Abstract
Background
Circular RNAs (circRNAs) are associated with propofol‐mediated inhibitory effect on non‐small cell lung cancer (NSCLC) progression. Circular hsa_circ_0003028 (circ_0003028) exerts a tumor‐promoting role in NSCLC. However, it is unclear whether propofol can mediate NSCLC progression via regulating circ_0003028 expression.
Methods
A total of 36 NSCLC patients were recruited in the study. Cell viability, proliferation, apoptosis, migration, and invasion were determined by 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT), colony formation, flow cytometry, and transwell assays. Relative expression of circ_0003028 in NSCLC samples and cells was detected by quantitative real‐time polymerase chain reaction (RT‐qPCR). Analysis of the latent binding of circ_0003028 to miR‐1305 was done by bioinformatic analysis and confirmed by luciferase reporter and RNA immunoprecipitation (RIP) assays. Xenografting in mice was done to verify the relationship between propofol and circ_0003028.
Results
Significant upregulation of circ_0003028 was detected in NSCLC samples and cells. Functionally, propofol treatment reduced circ_0003028 expression in NSCLC cells, and circ_0003028 overexpression impaired propofol‐mediated inhibitory effect on NSCLC cell proliferation, migration, and invasion. Interestingly, circ_0003028 could compete with miR‐1305 as a competing endogenous RNA and upregulate CORO1C expression in NSCLC cells.
Conclusion
Propofol‐mediated inhibiting effect on NSCLC growth partly depended on the circ_0003028/miR‐1305/CORO1C axis.
Keywords: circ_0003028, CORO1C, miR‐1305, NSCLC, propofol
Propofol‐mediated inhibiting effect on NSCLC progression partly depended on the circ_0003028/miR‐1305/CORO1C axis.
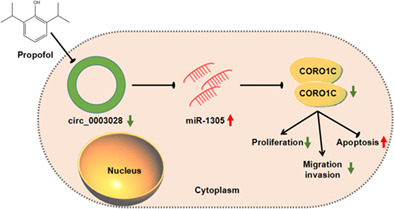
INTRODUCTION
Propofol (2,6‐diisopropyl phenol), an intravenous anesthetic with fast onset, strong action, and quick recovery, is widely used in clinical applications for induction and maintenance of anesthesia and sedation of critically ill patients in ICU. 1 In addition to its advantages in anesthesia, propofol also has a variety of non‐narcotic effects, such as analgesia, immune regulation, antiemetic, antioxidant, and neuroprotective effects. 2 , 3 Studies have shown that propofol can suppress host immunity to a lower degree compared with volatile anesthetics. 4 Accumulated evidence shows that the prognosis of cancer is related to the use of propofol during the perioperative period. 5 , 6 , 7 , 8 At present, the mechanism by which propofol exerts its antitumor activity is still unclear.
Non‐small cell lung cancer (NSCLC), which is an aggressive disease, accounts for more than 85% of all lung cancers. 9 Surgical resection is the main treatment for potentially curable NSCLC, but patients also experience postoperative recurrence. 10 , 11 Existing evidence shows that propofol anesthesia can reduce perioperative inflammation in patients undergoing lung cancer resection, protect patients' lung function, and shorten postoperative recovery time. 7 Moreover, propofol‐based intravenous anesthesia for patients with stage I NSCLC has a better prognosis than inhalation anesthesia. 12 At present, the mechanism by which propofol inhibits NSCLC needs to be further elucidated.
Circular RNAs (circRNAs), a new class of functional molecules, have a circular structure formed by typical 5′ to 3′‐phosphodiester bonds. This unique circular structure does not have 5′ or 3′ free ends, so it is not easily affected by exonuclease. 13 Because of their high stability, circRNAs are regarded as potential biomarkers in human diseases, including cancer. 14 Growing reports have highlighted the roles of circRNAs in NSCLC. For instance, circ_0084003, 15 circ‐SATB2, 16 and circ‐CPA4 17 are carcinogenic RNAs in NSCLC, while circ_0008305 18 and circ_0007518 19 exert an antitumor role in NSCLC. Recent evidence shows that the inhibitory effect of propofol on NSCLC may depend on the regulatory mechanism mediated by circRNAs. 20 , 21 One example is that propofol reduces lung cancer progression by downregulating circTADA2A. 20 Hsa_circ_0003028 (circ_0003028) has been reported as an oncogene in NSCLC. 22 However, whether propofol can restrain NSCLC progression via mediating circ_0003028 expression is unclear.
Some circRNAs located in the cytoplasm become important because they can act as decoys to regulate the expression of genes involved in cell function. 23 By sponging microRNAs (miRNAs), circRNAs can immobilize and inactivate their activity, thereby changing the stability of miRNA‐regulated linear mRNAs. 24 For instance, circ‐FLI1 drove NSCLC progression via upregulating ROCK1 by interacting with miR‐584. 25 Based on this function, we found that circ_0003028 may be a potential sponge for miR‐1305 through bioinformatic prediction. miR‐1305 expression is universally reduced in tumor samples, such as bladder cancer, 26 NSCLC, 27 cervical cancer, 28 and esophageal cancer. 29 Nevertheless, whether circ_0003028 can interact with miR‐1305 in NSCLC is unclear.
Coronin‐like actin‐binding protein 1C (CORO1C, also termed coronin 3) is one of the coronin family, which is a conserved family of actin cytoskeleton regulators that facilitate cell motility and regulate other actin‐dependent processes. 30 CORO1C is singularly upregulated in many tumors and has been shown to play a promoting effect on cancer cell migration. 31 , 32 , 33 In this context, CORO1C was predicted as a possible target of miR‐1305. However, the regulatory mechanisms between them are unclear.
In this study, we hypothesized that propofol treatment lessened NSCLC progression by the circ_0003028/miR‐1305/CORO1C axis. The study may provide novel insights into the underlying mechanism of propofol‐mediated NSCLC.
METHODS
Cell culture
16HBE (CL‐0249, Procell), H1299 (CL‐0165, Procell) and A549 (CL‐0016, Procell) cells were cultured in RPMI‐1640 (Thermo) or Ham's F‐12 K (Thermo) medium supplemented with 10% FBS (Sigma‐Aldrich) and 1% P/S (SA) in an incubator with 95% air and 5% carbon dioxide at 37°C.
For propofol exposure, NSCLC cells were seeded into media with different concentrations of propofol (SA), which was dissolved with dimethyl sulfoxide (Yesen). The same amount of dimethyl sulfoxide was also used as a control.
Cell viability analysis
About 5 × 103 cells were seeded into 96‐well plates and cultured for 24–72 h. After replacing the fresh medium, 10 μL of MTT working solution (5 mg/mL) was added to each well and mixed. After 4 h of incubation, the culture medium was removed from each well and supplemented with 100 μL of formazan solution. After another 4 h of incubation, measurement of absorbance was performed with a microplate reader (Bio‐Tek) at 570 nm.
Cell proliferation analysis
The clonogenic ability of the cells was used for proliferation analysis. Briefly, approximately 1 × 103 cells were allowed to form colonies onto six‐well plates. Once visible colonies had formed, the cells were fixed (4% paraformaldehyde, Beyotime) and stained (0.01% crystal violet, Beyotime), followed by imaging with a microscope using imaging software (Olympus).
Cell apoptosis analysis
An annexin V‐FITC/PI apoptosis detection kit (Yesen) was used for apoptosis analysis. After digestion with trypsin without EDTA, the cells were collected by centrifugation. The cells were washed and resuspended in 100 μL of 1 × binding buffer, followed by adding 5 μL of annexin V‐FITC and 10 μL of propidium iodide (PI) staining solution and mixing. After reacting for 10–15 min in the dark, the samples were detected by flow cytometry (Becton Dickinson) within 1 h.
Western blotting
The determination of protein concentration was performed using the BCA protein assay kit (Thermo) after preparing the total extract with RIPA buffer (Thermo). Western blotting was carried out as previously described using the following primary antibodies: cleaved caspase‐3 with a dilution of 1:500 (ab2302, Abcam), proliferating cell nuclear antigen (PCNA) with a dilution of 1:1000 (ab280088, Abcam), CORO1C with a dilution of 1:1000 (ab96266, Abcam), and glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) with a dilution of 1:5000 (ab8245, Abcam). Detection of protein signals was performed using chemiluminescence (Thermo). Densitometry was performed to quantify the changes in protein levels using ImageJ software (NIH).
Cell migration and invasion analysis
Transwell migration and invasion experiments were performed using 24‐well transwell chambers (Corning) without or with Matrigel (Corning) as previously described. 34 The number of migrating and invading cells was counted under a microscope (Olympus) in five randomly chosen fields.
Collection of clinical samples
Patients diagnosed with operable NSCLC at The First People's Hospital of Lianyungang were enrolled in the study (n = 36). The clinical samples of the enrolled patients were obtained during the operation in the hospital where the disease was confirmed. Participants were aware of the research and signed a personal consent form. The research was carried out with a permit authorized by the Ethics Committee of The First People's Hospital of Lianyungang.
Real‐time polymerase chain reaction (RT‐qPCR)
RNA isolation was conducted using the RNAeasy Plus animal RNA isolation kit with spin column (Beyotime) according to the manufacturer's instructions. RNase R digestion was performed by incubating total RNA from NSCLC cells with 3 U/μg RNase R (Epicenter). The synthesis of complementary DNA was done using 1 ng of total RNA along with an iScript cDNA synthesis kit (Bio‐Rad) or miRCURY LNA Universal RT microRNA PCR system (Exiqon, Euroclone). Amplifications were run on the 7900HT fast real‐time PCR system with an SYBR Master Mix (Bio‐Rad). Data were reported as relative quantity with respect to a calibrator sample using the equation 2−ΔΔCt. The primers used are listed in Table 1.
TABLE 1.
Primer sequences used for real‐time polymerase chain reaction (RT‐qPCR).
| Genes | Primer sequences (5'‐3') |
|---|---|
| circ_0003028 | Forward (F): 5'‐GCCGAATCTCTCCGCATGTA‐3′ |
| Reverse (R): 5'‐CTCACTTCCCTGGAGTCCTGGA‐3′ | |
| circ_0084003 | F: 5'‐CTTGACCTCCAACCAGCCG‐3′ |
| R: 5'‐GTGGCATAACGGACCTTGTAG‐3′ | |
| circ_0007518 | F: 5'‐TCACTCTGTACAATGCCGGT‐3′ |
| R: 5'‐TCCAGAACCACATGAGTCCG‐3′ | |
| circ_0008305 | F: 5'‐TCTCTGTGTCAGAAAAGATGTTGGT‐3' |
| R: 5'‐GCCGATTGCCAGTTCCAC‐3' | |
| CORO1C | F: 5'‐CTGGCCACGAATCATTTGCC‐3' |
| R: 5'‐CAGGCAATTGAGCATCCACG‐3' | |
| FUT8 | F: 5'‐TATGCTCACCAACCCCGAAC‐3' |
| R: 5'‐AGCCTCAGGATATGTGGGGT‐3' | |
| miR‐1305 | F: 5'‐GCCGAGTTTTCAACTCTAATGG‐3' |
| R: 5'‐AGTGCAGGGTCCGAGGTATT‐3' | |
| GAPDH | F: 5'‐GACAGTCAGCCGCATCTTCT‐3' |
| R: 5'‐GCGCCCAATACGACCAAATC‐3' | |
| U6 | F: 5'‐CTCGCTTCGGCAGCACA‐3' |
| R: 5'‐AACGCTTCACGAATTTGCGT‐3' |
Oligonucleotides and construction plasmids
Oligonucleotides used in the research were synthesized by GenePharma, including si‐circ_0003028, si‐CORO1C, si‐NC, miR‐1305 inhibitor (anti‐miR‐1305), anti‐miR‐NC, miR‐1305 mimic, and miR‐NC. Construction of circ_0003028 overexpressed plasmid (circ_0003028) was executed using the pCD‐ciR vector (Geneseed).
Transfection
NSCLC cells were transiently transfected with the above mentioned oligonucleotides separately using TransIT‐X2 (Mirus Bio) according to the manufacturer's protocol. For plasmid transfection, 2–3 μg plasmid was transfected using FuGENE HD transfection reagent (Promega) as per the manufacturer's protocol.
Dual‐luciferase reporter assay
Recombinant luciferase reporter plasmids were created by ligating the fragments of circ_0003028WT, circ_0003028MUT, CORO1C 3'UTRWT, or CORO1C 3'UTRMUT into the psiCHECK‐2 vector (Promega), respectively. TransIT‐X2 (Mirus Bio) was used to cotransfect miR‐1305 mimic or miR‐NC with a recombinant luciferase plasmid into NSCLC cells. Luciferase activity was measured using the dual‐luciferase assay system (Promega) and then normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
RIP analysis was carried out using an antibody against Ago2/IgG according to the manufacturer's instructions of the Magna RIP RNA‐binding protein immunoprecipitation kit (Millipore). Coprecipitated RNA complexes were purified and then analyzed by RT‐qPCR.
Animal experiments
Twelve male BALB/c nude mice (4 weeks old) (Vital River, Beijing, China) were allowed to acclimatize to the environment for 1 week in a specific pathogen‐free environment and then administered as follows: (1) control and (2) propofol (6 mice/group). Specifically, A549 cells (1 × 107) were injected subcutaneously into the armpits of mice. When the tumor volume grew to 100 mm3, the mice were treated with dimethyl sulfoxide (DMSO) or propofol (45 mg/kg) every 3 days. Tumor volume was monitored every 6 days from the administration of DMSO or propofol and calculated with the equation (volume = [length ×width2]/2). Then, 25 days later, the mice were killed and their tumors excised. Xenograft experiments were conducted with a permit authorized by the Animal Care Committee of The First People's Hospital of Lianyungang.
Immunohistochemical (IHC) analysis
IHC analysis was performed using 5 μm thick paraformaldehyde‐fixed de‐paraffinized sections as previously described. 35 Xenograft tumor tissue sections were probed with the antibody against Ki67 (ab243878, 1:300, Abcam).
Statistical analysis
Data were analyzed with GraphPad Prism software (version 6.01, GraphPad Software), and the data of three biological replicates are exhibited as mean ± standard deviation. Significance was evaluated by Student's t‐test or ANOVA. Differences were considered statistically significant at p < 0.05.
RESULTS
Propofol exposure decreased NSCLC cell malignant behaviors
We first analyzed the effects of different concentrations of propofol on tumor cells. Data in Figure 1a,b showed that propofol exposure led to a decrease in NSCLC cell viability in a concentration‐dependent manner and 10 μg/mL of propofol, which achieved a 50% inhibitory effect, was used for further investigation. Moreover, propofol exposure decreased NSCLC cell proliferation and induced NSCLC cell apoptosis in colony formation and flow cytometry assays (Figure 1c,d). Consistent with the above experiments, the proliferation‐related PCNA protein levels were reduced and the apoptosis‐related cleaved caspase‐3 protein levels were increased in propofol‐treated NSCLC cells (Figure 1e,f). In addition, propofol exposure restrained the migrating and invading capabilities of NSCLC cells in transwell experiments (Figure 1g,h). Together, propofol exposure decreased cell malignant behaviors in NSCLC cells.
FIGURE 1.
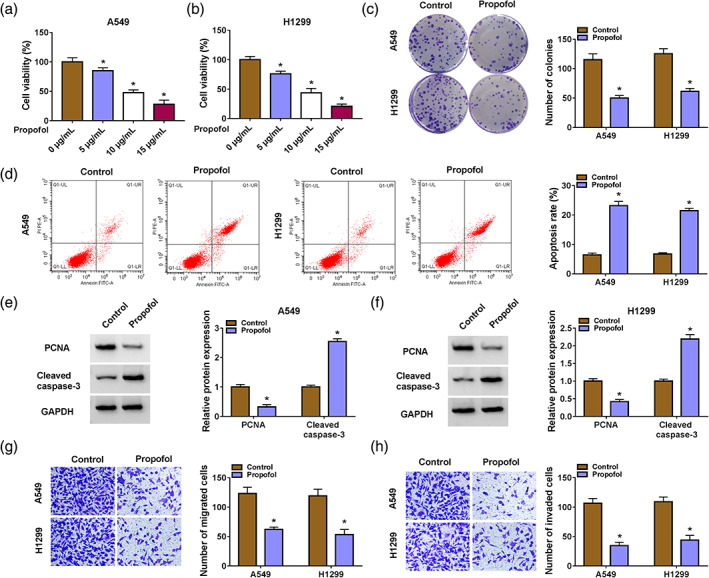
Propofol exposure reduced cell malignant behaviors in non‐small cell lung cancer (NSCLC) cells. (a and b) 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide (MTT) assays were used to investigate the effects of different concentrations of propofol on NSCLC cell viability. (c and d) Colony formation and flow cytometry assays were used to determine cell proliferation and apoptosis in NSCLC cells with propofol (10 μg/mL) exposure. (e and f) Western blotting was used to analyze cleaved caspase‐3 and proliferating cell nuclear antigen (PCNA) protein levels in NSCLC cells with propofol (10 μg/mL) exposure. (g and h) Transwell assays were used to detect cell migration and invasion in NSCLC cells with propofol (10 μg/mL) exposure. *p < 0.05.
Increased circ_0003028 expression was obtained in NSCLC
Next, we explored propofol‐mediated molecular mechanisms associated with circRNAs in NSCLC. By reading some reports, 15 , 18 , 19 , 22 we screened four circRNAs (circ_0003028, circ_0084003, circ_0008305, and circ_0007518) associated with NSCLC progression for further analysis. Propofol treatment caused a significant downregulation of circ_0003028, so circ_0003028 was selected for subsequent analysis (Figure 2a,b). Circ_0003028 expression levels in NSCLC samples and cells were detected to verify the function of circ_0003028. We observed a prominent elevation in circ_0003028 expression in NSCLC samples and cell lines compared with their corresponding controls (Figure 2c,d). Moreover, circ_0003028 was hardly digested by RNase R treatment, but its parental gene FUT8 was severely digested (Figure 2e,f). These results demonstrated that upregulated circ_0003028 might be related to NSCLC.
FIGURE 2.
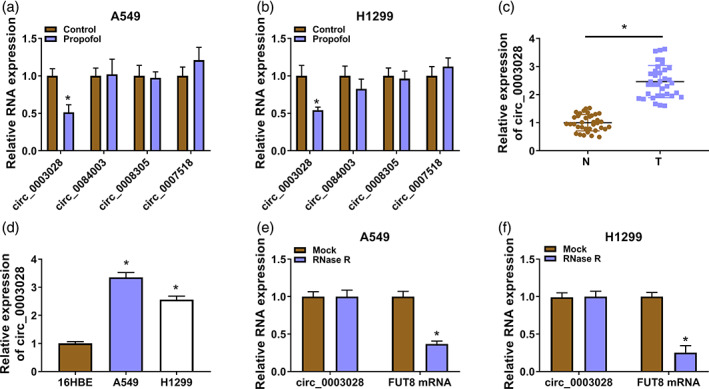
Propofol treatment decreased circ_0003028 expression in non‐small cell lung cancer (NSCLC). (a and b) Real‐time polymerase chain reaction (RT‐qPCR) analysis of the expression levels of four circRNAs (circ_0003028, circ_0084003, circ_0008305, and circ_0007518) in NSCLC cells with or without propofol (10 μg/mL) exposure. (c and d) RT‐qPCR was used to detect the relative levels of circ_0003028 in NSCLC samples and cell lines. (e and f) RT‐qPCR was used to analyze the relative levels of circ_0003028 and FUT8 mRNA in total RNA of NSCLC cells with or without RNase R treatment. *p < 0.05.
Overexpression of circ_0003028 weakened propofol‐mediated suppression on NSCLC cell malignant behaviors
We then examined whether circ_0003028 is involved in propofol‐mediated effects on NSCLC cell malignant behaviors. The circ_0003028 overexpression plasmid was then constructed, and the transfection efficiency was exhibited in Figure 3a. Further experiments showed that circ_0003028 overexpression impaired propofol‐mediated effects on cell viability, colony formation, and apoptosis in NSCLC cells (Figure 3b–d). Moreover, elevated cleaved caspase‐3 and reduced PCNA protein levels in propofol‐treated NSCLC cells were partly reversed after circ_0003028 overexpression (Figure 3e,f). In addition, the decline of NSCLC cell migrating and invading abilities caused by propofol treatment was attenuated after circ_0003028 overexpression (Figure 3g,h). These findings indicated that circ_0003028 was involved in propofol‐mediated inhibiting effect on NSCLC cell malignant behaviors.
FIGURE 3.
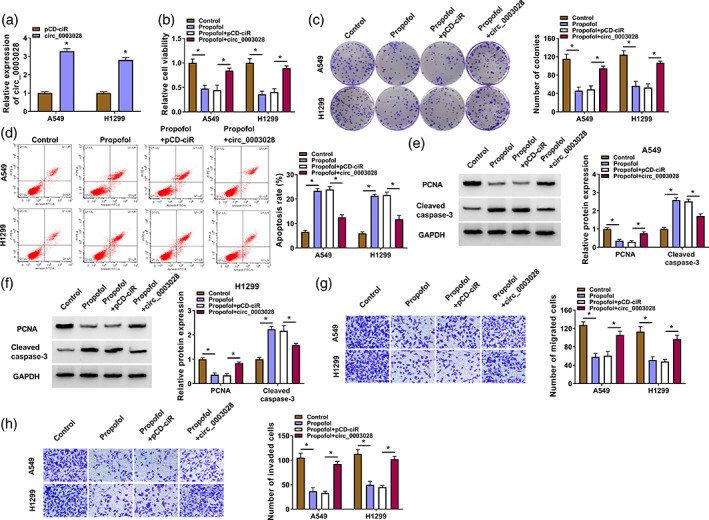
Upregulated circ_0003028 weakened propofol‐mediated suppression on non‐small cell lung cancer (NSCLC) cell malignant behaviors. (a) Real‐time polymerase chain reaction (RT‐qPCR) analysis of the transfection efficiency of the circ_0003028 overexpression plasmid. (b–h) NSCLC cells were treated as follows: control, propofol, propofol + pCD‐ciR, or propofol + circ_0003028. (b–d) The viability, colony formation, and apoptosis of NSCLC cells were determined. (e and f) Protein levels of cleaved‐caspase‐3 and proliferating cell nuclear antigen (PCNA) in NSCLC cells were detected. (g and h) The migration and invasion of NSCLC cells were evaluated. *p < 0.05.
Circ_0003028 acted as a miR‐1305 sponge
Since an important function of circRNAs is to act as ceRNA to antagonize miRNAs, we searched for miRNAs that might interact with circ_0003028. Circular RNA interactome predicted that circ_0003028 contained the binding sites for miR‐1305 (Figure 4a). To test this prediction, miR‐1305 mimic was used to overexpress miR‐1305 in NSCLC cells (Figure 4b). Moreover, miR‐1305 overexpression decreased the luciferase activity of the circ_0003028WT luciferase plasmid but not the circ_0003028MUT luciferase plasmid (Figure 4c,d). Circ_0003028 and miR‐1305 were also more enriched in RNA complexes precipitated by the anti‐Ago2 antibody than with the anti‐IgG antibody (Figure 4e,f). There was a conspicuous decrease in miR‐1305 expression in NSCLC samples and cells (Figure 4g,h). In contrast, a significant elevation was obtained in propofol‐treated NSCLC cells (Figure 4i). We also designed a siRNA against circ_0003028 in NSCLC cells, and its interference efficiency was shown in Figure 4j. In addition, circ_0003028 silencing led to an elevation in miR‐1305 expression in propofol‐treated NSCLC cells, while circ_0003028 overexpression caused a reduction in miR‐1305 expression in propofol‐treated NSCLC cells (Figure 4k,l). Together, these findings indicated that circ_0003028 acted as a sponge to miR‐1305.
FIGURE 4.
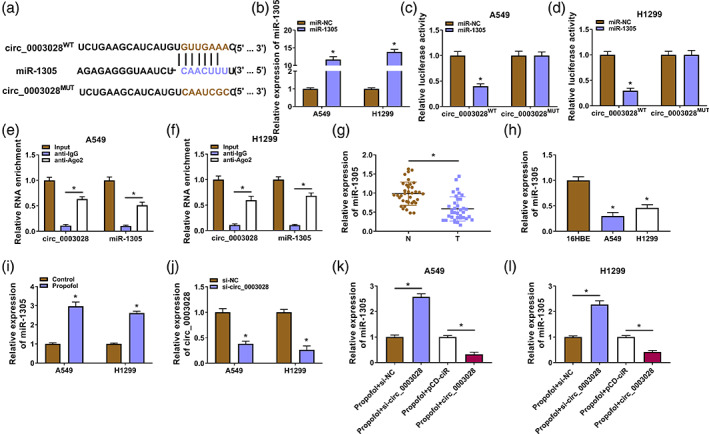
Circ_0003028 served as a miR‐1305 sponge. (a) Schematic diagram of the binding sites of circ_0003028 and miR‐1305. (b) The transfection efficiency of miR‐1305 mimic. (c and d) Luciferase reporter assay was used to analyze the luciferase activity of the circ_0003028WT/circ_0003028MUT luciferase plasmid. (e and f) Enrichment of circ_0003028 and miR‐1305 was detected by real‐time polymerase chain reaction (RT‐qPCR) after RNA immunoprecipation (RIP) assay. (g–i) Relative expression levels of miR‐1305 in non‐small cell lung cancer (NSCLC) samples and cells, as well as propofol‐treated NSCLC cells. (j) The interference efficiency of si‐circ_0003028 on circ_0003028 in NSCLC cells. (k and l) Relative expression levels of miR‐1305 in circ_0003028‐overexpressed/−silenced NSCLC cells with propofol treatment. *p < 0.05.
Circ_0003028 mediated propofol‐treated NSCLC cell malignant behaviors through miR‐1305
To verify that circ_0003028 mediates propofol‐treated NSCLC cell malignant behaviors through miR‐1305, we assessed whether miR‐1305 overexpression weakened the promoting effect of circ_0003028 upregulation on the malignant behaviors of propofol‐treated NSCLC cells. The decreased miR‐1305 in propofol‐treated NSCLC cells caused by circ_0003028 overexpression was partly overturned by the introduction of miR‐1305 mimic (Figure 5a). Furthermore, circ_0003028 upregulation elevated cell viability, facilitated cell proliferation, and decreased cell apoptosis in propofol‐treated NSCLC cells, while these effects were weakened by cointroduction of the circ_0003028 overexpression plasmid and miR‐1305 mimic (Figure 5b–d). Exogenetic expression of miR‐1305 also impaired the elevated PCNA and the decreased cleaved caspase‐3 protein levels in propofol‐treated NSCLC cells caused by circ_0003028 overexpression (Figure 5e,f). In addition, miR‐1305 overexpression partially counteracted the promoting effects of circ_0003028 overexpression on propofol‐treated NSCLC cell migration and invasion (Figure 5g,h). Collectively, these findings suggested that circ_0003028 regulated propofol‐treated NSCLC cell malignant behaviors through adsorbing miR‐1305.
FIGURE 5.
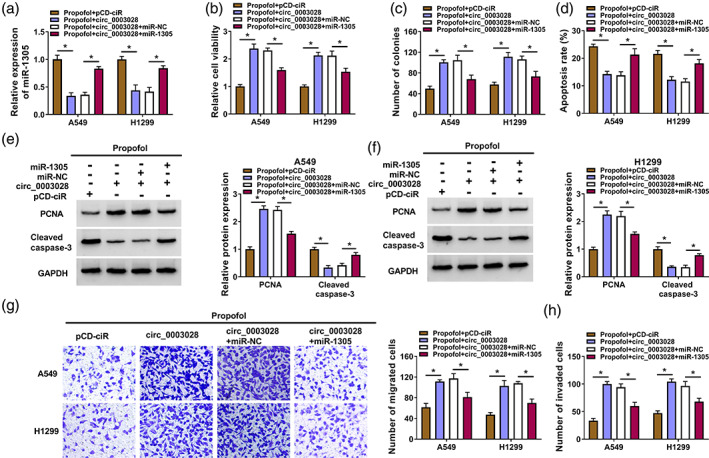
Circ_0003028 regulated cell malignant behaviors through adsorbing miR‐1305 in propofol‐treated non‐small cell lung cancer (NSCLC) cells. (a–h) NSCLC cells were treated as follows: propofol + pCD‐ciR, propofol + circ_0003028, propofol + circ_0003028 + miR‐NC, or propofol + circ_0003028 + miR‐1305. (a) Relative expression levels of miR‐1305 in NSCLC cells. (b–d) The viability, colony formation, and apoptosis of NSCLC cells were determined. (e and f) Relative protein levels of cleaved caspase‐3 and proliferating cell nuclear antigen (PCNA) in NSCLC cells were detected. (g and h) The migration and invasion of NSCLC cells were evaluated. *p < 0.05.
CORO1C was a downstream target of miR‐1305
TargetScan was used to predict the targets of miR‐1305. A tumor‐associated gene, CORO1C, was predicted as a potential target for miR‐1305 (Figure 6a). Also, exogenetic expression of miR‐1305 decreased the luciferase activity of the CORO1C 3'UTRWT luciferase plasmid instead of the CORO1C 3'UTRMUT luciferase plasmid (Figure 6b,c). Moreover, miR‐1305 and CORO1C mRNA were overtly enriched in the anti‐Ago2 group (Figure 6d,e). In NSCLC samples, CORO1C mRNA and protein levels were observably elevated (Figure 6f,g). Similar protein trends were observed in NSCLC cells (Figure 6h). In contrast, propofol treatment decreased CORO1C protein levels in NSCLC cells (Figure 6i). Furthermore, we silenced miR‐1305 expression in NSCLC cells using miR‐1305 inhibitor, and the transfection efficiency was exhibited in Figure 6j. MiR‐1305 mimic also decreased CORO1C protein levels in propofol‐treated NSCLC cells, but miR‐1305 inhibitor had the opposing effect (Figure 6k,l). Additionally, circ_0003028 overexpression resulted in an elevation in CORO1C protein levels in propofol‐treated NSCLC cells, while this effect mediated by circ_0003028 overexpression was partly counteracted after miR‐1305 mimic introduction (Supplementary Figure S1a,b). Together, circ_0003028 regulated CORO1C, which acted as a miR‐1305 target, through sponging miR‐1305.
FIGURE 6.
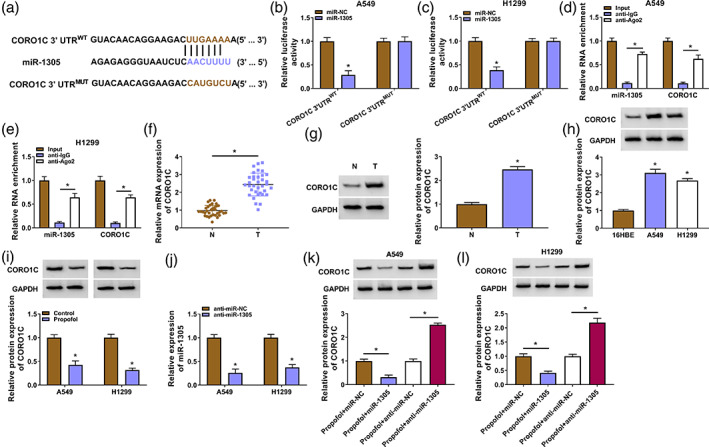
Coronin‐like actin‐binding protein 1C (CORO1C) was a target for miR‐1305. (a) Schematic diagram of the binding sites of miR‐1305 and the 3'UTR of CORO1C. (b and c) Luciferase reporter assay analyzed the luciferase activity of the CORO1C 3'UTRWT/CORO1C 3'UTRMUT luciferase plasmid. (d and e) Enrichment of miR‐1305 and CORO1C mRNA was detected after RNA immunoprecipitation (RIP) assay. (f and g) Relative levels of CORO1C mRNA and protein in NSCLC samples were evaluated. (h and i) Relative protein levels of CORO1C in NSCLC cells and propofol‐treated NSCLC cells. (j) The interference efficiency of miR‐1305 inhibitor on miR‐1305. (k and l) Relative protein levels of CORO1C in miR‐1305‐inhibited/−overexpressed NSCLC cells with propofol treatment. *p < 0.05.
miR‐1305 regulated propofol‐treated NSCLC cell malignant behaviors through targeting CORO1C
We further assessed whether miR‐1305 mediated the malignant behaviors of propofol‐treated NSCLC cells via CORO1C. A siRNA against CORO1C was used to knock down CORO1C (Figure 7a). Transfection of si‐CORO1C partly reversed the increased protein levels of CORO1C in propofol‐treated NSCLC cells caused by miR‐1305 knockdown (Figure 7b). Furthermore, miR‐1305 silencing increased cell viability and colony formation, as well as reduced cell apoptosis in propofol‐treated NSCLC cells, but these changes were alleviated by CORO1C knockdown (Figure 7c–e). The increased PCNA protein levels and the reduced cleaved caspase‐3 protein levels in propofol‐treated NSCLC cells mediated by miR‐1305 knockdown were also weakened after CORO1C inhibition (Figure 7f,g). In addition, miR‐1305 downregulation‐mediated promotion on propofol‐treated NSCLC cell migration and invasion were reversed by CORO1C silencing (Figure 7h,i). In summary, miR‐1305 regulated propofol‐treated NSCLC cell malignant behaviors via targeting CORO1C.
FIGURE 7.
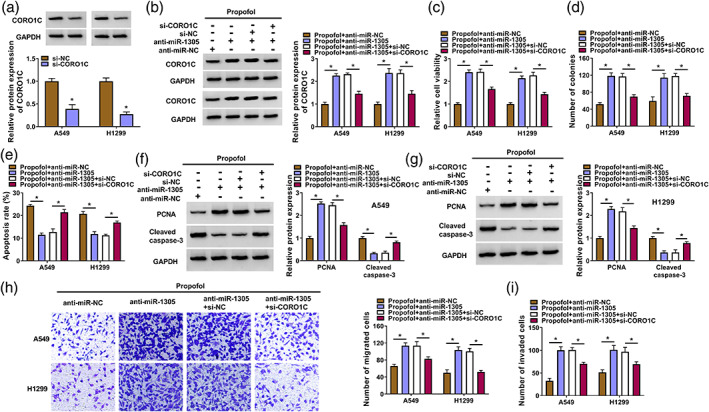
miR‐1305 regulated non‐small cell lung cancer (NSCLC) cell malignant behaviors through coronin‐like actin‐binding protein 1C (CORO1C) under propofol treatment. (a) Analysis of the CORO1C protein levels in NSCLC cells carrying si‐NC or si‐CORO1C. (b–i) NSCLC cells were treated as follows: propofol + anti‐miR‐NC, propofol + anti‐miR‐1305, propofol + anti‐miR‐1305 + si‐NC, or propofol + anti‐miR‐1305 + si‐CORO1C. (b) Detection of the CORO1C protein levels in NSCLC cells. (c–e) Evaluation of cell viability, colony formation, and apoptosis in NSCLC cells was carried out. (f and g) Detection of cleaved caspase‐3 and proliferating cell nuclear antigen (PCNA) protein levels in NSCLC cells was performed by western blotting. (h and i) Determination of cell migration and invasion in NSCLC cells was performed. *p < 0.05.
Propofol treatment reduced tumor growth partly by downregulating circ_0003028
Whether propofol treatment reduced tumor growth through downregulating circ_0003028 was validated through mice models. Propofol treatment decreased tumor volume and weight (Figure 8a,b). Moreover, circ_0003028 expression was significantly reduced in tumor tissues from mice treated with propofol, but miR‐1305 expression was markedly increased (Figure 8c,d). IHC showed a low number of Ki67/CORO1C‐positive cells in tumor tissues from mice treated with propofol than those in tumor tissues from the control group (Figure 8e). Collectively, propofol treatment reduced NSCLC growth partly by downregulating circ_0003028.
FIGURE 8.
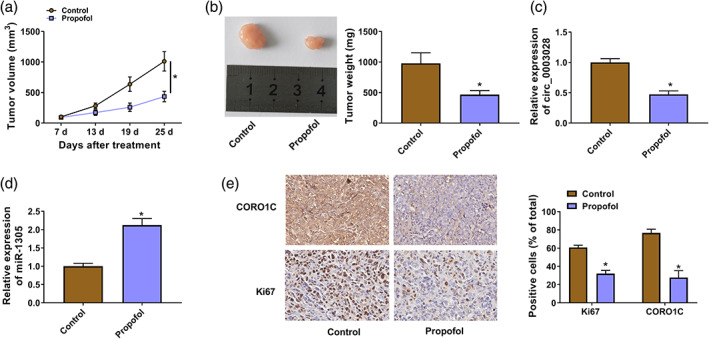
Propofol reduced tumor growth partly by downregulating circ_0003028. (a) Tumor volume in different groups. (b) Schematic representation of tumor images from different groups of mice (left), tumor weight in different groups (right). (c and d) Levels of circ_0003028 and miR‐1305 in tumor tissues from different groups of mice were detected. (e) Immunohistochemical (IHC) analysis of the number of Ki67/CORO1C‐positive cells in tumor tissues from different groups of mice. *p < 0.05.
DISCUSSION
Increasing evidence shows the inhibitory effect of propofol on NSCLC progression. Furthermore, the existing evidence exhibits that the deregulation of circRNAs disrupts the tightly regulated RNA network in NSCLC cells. 36 Therefore, it is meaningful to explore the mechanism by which propofol regulates circRNAs to inhibit NSCLC. Our findings uncovered that propofol decreased NSCLC progression through regulation of the circ_0003028/miR‐1305/CORO1C axis.
We validated the upregulation of circ_0003028 in NSCLC samples and cells, which was in line with a previous report. 22 Interestingly, propofol exposure overtly decreased circ_0003028 expression in NSCLC cells, and circ_0003028 overexpression impaired propofol‐mediated suppression on NSCLC cell proliferation, migration, and invasion, as well as promotion on NSCLC cell apoptosis. The combination of circ_0003028 silencing and propofol also further repressed xenograft tumor growth than silencing circ_0003028 alone, and circ_0003028 expression was lower in xenograft tumors derived from the combination of circ_0003028 silencing and propofol than silencing circ_0003028 alone. These results prompted us to conclude that propofol repressed NSCLC progression by decreasing circ_0003028 expression. In addition, our findings supported the oncogenic activity of circ_0003028 in NSCLC reported by Guan et al. Nevertheless, circ_0003028 has been reported to exert an inhibitory role in bladder cancer, 37 which might be related to cell‐specific expression.
The sponge function of circRNAs prompted us to further investigate whether circ_0003028 participates in the effects of propofol on NSCLC cells as an endogenous competitive RNA. Bioinformatic prediction showed that circ_0003028 might function as a miR‐1305 sponge, and this prediction was verified by luciferase reporter and RIP assays. Consistent with previous studies, 27 , 38 miR‐1305 was lowly decreased in NSCLC samples and cells. Moreover, propofol exposure overtly increased miR‐1305 expression, and exogenetic expression of miR‐1305 weakened the effects of circ_0003028 overexpression on propofol‐treated NSCLC cell malignant behaviors. These findings manifested that propofol‐mediated suppression on NSCLC progression depended on the circ_0003028/miR‐1305 axis.
Considering that circRNAs can modulate gene expression via adsorbing miRNAs, we further investigated the downstream target of miR‐1305. CORO1C, a cytoskeletal protein implicated in metastasis in many cancer types, was validated as a miR‐1305 in the research. The increased expression of CORO1C in NSCLC was also consistent with previous studies. 39 , 40 In addition, propofol treatment reduced CORO1C expression, and CORO1C knockdown mitigated the impacts of miR‐1305 inhibitor on cell malignancy in NSCLC cells under propofol treatment. Notably, circ_0003028 could adsorb miR‐1305 to modulate CORO1C expression. Thus, we inferred that propofol decreased CORO1C expression via the circ_0003028/miR‐1305 axis, thus repressing NSCLC cell malignant behaviors.
In conclusion, we herein identified that the circ_0003028/miR‐1305/CORO1C competitive endogenous RNA network participates in the inhibitory effect of propofol on NSCLC. These findings support the potential function of circ_0003028 as a target for NSCLC treatment.
AUTHOR CONTRIBUTIONS
Conceptualization and Methodology: Dongzhi Liu and Ping Wang; Formal analysis and Data curation: Ping Wang, and Naihe Liu; Validation and Investigation: Xiuli Zhang and Dongzhi Liu; Writing – original draft preparation and Writing – review and editing: Xiuli Zhang, Dongzhi Liu and Ping Wang; Approval of final manuscript: all authors.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
CONSENT FOR PUBLICATION
Patients agree to participate in this work.
Supporting information
Figure S1. circ_0003028 adsorbed miR‐1305 to regulate CORO1C expression. (a and b) Relative protein levels of CORO1C in NSCLC cells treated with propofol + pCD‐ciR, propofol + circ_0003028, propofol + circ_0003028 + miR‐NC, or propofol + circ_0003028 + miR‐1305. *p < 0.05.
Zhang X, Liu D, Wang P, Liu N. Propofol mediates non‐small cell lung cancer growth in part by regulating circ_0003028‐related mechanisms. Thorac Cancer. 2023;14(17):1606–1617. 10.1111/1759-7714.14906
DATA AVAILABILITY STATEMENT
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
REFERENCES
- 1. Dinis‐Oliveira RJ. Metabolic profiles of propofol and Fospropofol: clinical and forensic interpretative aspects. Biomed Res Int. 2018;2018:6852857–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vasileiou I, Xanthos T, Koudouna E, Perrea D, Klonaris C, Katsargyris A, et al. Propofol: a review of its non‐anaesthetic effects. Eur J Pharmacol. 2009;605:1–8. [DOI] [PubMed] [Google Scholar]
- 3. Irwin MG, Chung CKE, Ip KY, Wiles MD. Influence of propofol‐based total intravenous anaesthesia on peri‐operative outcome measures: a narrative review. Anaesthesia. 2020;75(Suppl 1):e90–e100. [DOI] [PubMed] [Google Scholar]
- 4. Longhini F, Bruni A, Garofalo E, De Sarro R, Memeo R, Navalesi P, et al. Anesthetic strategies in oncological surgery: not only a simple sleep, but also impact on immunosuppression and cancer recurrence. Cancer Manag Res. 2020;12:931–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu XL, Tang HH, Zhou ZG, Tang HH, Zhou ZG, Yin F, et al. The effect of sevoflurane inhalation anesthesia only and propofol total intravenous anesthesia on perioperative cytokine balance in lung cancer patients. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2011;27:659–61. [PubMed] [Google Scholar]
- 6. Chen X, Lu P, Chen L, Yang SJ, Shen HY, Yu DD, et al. Perioperative propofol‐paravertebral anesthesia decreases the metastasis and progression of breast cancer. Tumour Biol. 2015;36:8259–66. [DOI] [PubMed] [Google Scholar]
- 7. Tian HT, Duan XH, Yang YF, Wang Y, Bai QL, Zhang X. Effects of propofol or sevoflurane anesthesia on the perioperative inflammatory response, pulmonary function and cognitive function in patients receiving lung cancer resection. Eur Rev Med Pharmacol Sci. 2017;21:5515–22. [DOI] [PubMed] [Google Scholar]
- 8. Guerrero Orriach JL, Raigon Ponferrada A, Malo Manso A, Herrera Imbroda B, Escalona Belmonte JJ, Ramirez Aliaga M, et al. Anesthesia in combination with propofol increases disease‐free survival in bladder cancer patients who undergo radical tumor cystectomy as compared to inhalational anesthetics and opiate‐based analgesia. Oncology. 2020;98:161–7. [DOI] [PubMed] [Google Scholar]
- 9. Socinski MA, Obasaju C, Gandara D, Hirsch FR, Bonomi P, Bunn P, et al. Clinicopathologic features of advanced squamous NSCLC. J Thorac Oncol. 2016;11:1411–22. [DOI] [PubMed] [Google Scholar]
- 10. Kawachi R, Tsukada H, Nakazato Y, Takei H, Furuyashiki G, Koshi‐ishi Y, et al. Early recurrence after surgical resection in patients with pathological stage I non‐small cell lung cancer. Thorac Cardiovasc Surg. 2009;57:472–5. [DOI] [PubMed] [Google Scholar]
- 11. Schuchert MJ, Normolle DP, Awais O, Pennathur A, Wilson DO, Luketich JD, et al. Factors influencing recurrence following anatomic lung resection for clinical stage I non‐small cell lung cancer. Lung Cancer. 2019;128:145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayasaka K, Shiono S, Miyata S, Takaoka S, Endoh M, Okada Y. Prognostic significance of propofol‐based intravenous anesthesia in early‐stage lung cancer surgery. Surg Today. 2021;51:1300–8. [DOI] [PubMed] [Google Scholar]
- 13. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36. [DOI] [PubMed] [Google Scholar]
- 15. Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu J, et al. Circular RNA circFGFR1 promotes progression and anti‐PD‐1 resistance by sponging miR‐381‐3p in non‐small cell lung cancer cells. Mol Cancer. 2019;18:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 promotes progression of non‐small cell lung cancer cells. Mol Cancer. 2020;19:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ‐CPA4/let‐7 miRNA/PD‐L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non‐small cell lung cancer (NSCLC). Nat Commun. 2020;39:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L, Tong X, Zhou Z, Wang S, Lei Z, Zhang T, et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF‐β‐induced epithelial‐mesenchymal transition and metastasis by controlling TIF1γ in non‐small cell lung cancer. Mol Cancer. 2018;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H, et al. circNDUFB2 inhibits non‐small cell lung cancer progression via destabilizing IGF2BPs and activating anti‐tumor immunity. Mol Cancer. 2021;12:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao H, Wei H, He J, Wang D, Li W, Wang Y, et al. Propofol disrupts cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR‐455‐3p/FOXM1 axis in lung cancer. Cell Cycle. 2020;19:2538–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao J, Ding C, Zhou J, Wu G, Han Z, Li J, et al. Propofol suppresses lung cancer tumorigenesis by modulating the circ‐ERBB2/miR‐7‐5p/FOXM1 axis. Thorac Cancer. 2021;12:824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan H, Sun C, Gu Y, Li J, Ji J, Zhu Y. Circular RNA circ_0003028 contributes to tumorigenesis by regulating GOT2 via miR‐1298‐5p in non‐small cell lung cancer. Bioengineered. 2021;12:2326–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franco‐Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio‐Somoza I, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–7. [DOI] [PubMed] [Google Scholar]
- 24. Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75:1071–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, et al. FLI1 Exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res. 2019;25:1302–17. [DOI] [PubMed] [Google Scholar]
- 26. Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. CircRIP2 accelerates bladder cancer progression via miR‐1305/Tgf‐β2/smad3 pathway. Mol Cancer. 2020;19:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai Y, Hao Y, Ren H, Dang ZG, Xu H, Xue X, et al. miR‐1305 inhibits the progression of non‐small cell lung cancer by regulating MDM2. Mol Cancer. 2019;11:9529–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu W, Zhuang R, Feng S, Bai X, Jia Z, Kapora E, et al. Long non‐coding RNA ASB16‐AS1 enhances cell proliferation, migration and invasion via functioning as a ceRNA through miR‐1305/Wnt/β‐catenin axis in cervical cancer. Biomed Pharmacother. 2020;125:109965. [DOI] [PubMed] [Google Scholar]
- 29. Li X, Song L, Wang B, Tao C, Shi L, Xu M. Circ0120816 acts as an oncogene of esophageal squamous cell carcinoma by inhibiting miR‐1305 and releasing TXNRD1. Cancer Manag Res. 2020;20:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan KT, Creed SJ, Bear JE. Unraveling the enigma: progress towards understanding the coronin family of Actin regulators. Trends Cell Biol. 2011;21:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang J, Tsouko E, Jonsson P, Bergh J, Hartman J, Aydogdu E, et al. Mir‐206 inhibits cell migration through direct targeting of the Actin‐binding protein coronin 1C in triple‐negative breast cancer. Onco Targets Ther. 2014;8:1690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fan L, Wei Y, Ding X, Li B. Coronin3 promotes nasopharyngeal carcinoma migration and invasion by induction of epithelial‐to‐mesenchymal transition. Onco Targets Ther. 2019;12:9585–98. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Li J, Tian L, Jing Z, Guo Z, Nan P, Liu F, et al. Cytoplasmic RAD23B interacts with CORO1C to synergistically promote colorectal cancer progression and metastasis. Cancer Lett. 2021;516:13–27. [DOI] [PubMed] [Google Scholar]
- 34. Ren M, Xu W, Xu T. Salidroside represses proliferation, migration and invasion of human lung cancer cells through AKT and MEK/ERK signal pathway. Artif Cells Nanomed Biotechnol. 2019;47:1014–21. [DOI] [PubMed] [Google Scholar]
- 35. Liu T, Zhu J, Du W, Ning W, Zhang Y, Zeng Y, et al. AKT2 drives cancer progression and is negatively modulated by miR‐124 in human lung adenocarcinoma. Respir Res. 2020;21:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li C, Zhang L, Meng G, Wang Q, Lv X, Zhang J, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non‐small cell lung cancer. J Cancer Res Clin Oncol. 2019;145:2875–89. [DOI] [PubMed] [Google Scholar]
- 37. He Q, Yan D, Dong W, Bi J, Huang L, Yang M, et al. circRNA circFUT8 upregulates Krüpple‐like factor 10 to inhibit the metastasis of bladder cancer via sponging miR‐570‐3p. Mol Ther Oncolytics. 2020;16:172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao W, Yang X, Dong Y, Quan J, Huang L. Effects of miR‐1305 on proliferation and apoptosis of non‐small cell lung cancer cells via regulating high mobility group Box‐1 level. J Biomater Tissue Eng. 2020;10:1837–42. [Google Scholar]
- 39. Liao M, Peng L. MiR‐206 may suppress non‐small lung cancer metastasis by targeting CORO1C. Cell Mol Biol Lett. 2020;25:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang W, Song C, Ren X. Circ_0003998 regulates the progression and docetaxel sensitivity of DTX‐resistant non‐small cell lung cancer cells by the miR‐136‐5p/CORO1C Axis. Technol Cancer Res Treat. 2021;20:1533033821990040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. circ_0003028 adsorbed miR‐1305 to regulate CORO1C expression. (a and b) Relative protein levels of CORO1C in NSCLC cells treated with propofol + pCD‐ciR, propofol + circ_0003028, propofol + circ_0003028 + miR‐NC, or propofol + circ_0003028 + miR‐1305. *p < 0.05.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.


