with STS and Z-VAD-FMK, apoptosis dropped from (15.2±2.3)% to (11.7±2.7)% (P=0.035) in AIF-WT cells, but the p.T260A, p.R422W, or p.R451Q variant cells showed no significant decrease (Figs. 2a and 2b). These results suggested that the increased cell death in the variant cell lines occurs predominantly via caspase-independent and AIF-mediated apoptosis.
Fig. 2. Activated caspase-independent apoptosis in the variants. (a) Representative flow cytometry dot-plots in AIF-WT and AIF variant cells. Results are shown in three groups: Blank; STS (1 µmol/L for 1.5 h) in absence of Z-VAD; and STS (1 µmol/L for 1.5 h) in the presence of Z-VAD (50 µmol/L for pretreatment of 0.5 h). (b) Quantification analysis of flow cytometry dot-plots. Data are represented as mean±SEM (n=3). ns P ≥0.05, * P<0.05, *** P<0.001. (c) Western blotting of caspases 3, 7, and 9. Caspase refers to caspase proteins without activity, and cleaved-caspase refers to activated caspase proteins. AIF: apoptosis-inducing factor; WT: wild type; STS: staurosporine; Z-VAD: benzyloxycarbonyl-Val-Ala-Asp; SEM: standard error of the mean; ns: not significant; PE: phycoerythrin; FITC: fluorescein isothiocyanate; UL: upper left; UR: upper right; LL: lower left; LR: lower right; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
2.3. Impaired AIF dimerization and NADH-binding affinity in AIF variants
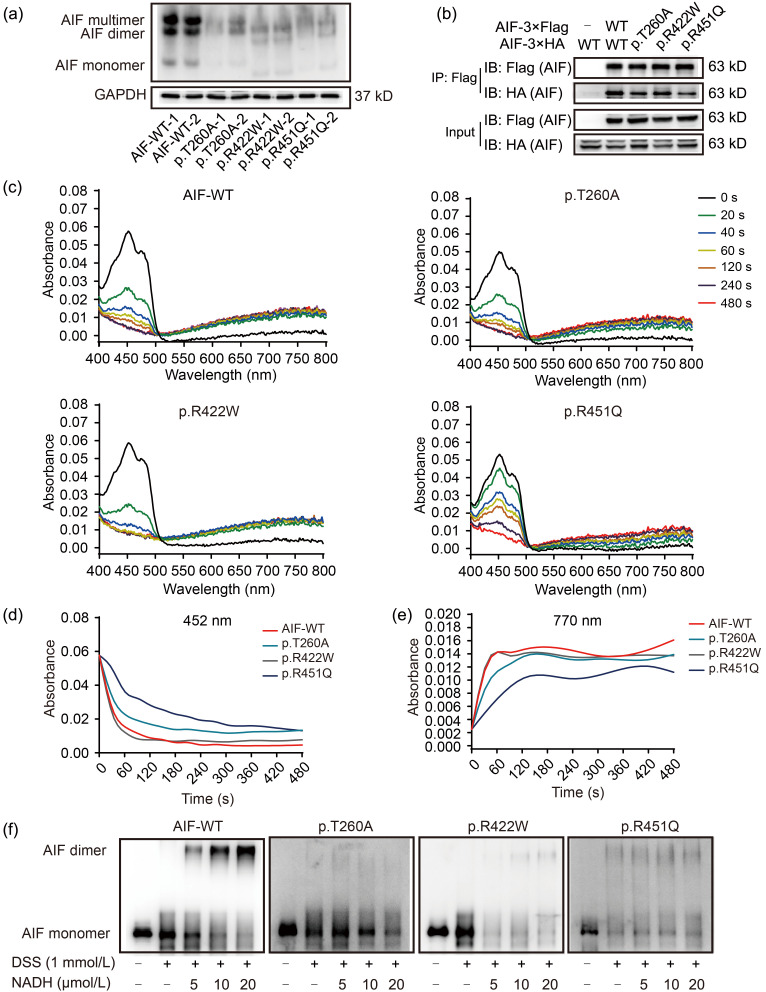
AIF is in a monomer–dimer equilibrium in mitochondria and CTC formations are vital to its function (Ferreira et al., 2014). To test whether the AIF dimerization was impaired in AIF variant cell lines, the proteins were extracted natively and further separated by native-polyacrylamide gel electrophoresis (PAGE). Results showed that AIF dimers were decreased in AIF variants compared to WT cell lines (Fig. 3a). To directly confirm the ability of AIF dimerization in AIF variants, transitory over-expression cell lines were generated by transforming p3×Flag-AIF (WT, p.T260A, p.R422W, or p.R451Q) and p3×HA-AIF (WT, p.T260A, p.R422W, or p.R451Q). Co-immunoprecipitation (Co-IP) results indicated that the interaction strength of the AIF p.T260A, p.R422W, and p.R451Q variants was significantly weakened compared to that of WT AIF (Fig. 3b). These results showed that the interaction of the two AIF monomers was impaired, and AIF monomer–dimer equilibrium disrupted due to these variants. To detect the NADH-binding affinity of AIF variants, the hAIFΔ1-102 proteins were purified in Escherichia coli. The results of kinetics showed that AIF proteins (whether WT or variant) could react with NADH. After adding NADH, the AIF monomer, as indicated by absorbance at 452 nm, was reduced to the AIF dimer, as indicated by absorbance at 770 nm (Fig. 3c). The absorbance changes at 452 nm over time showed that, compared to WT AIF, the reduction of p.T260A and p.R451Q variants proceeded at a far slower pace, while the p.R422W variant showed no difference (Fig. 3d). The absorbance changes at 770 nm over time showed that all AIF variants showed a decreased formation of dimer in contrast to WT AIF (Fig. 3e). We concluded that whilst the p.R422W variant had a mild impact on AIF reduction, the reduction for the p.T260A and p.R451Q variants had proceeded extremely slowly. However, the N-terminal of AIF precursor will be cleaved to form AIFΔ1-54 proteins located mainly in mitochondria in eukaryotic cells. As reported, the dimerization of AIFΔ1-54 proteins is vital to mitochondrial function (Hangen et al., 2015). To confirm the NADH-binding affinity of AIF variants in the normal physiological state, the AIF full-length proteins were expressed, and then AIFΔ1-54 proteins were purified in eukaryotic cells. After mixing AIF with NADH, cross-linking and western blotting were performed to detect the level of AIF dimer. Our results showed that only a 5-fold excess of NADH was required to form AIF dimer in AIF-WT, but AIF variants needed a 10-fold or more excess of NADH to convert the oxidized form to the reduced form (Fig. 3f). The AIFΔ1-102 proteins were also expressed and purified in eukaryotic cells to confirm the reduction rate of AIFΔ1-102 proteins. The reduction of the variants required more excess of NADH compared with WT AIFΔ1-102 both in eukaryotic cells and E. coli (Figs. S2a and S2b). These results demonstrated that AIF variants display an impaired ability to form CTCs with NADH.
Fig. 3. Impaired AIF dimerization and coenzyme-binding affinity in AIF variants. (a) Native-PAGE detection for AIF dimers in a native state. There were three forms of AIF, namely AIF monomer, dimer, and multimer. GAPDH in SDS-PAGE was indicated as the loading control. (b) Flag-tagged AIF or its variant was co-expressed with HA-tagged AIF or its variants in AIF-null cells. Direct interaction was determined by Co-IP and western blotting. The input bands show successful transfection. IP-flag bands are shown as the loading control. The IP-HA bands indicated the amounts of HA-AIF dragged down by Flag-AIF. (c) Absorbance changes during the reduction of AIFΔ1-102 (10 μmol/L) by NADH (100 μmol/L) scanned from 400 to 800 nm at 0, 20, 40, 60, 120, 240, and 480 s after mixing. Spectra were recorded for the reaction of NADH with AIF-WT, p.T260A, p.R422W, or p.R451Q. (d) The kinetics of the FAD reduction was recorded at 452 nm upon mixing 10 μmol/L AIFΔ1-102 with 100 μmol/L NADH in PBS buffer (pH 7.4). (e) The kinetics of the CTC formation was recorded at 770 nm upon mixing 10 μmol/L AIFΔ1-102 with 100 μmol/L NADH in PBS buffer (pH 7.4). (f) Western blotting analysis after mixing 1 μmol/L AIFΔ1-54 with various concentrations of NADH (5, 10, and 20 μmol/L) for 15-min incubation. DSS was added to crosslink the AIF dimer. AIF: apoptosis-inducing factor; PAGE: polyacrylamide gel electrophoresis; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; SDS: sodium dodecyl sulfate; HA: hemagglutinin; Co-IP: co-immunoprecipitation; NADH: nicotinamide adenine dinucleotide; WT: wild type; FAD: flavin adenine dinucleotide; PBS: phosphate-buffered saline; CTC: charge transfer complex; DSS: disuccinimidyl suberate.
2.4. Disrupted AIF dimer structure in AIF variants
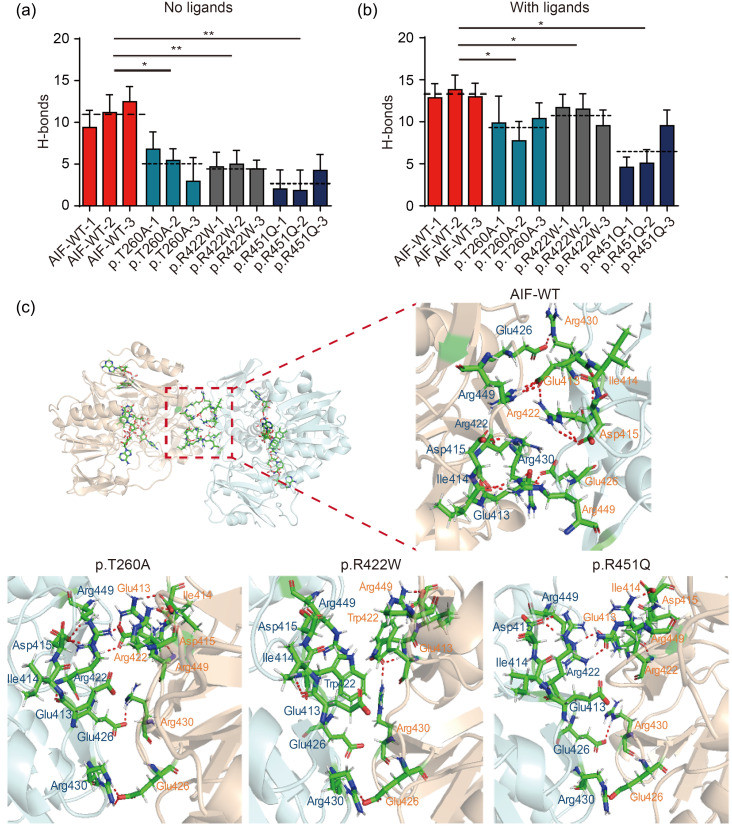
To test the stability of the AIF dimer, molecular dynamics simulations were performed using free AIF and AIF-ligands as starting structures. The root-mean-square deviations (RMSDs) of free AIF showed that the WT system remained relatively stable, but all of the variant systems showed significantly elevated fluctuations (Fig. S3a). In the system with ligands, the RMSDs of WT AIF were higher than those of the system without ligands. However, the WT systems remained relatively stable, unlike the AIF variant systems (Fig. S3b). These results showed that, compared to the WT AIF, AIF variants showed larger structural deviations, indicating AIF dimer instability. We further analyzed the number of H-bonds along the trajectories. Results showed that H-bonds were less frequent in AIF variants compared to AIF-WT, both in free AIF and AIF-ligand systems (Figs. 4a and 4b). To directly analyze the bonds at the dimer interface, the most stable snapshots from trajectories were selected. These conformations were analyzed in PyMOL (PyMOL Molecular Graphics System, Version 2.3.0). The result from AIF-WT indicated that H-bonds in Glu413-Arg449 and Glu426-Arg430 led to stabilization of the AIF dimer (Fig. 4c). However, when Thr260 was converted to Ala260, Trp422 replaced Arg422, or Gln451 replaced Arg451, the dimers became loose and the H-bonds in the dimerization interface were disrupted (Fig. 4c). Such results indicated an unstable AIF dimer in AIF variants. Surprisingly, Ala260 or Gln451 was not at the dimer interface. Thus, we inspected the H-bond network in the redox active site where the ligands integrated. The p.T260A variant was seen to impair binding with FAD, via a reduction from seven to three H-bonds (Figs. S3c and S3d). In the p.R422W variant, binding with NADH(A) was impaired (Fig. S3e). However, we could not detect any severe changes in the p.R451Q variant (Fig. S3f), and this finding requires further exploration.
Fig. 4. Disrupted AIF dimer structures. (a) The number of H-bonds between residues at the dimer interface across the trajectory time course without ligand. (b) The number of H-bonds between residues at the dimer interface across the trajectory time course with NADH and FAD ligands. The dotted line represents the average value of H-bonds. (c) Structures of AIF-WT and AIF variant dimers. These selected snapshots were the most stable structure from the trajectories. These conformations were then analyzed, and the figures were produced using PyMOL. The H-bonds in the dimerization interface were reduced. The residues were colored by element with the C atom in green, H atom in silver, N atom in blue, O atom in red, and the S atom in yellow. Dashed red lines represent H-bonds. Data are represented as mean±SEM (n=3). * P<0.05, ** P<0.01. AIF: apoptosis-inducing factor; NADH: nicotinamide adenine dinucleotide; FAD: flavin adenine dinucleotide; WT: wild type; SEM: standard error of the mean.
2.5. Improved AIF dimerization by NADH treatment
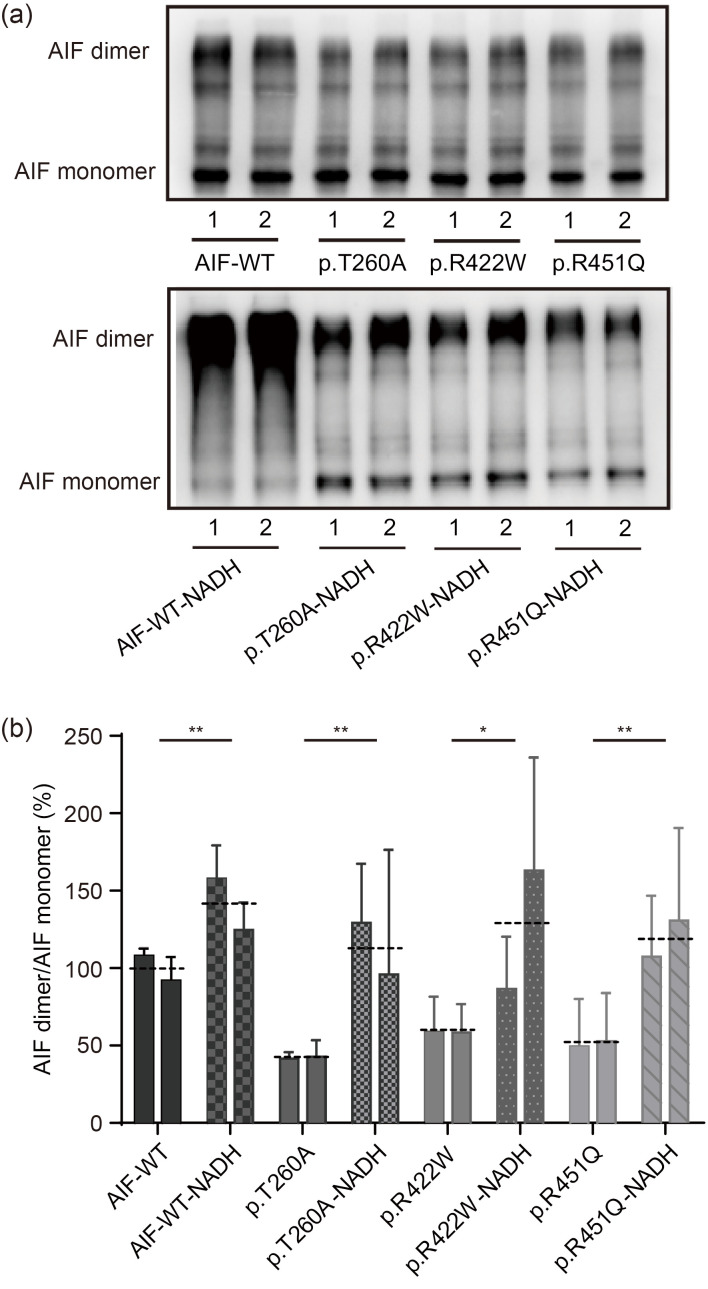
We next assessed whether NADH (200 μmol/L for 24 h) could restore AIF dimerization in the AIF variant cell lines. Western blotting analysis showed that the relative AIF dimer levels were decreased to (42.1±7.5)% (P<0.001) in p.T260A cells, (58.7±19.1)% (P<0.001) in p.R422W cells, and (51.1±28.6)% (P<0.001) in p.R451Q cells, compared to 100.0% in AIF-WT cells (Figs. 5a and 5b). The relative AIF dimer levels were increased to (112.6±59.2)% (P=0.0055) in p.T260A cells, (130.3±68.6)% (P=0.0139) in p.R422W cells, and (119.0±48.5)% (P=0.0042) in p.R451Q cells upon NADH treatment, compared to 100.0% in AIF-WT cells (Figs. 5a and 5b). Our results have provided the first in vivo evidence that NADH can rescue the disrupted dimerization in AIF variants.
Fig. 5. Improved AIF dimerization by NADH treatment. (a) Western blotting analysis of AIF dimers in AIF-WT and AIF variant stable transfection cell lines with NADH administration (200 µmol/L for 24 h). The cell pellets were treated with 4 µmol/L DSS for cross-linking before lysis. (b) Quantification analysis of AIF dimer in AIF-WT and AIF variants. Data are represented as mean±SEM (n=3). * P<0.05, ** P<0.01. The dotted line represents the average value of two clones. AIF: apoptosis-inducing factor; NADH: nicotinamide adenine dinucleotide; WT: wild type; DSS: disuccinimidyl suberate; SEM: standard error of the mean.
2.6. Decreased apoptosis after NADH treatment
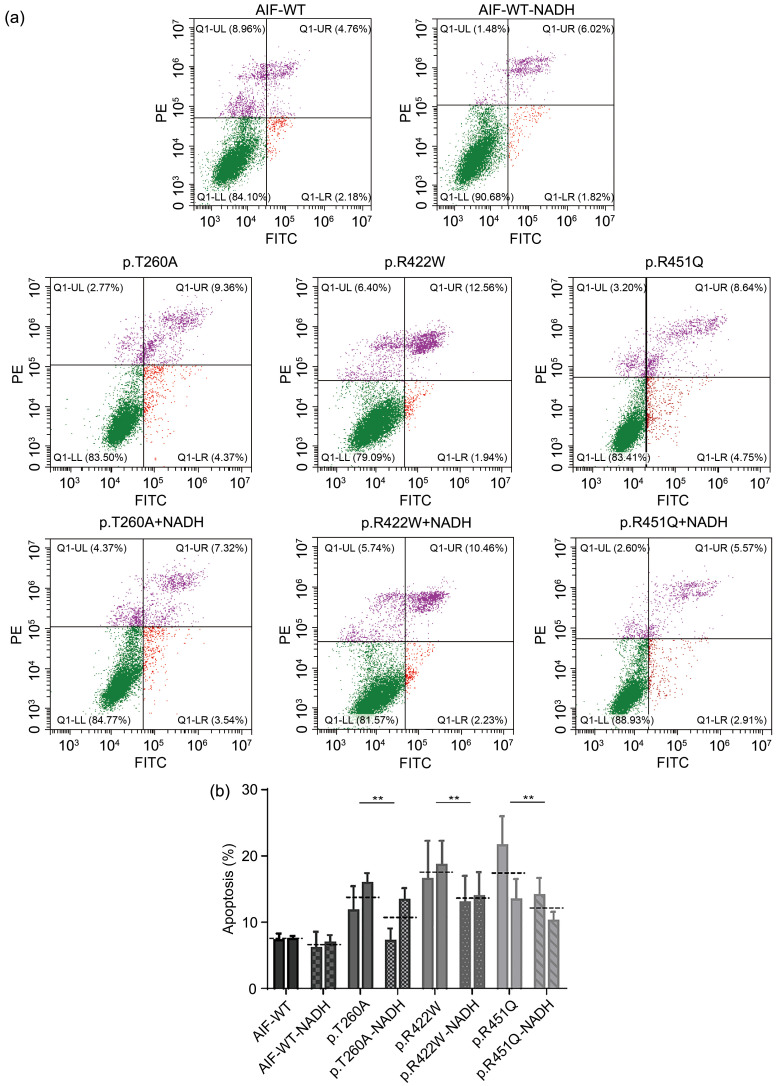
To determine whether restoration of AIF dimerization could reduce apoptosis of AIF variant cells, flow cytometry was performed. The percentage of apoptotic cells was reduced from (13.7±3.5)% to (10.0±3.7)% (P=0.027) in p.T260A cells, from (17.9%±4.2)% to (13.6±3.5)% (P=0.032) in p.R422W cells, and from (17.1%±5.5)% to (12.0±2.7)% (P=0.025) in p.R451Q cells (Figs. 6a and 6b). Collectively, NADH, as a coenzyme of AIF, can improve AIF dimerization and ultimately reduce the apoptosis of AIF variant cells.
Fig. 6. Decreased apoptosis after NADH treatment. (a) Flow cytometry analysis of apoptosis in AIF-WT and AIF variant stable transfection cell lines with NADH administration (200 µmol/L for 24 h). (b) Quantification analysis of apoptosis in AIF-WT and AIF variant stable transfection cell lines. Data are represented as mean±SEM (n=3). ** P<0.01. The dotted line represents the average value of two clones. AIF: apoptosis-inducing factor; WT: wild type; NADH: nicotinamide adenine dinucleotide; SEM: standard error of the mean; PE: phycoerythrin; FITC: fluorescein isothiocyanate; UL: upper left; UR: upper right; LL: lower left; LR: lower right.
3. Discussion
We previously reported 11 novel AIFM1 variants in a Chinese cohort of familial and sporadic cases of ANSD (Zong et al., 2015). Here, using functional assays and pharmacological restoration, we investigated the molecular mechanisms underlying the identified variants (p.T260A, p.R422W, and p.R451Q) in the familial cases. In the current study, our focus was on AIF dimerization, apoptosis, and the rescuing effect of NADH administration. We found that AIF variant cells had poor growth rates, likely resulting from the activation of apoptosis. Using STS and Z-VAD-FM, we characterized the increased apoptosis of AIF variant cells as AIF-dependent, but not caspase-dependent. This was further confirmed using western blotting for the caspase active form. These results could, in part, explain the poor physiological status of AIF variant cells. Other research has also shown relationship between AIF and caspase-independent apoptosis (Yu et al., 2006; Tang et al., 2019), suggesting a critical role of AIF in apoptosis. Beyond apoptosis, the poor physiological status of AIF variant cells may also be related to the impaired functions of AIF in mitochondria. For example, recent studies have shown AIF dimerization as indispensable for the assembly of coiled-coil-helix-coiled-coil-helix domain containing 4 (CHCHD4)-mediated respiratory complexes (Hangen et al., 2015; Reinhardt et al., 2020). How these AIF variants may further influence mitochondrial function will be a focus of a further study.
AIF dimerization is recognized as key to its function including modulating its proapoptotic function (Sevrioukova, 2011). To explore why apoptosis is active in AIF variants, we studied AIF dimerization. We detected a reduction in the content of AIF dimers and a weakened interaction between the two AIF monomers in AIF variants compared to AIF-WT. These results demonstrated that the AIF variants impaired dimerization. To complement our experimental data, molecular dynamics simulations were performed to test AIF dimer stability. The RMSDs showed increased fluctuation and a reduction in the number of H-bonds between two AIF monomers in AIF variant systems, in contrast to the WT systems. Collectively, these observations support the conclusion that the AIF variants caused AIF dimer instability. Similarly, AIF mutations have been reported elsewhere as causing disease via damaged dimerization. For example, the p.Gly262Ser mutant led to early-onset slowly progressive mitochondrial disease by affecting the formation/stability of CTCs (Ardissone et al., 2015; Sevrioukova, 2016). Similarly, the p.Gly308Glu mutant was unable to produce sufficient amounts of CTCs and effectively catalyze redox reactions (Berger et al., 2011; Sevrioukova, 2016). The Arg201del mutant produced a shorter-lived CTCs due to FAD cofactor release in the first transition, but NADH could stabilize this FAD cofactor (Ghezzi et al., 2010). Here, we first proposed the roles of AIF dimers in ANSD.
As reported, the interaction between AIF and NADH was key to CTC formation (Ferreira et al., 2014). We found that the AIF variants required a larger excess of NADH and longer time periods to reduce the monomer form to the CTC form. These results revealed that AIF variants impaired the NADH-binding affinity and led to the impaired dimerization. Some other published data have also showed impaired coenzyme-binding affinity for other AIF variants. These include the variants (p.E413A/R422A/R430A) in the dimer interface (Ferreira et al., 2014), the pathologic variants (p.V243L, p.G262S, p.G308E, and p.G338E) in neurodegenerative disorders (Sevrioukova, 2016), and the variants in active sites (p.H454A) (Brosey et al., 2016). Another recent study showed that W196 could stabilize the CTC form by enhancing the affinity for NADH (Romero-Tamayo et al., 2021). In the present study, we first elaborated on the impaired coenzyme-binding affinity of AIF variants in ANSD. Interestingly, the p.R422W variant showed no impact on the FAD reduction, but decreased dimer formation. This result may be related to the unstable AIF dimer. In addition, unlike Trp422, the variants Ala260 and Gln451 are not located at the dimer interface. Thus, we further inspected the H-bond network in the redox active site, where the ligands integrated. We found that the p.T260A variant impaired binding with FAD, which may affect dimerization. However, we could not detect any severe changes in the p.R451Q variant other than noting the slow pace of the reduction reaction. Clearly, the mechanism requires further study for clarification. In summary, AIF variants related to AIF dimerization are distributed throughout AIF, including the dimer interface (p.R422W, p.E413A/R422A/R430A (Ferreira et al., 2014)), the NADH-binding site (p.G308E (Berger et al., 2011) and p.G338E (Diodato et al., 2016)), C-loop salt bridges (p.ΔR201 (Ghezzi et al., 2010)), the cavity shielded by the Cβ-clasp (p.E493V (Rinaldi et al., 2012)), and other locations (p.T260A and p.R451Q in the present study). The reduction in AIF dimer may also be related to the impaired stability of the AIF monomer, such as for the p.ΔR201. Our results of native-PAGE showed a degree of AIF monomer degradation in variants, which may partly contribute to impaired dimerization. We further showed that AIF variants in ANSD cause caspase-independent apoptosis due to the impaired AIF dimerization. However, the detailed mechanisms of this increased apoptosis await further study.
NADH is a major electron donor in the electron transport chain (ETC) and plays important roles in mitochondrial oxidative phosphorylation (OXPHOS) (Ying, 2008). It has been known as a potential therapeutic drug for degenerative disorders, especially mitochondrial dysfunction-induced diseases, including Parkinson’s disease (PD) (Birkmayer et al., 1993), Alzheimer’s disease (AD) (Demarin et al., 2004), and chronic fatigue syndrome (Castro-Marrero et al., 2016). Here, we observed that apoptosis was significantly decreased in AIF variant cells upon NADH treatment, demonstrating that NADH has a therapeutic potential for ANSD treatment. Similarly, previous studies have reported that NADH can block poly(ADP-ribose) polymerase-1 (PARP-1)-mediated astrocyte death (Zhu et al., 2005), and protect cells from apoptosis by inhibiting mitochondrial permeability transition pore (mPTP) opening (Pittelli et al., 2011). It was reported that each AIF dimer has one FAD and two NADHs per AIF monomer, and the proportion of AIF dimers increases considerably upon the binding of NADH (Ferreira et al., 2014). Here, our results showed that NADH can rescue the disrupted dimerization in AIF variants and suggested that NADH treatment decreased cell apoptosis due mainly to the effect of NADH on AIF dimerization. Previous findings suggesting that NADH enhanced AIF dimerization were performed on purified proteins. Our results provided the first in vivo evidence that NADH can rescue the disrupted dimerization in AIF variant cells. The degree of impairment of the NADH-binding affinity of AIF variants is variable. Thus, we proposed that various concentrations and medication time-intervals may be required for personalized treatment of patients.
4. Conclusions
We suggest that the AIFM1 variants (p.T260A, p.R422W, and p.R451Q) inhibit AIF dimer conformation, disrupt the monomer–dimer equilibrium, and ultimately cause caspase-independent apoptosis. Pharmacological restoration of AIF dimerization by NADH treatment could markedly reduce apoptosis in AIF variant cells. We therefore suggest that NADH is a potential drug for ANSD treatment. Our findings provide an innovative insight into the molecular mechanisms of ANSD and provide evidence of novel therapies for ANSD treatment.
Materials and methods
The detailed methods are provided in the electronic supplementary materials of this paper.
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 32070584, 81830028, 31771398, 82222016, and 8207040100), the Zhejiang Provincial Natural Science Foundation of China (No. LZ19C060001), and the Fundamental Research Funds for the Central Universities (No. 2019QNA6001). We thank Chris WOOD from the College of Life Sciences, Zhejiang University for the proof reading of this manuscript. We thank Fangliang HUANG and Shelong ZHANG from the Instrument and Technical Service Platform, College of Life Sciences, Zhejiang University for their technical support.
Author contributions
Qingfeng YAN and Qiuju WANG conceived and designed the research. Yue QIU, Hongyang WANG, and Huaye PAN performed the experiments and analyzed the data. Lei YAN and Dong ZHANG performed molecular dynamics analysis. Hongyang WANG, Jing GUAN, Mingjie FAN, Hui ZHOU, Kaiwen WU, and Zexiao JIA provided technical support. Xuanhao ZHOU, Qianqian ZHUANG, Zhaoying LEI, Mengyao LI, and Xue DING verified the reproducibility of results. Aifu LIN, Yong FU, and Dong ZHANG contributed to discussion and data interpretation. Qingfeng YAN, Qiuju WANG, Yue QIU, and Huaye PAN wrote the manuscript. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Yue QIU, Hongyang WANG, Huaye PAN, Jing GUAN, Lei YAN, Mingjie FAN, Hui ZHOU, Xuanhao ZHOU, Kaiwen WU, Zexiao JIA, Qianqian ZHUANG, Zhaoying LEI, Mengyao LI, Xue DING, Aifu LIN, Yong FU, Dong ZHANG, Qiuju WANG, and Qingfeng YAN declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Al-Wardy NM, Al-Kindi MN, Al-Khabouri MJ, et al. , 2016. A novel missense mutation in the C2C domain of otoferlin causes profound hearing impairment in an Omani family with auditory neuropathy. Saudi Med J, 37(10): 1068-1075. 10.15537/smj.2016.10.14967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardissone A, Piscosquito G, Legati A, et al. , 2015. A slowly progressive mitochondrial encephalomyopathy widens the spectrum of AIFM1 disorders. Neurology, 84(21): 2193-2195. 10.1212/wnl.0000000000001613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bano D, Prehn JHM, 2018. Apoptosis-inducing factor (AIF) in physiology and disease: the tale of a repented natural born killer. eBioMedicine, 30: 29-37. 10.1016/j.ebiom.2018.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I, Ben-Neriah Z, Dor-Wolman T, et al. , 2011. Early prenatal ventriculomegaly due to an AIFM1 mutation identified by linkage analysis and whole exome sequencing. Mol Genet Metab, 104(4): 517-520. 10.1016/j.ymgme.2011.09.020 [DOI] [PubMed] [Google Scholar]
- Birkmayer JGD, Vrecko C, Volc D, et al. , 1993. Nicotinamide adenine dinucleotide (NADH)—a new therapeutic approach to Parkinson’s disease. Comparison of oral and parenteral application. Acta Neurol Scand, 87(S146): 32-35. 10.1111/j.1600-0404.1993.tb00018.x [DOI] [PubMed] [Google Scholar]
- Brosey CA, Ho C, Long WZ, et al. , 2016. Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure, 24(12): 2067-2079. 10.1016/j.str.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Marrero J, Sáez-Francàs N, Segundo MJ, et al. , 2016. Effect of coenzyme Q10 plus nicotinamide adenine dinucleotide supplementation on maximum heart rate after exercise testing in chronic fatigue syndrome—a randomized, controlled, double-blind trial. Clin Nutr, 35(4): 826-834. 10.1016/j.clnu.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Cheng X, Li L, Brashears S, et al. , 2005. Connexin 26 variants and auditory neuropathy/dys-synchrony among children in schools for the deaf. Am J Med Genet, 139A(1): 13-18. 10.1002/ajmg.a.30929 [DOI] [PubMed] [Google Scholar]
- Cheung ECC, Joza N, Steenaart NAE, et al. , 2006. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J, 25(17): 4061-4073. 10.1038/sj.emboj.7601276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Wu CC, Lu YC, et al. , 2010. Mutations in the OTOF gene in Taiwanese patients with auditory neuropathy. Audiol Neurootol, 15(6): 364-374. 10.1159/000293992 [DOI] [PubMed] [Google Scholar]
- Delmaghani S, del Castillo FJ, Michel V, et al. , 2006. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet, 38(7): 770-778. 10.1038/ng1829 [DOI] [PubMed] [Google Scholar]
- Demarin V, Podobnik SS, Storga-Tomic D, et al. , 2004. Treatment of Alzheimer’s disease with stabilized oral nicotinamide adenine dinucleotide: a randomized, double-blind study. Drugs Exp Clin Res, 30(1): 27-33. [PubMed] [Google Scholar]
- Diodato D, Tasca G, Verrigni D, et al. , 2016. A novel AIFM1 mutation expands the phenotype to an infantile motor neuron disease. Eur J Hum Genet, 24(3): 463-466. 10.1038/ejhg.2015.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P, Villanueva R, Martínez-Júlvez M, et al. , 2014. Structural insights into the coenzyme mediated monomer-dimer transition of the pro-apoptotic apoptosis inducing factor. Biochemistry, 53(25): 4204-4215. 10.1021/bi500343r [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Sevrioukova I, Invernizzi F, et al. , 2010. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am J Hum Genet, 86(4): 639-649. 10.1016/j.ajhg.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangen E, Blomgren K, Bénit P, et al. , 2010. Life with or without AIF. Trends Biochem Sci, 35(5): 278-287. 10.1016/j.tibs.2009.12.008 [DOI] [PubMed] [Google Scholar]
- Hangen E, Féraud O, Lachkar S, et al. , 2015. Interaction between AIF and CHCHD4 regulates respiratory chain biogenesis. Mol Cell, 58(6): 1001-1014. 10.1016/j.molcel.2015.04.020 [DOI] [PubMed] [Google Scholar]
- Heimer G, Eyal E, Zhu X, et al. , 2018. Mutations in AIFM1 cause an X-linked childhood cerebellar ataxia partially responsive to riboflavin. Eur J Paediatr Neurol, 22(1): 93-101. 10.1016/j.ejpn.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J, 2020. Apoptosis inducing factor and mitochondrial NADH dehydrogenases: redox-controlled gear boxes to switch between mitochondrial biogenesis and cell death. Biol Chem, 402(3): 289-297. 10.1515/hsz-2020-0254 [DOI] [PubMed] [Google Scholar]
- Hu B, Wang M, Castoro R, et al. , 2017. A novel missense mutation in AIFM1 results in axonal polyneuropathy and misassembly of OXPHOS complexes. Eur J Neurol, 24(12): 1499-1506. 10.1111/ene.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang-Roth R, Fischer-Krall E, Kornblum C, et al. , 2017. AUNA2: a novel type of non-syndromic slowly progressive auditory synaptopathy/auditory neuropathy with autosomal-dominant inheritance. Audiol Neurootol, 22(1): 30-40. 10.1159/000474929 [DOI] [PubMed] [Google Scholar]
- Li C, Brant E, Budak H, et al. , 2021. CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 22(4): 253-284. 10.1631/jzus.B2100009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka AJ, Morrissey ZD, Zhang CY, et al. , 2017. Directed differentiation of human embryonic stem cells toward placode-derived spiral ganglion-like sensory neurons. Stem Cells Transl Med, 6(3): 923-936. 10.1002/sctm.16-0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzewska H, Rydzanicz M, Biegański T, et al. , 2017. Spondyloepimetaphyseal dysplasia with neurodegeneration associated with AIFM1 mutation—a novel phenotype of the mitochondrial disease. Clin Genet, 91(1): 30-37. 10.1111/cge.12792 [DOI] [PubMed] [Google Scholar]
- Penido RC, Isaac ML, 2013. Prevalence of auditory neuropathy spectrum disorder in an auditory health care service. Braz J Otorhinolaryngol, 79(4): 429-433. 10.5935/1808-8694.20130077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittelli M, Felici R, Pitozzi V, et al. , 2011. Pharmacological effects of exogenous NAD on mitochondrial bioenergetics, DNA repair, and apoptosis. Mol Pharmacol, 80(6): 1136-1146. 10.1124/mol.111.073916 [DOI] [PubMed] [Google Scholar]
- Reinhardt C, Arena G, Nedara K, et al. , 2020. AIF meets the CHCHD4/Mia40-dependent mitochondrial import pathway. Biochim Biophys Acta Mol Basis Dis, 1866(6): 165746. 10.1016/j.bbadis.2020.165746 [DOI] [PubMed] [Google Scholar]
- Rinaldi C, Grunseich C, Sevrioukova IF, et al. , 2012. Cowchock syndrome is associated with a mutation in apoptosis-inducing factor. Am J Hum Genet, 91(6): 1095-1102. 10.1016/j.ajhg.2012.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanos J, Kimura L, Fávero ML, et al. , 2009. Novel OTOF mutations in Brazilian patients with auditory neuropathy. J Hum Genet, 54(7): 382-385. 10.1038/jhg.2009.45 [DOI] [PubMed] [Google Scholar]
- Romero-Tamayo S, Laplaza R, Velazquez-Campoy A, et al. , 2021. W196 and the β-hairpin motif modulate the redox switch of conformation and the biomolecular interaction network of the apoptosis-inducing factor. Oxid Med Cell Longev, 2021: 6673661. 10.1155/2021/6673661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Martínez A, Benito-Orejas JI, Tellería-Orriols JJ, et al. , 2017. Autosomal dominant auditory neuropathy and variant DIAPH3 (c. -173C>T). Acta Otorrinolaringol Esp, 68(3): 183-185. 10.1016/j.otorri.2016.06.004 [DOI] [PubMed] [Google Scholar]
- Sancho P, Sánchez-Monteagudo A, Collado A, et al. , 2017. A newly distal hereditary motor neuropathy caused by a rare AIFM1 mutation. Neurogenetics, 18(4): 245-250. 10.1007/s10048-017-0524-6 [DOI] [PubMed] [Google Scholar]
- Santarelli R, Cama E, Scimemi P, et al. , 2008. Audiological and electrocochleography findings in hearing-impaired children with connexin 26 mutations and otoacoustic emissions. Eur Arch Otorhinolaryngol, 265(1): 43-51. 10.1007/s00405-007-0412-z [DOI] [PubMed] [Google Scholar]
- Schoen CJ, Emery SB, Thorne MC, et al. , 2010. Increased activity of Diaphanous homolog 3 (DIAPH3)/diaphanous causes hearing defects in humans with auditory neuropathy and in Drosophila. Proc Natl Acad Sci USA, 107(30): 13396-13401. 10.1073/pnas.1003027107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, 2011. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid Redox Signal, 14(12): 2545-2579. 10.1089/ars.2010.3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, 2016. Structure/function relations in AIFM1 variants associated with neurodegenerative disorders. J Mol Biol, 428(18): 3650-3665. 10.1016/j.jmb.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Starr A, Sininger YS, Pratt H, 2000. The varieties of auditory neuropathy. J Basic Clin Physiol Pharmacol, 11(3): 215-230. [DOI] [PubMed] [Google Scholar]
- Starr A, Isaacson B, Michalewski HJ, et al. , 2004. A dominantly inherited progressive deafness affecting distal auditory nerve and hair cells. J Assoc Res Otolaryngol, 5(4): 411-426. 10.1007/s10162-004-5014-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, et al. , 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature, 397(6718): 441-446. 10.1038/17135 [DOI] [PubMed] [Google Scholar]
- Tang DL, Kang R, Berghe TV, et al. , 2019. The molecular machinery of regulated cell death. Cell Res, 29(5): 347-364. 10.1038/s41422-019-0164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin M, Akcayoz D, Incesulu A, 2005. A novel missense mutation in a C2 domain of OTOF results in autosomal recessive auditory neuropathy. Am J Med Genet, 138A(1): 6-10. 10.1002/ajmg.a.30907 [DOI] [PubMed] [Google Scholar]
- Tranebjærg L, Strenzke N, Lindholm S, et al. , 2018. The CAPOS mutation in ATP1A3 alters Na/K-ATPase function and results in auditory neuropathy which has implications for management. Hum Genet, 137(2): 111-127. 10.1007/s00439-017-1862-z [DOI] [PubMed] [Google Scholar]
- Varga R, Kelley PM, Keats BJ, et al. , 2003. Non-syndromic recessive auditory neuropathy is the result of mutations in the otoferlin (OTOF) gene. J Med Genet, 40(1): 45-50. 10.1136/jmg.40.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QJ, Li RH, Zhao H, et al. , 2005. Clinical and molecular characterization of a Chinese patient with auditory neuropathy associated with mitochondrial 12S rRNA T1095C mutation. Am J Med Genet, 133A(1): 27-30. 10.1002/ajmg.a.30424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QJ, Li QZ, Rao SQ, et al. , 2006. AUNX1, a novel locus responsible for X linked recessive auditory and peripheral neuropathy, maps to Xq23‒27. 3. J Med Gente, 43(7): e33 10.1136/jmg.2005.037929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Li SS, Zhang X, et al. , 2015. Gene editing and cell therapy based on induced pluripotent stem cells. Chin J Cell Biol, 37(1): 90-99. [Google Scholar]
- Ying WH, 2008. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal, 10(2): 179-206. 10.1089/ars.2007.1672 [DOI] [PubMed] [Google Scholar]
- Yu SW, Wang HM, Poitras MF, et al. , 2002. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science, 297(5579): 259-263. 10.1126/science.1072221 [DOI] [PubMed] [Google Scholar]
- Yu SW, Andrabi SA, Wang HM, et al. , 2006. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA, 103(48): 18314-18319. 10.1073/pnas.0606528103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu KQ, Swanson RA, Ying WH, 2005. NADH can enter into astrocytes and block poly(ADP-ribose) polymerase-1-mediated astrocyte death. NeuroReport, 16(11): 1209-1212. 10.1097/00001756-200508010-00015 [DOI] [PubMed] [Google Scholar]
- Zong L, Guan J, Ealy M, et al. , 2015. Mutations in apoptosis-inducing factor cause X-linked recessive auditory neuropathy spectrum disorder. J Med Genet, 52(8): 523-531. 10.1136/jmedgenet-2014-102961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.