Abstract
Gut-enriched Krüppel-like factor (GKLF or KLF4) is a pleiotropic (activating and repressive) transcription factor. This study characterizes the mechanisms of transactivation by GKLF. Using a GAL4 fusion assay, the activating domain of murine GKLF was localized to the 109 amino acid residues in the N-terminus. Site-directed mutagenesis showed that two adjacent clusters of acidic residues within this region are responsible for the activating effect. Transactivation by GKLF involves intermolecular interactions as demonstrated by the ability of wild-type, but not mutated, GKLF to compete with the N-terminal activation domain. In addition, wild-type adenovirus E1A, but not a mutated E1A that failed to bind p300/CBP, inhibited transactivation by the N-terminal 109 amino acids of GKLF, suggesting that p300/CBP are GKLF’s interacting partners. A physical interaction between GKLF and CBP was demonstrated by glutathione-S-transferase pull-down and by in vivo co-immunoprecipitation experiments. We also showed that the two acidic amino acid clusters are essential for this interaction, since GKLF with mutations in these residues failed to co-immunoprecipitate with CBP. Importantly, the same mutations abrogated the ability of GKLF to suppress cell growth as determined by a colony suppression assay. These studies therefore provide plausible evidence for a structural and functional correlation between the transactivating and growth-suppressing effects of GKLF.
INTRODUCTION
Eukaryotic transcription factors are modular proteins that depend on distinct domains for their functions such as DNA binding and modulation of transcription (1). Various structural motifs have been described that are involved in DNA binding and/or protein dimerization. Examples include the zinc finger, basic helix–loop–helix and leucine zipper motifs (2–6). Many transcription factors also contain distinct domains for the transcriptional activation or repression of target genes. Activation domains, for example, can be rich in glutamine (7,8), proline (9,10) or acidic residues (11–13). Adding to the complexity of transcriptional regulation is the recent identification of a group of co-activator proteins, such as p300 (14,15), CBP (16–18) and P/CAF (19). Collectively, these proteins exhibit a broad spectrum of activities including interaction with sequence-specific DNA-binding proteins and the basal transcriptional machinery, as well as chromatin remodeling (20,21).
The gut-enriched Krüppel-like factor (GKLF; also called Krüppel-like factor 4 or KLF4; 22) is a recently identified Krüppel-type transcription factor with 3 C2H2 zinc fingers. Expression of GKLF is enriched in epithelial cells of the gastrointestinal tract (23–25) and the skin (25,26), and in vascular endothelial cells (27). The in vitro expression of GKLF is temporally associated with a growth-arrested state, such as that induced by contact inhibition or serum deprivation (23). In addition, constitutive GKLF production in transfected cells resulted in the inhibition of DNA synthesis (23). These results suggest that GKLF is a negative regulator of cell growth. The significance of the in vivo function of GKLF is heightened by the fact that expression of its gene is developmentally regulated and is down-regulated in a rodent model of intestinal tumorigenesis (28). Taken together, these studies suggest that GKLF may have a potentially important function in regulating proliferation and differentiation of specific epithelial and endothelial tissues.
The amino acid sequences of the zinc fingers (23) and the nuclear localization signals (29) of GKLF are closely related to two other Krüppel-like transcription factors, LKLF (30) and EKLF (31). Among the three, EKLF is the most extensively characterized. EKLF is essential for expression of the β-globin gene (32,33) and for erythropoeisis (34,35). It activates the β-globin gene by binding to a CACCC element in the β-globin promoter (32,33). Because of the high degree of homology between the zinc finger regions of GKLF and EKLF, GKLF has been shown to bind to the same or similar elements (24,25,27,36). However, GKLF also binds DNA sequences other than the CACCC element (36). The basic transcription element (BTE), found in the promoter of a highly conserved family of genes encoding the cytochrome P450 drug-metabolizing enzymes including CYP1A1 (37,38), for example, is a high-affinity binding site for GKLF (36,39).
A recent examination of the relationship between GKLF and the promoter activity of the CYP1A1 gene indicates that GKLF is a suppressor of the CYP1A1 promoter in a BTE-dependent fashion (39). It does so by competing with the binding of Sp1 to BTE and by physically interacting with Sp1, which is a potent activator of CYP1A1 (39). Both effects seem to be mediated by the zinc fingers of GKLF. Another study showed that GKLF possesses an intrinsic repression domain in a region of protein preceding the zinc fingers (27). In different situations, however, GKLF can be a potent activator of transcription (24,25,36). Moreover, the N-terminal region of GKLF was shown to confer the activating function (25,27). These observations indicate that GKLF is a pleiotropic protein with a dual activity in modulating transcription. The present study examines in detail the mechanisms by which GKLF activates transcription and shows that such mechanisms are complex, requiring two clusters of acidic amino acid residues within GKLF for full activity. The same amino acid residues proved to be indispensable for the interaction of GKLF with the co-activator CBP and for the ability of GKLF to suppress cell growth. Our study therefore demonstrates an important association between two primary biological activities of GKLF, that of transcription activation and that of growth suppression.
MATERIALS AND METHODS
Plasmid DNA constructs
The expression construct encoding full-length murine GKLF cloned in the mammalian expression vector PMT3 (40), PMT3-GKLF(1–483), was described previously (23,29,36,39). Expression constructs with truncation mutations were generated by deleting various segments of the GKLF reading frame with the appropriate restriction endonucleases. They included PMT3-GKLF(1–401), containing the N-terminal region including the nuclear localization signal but excluding the zinc fingers (29,36,39); PMT3-GKLF(350–483), containing the nuclear localization signal and the three zinc fingers (29,36,39); and PMT3-GKLF(145–483), which had a deletion of the N-terminal 144 amino acid residues of GKLF. In addition, a mutant construct with an internal deletion between amino acid residues 158 and 349 was generated by deleting an internal SmaI–ApaI fragment of the GKLF cDNA (23).
Constructs of GKLF with point mutations were generated from PMT3-GKLF(1–483) using the QuickChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). Two such mutants were generated (Fig. 1C). One substituted valine residues for three glutamate residues at amino acid positions 93, 95 and 96 [PMT3-GKLF(E93/95/96V)] and the other substituted valine residues for three aspartate residues at positions 99, 102 and 104 [PMT3-GKLF(D99/102/104V)]. The reporter plasmid, TDAx2-E1bTATALUC, was described previously (36). This construct contained a luciferase (LUC) reporter driven by the adenovirus E1b TATA box (41) linked to two tandem copies of the empirically derived GKLF-binding site (36).
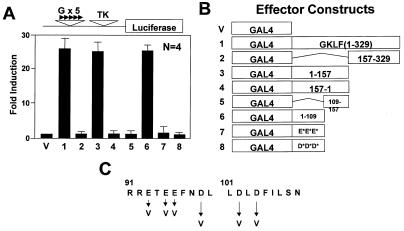
Figure 1.
Localization of the transactivation domain of GKLF by the GAL4 fusion assay. Chinese hamster ovary (CHO) cells were co-transfected with 5 µg/10 cm dish each of the indicated effector construct and the pG5TKLUC reporter. (A) Mean fold-induction in luciferase activity by the respective effector over that by the control vector containing only the DNA-binding domain of GAL4 (effector V). Bars represent standard deviations. (B) Schematic presentation of the various GAL4 fusion effector constructs. The point mutants involving the two clusters of acidic residues, effectors 7 and 8, are drawn as E*E*E* and D*D*D*, respectively. (C) Amino acid sequence between residues 91 and 110 of GKLF and identifies the mutagenized residues. All luciferase activities were standardized to the internal control β-galactosidase activities. Data represent the means of four independent experiments, each performed in duplicate.
Chimeric plasmids fusing the DNA-binding domain (amino acids 1–147) of the yeast transcription factor GAL4 to various regions of GKLF were generated in the plasmid pGALO (42,43), a kind gift of Dr Chi V. Dang. The junctions between the GAL4 and GKLF sequences in all constructs were confirmed to be in-frame by sequencing. The reporter construct, pG5TKLUC (44), also from Dr Dang, contains the luciferase gene driven by the herpes simplex virus TK promoter linked to five tandem copies of the GAL4 binding site. Two point mutants affecting the acidic domains were generated from the GAL4(1–147) and GKLF(1–109) fusion construct using the QuickChange mutagenesis kit as described above. The wild-type adenovirus E1A-containing construct, RSV-E1A12S, and the two mutant constructs, RSV-E1AΔCR1 and RSV-E1AΔCR2, were generously provided by Dr Tony Kouzarides (45). The expression plasmids containing p300/CBP, CMV-p300 and RSV-CBP were kindly provided by Drs Andrew B. Leiter (46) and Richard H. Goodman (47), respectively. Fusion constructs between glutathione-S-transferase (GST) and various regions of CBP including GST-CBP1 (amino acids 461–662), GST-CBP2 (amino acids 1621–1877) and GST-CBP3 (amino acids 1990–2441) were kindly provided by Drs Tony Kouzarides (45) and Robert G. Roeder (47).
Transfection and reporter assays
Transient transfection of Chinese hamster ovary (CHO) cells by lipofection was performed with CsCl-purified plasmid DNA as described previously (23,29,36,39). A constant amount of DNA of an internal standard, pCMV-SPORT-β-galactosidase (Life Technologies, Gaithersburg, MD), was used in all reactions. Assays to measure luciferase and β-galactosidase activities were described previously (36). All luciferase activities were standardized to β-galactosidase activities in transfected cells.
Production of recombinant proteins
Fusion proteins between GST and various segments of CBP (GST-CBP1, GST-CBP2 and GST-CBP3), or GST alone, were produced as recombinant proteins in bacteria. Single colonies of the DH5α strain of Escherichia coli (Life Technologies) were transformed by the respective plasmid construct and expanded in LB broth containing ampicillin to an optical density of 0.5 at 600 nm. Isopropyl-β-d-thiogalactopyranodise (1 mM) was then added and incubation continued for an additional 4 h at which time the bacterial pellets were collected by centrifugation. The pellets were resuspended in a lysis buffer containing 20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 1% glycerol, 1 µg/ml each of leupeptin, pepstatin and aprotinin, and sonicated with a Fisher Scientific 50 Sonic Dismembrator at a setting of 5 for 15 s at a time for 10 cycles. After clearing debris by centrifugation, solubilized proteins were stored in aliquots at –80°C.
Synthesis of GKLF protein by in vitro transcription and translation
Full-length GKLF labeled with [35S]methionine was synthesized by the TNT T7 Coupled Reticulocyte Lysate System manufactured by Promega (Madison, WI). Two micrograms of template DNA of GKLF in pBluescript (Stratagene, LaJolla, CA; 23) were incubated in a 50 µl reaction containing 25 µl TNT rabbit reticulocyte lysates, 2 µl TNT reaction buffer, 1 µl amino acid mixture minus methionine, 1 µl RNasin (a ribonuclease inhibitor), 40 µCi [35S]methionine (>1000 Ci/mmol; NEN Life Science Products, Boston, MA) and 1 µl TNT T7 RNA polymerase at 30°C for 90 min. An aliquot of the reaction was then resolved by denaturing polyacrylamide gel electrophoresis (PAGE) followed by autoradiography to verify the quality of the synthesized product.
GST pull-down experiments
GST pull-down experiments were performed using GST-CBP fusion proteins or GST alone and in vitro synthesized GKLF. One milligram of solubilized recombinant GST-CBP fusion proteins or GST was mixed with 500 µl glutathione–Sepharose 4B beads (Amersham Pharmacia Biotech, Piscataway, NJ) supplied as a 50% suspension and gently rocked at room temperature for 40 min. The beads were then collected by centrifugation and washed three times with 1.5 ml phosphate-buffered saline (PBS). A binding buffer containing 10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 20 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 10 µM ZnCl2 and 1.2% glycerol was then added to the washed beads to a final volume of 600 µl, followed by the addition of 15 ml of [35S]methionine-labeled GKLF synthesized in vitro. The mixture was gently rocked at 4°C for 1 h following which the beads were collected by centrifugation and washed three times with 1 ml PBS containing 0.025% NP-40. Bound proteins were eluted by incubating the washed beads with 10 mM reduced glutathione in 50 mM Tris–HCl, pH 8.0, at 4°C for 30 min. The eluant was concentrated by drying in a Speed Vac, resuspended in 3× sample buffer (0.18 M Tris–HCl, pH 6.8, 70 mM EDTA, 6% SDS, 30% glycerol and 0.015% bromophenol blue) and resolved by denaturing SDS–PAGE followed by autoradiography.
In vivo co-immunoprecipitation experiments
COS-1 cells were transiently co-transfected by lipofection with 10 µg/10 cm dish each of PMT3-GKLF(1–483), PMT3-GKLF(E93/95/96V), PMT3-GKLF(D99/102/104V) or PMT3 and RSV-CBP tagged with the influenza hemagglutinin (HA) epitope (47) followed by an overnight incubation in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). Cells were then washed three times with 10 ml methionine-free DMEM and incubated in 3 ml methionine-free DMEM containing 10% FBS and 500 µCi [35S]methionine (>1000 Ci/mmol; NEN Life Science Products) at 37°C for 3 h. Cells were then washed three times with 10 ml ice cold PBS and collected by scraping and centrifugation. A modified RIPA buffer (1% NP-40, 0.1% SDS, 0.5% Na deoxycholate, and 1 µg/ml each of leupeptin, pepstatin and aprotinin in PBS) at a volume of 400 µl/10 cm dish was then used to lyse the cells. The metabolically labeled cell extracts were divided into three 125 µl aliquots and incubated with pre-immune serum, anti-GKLF serum or anti-HA IgG (sc-805; Santa Cruz Biotechnology, Santa Cruz, CA) at 4°C for 2 h. Sixty microliters of a 50% suspension of protein A–Sepharose CL-4B (Amersham Pharmacia Biotech) were then added and the incubation continued at 4°C for 1 h. The beads were collected by centrifugation and washed three times with 1 ml modified RIPA buffer before being resuspended in 3× sample buffer for electrophoresis.
Colony suppression assays
Colony suppression assays were performed based on a modification of a previously published protocol (48). Rat1a cells (44) were transfected with PMT3, PMT3-GKLF(1–483), PMT3-GKLF(E93/95/96V) or PMT-GKLF(D99/102/104V) and pBabe Puro (48) which contained a puromycin-resistant gene, at a molar ratio of 40:1. Two days following transfection cells were fed DMEM containing 10% FBS and 0.75 µg/ml puromycin. Two weeks into the puromycin selection resistant colonies were visualized by staining with a 1% methylene blue solution.
RESULTS
Transactivation by GKLF depends on two clusters of acidic amino acid residues in its N-terminus
As an initial attempt to localize the domain responsible for the transactivating activity of GKLF, we adopted the previously established GAL4 fusion assay (42,43). We generated eight effector constructs by fusing various regions of the GKLF reading frame outside its zinc fingers to the N-terminal 147 amino acid DNA-binding domain of the yeast transcription factor GAL4 (Fig. 1B). A luciferase reporter driven by the herpes virus thymidine kinase (TK) promoter linked to five tandem copies of the GAL4 binding site [pG5TKLUC (44); Fig. 1A] was used to read out the activating potential of the various regions of GKLF upon co-transfection. The results in Figure 1A show that the N-terminal 329 amino acids of GKLF (effector 1) harbors an activation domain, as evidenced by the significant induction in the luciferase activity in cells co-transfected with this effector when compared to those with the vector (effector V) alone. A deletion in the N-terminal 156 amino acids of GKLF (effector 2) resulted in a complete loss of activating potentials. In contrast, the N-terminal 157 amino acids of GKLF maintained its inducing effect when present in the sense (effector 3) but not in the antisense (effector 4) orientation. Further deletion analysis revealed that the activation domain of GKLF is solely localized to the N-terminal 109 amino acids (effector 6).
The N-terminal 109 amino acid region of GKLF contains several closely clustered acidic amino acids (23). As acidic residues have previously been shown to be involved in activation of transcription (11–13), we selectively mutagenized two neighboring clusters of acidic residues, one of three glutamates and the other of three aspartates, in the region between amino acid residues 93–104 (Fig. 1C). As seen in Figure 1A, mutation in either cluster in the context of the fusion between GAL4(1–147) and GKLF(1–109) completely abolished the ability of the N-terminal 109 amino acids of GKLF to activate reporter expression. These results strongly suggest that both clusters of acidic residues are involved in mediating transactivation by GKLF.
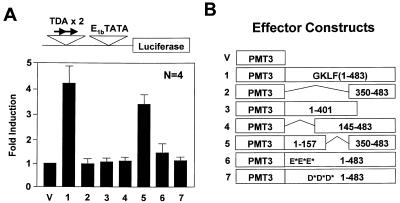
The activation domains of GKLF were verified by co-transfection experiments involving full-length or deleted GKLF constructs cloned in the mammalian expression vector, PMT3, and a luciferase reporter containing two tandem copies of the empirically determined GKLF-binding site (TDA; 36) linked to the adenovirus E1b TATA box (TDAx2-E1bTATALUC). As shown in Figure 2A, full-length GKLF (effector 1) induced the luciferase activity by a mean of 4.3-fold as compared to the PMT3 vector (effector V). The activating effect was abolished in a construct that contained only the zinc fingers (effector 2) or one that had the zinc fingers deleted (effector 3). Moreover, the activating effect was also abolished when the N-terminal 144 amino acids (effector 4) were deleted. In contrast, a construct with an internal deletion between amino acid residues 158 and 349 (effector 5) retained ∼75% of the activity of the full-length construct. These results are therefore consistent with those of the GAL4 fusion assays in Figure 1.
Figure 2.
Localization of the transactivation domain of GKLF using its cognate binding site. CHO cells were co-transfected with 5 µg/10 cm dish each of the TDAx2-E1bTATALUC reporter and the respective effector constructs containing various regions or mutations of GKLF (effectors 1–7) or the vector PMT3 (V) alone. (A) Means of fold-induction in reporter activity over that of the vector (n = 4). (B) Schematic of the various effector constructs. The three E*s in effector 6 indicate the mutated glutamate residues and the D*s in effector 7 indicate the three mutated aspartate residues.
To verify the preceding findings from the GAL4 fusion assay that the two clusters of acidic residues in the N-terminal 109 amino acids are indeed crucial for GKLF’s ability to activate transcription, we generated two point mutants that selectively changed either the three glutamates or three aspartates to valines in the context of full-length GKLF (Fig. 2B). As seen in Figure 2A, conversion of the three glutamates to valines (effector 6) resulted in a significant (∼75%) reduction in the ability of GKLF to activate transcription. In comparison, mutation of the three aspartates (effector 7) resulted in a total loss of activity. These findings suggest that both clusters of acidic residues are involved in mediating transactivation by GKLF although mutation of the aspartates seems more deleterious than that of the glutamates.
Transactivation by GKLF involves intermolecular interaction
Among members of the subfamily of Krüppel proteins closely related to GKLF (29), EKLF was the earliest identified (31) and probably best characterized. Like GKLF, the transactivation domain of EKLF has previously been localized to its N-terminal 124 amino acid (49). Although it has a different primary amino acid sequence from that of GKLF, this region of EKLF is also rich in acidic residues (31). Moreover, the mechanism of transactivation by EKLF appears to be complex and involves intermolecular interaction (49). We therefore asked the question whether a similar mechanism is also involved in transactivation by GKLF.
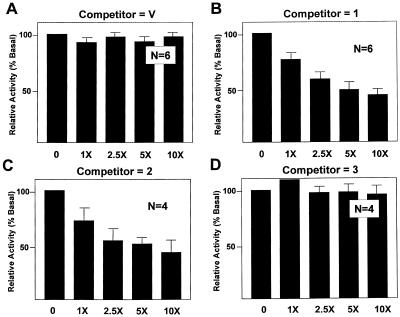
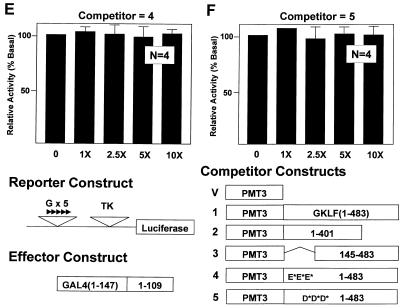
The possibility of intermolecular interaction as part of the mechanism for transactivation of GKLF was examined by an in vivo competition assay similar to that described previously (49). In this assay an expression construct containing full-length GKLF was introduced by transfection in gradually increasing amounts relative to the GAL4 fusion construct containing the N-terminal 109 amino acids of GKLF along with the pG5TKLUC reporter. If transactivation by GKLF requires the participation of interacting molecules, the presence of full-length GKLF in excess might ‘squelch’ the activating ability of its own transactivation domain. As shown in Figure 3, increasing amounts of full-length GKLF (competitor 1) competed for the activating effect of GAL4-GKLF(1–109) on the reporter (Fig. 3B) in a dose-dependent fashion, whereas the vector alone (competitor V) failed to do so (Fig. 3A). Moreover, the competing effect of GKLF was mediated by the N-terminal 401 amino acids (competitor 2; Fig. 3C) but not by the sequence between amino acids 145 and 483 (competitor 3; Fig. 3D). This indicates that the N-terminal 144 amino acids of GKLF compete for an interacting molecule(s) involved in transactivation. Significantly, both mutant constructs with substitution in the two acidic clusters within the N-terminus of GKLF (competitors 4 and 5) failed to compete for the activating effect of the N-terminal transactivation domain (Fig. 3E and F). These results suggest that intermolecular interaction is responsible, at least in part, for the transactivation by GKLF. Moreover, the same two clusters of acidic amino acids in the N-terminus necessary for GKLF’s transactivating effect are involved in such interaction.
Figure 3.
In vivo competition assays of transactivation by GKLF. CHO cells were transfected with 2 µg/10 cm dish of pG5TKLUC, 0.5 µg/10 cm dish of GAL4-GKLF(1–109) and increasing amounts of the various competitor PMT3 constructs in fold-excess relative to that of GAL4-GKLF(1–109) as indicated. The relative luciferase activity of cells that received no competitor DNA was chosen as 100%. n = 6 in experiments (A) and (B), and n = 4 in experiments (C)–(F).
Transactivation by GKLF involves p300/CBP
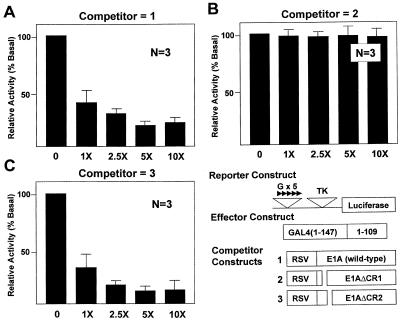
The recently described p300/CBP family of co-activators have been implicated in the regulation of transcription by modifying chromatin structure and by physically interacting with a host of transcription factors (50–52), including EKLF (53). An often employed strategy to study whether p300/CBP are involved in mediating the function of a transcription factor is to examine the effect of the adenovirus E1A oncoprotein on the activity of that factor. E1A is a multifunctional viral protein that modulates transcription of cellular genes by interacting with several key cellular proteins that include p300/CBP (14,15). To determine whether E1A also modulates the transactivating effect of GKLF, we performed in vivo competition experiments using expression constructs containing either wild-type or mutated E1A protein. As shown in Figure 4A, wild-type E1A (competitor 1) potently inhibited transactivation by the GAL4-GKLF(1–109) construct in a dose-dependent manner. Similarly, a mutated E1A construct that contained a deletion in its conserved region 2 (E1AΔCR2; competitor 3) was able to inhibit transactivation by the N-terminal 109 amino acids of GKLF (Fig. 4C). In contrast, another mutated E1A that contained a deletion in the conserved region 1 (E1AΔCR1; competitor 2) failed to inhibit GKLF’s ability to transactivate (Fig. 4B). These results indicate that E1A is a potent inhibitor of transactivation by GKLF and that the conserved region 1 of E1A exerts this inhibitory effect.
Figure 4.
In vivo competition of GKLF’s transactivation by E1A-expressing constructs. CHO cells were transfected with 2 µg/10 cm dish of pG5TKLUC, 0.5 µg/10 cm dish of GAL4-GKLF(1–109) and increasing amounts of a competitor RSV construct containing either wild-type or mutated E1A in fold-excess relative to that of GAL4-GKLF(1–109). The relative luciferase activity of cells that received no competitor DNA was chosen as 100%. n = 3 in all experiments. CR indicates conserved region.
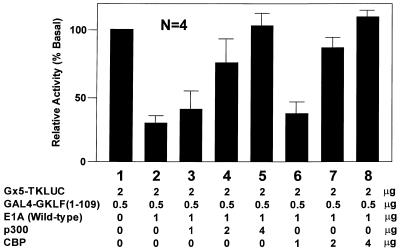
The conserved region 1 of E1A has previously been shown to bind p300/CBP (45,54,55). It is therefore possible that p300/CBP may mediate transactivation by GKLF. To determine this, we performed co-transfection experiments using expression constructs containing p300 or CBP (46,47). Figure 5 shows that both p300 and CBP were able to reverse the inhibition of GKLF’s transactivation by E1A in a dose-dependent fashion. These findings suggest that p300/CBP are indeed involved in mediating the transactivating activity by GKLF.
Figure 5.
p300/CBP reverses the inhibitory effect of E1A on transactivation by GKLF. CHO cells were transfected with 2 µg/10 cm dish of pG5TKLUC, 0.5 µg/10 cm dish of GAL4-GKLF(1–109), 1 µg/10 cm dish of RSV-E1A except for lane 1, and increasing amounts of RSV-p300 (lanes 3–5) or CMV-CBP (lanes 6–8) at the indicated concentrations. The relative luciferase activity of lane 1 was taken as 100%. n = 4.
GKLF and CBP physically interact with each other
E1A binds to p300/CBP. This raises the possibility that GKLF may also physically interact with p300/CBP. We therefore performed GST pull-down experiments using [35S]methionine-labeled, in vitro synthesized GKLF and GST fusion proteins containing various segments of the CBP protein. As shown in Figure 6, GKLF was recovered from the eluants when it was first incubated with GST-CBP2 (lane 5) and GST-CBP3 (lane 6), with CBP2 exhibiting a higher binding affinity. In contrast, GKLF failed to interact with GST-CBP1 (lane 4) or GST alone (lane 3). Similarly, no activity was recovered from the beads when no GST proteins were present in the initial incubation (lane 2). These results indicate that GKLF specifically binds to CBP and that this binding occurs in at least two different regions of CBP.
Figure 6.
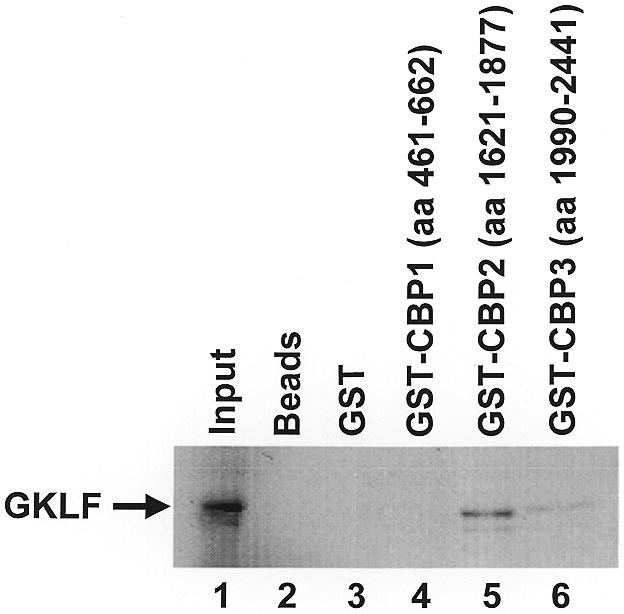
GST pull-down of GKLF by CBP. In vitro [35S]methionine-labeled GKLF was incubated with recombinant GST fusion proteins containing three different segments of CBP (lanes 4–6) or GST only (lane 3), followed by the addition of glutathione–Sepharose 4B beads. After thoroughly washing the beads with a detergent-containing solution, the bound proteins were eluted with reduced glutathione, resolved by denaturing PAGE and visualized by autoradiography. Lane 1, input GKLF, which represents 10% of the protein used in the pull-down experiments; lane 2, no input GST proteins.
The interaction between GKLF and CBP was further examined by co-immunoprecipitation of in vivo labeled proteins. COS-1 cells were co-transfected with RSV-CBP tagged with the HA epitope (47) and PMT3 or PMT3-GKLF, and then metabolically labeled with [35S]methionine. Lysates from transfected cells were immunoprecipitated with pre-immune serum, anti-GKLF serum or anti-HA antibodies, and the precipitated materials resolved by autoradiography. As shown in Figure 7, the anti-GKLF serum precipitated GKLF only in cells transfected with PMT3-GKLF (lane 5) but not in those with PMT3 (lane 2). Similarly, anti-HA antibodies precipitated a high-molecular-weight protein likely representing the HA-tagged CBP in transfected cells (lanes 3 and 6). Importantly, anti-HA antibodies also precipitated [35S]methionine-labeled GKLF in PMT3-GKLF-transfected cells (lane 6) but not in PMT3-transfected cells (lane 3). Moreover, when similar co-immunoprecipitation experiments were performed on the two point mutant proteins that involved the acidic residues of GKLF, anti-HA antibodies could no longer precipitate either GKLF mutant (lanes 9 and 11). These results strongly suggest that a physical interaction between GKLF and CBP occurs in vivo, and that the two clusters of acidic residues in the N-terminus of GKLF are crucial for this interaction.
Figure 7.
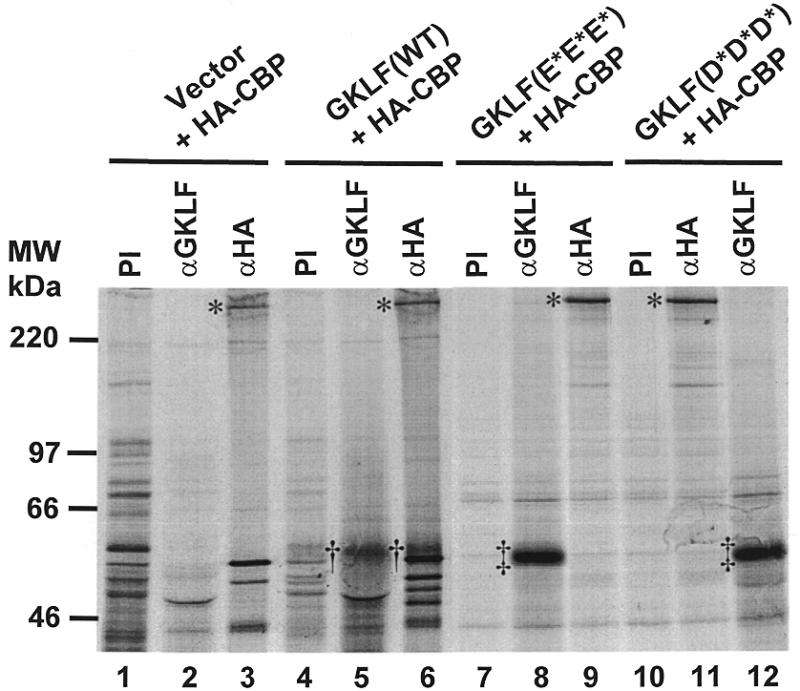
Co-immunoprecipitation of GKLF and CBP. COS-1 cells were transfected with 10 µg/10 cm dish each of RSV-HA-CBP and PMT3 (lanes 1–3), wild-type (WT) PMT3-GKLF (lanes 4–6), PMT3-GKLF(E93/95/96V) (E*E*E*; lanes 7–9), or PMT3-GKLF(D99/102/104V) (D*D*D*; lanes 10–12). Twenty-four hours later, proteins were metabolically labeled with [35S]methionine in vivo. Cells were then lysed and the lysates incubated with pre-immune (PI) serum, anti-GKLF serum (αGKLF) or anti-HA antibodies (αHA). The immune complex was precipitated with protein A–Sepharose beads and resolved by gel electrophoresis following repeated washings of the beads. The asterisks identify HA-CBP. †, Wild-type GKLF; ‡, the mutant GKLF.
Acidic residues are required for the growth-suppressing effect of GKLF
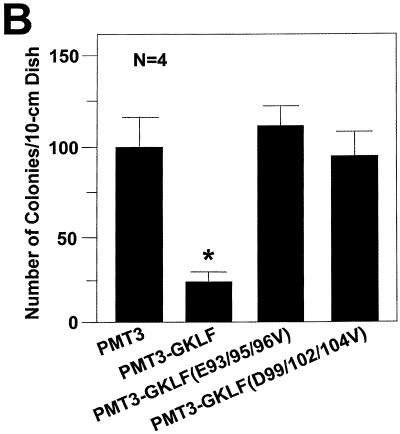
GKLF is a potent inhibitor of cell growth as demonstrated by previous studies involving transient transfection (23). In the present study we used an independent method to corroborate the growth-suppressing effect of GKLF—the colony suppression assay (48) that is based on stable transfection. Figure 8A shows that while cells transfected with the PMT3 control plasmid and a puromycin resistance-containing plasmid led to the formation of a large number of puromycin-resistant colonies, those transfected with PMT3-GKLF and the resistant marker had far fewer. The degree of colony suppression by GKLF was slightly over 75% as compared to vector alone, which represents a significant reduction (P < 0.005 by two-tailed Students’ t-test) (Fig. 8B). These results suggest that expression of GKLF hindered cell growth, therefore preventing the formation of colonies. Importantly, the two point mutants of GKLF that abolished the transactivating and interacting abilities of GKLF completely failed to suppress colony formation (Fig. 8A and B). We conclude that the same acidic residues in the N-terminus of GKLF are implicated in at least three of its biological activities including transactivation, protein–protein interaction and growth suppression.
Figure 8.
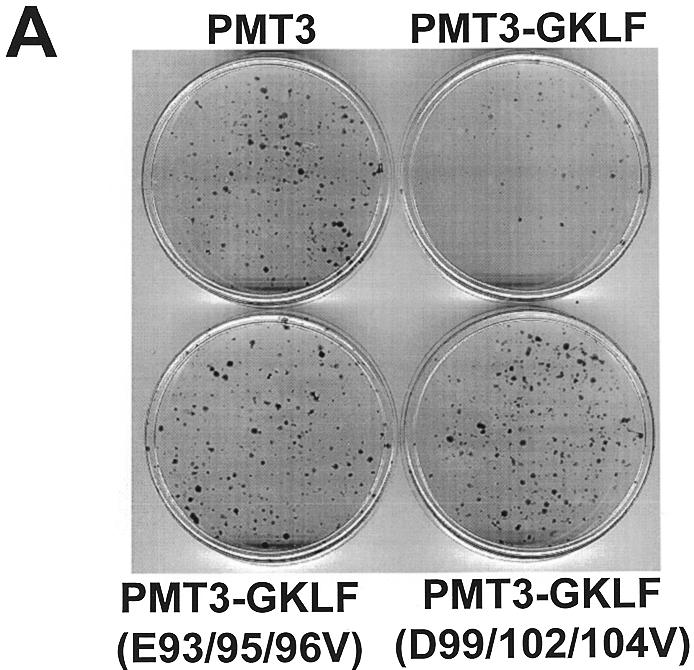
Colony suppression assays of GKLF. Rat1a cells were transfected with 10 µg/10 cm dish of the stated PMT3 constructs and 0.25 µg/10 cm dish of pBabe Puro (48). Two days following transfection, cells were fed media containing puromycin for 2 weeks. Resistant colonies of cells were visualized by staining with methylene blue (A). (B) Mean number of colonies for four dishes of cells transfected with each construct. *P < 0.005 by two-tailed Students’ t-test between PMT3- and PMT3-GKLF-transfected cells.
DISCUSSION
GKLF was first identified 3 years ago based on low-stringency cDNA library screening (23) and polymerase chain reaction amplification of mouse embryonic cDNA with degenerate oligonucleotides (25). Although the definitive physiological functions of GKLF have yet to be established, studies suggest that it is involved in the regulation of proliferation and differentiation of certain epithelial tissues (23–27). One important role that GKLF plays is to maintain a barrier function of the skin as demonstrated by recent experiments involving gene targeting (26). Recent studies have also delineated some of the biochemical properties of GKLF including the DNA sequences to which it binds (24,25,27,36,39), the potential target genes subject to its regulation (24,26,39) and the signals required for its nuclear localization (29). In addition, GKLF was shown to exhibit a dual mechanism in modulating transcription, acting as a repressor under certain circumstances (26,39) and an activator in others (24–26,36). Moreover, studies involving deletional mutagenesis showed that GKLF contains distinct repressive and activating domains (27). Taken together, these findings suggest that the mechanism of action of GKLF is complex. The current study thus provides new and detailed information on the mechanism of transactivation by GKLF from a structural–functional perspective.
Various mechanisms have been shown to be involved in the activation of transcription by eukaryotic transcription factors. The domains responsible for transactivation often belong to one or more of three sequences that are rich in glutamates (7,8), prolines (9,10) or acidic residues (11–13). Among the Krüppel-related proteins, Sp1 uses glutamate-rich domains for activation (7) while NGFI-A uses all three types of domains for activation (8). The transactivation domain of BTEB2, which exhibits significant homology to GKLF in the zinc finger region, is rich in proline residues (56). Although the primary amino acid sequence of GKLF is also very rich in prolines (12.8%) (23), our results indicate that these residues are not essential for transactivation by GKLF. Instead, certain acidic amino acid residues are required. These studies therefore demonstrate a functional heterogeneity in the mechanisms of transactivation by the Krüppel family of related proteins.
Be that as it may, there is a remarkable degree of similarity in the mechanism of transactivation between GKLF and EKLF, a member of the GKLF subfamily of Krüppel proteins (23,29). The activation domain of EKLF has been localized to between amino acid residues 20 and 124, a region that is rich in both acidic and proline residues (49). Protein–protein interaction has also been shown to be important in the ability of EKLF to activate transcription and evidence suggests that such interaction is mediated by acidic residues localized within the N-terminus of EKLF (49). EKLF physically interacts with p300 and CBP (53). In transient transfection assays the addition of p300/CBP increased transactivation by EKLF (53). Thus, despite a difference in the primary sequence between the activation domains of GKLF and EKLF, there appears to be a significant degree of functional conservation in the mechanism by which the two proteins activate transcription.
p300 and CBP belong to the family of proteins that exhibit histone acetyltransferase (HAT) activity (57). They function to stimulate transcription of specific promoters following their recruitment by specific DNA-binding proteins (50,51). The HAT activity within p300/CBP is responsible, at least in part, for this transcriptional stimulation (58,59). Numerous studies have demonstrated that p300/CBP may play a key role in the regulation of cell proliferation and differentiation (50,57). For example, p300 has been shown to physically interact with the myogenic transcription factor MyoD in differentiating myocytes. This interaction is necessary for both MyoD-induced activation of muscle-specific gene transcription and cell cycle arrest due to an up-regulation of the cyclin-dependent kinase inhibitor p21 (60). p300/CBP have also been shown to interact with the tumor suppressor p53, thereby modulating the cell cycle regulatory activity of p53 (47,61). Moreover, there is considerable evidence to suggest that interaction between the adenoviral oncoprotein E1A and p300/CBP is important for the biological effects of E1A including regulation of transcription, suppression of differentiation and immortalization of cells in culture (15). In the context of gut epithelial differentiation, expression of CBP has been associated with terminal differentiation of a model enterocyte, Caco-2 (62). Furthermore, CBP interacts with Cdx2, a homeodomain protein purportedly implicated in effecting intestinal epithelial differentiation (63). Taking into view the potential role of GKLF in regulating cell proliferation and differentiation, it is very possible that p300/CBP are also an important component that mediates some of GKLF’s biological activities.
Using transient transfection assays, we previously showed that GKLF is a potent inhibitor of DNA synthesis (23). This inhibitory effect was substantiated in the present study by the colony suppression assay that is based on stable transfection. Importantly, point mutations in GKLF abolishing the transactivating activity of GKLF also abolished its growth-inhibitory effect. Thus, transactivation is an essential element of GKLF’s biological functions. The significance of this finding is further increased by the fact that the same acidic residues are crucial for interaction with CBP. Consequently, these domains likely represent the convergence of several important aspects of GKLF’s activities, including transactivation, protein–protein interaction and growth suppression. Our study therefore provides strong evidence for an important structural and functional correlation among the various biological effects of GKLF.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs C. V. Dang, A. B. Leiter, T. Kouzarides, R. H. Goodman and R. G. Roeder for plasmids, and Channing Mahatan for technical assistance. This work was in part supported by grants DK44484, DK52230 and CA84197 (to V.W.Y.) from the National Institutes of Health. H.T.-T. was a recipient of a National Research Service award from the National Institutes of Health (DK09492). J.M.J. was a participant of the Minority Summer Internship Program of the Johns Hopkins University School of Medicine and a recipient of a Student Research Fellowship Award from the American Digestive Health Foundation.
REFERENCES
- 1.Yang V.W. (1998) J. Nutr., 128, 2045–2051. [DOI] [PubMed] [Google Scholar]
- 2.Johnson P.F. and McKnight,S.L. (1989) Annu. Rev. Biochem., 58, 799–839. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P.J. and Tjian,R. (1989) Science, 245, 371–378. [DOI] [PubMed] [Google Scholar]
- 4.Frankel A.D. and Kim,P.S. (1991) Cell, 65, 717–719. [DOI] [PubMed] [Google Scholar]
- 5.McKnight S.L. and Yamamoto,K.R. (1992) Transcriptional Regulation. Cold Spring Harbor Laboratory, New York, NY.
- 6.Klug A. (1995) Ann. N. Y. Acad. Sci., 758, 143–160. [DOI] [PubMed] [Google Scholar]
- 7.Courey A.J., Holtzman,D.A., Jackson,S.P. and Tjian,R. (1989) Cell, 59, 827–836. [DOI] [PubMed] [Google Scholar]
- 8.Russo M.W., Matheny,C. and Milbrandt,J. (1993) Mol. Cell. Biol., 13, 6858–6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mermod N., O’Neill,F.A., Kell,T.J. and Tjian,R. (1989) Cell, 58, 741–753. [DOI] [PubMed] [Google Scholar]
- 10.Artandi S.E., Merrell,K., Avitahl,N., Wong,K.K. and Calame,K. (1995) Nucleic Acids Res., 23, 3865–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hope I.A. and Struhl,K. (1986) Cell, 46, 885–894. [DOI] [PubMed] [Google Scholar]
- 12.Ma J. and Ptashne,M. (1987) Cell, 51, 113–119. [DOI] [PubMed] [Google Scholar]
- 13.Sadowski I., Ma,J., Triezenberg,S. and Ptashne,M. (1988) Nature, 335, 563–564. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N. and Harlow,E. (1992) Cancer Surv., 12, 161–195. [PubMed] [Google Scholar]
- 15.Moran E. (1993) Curr. Opin. Genet. Dev., 3, 63–70. [DOI] [PubMed] [Google Scholar]
- 16.Chrivia J.C., Kwok,R.P., Lamb,N., Hagiwara,M., Montminy,M.R. and Goodman,R.H. (1993) Nature, 365, 855–859. [DOI] [PubMed] [Google Scholar]
- 17.Kwok R.P., Lundblad,J.R., Chrivia,J.C., Richards,J.P., Bachinger,H.P., Brennan,R.G., Roberts,S.G., Green,M.R. and Goodman,R.H. (1994) Nature, 370, 223–226. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R., Ewen,M.E., Newsome,D., Gerdes,M., DeCaprio,J.A., Lawrence,J.B. and Linvingston,D.M. (1994) Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- 19.Yang X.J., Ogryzko,V.V., Nishikawa,J.I., Howard,B.H. and Nakatani,Y. (1996) Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- 20.Shikama N., Lyon,J. and La Thangue,N.B. (1997) Trends Cell Biol., 7, 230–236. [DOI] [PubMed] [Google Scholar]
- 21.Wu C. (1997) J. Biol. Chem., 272, 28171–28174. [DOI] [PubMed] [Google Scholar]
- 22.Turner J. and Crossely,M. (1999) Trends Biochem. Sci., 24, 236–240. [DOI] [PubMed] [Google Scholar]
- 23.Shields J.M., Christy,R.J. and Yang,V.W. (1996) J. Biol. Chem., 271, 20009–20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins T.D., Opitz,O.G., Okano,J. and Rustgi,A.K. (1998) J. Biol. Chem., 273, 10747–10754. [DOI] [PubMed] [Google Scholar]
- 25.Garrett-Sinha L.A., Eberspaecher,H., Seldin,M.F. and de Crombrugghe,B. (1996) J. Biol. Chem., 271, 31384–31390. [DOI] [PubMed] [Google Scholar]
- 26.Segre J.A., Bauer,C. and Fuchs,E. (1999) Nature Genet., 22, 356–360. [DOI] [PubMed] [Google Scholar]
- 27.Yet S.F., McA’Nulty,M.M., Folta,S.C., Yen,H.W., Yoshizumi,M., Hsieh,C.M., Layne,M.D., Chin,M.T., Wang,H., Perrella,M.A., Jain,M.K. and Lee,M.E. (1998) J. Biol. Chem., 273, 1026–1031. [DOI] [PubMed] [Google Scholar]
- 28.Ton-That H., Kaestner,K.H., Shields,J.M., Mahatanankoon,C.S. and Yang,V.W. (1997) FEBS Lett., 419, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shields J.M. and Yang,V.W. (1997) J. Biol. Chem., 272, 18504–18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson K.P., Kern,C.B., Crable,S.C. and Lingrel,J.B. (1995) Mol. Cell. Biol., 15, 5957–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller I.J. and Bieker,J.J. (1993) Mol. Cell. Biol., 13, 2776–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartzog G.A. and Myers,R.M. (1993) Mol. Cell. Biol., 13, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bieker J.J. and Southwood,C.M. (1995) Mol. Cell. Biol., 15, 852–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuez B., Michalovich,D., Bygrave,A., Ploemacher,R. and Grosveld,F. (1995) Nature, 375, 316–318. [DOI] [PubMed] [Google Scholar]
- 35.Perkins A.C., Sharpe,A.H. and Orkin,S.H. (1995) Nature, 375, 318–322. [DOI] [PubMed] [Google Scholar]
- 36.Shields J.M. and Yang,V.W. (1998) Nucleic Acids Res., 26, 796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanagida A., Sogawa,K., Yasumoto,K.I. and Fujii-Kuriyama,Y. (1990) Mol. Cell. Biol., 10, 1470–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujii-Kuriyama Y., Imataka,H., Sogawa,K., Yasumoto,K.-I. and Kikuchi,Y. (1992) FASEB J., 6, 706–710. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W., Shields,J.M., Sogawa,K., Fujii-Kuriyama,Y. and Yang,V.W. (1998) J. Biol. Chem., 273, 17917–17925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swick A.G., Janicot,M., Cheneval-Kastelic,T., McLenithan,J.C. and Lane,M.D. (1992) Proc. Natl Acad. Sci. USA, 89, 1812–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potter J.J., Cheneval,D., Dang,C.V., Resar,L.M.S., Mezey,E. and Yang,V.W. (1991) J. Biol. Chem., 266, 15457–15463. [PubMed] [Google Scholar]
- 42.Kato G.J., Barrett,J., Villa-Garcia,M. and Dang,C.V. (1990) Mol. Cell. Biol., 10, 5914–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dang C.V., Barrett,J., Villa-Garcia,M., Resar,L.M.S., Kato,G.J. and Fearon,E.R. (1991) Mol. Cell. Biol., 11, 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen K.J., Hanna,J.S., Prescott,J.E. and Dang,C.V. (1996) Mol. Cell. Biol., 16, 5527–5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bannister A.J. and Kouzarides,T. (1995) EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mutoh H., Naya,F.J., Tsai,M.J. and Leiter,A.B. (1998) Genes Dev., 12, 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu W., Shi,X.L. and Roeder,R.G. (1997) Nature, 387, 819–823. [DOI] [PubMed] [Google Scholar]
- 48.Egan S.E., Giddings,B.W., Brooks,M.W., Buday,L., Sizeland,A.M. and Weinberg,R.A. (1993) Nature, 363, 45–51. [DOI] [PubMed] [Google Scholar]
- 49.Chen X. and Bieker,J.J. (1996) EMBO J., 15, 5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 50.Giordano A. and Avantaggiati,M.L. (1999) J. Cell Physiol., 181, 218–230. [DOI] [PubMed] [Google Scholar]
- 51.Struhl K. (1998) Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 52.Wu C. (1997) J. Biol. Chem., 272, 28171–28174. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W. and Bieker,J.J. (1998) Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stein R.W., Corrigan,M., Yaciuk,P., Whelan,J. and Moran,E. (1990) J. Virol., 64, 4421–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong H.K. and Ziff,E.B. (1994) J. Virol., 68, 4910–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kojima S., Kobayashi,A., Gotoh,O., Ohkuma,Y., Fujii-Kuriyama,Y. and Sogawa,K. (1997) J. Biochem., 121, 389–396. [DOI] [PubMed] [Google Scholar]
- 57.Kouzarides T. (1999) Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- 58.Marinez-Balbas M.A., Bannister,A.J., Martin,K., Haus-Seuffert,P., Meisterernst,M. and Kouzarides,T. (1998) EMBO J., 17, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney,E.M., Mullen,T., Glass,C.K. and Rosenfeld,M.G. (1998) Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- 60.Puri P.L., Avantaggiati,M.L., Balsano,C., Sang,N., Graessmann,A., Giordano,A. and Levrero,M. (1997) EMBO J., 16, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lill N.L., Grossman,S.R., Ginsberg,D., DeCaprio,J. and Livingston,D.M. (1997) Nature, 387, 823–827. [DOI] [PubMed] [Google Scholar]
- 62.Lorentz O., Suh,E.R., Taylor,J.K., Boudreau,F. and Traber,P.G. (1999) J. Biol. Chem., 274, 7196–7199. [DOI] [PubMed] [Google Scholar]
- 63.Suh E. and Traber,P.G. (1996) Mol. Cell. Biol., 16, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]