Abstract
The Bisulfite Genomic Sequencing technique has found wide acceptance for the generation of DNA-methylation maps with single-base resolution. The method is based on the selective deamination of cytosine to uracil (and subsequent conversion to thymine via PCR), whereas 5-methylcytosine residues remain unchanged. Methylation maps are created by the comparison of bisulfite converted sequences with the untreated genomic sequence. ‘MethTools’ is a collection of software tools that replaces the time-consuming manual comparison process, generates graphical outputs of methylation patterns and methylation density, estimates the systematic error of the experiment and searches for conserved methylated nucleotide patterns. The programs are written in Perl 5 and C, and the source code can be downloaded. All tools run independently but the programs are interfaced. Thus, a script can perform the entire analysis procedure automatically. In addition, a web-based remote analysis service is offered. Both the source code and the remote analysis are available at http://genome.imb-jena.de/methtools/
INTRODUCTION
For many years, evidence has been accumulating suggesting that methylation of cytosines in the DNA of animals, plants and fungi is involved in gene silencing. Transfection experiments with in vitro methylated reporter gene constructs have shown that methylation in certain regions of the gene inhibits transcription (1,2). Methylation either directly inhibits the binding of transcription factors (reviewed in 3) or methylcytosine binding proteins interact with other structural compounds of the chromatin making the DNA inaccessible to transcription factors through histone deacetylation and chromatin structure changes (4). Differential methylation was found to be a key element in the transcriptional regulation of genes whose expression state depends on the sex of the parent from whom they were inherited (imprinted genes) (5–8). Also, the initiation and/or maintenance of X-inactivation in female eutherians is associated with methylation (9,10). Likewise, methylation is important for the maintenance of transcriptional silencing in plants (11). The development of cancer in mammalia was found to be accompanied by genome wide demethylation and local hypermethylation of tumor suppressor genes (12–14). In vertebrates, methylation is probably restricted to cytosines that are followed by a guanine in the 5′ to 3′ direction (CpG) (15,16) but in plants, methylation was observed also at non-symmetric sites (17,18).
Several hypotheses about the origin and function of methylation were proposed ranging from a host defense mechanism that silences most of the parasitic sequence elements (19–21) to the function as a molecular instrument to lock developmental changes and to determine tissue-specific gene expression (22–24). However, it should be noted that most of the evidence that the occurrence of 5-methylcytosine (5mC) actually controls gene expression in vivo is of correlative nature.
Our knowledge about methylation has profited from investigations with isoschizomeric restriction enzymes. These enzymes recognize identical restriction sites but show different sensitivity towards methylation (25). However, only the introduction of the Bisulfite Genomic Sequencing technique made it possible to study small amounts of DNA and to identify methylation patterns with single base resolution (26). The technique is based on the selective deamination of cytosine to uracil. In contrast, 5mC remains unchanged. PCR amplification converts all uracils into thymine and comparison of the sequences of the PCR products with the original genomic sequence (mother-sequence) reveals which cytosine was methylated and which was not. Subcloning of the PCR products and sequencing of individual clones delivers the methylation pattern of single-strand DNA molecules.
Alignment and comparison of the sequences is usually done by manual inspection, a process which is labor intensive, time consuming and error prone. Thus, we present here a collection of software tools for the evaluation of bisulfite sequencing data, for the visualization of methylation patterns and for the search of common motives in these data.
DESCRIPTION OF SOFTWARE TOOLS
All software tools, except ‘logo5mC’, are written in Perl 5 (27) a programming language which is available for all common computer platforms. The source code can easily be transferred as a text file and can be adapted to local needs with basic programming knowledge. The programs are interfaced with each other and automation of the whole evaluation process is possible with a suitable script. A short help text is displayed when the program name is entered followed by parameter ‘-h’. The program ‘logo5mC’ for the calculation of generic and methylation sequence logos (28) is written in C++. Deliberately, the user interface of all programs was restricted to a simple command line in order to facilitate script-based automation and/or the integration into existing local graphical user interfaces. In the following, ‘usage: >’ indicates the beginning of the command line that has to be entered under UNIX and compatible operating systems. Parameters that have to be supplied by the users are written in italic.
Extraction of methylation patterns from bisulfite converted DNA sequences
• ‘convert_bisulfite’
usage: >convert_bisulfite.pl -f filename > filename.log
Since the IUB code provides no symbols for modified bases (29,30) we decided to symbolize 5mC by an uppercase ‘C’ and all unmodified bases by lowercase letters ‘c, a, t, g’. Sequences of this format are generated by the program ‘convert_bisulfite’. The input format for this program consists of a single file containing the mother-sequence followed by the bisulfite generated sequences in FASTA format (>name[new-line]-sequence[new-line]). All sequences have to be aligned and brought to equal length to generate this input file (‘filename’). The alignment can be done by hand or with a suitable program. We recommend ClustalW (31) using a modified conversion matrix with equal probability of [c to c] and [c to t] exchanges. The mother-sequence is supposed to have no internal gaps while ‘n’ is allowed to represent unknown bases.
The program compares the mother-sequence with the bisulfite generated sequences below in the file ‘filename’, and generates individual files carrying the FASTA-name extended by ‘.seq1’. These files follow the FASTA-standard but cytosines in the bisulfite sequences are converted into uppercase ‘C’, and thymines that used to be cytosines in the mother-sequence to lowercase ‘c’. All other bases are written in lowercase letters. In this paper, this format will be referred to as ‘seq1-format’. Except for cytosine/thymine exchanges, conflicts between the sequences are resolved in favor of the untreated original. In parallel, a second file is generated (FASTA-name.seq2) containing a symbolic representation of the seq1-file: 5mC is coded by ‘│’, unmethylated cytosine by ‘O’ and other bases by ‘–’. Unknown bases are identified by ‘?’ and mismatches between mother-sequence and PCR products by ‘C’ (for conflict). This type of representation can be used in many parsimony programs which might be useful to study clonal inheritance of methylation patterns within the investigated population of cells.
The program writes a log-file to the standard output. We suggest redirecting this output into a file for further use. The log-file contains the FASTA-names of the processed sequences, conversion statistics and the position and nature of differences (other than C→T exchanges) between the mother-sequence and the PCR products.
Finally, a table is written into a file ‘filename.tab’. The first column contains the positions from base 1 to the end of the sequence, the second the nucleotide symbol, the next column the average methylation at cytosine sites (in percent) and the fourth column the number of analyzed cytosines per position. This file can be imported into conventional data representation programs. Once the files in seq1-format are generated a number of additional independent analyses can be performed. It is recommended to generate a subdirectory for each analyzed DNA fragment, and to place both the log-file and the seq1-files there after the initial conversion procedure.
Estimation of the systematic error of the experiment
• ‘EvalLog’
usage: >EvalLog.pl -d path -n outfile
The fidelity of the PCR and the sequencing reaction has a strong impact on the accuracy of the bisulfite generated methylation data. It can be estimated from the number of base exchanges other than C→T conversions which are recorded in the log-file of ‘convert_bisulfite’. The ‘EvalLog’ program scans the log-files for these substitutions and determines in parallel the total number of the five bases, 5mC, c, t, a and g, in the corresponding seq1-files. These values are written to a text-file ‘outfile.err’ and can be used to estimate the error rate of the reactions. Both the log-files and the related seq1-files have to be in the same subdirectory ‘path’. Multiple log-files are possible (e.g. for repeated PCR reactions of one DNA fragment). Files without the ‘.log’ or ‘.seq1’ extension will be ignored.
Simple graphical representation of methylation patterns
• ‘Plot_CpGs_to_PS’
usage: >Plot_CpGs_to_PS.pl -d path -n outfile -s scale
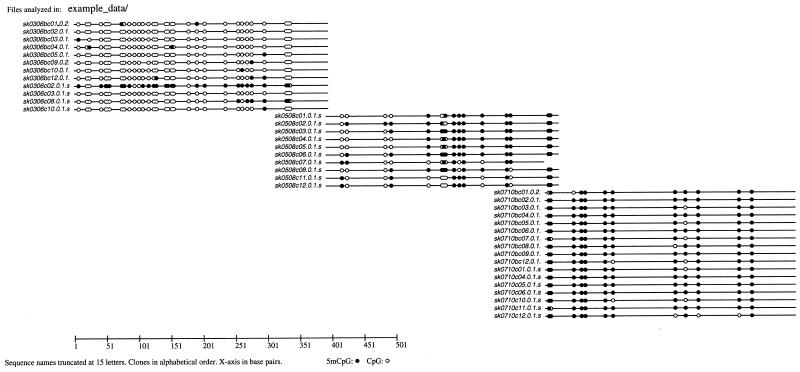
Representation of unmethylated CpGs as hollow circles and methylated CpG as full circles on a line representing the sequence itself enjoys common popularity in the related literature. ‘Plot_CpGs_to_PS’ generates this projection of methylation patterns. The program reads files in seq1-format carrying the extension ‘.seq1’ from the subdirectory ‘path’ indicated by option -d. It writes a postscript graphic into the file ‘outfile’ specified by -n. If -n is omitted the name of the subdirectory is used as name of the output file. Unless ‘outfile’ has already been entered with the extension ‘.ps’, this extension is added by the program. Depending on the total length of the sequences and the number of analyzed subclones, an appropriate numeric scale parameter has to be given with option -s (default = 1). An example of the graphical output generated with this program is shown in Figure 1.
Figure 1.
Methylation patterns in an example data set. This graphical representation of methylation patterns was generated with the program ‘Plot_CpGs_to_PS’ (scale parameter -s 0.7). The lengths of the aligned DNA fragments are indicated as black lines. Full black circles represent CpG pairs containing 5mC; hollow circles represent unmethylated CpG pairs. The FASTA-names of the sequences are written on the left-hand side and truncated at 15 characters. On the top, the full directory path of the analyzed sequences is printed. On the bottom, the scale of the figure is given in base pairs. The example data were taken from an unpublished study of the MSSK1 promotor region in human Xq28.
• ‘Plot_CpGs_to_Gif’ and ‘Plot_CpNpGs_to_Gif’
usage: >Plot_CpGs_to_Gif.pl -d path -n outfile
While postscript files are suitable for high resolution prints, they are less appropriate for on-screen display and web-publishing. ‘Plot_CpGs_to_Gif’ generates colored Gif-images which display 5mCpG as red circles and unmethylated CpG as blue circles. For organisms that show no restriction of cytosine methylation to CpGs the program ‘Plot_CpNpGs_to_Gif’ can be applied. It uses a set of red, blue, green, black and yellow circles to represent the most commonly methylated pairs and triplets CpG, CpApA, CpApG, CpTpG and CpTpA, respectively. The programs read files in seq1-format carrying the extension ‘.seq1’. These files are expected to be in a subdirectory ‘path’. Output is an image file in Gif-format (‘outfile.gif’). This format can be displayed with any image viewer (e.g. XView) or web-browser (e.g. Netscape).
Extended graphical representation and further analysis of methylation patterns
• ‘Plot_5mCpG_density_to_Gif’ and ‘Plot_5mC_density_to_Gif’
usage: >Plot_5mCpG_density_to_Gif.pl -d path -n outfile
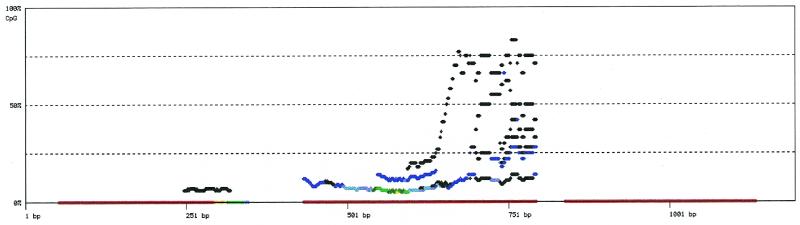
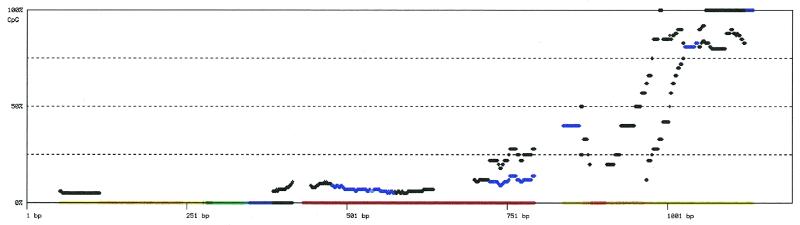
The methylation density, i.e. the number of methylated cyto-sines in a given sequence window, is believed to be important for the effect methylation has on gene expression. Accordingly, variations of the methylation density within the investigated population of PCR products (i.e. cells) will be interesting to screen for. ‘Plot_5mCpG_density_to_Gif’ displays a methylation density plot. Again, the program reads files in seq1-format carrying the extension ‘.seq1’ in a subdirectory ‘path’ specified by -d. The methylation density of individual clones is calculated as the ratio of the number of methylated CpGs to the total number of CpGs [5mCpG/(5mCpG + CpG)] in a 100 bp window shifted in 1 bp steps over the sequences. These default values can be changed in the source code. The number of clones with identical densities at a given position of the sequence is indicated by the color of the corresponding data point. The color spectrum ranges from black for one clone to red for >10 clones. A similar algorithm is implemented in ‘Plot_5mC_density_to_Gif’, but here the density of methylated C [5mC/(5mC + C)], and not CpGs, is calculated. We recommend this program for methylation data in non-CpG context. Density plots are written to a file ‘outfile.distr.gif’ in Gif-format.
Besides the information about the density distribution itself, we found this data representation useful for the evaluation of the quality of the raw data. Figures 2 and 3 show the methylation densities for clones of three different overlapping PCR fragments in two different tissues. Clearly, the methylation densities are different in the two tissues. Interestingly, the density of a few clones of the central fragment of Figure 2 increases gradually and drops suddenly when the end of the fragment is reached. Unexpectedly, no clones of similar density are found in the 3′ adjacent PCR fragment. This suggests data are missing there, probably due to the low overall methylation in this region.
Figure 2.
Methylation density distribution plot for example data set A. The shown example of a plot of the methylation density distribution was generated using the program ‘Plot_5mCpG_density_to_Gif’. On the x-axis the sequence length is given in base pairs; the y-axis represents the methylation density in percent. The sequences are displayed in 5′ (left) to 3′ (right) direction. The methylation density was calculated as the percentage of methylated CpGs in the total number of CpGs [100 × 5mCpG/(5mCpG + CpG)] in a 100 bp window which is shifted in 1 bp steps over the sequences. Methylation densities are computed for each individual PCR subclone of bisulfite treated DNA. Each clone represents the methylation pattern of a single DNA molecule. When multiple subclones deliver the same data point, the number of clones that contribute to this data point is indicated by the color of the point (black for one clone, dark blue to light blue for two to four clones, light and dark green for five and six, light and dark yellow for seven and eight, light and dark red for nine and >10 clones). No methylation density can be determined for the first and last 50 bp of each sequence (half window size); thus, gaps exist between neighboring fragments. The example data set consists of three overlapping bundles of 34 PCR fragments. It is part of an unpublished study of the SLC6A8 promotor region on human Xq28. The methylation density of a number of clones in the central fragment of the example tissue increases gradually and drops suddenly when the end of the fragment is reached. Unexpectedly, no clones with a similar density of methylation signals are found in the 3′ adjacent PCR fragment. Either data is missing there, or the clones with higher methylation represent a contamination.
Figure 3.
Methylation density plot for example data set B. The density plot was generated as described for Figure 2, but a set of 52 PCR products from a different tissue was used. In contrast to Figure 2, the changes in methylation density follow a general trend along the entire region. This indicates that the randomly chosen PCR subclones that were used to generate this data set B were probably more evenly distributed than those in example data set A. Both data sets were arbitrarily chosen to illustrate one potential use of this sort of data representation.
• ‘InfoScan’
usage: >InfoScan.pl -d path -n outfile
The calculation and visualization of the information content is a useful tool to disclose conserved sequence patterns (32) and has been suggested as an improvement to consensus sequences in general (28). In order to identify conserved sequence patterns around methylated cytosines, the program ‘InfoScan’ determines the frequency of each nucleotide for positions up to 300 bp up- and downstream of each 5mC in a given data set. The information content R of each position (32) is calculated using:
R(pos.) = H(max.) – H(pos.) – e(pos.),
with e being a correction factor, H(max.) the maximum uncertainty 2.32 (for equal frequency of each base), and H(pos.) the uncertainty at each position given as:
H(pos.) = –![]() f(base, pos.)·log2 f(base, pos.),
f(base, pos.)·log2 f(base, pos.),
where f is the base frequency at a position (pos.) and base corresponds to one of the five nucleotides, including 5mC. If the number of nucleotides (n) per position is >50, the correction factor e(pos.) can be approximated by:
(28,32) where N is the number of nucleotide bases (five in this case). Albeit in our own experiments the number of analyzed nucleotides per investigated position usually exceeded 50, we observed higher fluctuation in uncertainty values when the number of nucleotides dropped below 100 (data not shown).
To incorporate the overall distribution of the five nucleotides in the investigated region the relative entropy H′ (33) [or Shannon information (34)]:
is calculated. Here, q(base) is the global frequency of the base in the corresponding .seq1-files. The value of H′ will be zero when the frequency, f, of nucleotide bases found at a given position matches exactly with their global occurrence, g. H′ is expected to be more accurate than R since the actual distribution of the bases in the investigated region is incorporated into the calculation.
The output of the program consists of a table containing the relative positions to 5mC, the observed frequencies of g, a, t, c and 5mC, R and H′. The table is written into the file ‘outfile.inf’. A useful graphical representation of this type of data has been developed by Schneider and Stephens and is called ‘logo’ (28). The output file of ‘InfoScan’ can be used as input file for the program ‘logo5mC’.
• ‘logo5mC’
usage: >logo5mC -b -f start -l length infile > outfile.ps
The program constructs a sequence logo (28) by ranking nucleotide letters in terms of their positional frequency. The height of the stack of letters is determined by the total amount of relative entropy H′ at each position. The height of each letter represents the frequency of the respective nucleotide multiplied with H′ at this position. For frequencies <0.2 (i.e. roughly below equal distribution) the letters are written upside down. With option -b the logo5mC program reads the positional nucleotide frequencies and information from the file ‘infile’ and generates the sequence logo as a postscript file ‘outfile.ps’. Option -f determines the starting point ‘start’ of the region to be plotted, and -l the length ‘length’. Further options are specified in the enclosed ‘readme’ file. The probability table input file contains the relative position, the frequencies and the positional information of nucleotides in the vicinity of methylated nucleotides. It resembles the output of ‘InfoScan’ but can of course be generated by other means too. The sequence logo displays methylated nucleotides using the symbol ‘5’ as 5mC letter code (not uppercase ‘C’). Figure 4 shows an example of a 5mC sequence logo.
Figure 4.
Frequencies of nucleotides around methylated sites in an example data set displayed as 5mC-Logo. The shown graphic was produced using ‘logo5mC’ and is based on a table generated by ‘InfoScan’. The y-axis represents the amount of Shannon information H′ in bits, the x-axis the position relative to 5mC at position 0 in base pairs. The length of the letters of each stack reflects the frequency of the respective base multiplied by the total information at this position. The information content H′ at a given position increases with the increasing difference from the expected equal distribution of all nucleotides at this position. The information content H′ is 0 if all bases are equally distributed at a given position and occur with their global frequency. The information H′ is maximal if exclusively the rarest base occurs at a certain position. The example data used in this case are part of a methylation analysis of the human X-linked creatine transporter gene SLC6A8. 5mC at position 0 is usually followed by a guanine in 3′ direction, at positions 4 and 8 upstream and downstream of 5mC regular over-representation of cytosine, guanine and 5mC is observed, while adenine and thymine are under-represented. The significance of this observation remains to be investigated.
The software tools described in this paper will accelerate the evaluation of Bisulfite Genomic Sequencing data. The graphical representation of the data and the determination of the information content around methylated cytosines provide ways to search for patterns in DNA methylation. We hope our programs will contribute to finally deciphering the information encoded in these patterns.
AVAILABILITY OF SOFTWARE AND WEB-BASED REMOTE ANALYSIS
The MethTools package (UNIX/Linux and Macintosh versions), example data and a modified matrix for ClustalW (version 1.6 or greater) described in this paper can be downloaded from http://genome.imb-jena.de/methtools/ . The Macintosh versions of the programs do not use a command line interface and the usage is therefore slightly different from the UNIX version described in this paper. However, the website provides detailed installing instructions for both systems and gives a short example for an analysis. As an alternative to the download and installation of the programs, input files for ‘convert_bisulfite’ can also be submitted to our webserver under http://genome.imb-jena.de/methtools/ and a complete analysis is performed. The results of the analysis are returned by email. For the moment, the generation of LOGOs is excluded from this service. The Perl 5 software package is available as a public domain program for UNIX, Macintosh and other operating systems. The GD.lib (between versions 1.6 and 1.7.3) (35) and the GD.pm interface (version 1.19) (36) have to be installed for the ‘MethTools’ Perl-scripts that generate a graphical output. GD.lib and GD.pm as well as the latest version of ClustalW are available via anonymous ftp (see 31,35,36). ‘logo5mC’ has been programmed in C++. Under a UNIX environment it can be automatically compiled using the ‘make’ command. Currently, ‘logo5mC’ is not available for operating systems other than UNIX and compatibles. Parts of ‘logo5mC’ belong to the GENIO/logo project for a WWW-based generation of sequence logos (37).
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to T. Wiehe and M. Platzer for carefully reading the manuscript. This work was in part supported by a grant of the Klaus Tschira Foundation to C.G.
REFERENCES
- 1.Harbers K., Schnieke,A., Stuhlmann,H., Jahner,D. and Jaenisch,R. (1981) Proc. Natl Acad. Sci. USA, 78, 7609–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh C.L. (1997) Mol. Cell. Biol., 17, 5897–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jost J.P. and Saluz,H.P. (1993) DNA Methylation: Molecular Biology and Biological Significance. Birkhäuser Verlag, Basel, Boston, Berlin.
- 4.Nan X., Ng,H.H., Johnson,C.A., Laherty,C.D., Turner,B.M., Eisenman,R.N. and Bird,A. (1998) Nature, 393, 386–389. [DOI] [PubMed] [Google Scholar]
- 5.Michalowsky L.A. and Jones,P.A. (1989) Environ. Health Perspect., 80, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li E., Beard,C., Forster,A.C., Bestor,T.H. and Jaenisch,R. (1993) Cold Spring Harb. Symp. Quant. Biol., 58, 297–305. [DOI] [PubMed] [Google Scholar]
- 7.Gold J.D. and Pedersen,R.A. (1994) Curr. Top. Dev. Biol., 29, 227–280. [DOI] [PubMed] [Google Scholar]
- 8.Constancia M., Pickard,B., Kelsey,G. and Reik,W. (1998) Genome Res., 8, 881–900. [DOI] [PubMed] [Google Scholar]
- 9.Monk M. (1992) J. Inherit. Metab. Dis., 15, 499–513. [DOI] [PubMed] [Google Scholar]
- 10.Riggs A.D. (1975) Cytogenet. Cell Genet., 14, 9–25. [DOI] [PubMed] [Google Scholar]
- 11.Dieguez M.J., Vaucheret,H., Paszkowski,J. and Mittelsten Scheid,O. (1998) Mol. Gen. Genet., 259, 207–215. [DOI] [PubMed] [Google Scholar]
- 12.Schulz W.A. (1998) Int. J. Oncol., 13, 151–167. [PubMed] [Google Scholar]
- 13.Jones P.A. (1986) Cancer Res., 46, 461–466. [PubMed] [Google Scholar]
- 14.Holliday R. (1987) Mutat. Res., 181, 215–217. [DOI] [PubMed] [Google Scholar]
- 15.Keshet E. and Cedar,H. (1983) Nucleic Acids Res., 11, 3571–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy P.H. and Smith,H.O. (1973) J. Mol. Biol., 81, 427–444. [DOI] [PubMed] [Google Scholar]
- 17.Meyer P., Niedenhof,I. and ten Lohuis,M. (1994) EMBO J., 13, 2084–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruenbaum Y., Naveh-Many,T., Cedar,H. and Razin,A. (1981) Nature, 292, 860–862. [DOI] [PubMed] [Google Scholar]
- 19.Bestor T.H. (1998) Novartis Found. Symp., 214, 187–195. [DOI] [PubMed] [Google Scholar]
- 20.Woodcock D.M., Lawler,C.B., Linsenmeyer,M.E., Doherty,J.P. and Warren,W.D. (1997) J. Biol. Chem., 272, 7810–7816. [DOI] [PubMed] [Google Scholar]
- 21.Yoder J.A., Walsh,C.P. and Bestor,T.H. (1997) Trends Genet., 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 22.Siegfried Z. and Cedar,H. (1997) Curr. Biol., 7, R305–R307. [DOI] [PubMed] [Google Scholar]
- 23.Razin A. and Cedar,H. (1991) Microbiol. Rev., 55, 451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cedar H. (1988) Cell, 53, 3–4. [DOI] [PubMed] [Google Scholar]
- 25.Waalwijk C. and Flavell,R.A. (1978) Nucleic Acids Res., 5, 3231–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wall L. (1998) perl—Practical Extraction and Report Language. 5th version. http://www.perl.com
- 28.Schneider T.D. and Stephens,R.M. (1990) Nucleic Acids Res., 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nomenclature Committee of the International Union of Biochemistry (NC-IUB)(1985) Eur. J. Biochem., 150, 1–5. [DOI] [PubMed] [Google Scholar]
- 30.Nomenclature Committee of the International Union of Biochemistry (NC-IUB) (1985) Biochem. J., 229, 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. http://www.csc.fi/molbio/progs/clustalw/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider T.D., Stormo,G.D., Gold,L. and Ehrenfeucht,A. (1986) J. Mol. Biol., 188, 415–431. [DOI] [PubMed] [Google Scholar]
- 33.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis. Cambridge University Press, Cambridge.
- 34.Shannon C.E. and Weaver,W. (1949) The Mathematical Theory Communication. University of Illinois Press, Urbana, IL.
- 35.Boutell T. (1998) http://www.boutell.com/gd/gd.html
- 36.Stein L.D. (1995) http://stein.cshl.org/WWW/software/GD/GD.html
- 37.Mache N. (1998) http://ipvr2.informatik.uni-stuttgart.de/GENIO/logo