Abstract
Background
Circular RNAs (circRNAs) have been reported to play vital roles in the progression of diverse human cancers, including non‐small cell lung cancer (NSCLC). The purpose of this study was to explore the exact role and underlying mechanism of circ_PLXND1 in NSCLC progression.
Methods
Quantitative real‐time polymerase chain reaction (qRT‐PCR) assay was performed to determine the expression levels of circ_PLXND1, microRNA (miR)‐1287‐5p and human epidermal growth factor receptor 3 (ERBB3). The localization of circ_PLXND1 in NSCLC cells was tested by subcellular fractionation and localization assay. Cell angiogenesis, proliferation, apoptosis, migration and invasion were evaluated by tube formation assay, 5‐ethynyl‐2′‐deoxyuridine (EdU) incorporation assay, 3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyltetrazolium bromide (MTT) assay, flow cytometry and transwell assay. Dual‐luciferase reporter assay was utilized to confirm the interaction between miR‐1287‐5p and circ_PLXND1 or ERBB3. Western blot assay was exploited to examine the expression of proteins.
Results
Circ_PLXND1 and ERBB3 were upregulated while miR‐1287‐5p was downregulated in NSCLC tissues and cells. Circ_PLXND1 was a stable circRNA and mainly located in cytoplasm. Circ_PLXND1 silencing suppressed the proliferation, angiogenesis, migration and invasion of NSCLC cells in vitro. For mechanism analysis, circ_PLXND1 could positively regulate ERBB3 expression via sponging miR‐1287‐5p. The inhibitory impacts of circ_PLXND1 knockdown on the malignant behaviors of NSCLC cells were overturned by miR‐1287‐5p inhibitor. Overexpression of miR‐1287‐5p repressed the malignant phenotypes of NSCLC cells by targeting ERBB3. Furthermore, circ_PLXND1 interference inhibited tumor growth in vivo.
Conclusions
Circ_PLXND1 knockdown impeded NSCLC progression through modulating the miR‐1287‐5p/ERBB3 axis, indicating a promising molecular target for NSCLC treatment.
Keywords: circ_PLXND1, ERBB3, miR‐1287‐5p, NSCLC
Circ_PLXND1 was overexpressed in NSCLC tissues and cells, and contributed to NSCLC cell angiogenesis, proliferation, migration, invasion and suppressed cell apoptosis via upregulating ERBB3 through sequestering miR‐1287‐5p.
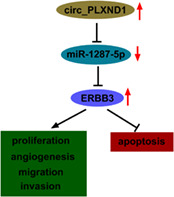
INTRODUCTION
Non‐small cell lung cancer (NSCLC), a major type of lung cancer, is a huge intimidation to human health with a dismal survival rate, leading to cancer‐associated morbidity and mortality. 1 , 2 Despite considerable progress having been made in treatment strategies for NSCLC, the prognosis of NSCLC patients remains unsatisfactory. 3 , 4 Therefore, there is a necessity to elucidate the pathogenic mechanism of NSCLC and find novel targeted curative strategies for NSCLC patients.
Circular RNAs (circRNAs) are a novel class of endogenous non‐coding RNAs (ncRNAs) with covalent closed loops. 5 , 6 Currently, diverse circRNAs have been demonstrated to be linked with the occurrence and advancement of diversiform tumors, including NSCLC. 7 For example, circ‐PITX1 expedited the tumorigenesis of NSCLC through the miR‐1248/CCND2 pathway. 8 Circ_0020123 exerted oncogenic property in NSCLC through the miR‐142‐3p/ZFX axis. 9 Moreover, circ_PLXND1 (hsa_circ_0067301), derived from the PLXND1 gene, is suggested to have elevated expression in NSCLC tissues through analyzing the GSE101684 data from the Gene Expression Omnibus database (GEO) public database. Nonetheless, the biological function and regulatory theory of circ_PLXND1 in NSCLC has not yet been explored.
MicroRNAs (miRNAs) are short ncRNAs that trigger translational repression and exert biological function in cancer via binding to the 3′ untranslated regions (3'UTRs) of targeted mRNAs. 10 , 11 , 12 Similarly, miRNAs have been reported to be novel biomarkers and promising targets for NSCLC treatment. 13 , 14 In addition, miR‐1287 has been shown to be involved in NSCLC cell proliferation and invasion via serving as a target of circ_0026134. 15 However, whether circ_PLXND1 might modulate NSCLC advancement by interacting with miR‐1287‐5p remains uninvestigated.
Human epidermal growth factor receptor 3 (ERBB3, HER3), a member of the ERBB receptor family, is relevant to the development and progression of cancers. 16 , 17 Furthermore, ERBB3 has been found to be upregulated in NSCLC thus promoting cancer development. 18 , 19 In this report, utilizing starBase v2.0, ERBB3 was forecasted to be targeted by miR‐1287‐5p. Thereby, further investigations on the role of ERBB3 in the functional effects mediated by miR‐1287‐5p on NSCLC progression were conducted in this study.
Herein, we examined the abundances of circ_PLXND1, miR‐1287‐5p and ERBB3 in NSCLC tissue samples and cells. In addition, the functional effects of circ_PLXND1 in NSCLC progression and the potential circ_PLXND1/miR‐1287‐5p/ERBB3 pathway were investigated.
METHODS
Tissue acquisition
A total of 45 pairs of NSCLC tissue samples and adjacent healthy tissues were collected from NSCLC patients who underwent surgery at Xi'an Hospital of Traditional Chinese Medicine. None of the subjects received other preoperative radiation treatment or chemical therapy. The study obtained approval from the Ethics Committee of Xi'an Hospital of Traditional Chinese Medicine. Before sample collection, written informed consent was also obtained from every patient.
Cell culture
The human normal bronchial epithelial cell line (16HBE) and NSCLC cell line H1299 were purchased from Procell. The NSCLC cell line PC9 was purchased from Cobioer Biosciences Co., Ltd. (Nanjing, China). Above cells were grown in RPMI‐1640 medium (Procell) plus 10% FBS (Procell) and 1% streptomycin/penicillin (P/S, Procell) in a moist atmosphere at 37°C with 5% CO2.
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
RNAiso Plus regent (Takara) was employed for total RNA separation. Then, cDNA was obtained through reverse transcription of RNA utilizing PrimeScript RT reagent kit (Takara) or miScript II RT kit (Qiagen), followed by qRT‐PCR assay on a PCR system exploiting a SYBR Premix Ex Taq II kit (Takara). The relative expression was computed via the 2−ΔΔCt method with glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) or U6 as inner contrast. The specific primers are shown in Table 1.
TABLE 1.
The list of primer sequences for PCR.
| Primers for PCR (5′‐3′) | ||
|---|---|---|
| circ_PLXND1 | Forward | CTCAGTGGTGGAAGACGG |
| Reverse | CAGGTCCCGAAGGATGTA | |
| PLXND1 | Forward | TGACCGTGAACGCCTCTAAGGA |
| Reverse | GAGCCTACATGGAGGTCATTCC | |
| miR‐1287‐5p | Forward | GCTGGATCAGTGGTTCG |
| Reverse | GAACATGTCTGCGTATCTC | |
| ERBB3 | Forward | CTATGAGGCGATACTTGGAACGG |
| Reverse | GCACAGTTCCAAAGACACCCGA | |
| GAPDH | Forward | GGAGCCAAAAGGGTCATC |
| Reverse | CCAGTGAGCTTCCCGTTC | |
| U6 | Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CGCTTCACGAATTTGCGTGTCAT | |
Subcellular fractionation and localization
The nuclear RNA and cytoplasmic fractions of H1299 and PC9 cells were isolated using the mirVana PARIS kit (Ambion). The proportion of circ_PLXND1 in the cytoplasm and nucleus was inspected via qRT‐PCR assay, with GAPDH and U6 as internal controls, respectively.
Actinomycin D assay
For the stability analysis of circ_PLXND1, transcription inhibitor actinomycin D (1 mg/mL; Abcam) was utilized to incubate with H1299 and PC9 cells for 0, 8, 16 or 24 h. The RNAs were isolated from H1299 and PC9 cells and then subjected to qRT‐PCR analysis for detecting the abundances of circ_PLXND1 and PLXND1.
Cell transfection
To silence circ_PLXND1, circ_PLXND1 small interference RNA (si‐circ_PLXND1) and short hairpin RNA (sh‐circ_PLXND1) were constructed by RiboBio, with si‐NC or sh‐NC as the negative control. For elevating or inhibiting miR‐1287‐5p level, miR‐1287‐5p mimic or inhibitor (miR‐1287‐5p or anti‐miR‐1287‐5p) and matched control (miR‐NC or anti‐miR‐NC) were also synthesized by RiboBio. For the acquisition of ERBB3 overexpression plasmid, ERBB3 full‐length sequence was introduced into pcDNA3.1 vector (pcDNA) (Invitrogen). H1299 and PC9 cells were transduced with oligonucleotides or vectors utilizing lipofectamine 3000 reagent (Invitrogen) referring to the specifications.
Tube formation assay
The serum‐free culture medium of assigned H1299 and PC9 cells was collected after centrifugation. Each well of the 96‐well plate was coated with 50 μL Matrigel (BD Biosciences) for 1 h. Human umbilical vein endothelial cells (HUVECs: Procell) were starved for 24 h and then seeded on the Matrigel gel with 200 μL collected culture medium plus 1% FBS. The tube formation of HUVECs was detected using a microscope.
5‐ethynyl‐2′‐deoxyuridine (EdU) incorporation assay
For assessing the capacity for DNA synthesis in proliferating cells, Cell‐Light EDU Apollo 567 In Vitro Imaging Kit (RiboBio) was utilized. Briefly, transfected H1299 and PC9 cells were cultivated for 48 h at 37°C in 96‐well plates, and subsequently immobilized using paraformaldehyde (4%; Solarbio) for 0.5 h and permeabilized by Triton X‐100 (0.5%; Solarbio) for 10 min in the indoor environment. Subsequently, cells were added with 10 μM EdU at 37°C for 2 h‐incubation, and 5 μg/mL DAPI solution (Solarbio) was utilized for the staining of cell nuclei for 10 min. A fluorescence microscope (100×; Olympus) was employed for counting the number of EdU‐positive cells in several randomly selected areas. Cell nuclei were stained blue, and EdU‐positive cells were stained red.
3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyltetrazolium bromide (MTT) assay
In brief, transfected H1299 and PC9 cells were seeded in 96‐well plates for the cultivation of varying time (1, 2, or 3 days). Thereafter, 20 μL MTT (5 mg/mL; Solarbio) was supplemented into the well for an additional 4 h‐incubation in the indoor environment. Later, cell supernatant was discarded and 150 μL DMSO (Solarbio) was utilized for melting the formazan crystals. Ultimately, a microplate reader was employed for the determination of absorbance at 570 nm.
Flow cytometry analysis
The detection of cell apoptosis was manipulated exploiting annexin V‐FITC Apoptosis Staining/Detection Kit (Abcam). After 48 h, transfected H1299 and PC9 cells were gathered and suspended in binding buffer (400 μL). Then, cells were dyed using annexin V‐FITC (10 μL) together with PI (5 μL) in darkness for 15 min. The apoptotic cells (annexin V‐FITC+ and PI+ or PI−) were identified and the apoptotic rate was analyzed by flow cytometry.
Transwell assay
Cell migratory and invasive capacities were respectively analyzed utilizing a transwell chamber (Corning Inc.) coated without and with Matrigel (Solarbio). Briefly, transfected H1299 and PC9 cells (1 × 104) were suspended in FBS‐free medium and then plated into the top compartment of the chamber. Meanwhile, 500 μL complete medium (plus FBS) was added to the bottom compartment. Following 24 h‐cultivation, migratory and invasive cells were fixed in paraformaldehyde (4%; Solarbio) for 0.5 h and dyed using 0.5% crystal violet (Solarbio). Then, a microscope (Olympus) was utilized for counting and photographing the migratory and invasive cells from five random areas at 100× magnification.
Dual‐luciferase reporter assay
The fragments of circ_PLXND1 or ERBB3 3'UTR possessing presumptive or mutant complementary sequence for miR‐1287‐5p were generated and introduced into pmirGLO vector (Promega) to construct circ_PLXND1‐wild type (WT), or ERBB3‐WT, circ_PLXND1‐mutant type (MUT) and ERBB3‐MUT luciferase reporter vectors, respectively. Afterwards, constructed plasmids and miR‐NC or miR‐1287‐5p were transfected into H1299 and PC9 cells using lipofectamine 3000.
Western blot assay
Total protein was segregated utilizing the RIPA lysis buffer (Solarbio). After quantification utilizing a BCA kit (Beyotime), protein (20 μg) was added on 10% SDS‐PAGE (Beyotime), and later transferred onto PVDF membrane (Solarbio). Thereafter, the membrane was sealed in 5% fat‐free milk for 1 h in the indoor environment. Afterward, the membrane was reacted with specific primary antibodies against proliferating cell nuclear antigen (PCNA) (ab152112, 1:2000, Abcam), twist family bHLH transcription factor 1 (Twist1) (ab175430, 1:1000, Abcam), E‐cadherin (ab133597; 1: 2000, Abcam), ERBB3 (ab131444, 1:1000, Abcam) or GAPDH (1:3000, ab8245, Abcam) at 4°C overnight, followed by incubation with secondary antibodies (1:3000, ab205718 or ab205719, Abcam) for 1 h in the indoor environment. Finally, the protein signals were examined via BeyoECL Plus kit (Beyotime).
Tumor xenograft assay
BALB/c nude mice were acquired from Vital River Laboratory Animal Co., Ltd (4–6 weeks old; Beijing, China) and randomly divided into two groups (n = 5/group). PC9 cells (2 × 106/0.2 mL PBS) stably introduced with sh‐NC or sh‐circ_PLXND1 were inoculated into the right flanks of the mice through subcutaneous injection. Then, 10 days after inoculation, tumor volume was detected every 5 days by the formula (length × width2)/2. At 30 days post inoculation, the mice were euthanized, and subcutaneous xenografts from the sacrificed mice were weighed and collected for further study. Permission was obtained from the Animal Care and Use Committee of XXXXXXXXX for the animal assay.
Immunohistochemistry (IHC) analysis
The paraffin‐embedded tissue sections from transplanted mice were divided into sections (4 μm) after dewaxing. After that, the sections were coreacted with primary antibodies against ERBB3 (ab131444, 1:50, Abcam) and Ki67 (1:200, ab16667, Abcam) at 4°C overnight, followed by incubation with secondary antibody (1:5000, ab205718, Abcam) for 1 h in the indoor environment. Afterwards, the sections were stained by 3, 3′‐diaminobenzidine solution (DAB; Beyotime), and the nuclei was counterstained with hematoxylin (Beyotime). Lastly, the sections were visualized using a microscope (Olympus).
Statistical analysis
The data from at least three duplicates are exhibited as mean ± standard deviation and were analyzed by Graphpad Prism 7 software. The differences were evaluated utilizing Student's t‐test or one‐way analysis of variance. The linear associations among circ_PLXND1, miR‐1287‐5p and ERBB3 in NSCLC tissues were assessed via Spearman's correlation coefficient analysis. p‐values < 0.05 were considered statistically significant.
RESULTS
Circ_PLXND1 was highly expressed in NSCLC tissues and cells
In this study, the public database (GSE101684) was used to inspect the expression change of circRNAs in NSCLC tumor tissues (T) versus adjacent normal tissues (N) (Figure 1a). Through analysis of large fold change (p < 0.05), we found a total of five differentially expressed circRNAs in NSCLC tumor tissues (Figure 1a,b). As shown in Figure 1c, circ_PLXND1 was the most upregulated circRNA among the above five differentially expressed circRNAs in NSCLC cells (H1299 and PC9) relative to that in normal 16HBE cells. Therefore, circ_PLXND1 was selected as the target for further investigation. The expression of circ_PLXND1 in 45 pairs of NSCLC tissues and neighboring normal tissues was detected by qRT‐PCR analysis. The results displayed that circ_PLXND1 expression was elevated in NSCLC tissues compared to that in neighboring normal tissues (Figure 1d). Next, the localization of circ_PLXND1 was determined in H1299 and PC9 cells through subcellular fractionation and localization assay. The qRT‐PCR data revealed that circ_PLXND1 was predominantly located in the cytoplasm (Figure 1e,f), implying its potential as the sponge of miRNAs. The characteristic of circ_PLXND1 in H1299 and PC9 cells was explored using transcription inhibitor actinomycin D. The result suggested that circ_PLXND1 was more stable than linear PLXND1 mRNA in H1299 and PC9 cells (Figure 1g,h), confirming that circ_PLXND1 was indeed a circRNA. These data indicated that circ_PLXND1 was a circular RNA and might be associated with the development of NSCLC.
FIGURE 1.
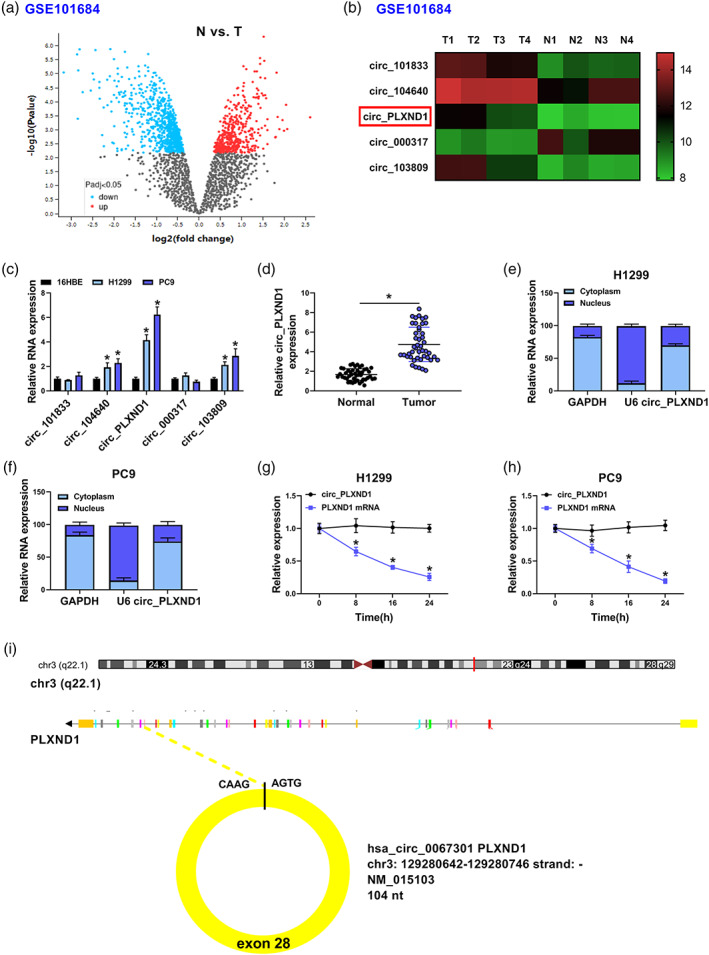
Circ_PLXND1 level was raised in non‐small cell lung cancer (NSCLC) tissues and cells. (a) Differentially expressed genes were identified via volcano plot analysis according to GSE101684 dataset. (b) The five most upregulated circRNAs according to GSE101684 dataset were shown. (c) The expression of above five upregulated circRNAs in NSCLC cells (H1299 and PC9) and normal 16HBE cells was determined by qRT‐PCR. (d) The expression of circ_PLXND1 in 45 pairs of NSCLC tissues and neighboring normal tissues was detected by qRT‐PCR. (e and f) The subcellular location of circ_PLXND1 in H1299 and PC9 cells was analyzed by qRT‐PCR analysis. (g and h) H1299 and PC9 cells were treated with actinomycin D at indicated time points, and qRT‐PCR assay was performed for the levels of circ_PLXND1 and PLXND1 mRNA. *p < 0.05.
Circ_PLXND1 knockdown suppressed cell angiogenesis, proliferation, migration, invasion and facilitated apoptosis in NSCLC cells
For investigating the precise role of circ_PLXND1 in the tumorigenesis of NSCLC, loss‐of‐function experiments were carried out through transfecting si‐circ_PLXND1 or si‐NC into H1299 and PC9 cells. The transfection of si‐circ_PLXND1 in H1299 and PC9 cells markedly decreased the level of circ_PLXND1 but not linear PLXND1 mRNA, indicating the successful transfection of si‐circ_PLXND1 (Figure 2a,b). Tube formation assay exhibited that the tube formation numbers of HUVECs were distinctly inhibited by circ_PLXND1 deficiency (Figure 2c). EdU assay showed that the downregulation of circ_PLXND1 drastically reduced the numbers of EdU‐positive cells in H1299 and PC9 cells (Figure 2d). MTT assay manifested that circ_PLXND1 silencing led to an obvious suppression in cell viability in H1299 and PC9 cells (Figure 2e,f). Meanwhile, the expression of proliferation‐related maker PCNA was markedly repressed by the introduction of si‐circ_PLXND1 (Figure 2g). All these experimental results confirmed that circ_PLXND1 knockdown impeded the proliferation of H1299 and PC9 cells. Furthermore, flow cytometry analysis demonstrated that circ_PLXND1 interference facilitated the apoptosis of H1299 and PC9 cells (Figure 2h). Transwell assay proved that circ_PLXND1 knockdown restrained cell migration and invasion capacities in H1299 and PC9 cells (Figure 2I,J). Additionally, the impact of circ_PLXND1 knockdown on metastasis was analyzed by detecting the levels of metastasis‐related proteins. As a result, circ_PLXND1 silencing obviously decreased the protein level of mesenchymal marker Twist1 and increased the protein level of epithelial marker E‐cadherin in H1299 and PC9 cells (Figure 2k,l). To summarize, circ_PLXND1 knockdown suppressed the malignant behaviors of NSCLC cells.
FIGURE 2.
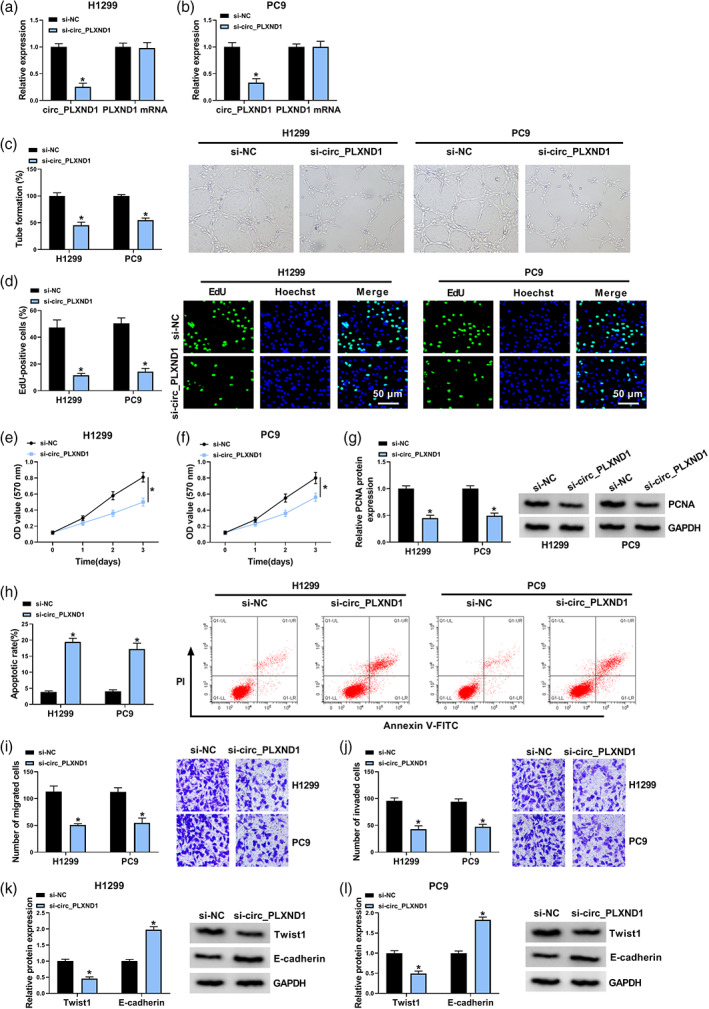
Circ_PLXND1 knockdown hampered cell angiogenesis, proliferation, migration, invasion and enhanced apoptosis in NSCLC cells. H1299 and PC9 cells were transfected with si‐circ_PLXND1 or si‐NC. (a and b) QRT‐PCR assay was conducted for the expression of circ_PLXND1 and linear PLXND1 mRNA in H1299 and PC9 cells. (c) Tube formation assay was performed to measure the number of formed tubes. (d) The number of EdU‐positive cells in H1299 and PC9 cells was assessed by EdU assay. (e and f) Cell viability was evaluated by MTT assay. (g) The expression of proliferation‐related maker PCNA was tested by western blot assay. (h) Flow cytometry analysis was employed to determine cell apoptosis rate. (i and j) Transwell assay was utilized to estimate the number of migration and invasion cells (×100). (k and l) Western blot assay was carried out for the protein levels of Twist1 and E‐cadherin in H1299 and PC9 cells. *p < 0.05.
Circ_PLXND1 acted as a sponge of miR‐1287‐5p
To trace out the action mechanism by which circ_PLXND1 affected the development of NSCLC cells, the target miRNAs of circ_PLXND1 were estimated using prediction software Circular RNA interactome (https://circinteractome.nia.nih.gov/). miR‐1287‐5p was also identified to bind with circ_PLXND1 through complementary sites (Figure 3a). Next, the relationship between circ_PLXND1 and miR‐1287‐5p was further confirmed by dual‐luciferase reporter assay. The data manifested that miR‐1287‐5p upregulation prominently reduced the luciferase activity of circ_PLXND1‐WT (more than 50%) in H1299 and PC9 cells, while there was no alteration in the luciferase activity of circ_PLXND1‐MUT (Figure 3b,c). Also, circ_PLXND1 silencing obviously elevated the expression of miR‐1287‐5p in H1299 and PC9 cells (Figure 3d). In addition, miR‐1287‐5p expression was decreased in NSCLC cells (H1299 and PC9) and tumor tissues compared to that in normal 16HBE cells and neighboring non‐tumor tissues (Figure 3e,f). In addition, Spearman's correlation analysis demonstrated that the expression of miR‐1287‐5p was negatively correlated with circ_PLXND1 expression in NSCLC tumor tissues (Figure 3g). Collectively, miR‐1287‐5p was a direct target of circ_PLXND1 and negatively regulated by circ_PLXND1 in NSCLC cells.
FIGURE 3.
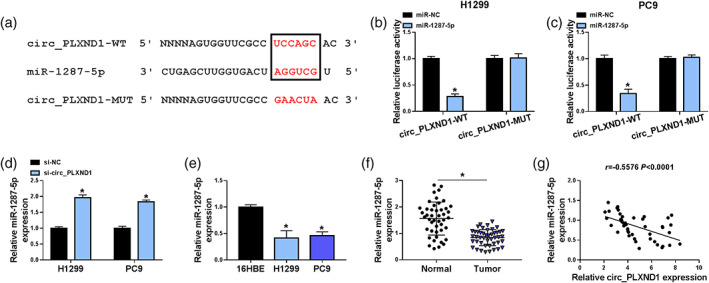
Circ_PLXND1 sponged miR‐1287‐5p to negatively regulate miR‐1287‐5p expression. (a) The binding sites between circ_PLXND1 and miR‐1287‐5p were presented. (b and c) The luciferase activity in H1299 and PC9 cells cotransfected with miR‐1287‐5p or miR‐NC and circ_PLXND1‐WT or circ_PLXND1‐MUT was determined by dual‐luciferase reporter assay. (d) The expression of miR‐1287‐5p in H1299 and PC9 cells transfected with si‐NC or si‐circ_PLXND1 was analyzed by qRT‐PCR. (e) The abundance of miR‐1287‐5p in NSCLC cells (H1299 and PC9) and normal 16HBE cells was detected by qRT‐PCR. (f) The expression of miR‐1287‐5p in 45 pairs of NSCLC tissues and neighboring normal tissues was examined by qRT‐PCR. (g) The correlation between miR‐1287‐5p and circ_PLXND1 expression in non‐small cell lung cancer (NSCLC) tissues was analyzed by spearman's correlation coefficient analysis. *p < 0.05.
MiR‐1287‐5p inhibitor alleviated the inhibitory effects of circ_PLXND1 knockdown on NSCLC progression
Subsequently, rescue experiments were carried out to confirm whether circ_PLXND1 regulated NSCLC progression via targeting miR‐1287‐5p. H1299 and PC9 cells were transfected with si‐NC, si‐circ_PLXND1, si‐circ_PLXND1 + anti‐miR‐NC or si‐circ_PLXND1 + anti‐miR‐1287‐5p. As exhibited in Figure 4a, circ_PLXND1 interference overtly enhanced the expression of miR‐1287‐5p in H1299 and PC9 cells, whereas the transfection of anti‐miR‐1287‐5p partly relieved the effect, showing that the transfection was effective. Furthermore, miR‐1287‐5p inhibition reversed the inhibition on the tube formation ability of HUVECs induced by si‐circ_PLXND1 (Figure 4b). The suppressive effect of circ_PLXND1 knockdown on the proliferation of H1299 and PC9 cells was effectively reversed by the downregulation of miR‐1287‐5p, reflected by a decrease in the number of EdU‐positive cells (Figure 4c), arrest in cell viability (Figure 4d,e) and reduced PCNA protein expression (Figure 4f). Moreover, si‐circ_PLXND1‐mediated proapoptosis (Figure 4g), antimigration (Figure 4h) and anti‐invasion (Figure 4i) effects were overturned by the silencing of miR‐1287‐5p. In addition, the effects of circ_PLXND1 knockdown on Twist1 and E‐cadherin levels in H1299 and PC9 cells were also reversed by miR‐1287‐5p suppression (Figure 4j,k). In summary, circ_PLXND1 deficiency repressed the malignancy of NSCLC cells via sponging miR‐1287‐5p.
FIGURE 4.
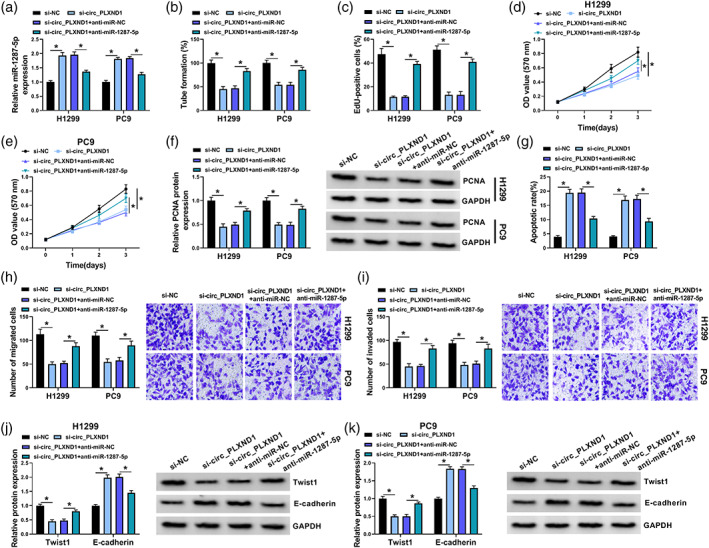
Knockdown of circ_PLXND1 suppressed non‐small cell lung cancer (NSCLC) cell development through interacting with miR‐1287‐5p. H1299 and PC9 cells were transfected with si‐NC, si‐circ_PLXND1, si‐circ_PLXND1 + anti‐miR‐NC or si‐circ_PLXND1 + anti‐miR‐1287‐5p. (a) The level of miR‐1287‐5p in H1299 and PC9 cells was examined by qRT‐PCR assay. (b) Tube formation assay was performed to measure the number of formed tubes. (c–e) The proliferation of H1299 and PC9 cells were analyzed by EdU assay (c) and MTT assay (d and e). (f) The protein level of PCNA in H1299 and PC9 cells was measured via Western blot assay. (g–i) The apoptosis (g), migration and invasion (h and i) of H1299 and PC9 cells were evaluated by flow cytometry analysis and transwell assay, respectively. (g and k) The protein levels of Twist1 and E‐cadherin in H1299 and PC9 cells were measured using western blot assay. *p < 0.05.
Circ_PLXND1 could regulate ERBB3 expression through competitively binding to miR‐1287‐5p
For deeper understanding of miR‐1287‐5p role in the progression of NSCLC cells, its molecular targets were searched utilizing starBase v2.0 (http://starbase.sysu.edu.cn/). Prediction results showed that ERBB3 3'UTR had a complementary binding position with miR‐1287‐5p (Figure 5a). Dual‐luciferase reporter assay revealed that elevated miR‐1287‐5p expression reduced the luciferase activity of ERBB3‐WT luciferase reporter in H1299 and PC9 cells, but there was no overt difference in the luciferase activity of ERBB3‐MUT reporter (Figure 5b,c). Next, miR‐1287‐5p or anti‐miR‐1287‐5p was successfully transfected into H1299 and PC9 cells to determine the effect of miR‐1287‐5p on ERBB3 expression, and the overexpression or knockdown efficiency was evaluated by qRT‐PCR assay (Figure 5d). Expectedly, miR‐1287‐5p overexpression visibly decreased the mRNA and protein levels of ERBB3 in H1299 and PC9 cells, while miR‐1287‐5p inhibition exhibited the opposite results (Figure 5e,f). Moreover, it was observed that the mRNA and protein levels of ERBB3 were strikingly enhanced in NSCLC tissues relative to neighboring normal tissues (Figure 5g,h). Furthermore, the expression of ERBB3 mRNA in NSCLC tissues was negatively correlated with miR‐1287‐5p expression (Figure 5i), and positively correlated with circ_PLXND1 expression (Figure 5j). Likewise, the mRNA and protein levels of ERBB3 in H1299 and PC9 cells were higher than in normal 16HBE cells (Figure 5k,l). Additionally, downregulation of circ_0043278 markedly reduced the mRNA and protein levels of ERBB3 in H1299 and PC9 cells, while this trend was effectively ameliorated by miR‐1287‐5p inhibition (Figure 5m,n). Together, these findings manifested that circ_PLXND1 positively regulated ERBB3 expression via sponging miR‐1287‐5p in NSCLC cells.
FIGURE 5.
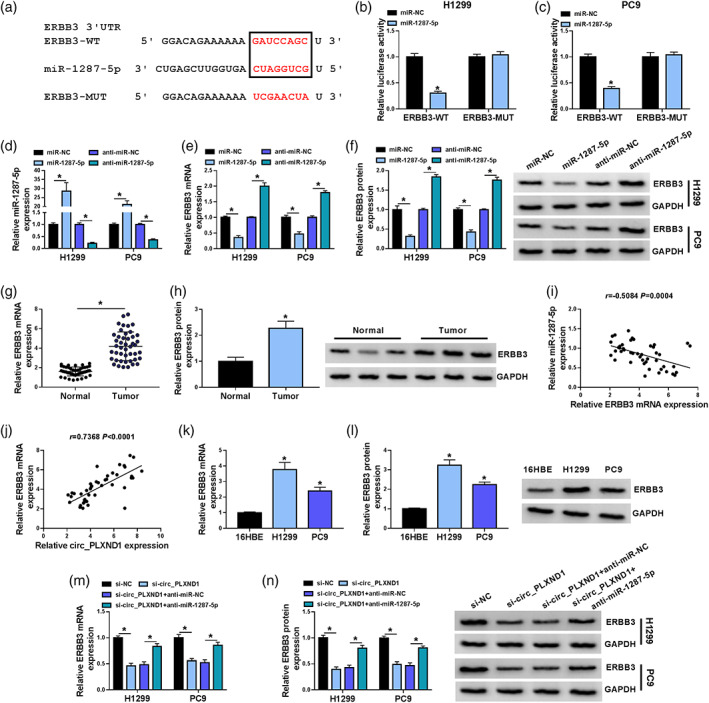
Circ_PLXND1 could regulate ERBB3 expression through targeting miR‐1287‐5p. (a) The putative binding sites between miR‐1287‐5p and ERBB3 3'UTR were predicted by starBase v2.0. (b and c) Dual‐luciferase reporter assay was conducted to verify the interaction between miR‐1287‐5p and ERBB3. (d–f) The expression of miR‐1287‐5p, ERBB3 mRNA and ERBB3 protein in H1299 and PC9 cells transfected with miR‐NC, miR‐1287‐5p, anti‐miR‐NC or anti‐miR‐1287‐5p was measured by qRT‐PCR assay or western blot assay. (g and h) The mRNA and protein levels of ERBB3 in NSCLC tissues and normal tissues were determined by qRT‐PCR assay and western blot assay, respectively. (i and j) The correlation between the level of ERBB3 mRNA and miR‐1287‐5p or circ_PLXND1 was analyzed by Spearman's correlation coefficient analysis. (k and l) The mRNA and protein levels of ERBB3 in 16HBE, H1299 and PC9 cells were examined by qRT‐PCR assay and western blot assay, respectively. (m and n) The mRNA and protein levels of ERBB3 in H1299 and PC9 cells transfected with si‐NC, si‐circ_PLXND1, si‐circ_PLXND1 + anti‐miR‐NC or si‐circ_PLXND1 + anti‐miR‐1287‐5p were tested by qRT‐PCR assay and western blot assay, respectively. *p < 0.05.
MiR‐1287‐5p overexpression‐mediated suppressed influences in the progression of NSCLC cells were partly overturned by ERBB3 accumulation
To further investigate the effects of miR‐1287‐5p and ERBB3 on NSCLC cell progression, H1299 and PC9 cells were transfected with miR‐NC, miR‐1287‐5p, miR‐1287‐5p + pcDNA or miR‐1287‐5p + ERBB3. As shown in Figure 6a,b, miR‐1287‐5p transfection resulted in a remarkable reduction in ERBB3 mRNA and protein enrichments in H1299 and PC9 cells, while the impact was largely rescued by ERBB3 addition. Moreover, the suppressive effects of miR‐1287‐5p upregulation on cell angiogenesis (Figure 6c), proliferation (Figure 6d–f), the expression of PCNA (Figure 6g), migration (Figure 6i) and invasion (Figure 6j) and promoting effect of miR‐1287‐5p upregulation on cell apoptosis (Figure 6h) were all abolished by the overexpression of ERBB3. In addition, western blot assay indicated that miR‐1287‐5p overexpression suppressed the protein expression of Twist1 and increased the expression of E‐cadherin, whereas the effects were restored by introduction of ERBB3 (Figure 6k,l). Thus, it was deduced that miR‐1287‐5p overexpression suppressed the progression of NSCLC cells by targeting ERBB3.
FIGURE 6.
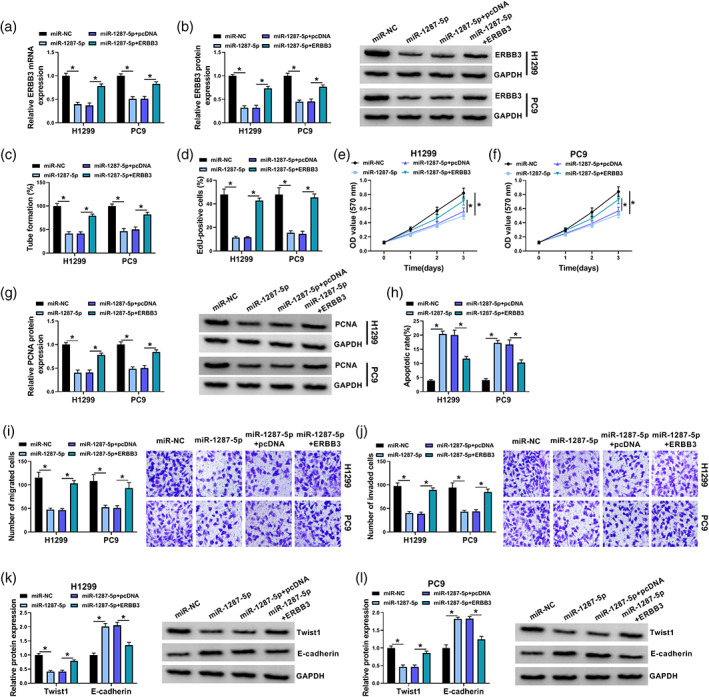
MiR‐1287‐5p inhibited the progression of NSCLC cells by targeting ERBB3. H1299 and PC9 cells were transfected with miR‐NC, miR‐1287‐5p, miR‐1287‐5p + pcDNA or miR‐1287‐5p + ERBB3. (a and b) The mRNA and protein expression of ERBB3 was measured by qRT‐PCR assay and western blot assay, respectively. (c) Tube formation assay was performed to measure the number of formed tubes. (d–f) EdU assay and MTT assay were applied to determine cell proliferation. (g) The level of PCNA in H1299 and PC9 cells was tested via western blot assay. (h) Cell apoptosis was evaluated by flow cytometry analysis. (i and j) Transwell assay was used to determine cell migration and invasion. (k and l) The protein expression of Twist1 and E‐cadherin in H1299 and PC9 cells was assessed by western blot assay. *p < 0.05.
Knockdown of circ_PLXND1 suppressed tumor growth in vivo
To clarify the function of circ_PLXND1 in tumor progression in vivo, PC9 cells (2 × 106/0.2 mL PBS) stably transfected with sh‐circ_PLXND1 or sh‐NC were injected into the nude mice. As illustrated in Figure 7a,b, the tumor volume and weight in the mice with circ_PLXND1 knockdown were markedly decreased compared to the sh‐NC group. Furthermore, the levels of circ_PLXND1, miR‐1287‐5p and ERBB3 in the harvested tumors were examined. The results showed that circ_PLXND1, as well as ERBB3 mRNA and protein levels were decreased, and miR‐1287‐5p level was enhanced in the tumors harvested from sh‐circ_PLXND1 group relative to sh‐NC group (Figure 7c–f). Moreover, IHC assay further confirmed the downregulated expression of ERBB3 and Ki67 (proliferation marker) in sh‐circ_PLXND1 group (Figure 7g). Hence, these results demonstrated that circ_PLXND1 knockdown suppressed the growth of NSCLC in vivo by upregulating miR‐1287‐5p and downregulating ERBB3.
FIGURE 7.
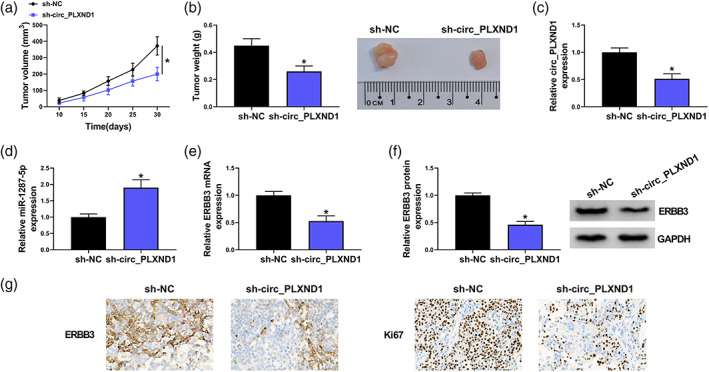
Knockdown of circ_PLXND1 restrained tumor growth in vivo. PC9 cells transfected with sh‐circ_PLXND1 or sh‐NC were injected subcutaneously into the nude mice. (a) Xenograft tumor volume was monitored every 5 days. (b) Xenograft tumor weight was examined on day 30. (c–e) The expression levels of circ_PLXND1, miR‐1287‐5p and ERBB3 mRNA in the collected tumor tissues were detected by qRT‐PCR assay. (f) Western blot assay was carried out to examine the protein expression of ERBB3 in harvested tumor tissues. (g) ERBB3‐positive and Ki67‐positive cells in tumor tissues were detected by immunohistochemistry (IHC) analysis. *p < 0.05.
DISCUSSION
There is currently growing evidence that circRNAs are intimately linked to the tumorigenesis and development of many tumors. 20 However, the role of circ_PLXND1 and underlying mechanism in NSCLC progression have not been explored. Herein, the role of circ_PLXND1 and a newfound circ_PLXND1/miR‐1287‐5p/ERBB3 network was exhibited in NSCLC.
Studies have manifested that various dysregulated circRNAs could act as crucial modulators during tumor progression. 21 , 22 Likewise, many key differentially expressed circRNAs in NSCLC have been identified, 23 and multiple circRNAs have been demonstrated to serve vital roles in the carcinogenesis of NSCLC. 7 For example, Zhang et al. unraveled that circDENND2A might act as a tumor promoter in NSCLC via miR‐34a/CCNE1 axis. 24 Cheng et al. suggested that CircSEC31A sponged miR‐376a to accelerate the malignant development of NSCLC by upregulating SEC31A. 25 Moreover, Zhang et al. pointed out that circ_PLXND1 was related to the epithelial‐mesenchymal transition process in endometriosis via the miR‐141‐5p/Notch‐1 axis. 26 However, the function of circ_PLXND1 in NSCLC progression remains unknown. Through the analysis of GSE101684 dataset, five differentially expressed circRNAs in NSCLC tumor tissues were identified, including circ_PLXND1. Also, in this study, circ_PLXND1 was verified to be most upregulated in NSCLC cells relative to that in normal 16HBE cells among the five dysregulated circRNAs. Consistent with the result of GSE101684 dataset, here, circ_PLXND1 abundance was also observed to be enhanced in NSCLC. Furthermore, the loss‐of‐function experiments showed that circ_PLXND1 deficiency caused obvious repressions in the growth and metastasis and an enhancement in apoptosis of NSCLC cells. These results illustrate that circ_PLXND1 might exert an oncogenic role in NSCLC progression.
It is widely accepted that circRNAs exert their biological functions through competitively binding to miRNAs and relieving the repressive effects of miRNAs on their targeted genes. 6 , 27 , 28 To explore whether circ_PLXND1 regulated NSCLC progression via binding to miRNAs, the potential targets of circ_PLXND1 were forecasted by circular RNA interactome. The results suggested that miR‐1287‐5p was sponged by circ_PLXND1. MiR‐1287‐5p has been reported to exert tumor‐inhibiting function to participate in the development of diversiform malignancies, including pancreatic carcinoma, 29 colorectal cancer 30 and cervical cancer. 31 Likewise, Li et al. uncovered that miR‐1287 was downregulated in NSCLC and concerned with the tumorigenesis of NSCLC through being targeted by circ_0016760. 32 Conformably, we attested the reduced level of miR‐1287‐5p in NSCLC, and the inverse relevance of miR‐1287‐5p and circ_PLXND1 expression in NSCLC tissues. Further investigation disclosed that miR‐1287‐5p introduction distinctly inhibited the development of NSCLC cells. More importantly, rescue experiments exhibited that miR‐1287‐5p inhibition relieved the impeditive impacts of circ_PLXND1 silencing in the vicious phenotypes of NSCLC cells. Therefore, we speculated that circ_PLXND1 could modulate the progression of NSCLC cells by interacting with miR‐1287‐5p.
MiRNAs have been expounded to take part in multifarious physiological and pathological processes by modulating the expression of targeted mRNAs. 33 Herein, ERBB3 was evidenced as a candidate target of miR‐1287‐5p. Research has shown that ERBB3 was upregulated in NSCLC and connected with the poor outcome of NSCLC patients. 18 , 34 In keeping with these reports, this study also proved the elevated level of ERBB3 in NSCLC tissues and cells. Meanwhile, ERBB3 expression was reversely related to miR‐1287‐5p expression and positively linked to circ_PLXND1 expression. Furthermore, we attested the positive regulation of circ_PLXND1 on ERBB3 expression via downregulating miR‐1287‐5p. Functionally, enforced expression of ERBB3 attenuated the antitumor role of miR‐1287‐5p in NSCLC cells, implying that miR‐1287‐5p could suppress the development of NSCLC cells by modulating ERBB3. For in vivo experiments, circ_PLXND1 downregulation blocked tumor growth via upregulating miR‐1287‐5p and downregulating ERBB3. Overall, circ_PLXND1 silencing inhibited the progression of NSCLC by elevating ERBB3 via miR‐1287‐5p.
In summary, our results revealed that circ_PLXND1 was overexpressed in NSCLC, and circ_PLXND1 knockdown efficiently suppressed the malignancy of NSCLC through modulating miR‐1287‐5p/ERBB3 axis. This study therefore offers new insights into the action pattern of circ_PLXND1 in NSCLC advancement and indicates that circ_PLXND1 might be a promising therapeutic target for NSCLC patients.
AUTHOR CONTRIBUTIONS
Jinzhou Wu designed and performed the research; Chenyang Liu and Guiping Yu analyzed the data; Jinzhou Wu wrote the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Wu J, Liu C, Yu G. Downregulation of circ_PLXND1 inhibits tumorigenesis of non‐small cell lung carcinoma via miR‐1287‐5p/ERBB3 axis. Thorac Cancer. 2023;14(17):1543–1555. 10.1111/1759-7714.14897
REFERENCES
- 1. Camps C, del Pozo N, Blasco A, Blasco P, Sirera R. Importance of quality of life in patients with non‐small‐cell lung cancer. Clin Lung Cancer. 2009;10:83–90. [DOI] [PubMed] [Google Scholar]
- 2. Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non‐small‐cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagano T, Tachihara M, Nishimura Y. Molecular mechanisms and targeted therapies including immunotherapy for non‐small cell lung cancer. Curr Cancer Drug Targets. 2019;19:595–630. [DOI] [PubMed] [Google Scholar]
- 4. Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non‐small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. [DOI] [PubMed] [Google Scholar]
- 5. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. [DOI] [PubMed] [Google Scholar]
- 6. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 7. Li S, Liu Y, Qiu G, Luo Y, Li X, Meng F, et al. Emerging roles of circular RNAs in nonsmall cell lung cancer (review). Oncol Rep. 2021;45:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yue Q, Xu Y, Deng X, Wang S, Qiu J, Qian B, et al. Circ‐PITX1 promotes the progression of non‐small cell lung cancer through regulating the miR‐1248/CCND2 Axis. Onco Targets Ther. 2021;14:1807–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu J, Ma X, Lin J, Hou P. Circ_0020123 increases ZFX expression to facilitate non‐small cell lung cancer progression by sponging miR‐142‐3p. Cancer Manag Res. 2021;13:1687–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng B, Zhang K, Wang R, Chen L. Non‐small‐cell lung cancer and miRNAs: novel biomarkers and promising tools for treatment. Clin Sci (Lond). 2015;128:619–34. [DOI] [PubMed] [Google Scholar]
- 14. Florczuk M, Szpechcinski A, Chorostowska‐Wynimko J. miRNAs as biomarkers and therapeutic targets in non‐small cell lung cancer: current perspectives. Target Oncol. 2017;12:179–200. [DOI] [PubMed] [Google Scholar]
- 15. Chang H, Qu J, Wang J, Liang X, Sun W. Circular RNA circ_0026134 regulates non‐small cell lung cancer cell proliferation and invasion via sponging miR‐1256 and miR‐1287. Biomed Pharmacother. 2019;112:108743. [DOI] [PubMed] [Google Scholar]
- 16. Sithanandam G, Anderson LM. The ERBB3 receptor in cancer and cancer gene therapy. Cancer Gene Ther. 2008;15:413–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hafeez U, Parslow AC, Gan HK, Scott AM. New insights into ErbB3 function and therapeutic targeting in cancer. Expert Rev Anticancer Ther. 2020;20:1057–74. [DOI] [PubMed] [Google Scholar]
- 18. Wang A, Zhang H, Wang J, Zhang S, Xu Z. MiR‐519d targets HER3 and can be used as a potential serum biomarker for non‐small cell lung cancer. Aging (Albany NY). 2020;12:4866–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scharpenseel H, Hanssen A, Loges S, Mohme M, Bernreuther C, Peine S, et al. EGFR and HER3 expression in circulating tumor cells and tumor tissue from non‐small cell lung cancer patients. Sci Rep. 2019;9:7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arnaiz E, Sole C, Manterola L, Iparraguirre L, Otaegui D, Lawrie CH. CircRNAs and cancer: biomarkers and master regulators. Semin Cancer Biol. 2019;58:90–9. [DOI] [PubMed] [Google Scholar]
- 21. Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J. 2021;19:910–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li L, Sun D, Li X, Yang B, Zhang W. Identification of key circRNAs in non‐small cell lung cancer. Am J Med Sci. 2021;361:98–105. [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y, Shan C, Chen Y, Sun S, Liu D, Zhang X, et al. CircDENND2A promotes non‐small cell lung cancer progression via regulating MiR‐34a/CCNE1 signaling. Front Genet. 2020;11:987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng F, Yu J, Zhang X, Dai Z, Fang A. CircSEC31A promotes the malignant progression of non‐small cell lung cancer through regulating SEC31A expression via sponging miR‐376a. Cancer Manag Res. 2020;12:11527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang M, Wang S, Tang L, Wang X, Zhang T, Xia X, et al. Downregulated circular RNA hsa_circ_0067301 regulates epithelial‐mesenchymal transition in endometriosis via the miR‐141/notch signaling pathway. Biochem Biophys Res Commun. 2019;514:71–7. [DOI] [PubMed] [Google Scholar]
- 27. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 28. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- 29. Zhang X, Xue C, Cui X, Zhou Z, Fu Y, Yin X, et al. Circ_0075829 facilitates the progression of pancreatic carcinoma by sponging miR‐1287‐5p and activating LAMTOR3 signalling. J Cell Mol Med. 2020;24:14596–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cui G, Zhao H, Li L. Long noncoding RNA PRKCQ‐AS1 promotes CRC cell proliferation and migration via modulating miR‐1287‐5p/YBX1 axis. J Cell Biochem. 2020;121:4166–75. [DOI] [PubMed] [Google Scholar]
- 31. Ji F, Du R, Chen T, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR‐1287‐5p/HOXA7 Axis. Mol Ther Nucleic Acids. 2020;19:413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X, et al. Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR‐1287/GAGE1 axis. Biochem Biophys Res Commun. 2018;503:2089–94. [DOI] [PubMed] [Google Scholar]
- 33. Felekkis K, Touvana E, Stefanou C, et al. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–40. [PMC free article] [PubMed] [Google Scholar]
- 34. Siegfried JM, Lin Y, Diergaarde B, Lin HM, Dacic S, Pennathur A, et al. Expression of PAM50 genes in lung cancer: evidence that interactions between hormone receptors and HER2/HER3 contribute to poor outcome. Neoplasia. 2015;17:817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]


