Abstract
The plastid ndhH–D operon produces several transcripts containing ndhA sequence with and without its group II intron. After sequencing an 8125 bp fragment of barley plastid DNA including the ndhH–D operon, we investigated the editing–splicing status of transcripts in the range 1.0–7.8 kb. Reverse transcription and sequencing of RNA bands separated by electrophoresis were used to determine C→U editing sites. Sites I, II and IV of ndhA and site V of ndhD were edited in all transcripts analysed and, probably, were edited before any splicing had taken place. In contrast, site III of ndhA (13 bp from the 5′-end base of the second exon) was not edited in transcripts containing the intron (including the 1.7 kb intermediary transcript consisting of the intron and the second exon) but was edited in all transcripts lacking the ndhA intron. Comparison of the secondary structures of the ndhA intron and intron–second exon intermediate suggests that G pairing prevents editing of site III in transcripts containing the intron and maintains the secondary structure required for splicing. Splicing of the ndhA intron releases the site III C from pairing and, probably, brings it close to cis- acting elements for editing upstream in the first exon.
INTRODUCTION
The plastid DNA of higher plants contains 11 reading frames (ndhA–ndhK) that are homologous to genes encoding subunits of mitochondrial NADH dehydrogenase (complex I, EC 1.6.5.3) (1). Transcripts of plastid ndh genes were first identified by northern blot hybridisation in tobacco (2) and later in maize (3), pea (4), rice (5) and many other higher plants. A 7–8 kb transcript (5), resulting from a transcriptional unit consisting of the ndhH to ndhD genes (the ndhH–D operon), includes ndh genes H, A, I, G, E and D and the gene psaC, which encodes the PsaC protein of photosystem I and maps between the ndhE and ndhD genes.
As with other plastid DNA operons, a number of transcripts containing different combinations of genes of the ndhH–D operon were found in addition to the longest polycistronic transcript (6,7). RT–PCR amplification and sequencing indicated that at least one transcript of the ndhH–D operon contains the intron of the ndhA gene, while another contains the message for the ndhA gene without the intron (8). Several nuclear mutations in Arabidopsis thaliana (6,7) have pleiotropic effects in the transcript pattern of different plastid operons, including ndhH–D, indicating that the nuclear mutated genes encode polypeptides of the splicing/processing machinery which are common for large primary transcripts of different plastid DNA operons. Similarly, nuclear mutations in maize affect splicing of the ndhA intron (9).
In addition, several primary transcripts of ndh genes are edited by changing certain cytosine (C) bases to uracil (U) bases in the RNA (10). Edited sites are identified by comparing genomic and complementary DNA (cDNA) sequences. Thus, ndhD requires editing in dicotyledons to restore the start codon (10,11), while in monocotyledons it is edited at nucleotide position 878 (12), changing the genomic UCA codon encoding serine to UUA, encoding a highly conserved leucine at position 293 of the amino acid sequence. The mature ndhA transcript has four edited sites (two in each exon) in barley and one to four edited sites in equivalent positions in many plants (8,13), but none in Arabidopsis (our unpublished results).
In general, editing creates a start AUG codon in mRNA from an ACG codon in primary transcripts (11) or modifies nucleotides within the message (13) rendering codons for conserved amino acids (10,14). However, silent editing of the third base of codons (not changing the encoded amino acid) and in non-coding regions near the initiation codon are sometimes found (10,15). In the latter cases, editing might change the secondary structure of RNA as a necessary step for its translation or splicing. Alternatively, in some cases splicing may be required for editing.
Despite some reports about editing in unspliced and polycistronic transcripts (16,17), the order of splicing and editing in plastid transcript processing is poorly known. Within the ndhH–D operon, questions such as which of the many transcripts are edited or contain the ndhA intron also remain unanswered.
In this work, we have sequenced the entire ndhH–D operon of barley (Hordeum vulgare L cv. Hassan) and have selected an electrophoresis system which, in the absence of formaldehyde, allowed reverse transcription, amplification and sequencing of isolated RNA bands. By comparing cDNA sequences of different bands and the deduced secondary structures, we have investigated the relationship between splicing and site III editing in ndhA transcripts.
MATERIALS AND METHODS
Plant materials
Greening leaves (from plants grown for 6 days in the dark followed by 18 h in light) of barley (Hordeum vulgare L cv. Hassan) were used as the source of chloroplast DNA and RNA.
Isolation of chloroplast DNA and RNA
Total DNA was isolated from barley leaves by a modification of the method described by Ausubel et al. (18). Leaves were ground in liquid nitrogen and the frozen powder was resuspended (8 ml buffer/g leaves) in 100 mM Tris–HCl, 50 mM EDTA, 10 mM NaCl, 100 µg/ml proteinase K and 2% (w/v) SDS (pH 8.5) and incubated for 30 min at 50°C. DNA was extracted twice with phenol:chloroform:isoamyl alcohol (25:24:1 v/v/v) saturated with 10 mM Tris–HCl pH 8.9 and twice with chloroform:isoamyl alcohol (24:1 v/v). A few times, chloroplast DNA was isolated by suspending the frozen leaf powder (8 ml buffer/g leaves) in 0.3 M sorbitol, 50 mM Na HEPES, 2 mM DTT (pH 8.0) as previously (19). The DNA was precipitated with isopropanol, washed with 70% ethanol and finally dissolved in water.
For RNA isolation, leaf segments (1 g) were ground in liquid nitrogen and RNA preparation was performed by phenol/SDS extraction and selective precipitation with LiCl (20,21). RNA yields were ~0.25 mg/g leaves.
RNA electrophoresis, isolation and northern blot hybridisation
Total RNA samples were treated with formaldehyde/formamide (22) and different electrophoresis systems were assayed for the separation of transcripts (80 µg RNA/lane): formaldehyde-containing gels (22), urea-containing gels and non-denaturing pre-cast (Reliant-Gel System) FMC gels. After electrophoresis, RNA bands were isolated using the RNAaid Kit from BIO101 for synthesis of the corresponding cDNAs, their amplification and sequencing. Most electrophoreses were carried out in non-denaturing pre-prepared FMC gels.
For northern blot hybridisation, after electrophoresis, separated RNA bands were immobilised on nylon membranes (Zeta-Probe; Bio-Rad). Hybridisations were carried out using the non-radioactive DNA labelling and detection kit from Boehringer Mannheim (Mannheim, Germany) and PCR-generated probes.
Reverse transcription, and amplification and sequencing of DNA and cDNA
Total RNA and RNA bands isolated from non-denaturing FMC gels were reverse transcribed and amplified using the Access RT-PCT System (Promega). Cycling conditions were: initial reverse transcription at 48°C for 45 min; one cycle of 94°C for 2 min; then 94°C for 30 s, 50°C for 90 s and 68°C for 3 min for 50 cycles; followed by a final extension at 68°C for 7 min. Primer pairs were based on the sequence of maize plastid DNA (1) and partial sequences of barley plastid DNA obtained in our laboratory.
Amplification products were subjected to electrophoresis through 1.5% agarose gels, purified with a DNA gel extraction kit (Qiaex II; Qiagen) and used for sequence analysis.
Genomic DNA and cDNAs were sequenced in an Applied Biosystems automatic sequencer. Three to five different RT–PCR sequencing assays were carried out for each specific RNA preparation to identify editing sites.
The Zuker and Stiegler program (23; GCG, Madison, WI) was used to determine secondary structures of RNA.
RESULTS
The ndhH–D operon of barley plastid DNA
An 8125 bp sequence (DDBJ/EMBL/GenBank accession no. AJ011848) mapped in the small single copy region of barley plastid DNA was determined by PCR amplification and sequencing using 49 different primers. The sequence included the ndh genes H, A, I, G, E and D (in that order) and the gene psaC (which encodes the PsaC protein of photosystem I), mapping between the ndhE and ndhD genes.
The gene organisation in this sequence is similar to that found in other plants and corresponds to a transcriptional unit of the ndhH to ndhD genes named the ndhH–D operon (1,24–28). The sequence from the first base of the ndhH gene to the last base of the ndhD gene is longer (7602 bp) in barley than in rice (7492 bp), maize (7500 bp), tobacco (7595 bp), Arabidopsis (7528 bp) and Marchantia (6716 bp).
ndhH–D operon transcripts containing ndhA and/or ndhD mRNAs
To investigate C→U editing sites in ndhH–D transcripts, we assayed the ability of different RNA electrophoresis systems to obtain isolated RNAs for cDNA synthesis, amplification and sequencing. Electrophoresis in non-denaturing pre-prepared FMC gels produced medium quality band separation and northern blot hybridisation patterns, but the isolated bands were suitable for synthesis of the corresponding cDNAs, their amplification and sequencing.
Figure 1 shows northern blot hybridisation patterns obtained after electrophoresis of RNA in pre-prepared FMC gel when PCR-produced probes specific for the ndhA gene (ndhA, S2), ndhA intron (S3) and ndhD gene (S5) were used. The transcript of the 7.8 kb band could contain the message of the seven genes, including the intron of ndhA. The wide 6.7 kb band detected with the ndhA probe could include several transcripts of very similar size. For instance, a transcript containing the sequence for the seven genes including the ndhA coding region without its intron and a transcript containing the sequence for the six genes from ndhA (including the intron) to ndhD (Fig. 2). Northern blots hybridised with other probes (not shown) and sequence analysis described below confirmed the heterogeneity of the 6.7 kb band. Similar assays suggested that the 5.4 kb band might contain the sequence for the six genes from ndhA (without the intron) to ndhD. Bands of 3.5 and 2.8 kb could include transcripts containing at least the ndhA sequence with the intron, while the 2.3 and 1.2 kb bands probably contain a spliced ndhA sequence (without the intron). The 1.0 kb band might correspond to the ndhA intron. Figure 2 shows the proposed map (deduced from the northern blots of Figure 1 and others not shown with probes S1 and S4) of the most abundant transcripts containing ndhA mRNA with or without its intron and/or ndhD mRNA.
Figure 1.
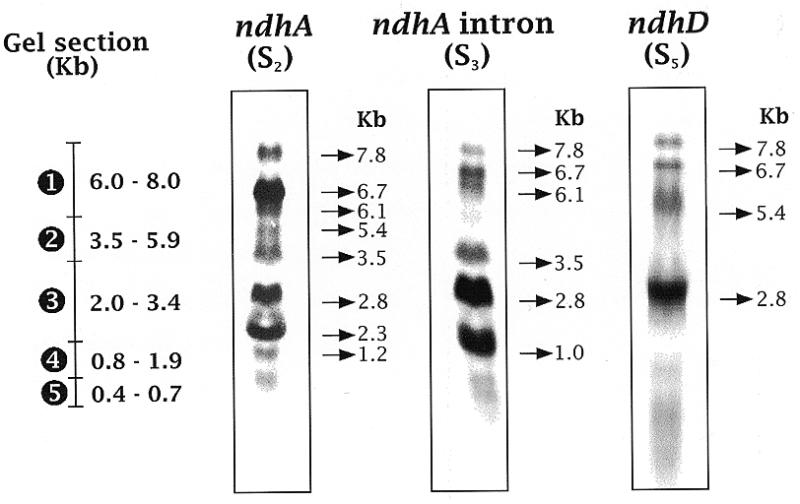
Northern blots of transcripts of the ndhH–D operon and gel sections used for transcript elution. After electrophoresis of barley RNA, lanes were northern blotted with probes S2 (the exons of ndhA, generated by RT–PCR from spliced ndhA transcripts), S3 (the intron of ndhA) and S5. The gel sections used to isolate RNA are indicated at the left of the northern lanes. Sizes (kb) of the main transcripts detected by northern blot and of the RNAs from the gel section are also indicated.
Figure 2.
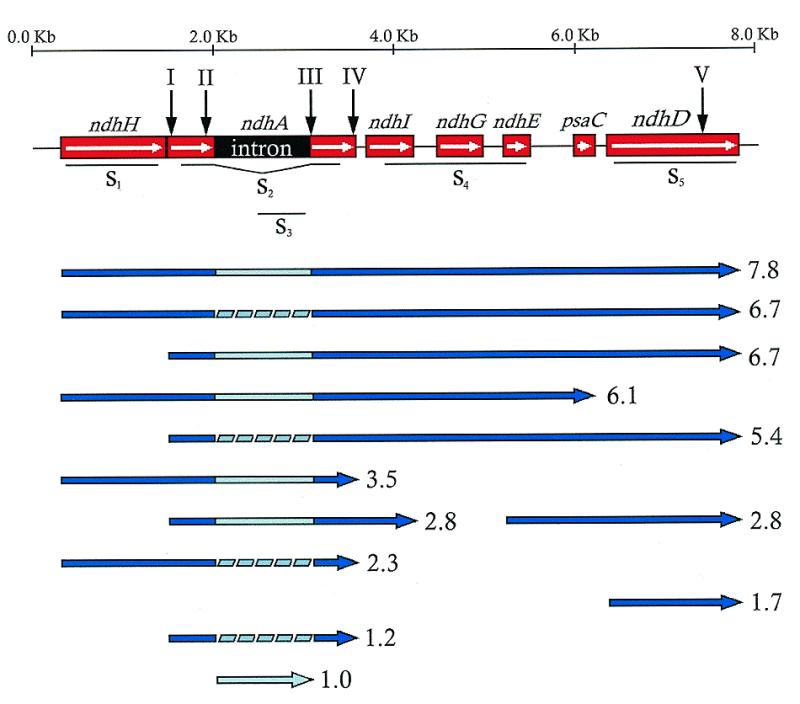
Gene and transcript maps of the ndhH–D operon of barley. Gene map of the 8125 bp sequenced DNA region of barley (up) shows with roman numbers on vertical arrows the location of the edited sites. Probes used in northern assays are represented under the gene map. The map of transcripts (down) was deduced from northern assays and RT–PCR amplifications and sequencing with specific primer pairs. The estimated sizes are indicated (kb) at the right of each transcript.
Western assays with a specific antibody (29) showed that polypeptide NDH-A, encoded by the ndhA gene, was present in barley leaves at different developmental stages. The level of NDH-A polypeptide increased with leaf age and after photo-oxidative treatment of the leaves. One possibility is that changes in the relative level of each transcript (due to control of specific transcript processing steps) together with differential translatability of each transcript containing ndhA could control the synthesis and level of NDH-A. As the ndhA gene without the intron has 1089 bases, the 1.2 kb band (Figs 1 and 2) could contain the shorter transcript that translates as NDH-A protein.
Processing of the large primary 7.8 kb transcript of the ndhH–D operon (Fig. 2) may originate the other transcripts. However, from our experiments, the possibility that there is more than one primary transcript cannot be excluded. In any case, the order in which splicing and editing of specific sites takes place are questions to be investigated. To advance in this direction, we have isolated and sequenced specific transcript bands.
Editing/splicing of different transcripts of the ndhH–D operon
Comparison of cDNA sequences obtained from total RNA preparations with genomic sequences revealed that in barley four C sites are edited to U in the mature ndhA transcript (two in each exon) (8) and one in ndhD (28). Editing changes the serines encoded at the genome level to leucines (sites I, II, III and V) or phenylalanine (site IV), which are highly conserved when the sequences of homologous proteins are compared in different plants. The map of the barley ndhH–D operon in Figure 2 shows the five editing sites and Table 1 gives the sequences around them.
Table 1. Sequences flanking the five sites edited in transcripts of the ndhH–D operon.
Edited C residues are indicated in bold. Numbers indicate the positions from the start codon of mature ndhA mRNA (sites I, II, III and IV) or ndhD (site V). The positions of intron ends are also indicated.
After electrophoresis of RNA in non-denaturing pre-prepared FMC gels, RNA bands of gel slices (of two contiguous lanes) were isolated. Lanes with size markers and with leaf RNA that was later northern blotted and hybridised with ndhA probe were run in parallel. After cDNA synthesis, specific DNA amplification primers were used to identify sequences contained in the RNAs isolated from different slices. According to size markers, five agarose gel sections (1–5 indicated in Fig. 1), which mainly included transcripts of 6.0–8.0, 3.5–5.9, 2.0–3.4, 0.8–1.9 and 0.4–0.7 kb, respectively, were sliced. It must be noted that many transcripts of the ndhH–D operon not containing ndhA message must be present in different gel slices.
The amplification of specific cDNAs synthesised from RNA eluted from each gel section with specific primer pairs further confirmed the transcript map deduced from northern blot hybridisation and shown in Figure 2.
The genomic C of site V (in ndhD-containing transcripts) was found to be always edited to U in cDNAs amplified from gel sections 1–4 (see Fig. 1). As expected by the size of the ndhD gene (1.5 kb), none of the above cited primer pairs could amplify any cDNA from RNA eluted from gel section 5. Controls with primer pairs within the short ndhG gene (531 bases) successfully amplified the corresponding cDNA from RNA eluted from section 5. Additional controls with RNA eluted from gel sections above 1, which would include RNAs longer than 8.0 kb, did not amplify with any primer pair, which indicated the validity of the approach to synthesise and amplify cDNA from RNAs separated by electrophoresis. The results on site V of ndhD indicated that it is probably edited before splicing or other processes modify the primary transcripts.
The C→U editing of sites I–IV was investigated by comparing genomic sequences with those of cDNAs synthesised from RNA isolated from different gel sections. After cDNA synthesis, a large number of primer pair combinations from the 5′-end of ndhH (P11) to the 3′-end of ndhG (P33) were used for primary and secondary amplifications and for sequencing. This approach allowed distinction between ndhA intron-containing transcripts and those not containing the ndhA intron. Thus, as shown in Figure 3, primers P21 (in the ndhA intron) and P30 (in ndhI) amplified the expected 1226 bp fragment (lane 1) including approximately one-third of the intron, the second exon of ndhA, the ndhA–ndhI intergenic region and approximately two-thirds of ndhI. From the same RNA sample, primers P16 (close to the 5′-end of the first exon of ndhA) and P30 (in ndhI) amplified a 1048 bp fragment (lane 3) including the end of the first exon, the entire second exon of ndhA, the ndhA–ndhI intergenic region and approximately two-thirds of ndhI. With the latter primer pair, a hypothetical fragment of 2082 bp, which would include the intron of ndhA, could not be amplified (see Fig. 3, lane 3).
Figure 3.
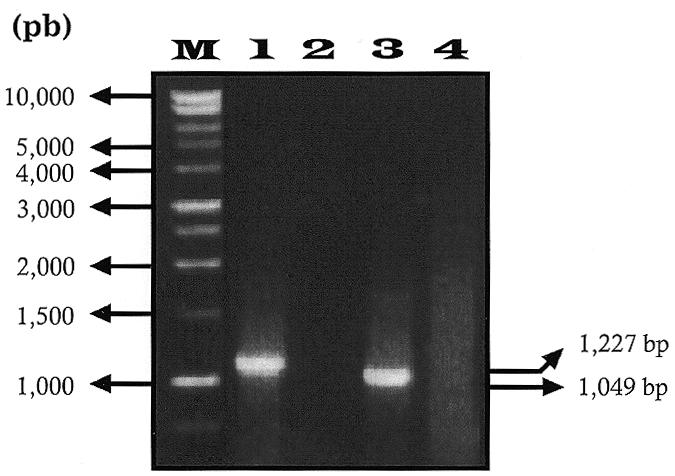
Agarose gel electrophoresis of RT–PCR amplification products with and without sequences of the ndhA intron. RNA isolated from gel section 1 (Fig. 1) was RT–PCR amplified with primer pairs P21 (bases 798–819 of the ndhA intron) and P30 (bases 331–352 of ndhI) (lane 1) and P16 (bases 490–515 of the first exon of ndhA) and P30 (lane 3). Lanes 2 and 4 are RT–PCR controls without eluted RNA of lanes 1 and 3, respectively. Lane M contains size markers (bp) indicated at the left. Sizes (bp) of amplified bands are indicated at the right.
When possible, amplifications with appropriate primer pairs were also used to confirm the polycistronic nature of large sized bands containing the ndhA message (Fig. 2).
All ndhA-containing transcripts, with sizes ranging from 0.8 to 8.0 kb, with or without the ndhA intron, underwent C→U editing at sites I, II and IV. As in the case of site V in ndhD, transcripts not edited at sites I, II and IV, if present, were at concentrations too low to be detected by reverse transcription and amplification. Thus, sites I, II and IV are probably edited early during the processing of transcription of the ndhH–D operon.
The transcript map (Fig. 2) indicates that each gel section (1–4 in Fig. 1) included at least one transcript containing the ndhA intron. In fact, the 1.0 kb band seems to contain only the intron. Accordingly, primer pairs containing sequences of the intron amplified the corresponding cDNA of RNA isolated from all of these gel sections but not from section 5 (Fig. 1). Pairs consisting of one primer from the intron (P18, P20 or P21) and one primer from the second exon (P23 or P25) of ndhA also amplified cDNA synthesised from RNA isolated from sections 1–3 and (surprisingly) 4. P27 is 47–71 bases downstream of the 3′-end of the second exon (in the intergenic region between ndhA and ndhI; see Fig. 2). Accordingly, P20/P27 and P21/P27 amplified cDNA of a 1.7 kb transcript of section 4 which includes the ndhA intron and the second exon. Although probes S2 and S3 (Fig. 2) recognised some transcripts in section 4 (Fig. 1), no obvious common transcript was recognised by both probes in section 4. The cDNA amplification of RNA from section 4 with primer pairs P20/P27 and P21/P27 suggested that a low level ndhA transcript, not detected by northern blot, was present in gel section 4. This transcript is probably an intermediate of intron splicing of the monocistronic ndhA transcript.
Comparison of the different cDNA sequences with genomic sequences showed that, regardless of transcript size, site III is edited only in spliced ndhA transcripts lacking the intron. Results obtained from sequencing cDNA regions (not genomic DNA) amplified with primer pairs P20/P27 and P21/P27 to include sites III and IV showed that only the latter was edited. Either intron splicing is required to edit site III or editing of site III is required for splicing of the ndhA intron. Northern blots (Fig. 1) indicated that chloroplasts contain significant levels of many transcripts that include the ndhA intron, which leads us to conclude that editing of site III and splicing of the ndhA intron do not immediately follow transcription.
Secondary structure of the ndhA intron–second exon transcript region
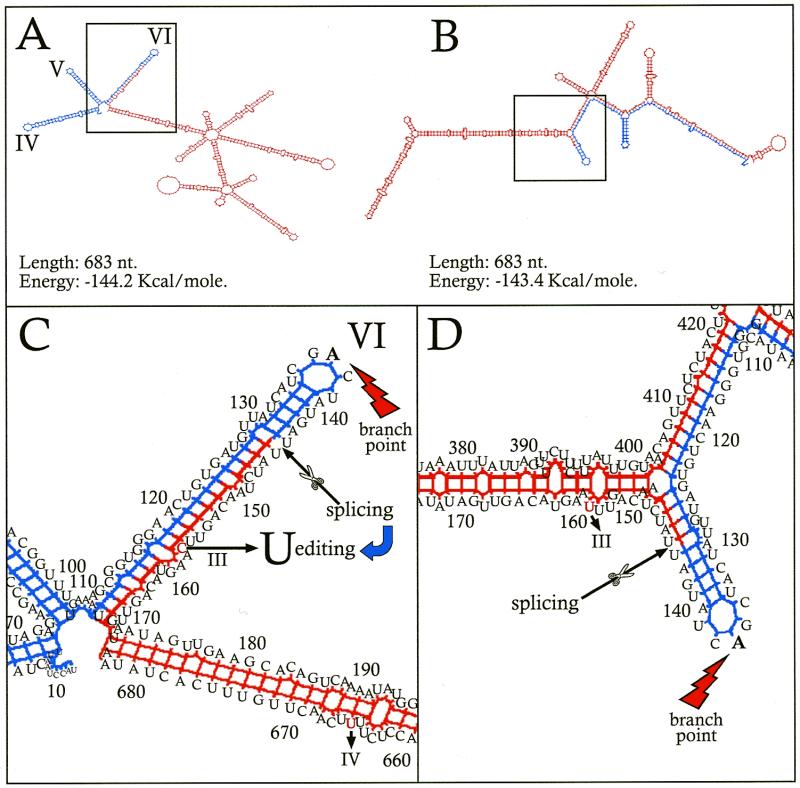
The secondary structure predicted for the RNA of the ndhA intron of barley has the typical features of group II introns (30) (not shown). At the 3′-end, in domain VI, an A base (eighth base from the 3′-end) forms a transitory 2′-phosphodiester bond with the intron 5′-end during splicing. When C remains unedited at site III in intron-containing ndhA transcripts, the secondary structure around site III conserves the domain structure of the intron (Fig. 4A and C) (energy –144.2 kcal/mol for the 683 base sequence including domains IV–VI of the intron and the entire second exon). Although domain VI is enlarged with 24 bases of the second exon 5′-end, the A base forming a 2′-phosphodiester bond remains unpaired in the central loop. Probably, the intron may be removed from this intermediate structure in which the unedited C (13th base of the exon 5′-end) is paired with the 29th base from the intron 3′-end. Surprisingly, the predicted secondary structure of the intron–second exon region when C is edited to U at site III is completely different (Fig. 4B and D) (energy –143.4 kcal/mol). It lacks the typical domains of group II introns. Probably, the intron cannot be removed from this structure, i.e. splicing could not occur if site III was previously edited. Hence, editing of site III must take place after the intron is removed from the intron–second exon intermediate.
Figure 4.
Predicted secondary structures flanking editing site III in intron-containing ndhA transcripts of barley. The secondary structures were determined (35) for a RNA region including the 144 bases of the intron 3′-end (blue) and the entire second exon (539 bases) (red) with site IV edited to U and site III unedited (A) and edited (B). (C and D) Expanded details around site III of (A) and (B), respectively. Domains IV, V, VI and A of the maturation branching point are indicated. Numbers correspond to nucleotide positions starting from the intron 5′-end. Other details are given in the text.
Site III of ndhA does not require editing in Arabidopsis, maize and tobacco, where it is a T at the genome level. This poses the question of the secondary structure of their presumed intron–second exon intermediates, which would contain U at site III. In contrast to barley, the secondary structures (not represented) predicted for Arabidopsis, maize and tobacco sequences equivalent to that shown in Figure 4A (energies –136.6, –141.0 and –160.6 kcal/mol, respectively) conserve the typical structure of group II introns. They are similar to that shown in Figure 4A for the unedited barley transcript. Apparently, other features of the second exon and/or intron sequences in these plants maintain the group II intron structure in the intron–second exon intermediate. After intron splicing, the predicted secondary structure in barley (not represented) shows the C base of site III unpaired in a loop, presumably exposed for editing.
DISCUSSION
The possibility of agarose gel extraction of specific RNA bands that can be reverse transcribed, amplified and sequenced allows the investigation of specific transcripts in mixtures containing different transcripts of a gene. This problem arises when transcripts of plastid polycistronic operons are analysed, since both primary and processed products containing different gene combinations are usually present (5). Questions such as which transcripts are edited, which are the primary transcripts and the processed products and which are translated by in vitro systems, as well as many other aspects, could probably be answered with this technique.
Here, we have applied the technique to identify editing sites in specific transcripts. Obviously, the conclusion that site III is not edited in transcripts containing the ndhA intron may also be reached by using appropriate primer pairs for the amplification of cDNA synthesised from total RNA. However, only cDNA synthesis and amplification of size-determined gel bands allows determination of whether a cDNA fragment amplified with appropriate primers proceeds from short or long transcripts. Since the efficiencies of reverse transcription and cDNA amplification decrease drastically for RNAs longer than 2.0 kb (22), previous size separation by electrophoresis has also allowed us to demonstrate that the ndhA intron is present in both long and short transcripts.
Apparently, the high formaldehyde concentrations used in conventional electrophoresis, in contrast to native electrophoresis, produce changes in RNA that make it unsuitable for cDNA synthesis. Gel sections shorter than those used in this work, combined with long run electrophoresis gels, would probably allow the identification of sequence characteristics of RNAs of more specific bands. The 5′- and 3′-end sequences could eventually be determined, although the large number of transcripts and indirect evidences for the existence, in chloroplasts, of multiple transcripts differing by a few end nucleotides leaves those determinations outside the scope of this work.
Clearly, at least for site III, editing does not immediately follow transcription. Site III, edited in ndhA transcripts lacking the ndhA intron but not in those unspliced, is located at the 13th base from the second exon 5′-end. The amplification of cDNA synthesised from RNA of section 4 with primer pairs based on the intron and second exon sequences indicated the existence of a low level transcript, of ~1.7 kb, consisting of the ndhA intron and its second exon, in which site III is not edited. This transcript would be the intron splicing intermediate of the monocistronic ndhA transcript which has been described (31), typical of group II introns such as that of ndhA (32). In the case of site III, the lack of editing in the short intron–second exon transcript suggests that splicing of the ndhA intron occurs prior to editing of site III. The comparison of secondary structure predictions with the mechanism (31–33) of splicing of group II introns supports this suggestion.
The pairing of the site III C in Figure 4C suggests that it can hardly be deaminated to U before removing the intron, thus complete splicing of the intron would be required for editing of site III. On the other hand, the secondary structure predicted for the 1.7 kb intron–edited second exon intermediate (Fig. 4B) suggests that the intron could hardly be completely removed if transcripts of ndhA are previously edited at site III. In plants such as Arabidopsis, maize and tobacco, in which site III is a U because T (not C) is present at the genome level, other features of the second exon and/or intron sequences maintain the appropriate structure of the group II intron and, consequently, proper splicing of the intron. If not accompanied by other sequence changes, the correction of site III at the genome level in barley would produce ndhA transcripts that cannot be spliced out. In barley plastids, the site III C would be edited to U only after the intron is spliced out because the C base becomes unpaired and/or linking of the first and second exons permits the approximation of upstream sequences which could favour editing (34,35).
The results described here are compatible with transcript processing of the ndhH–D operon in which a primary transcript of ~7.8 kb (containing the sequence of the seven genes of the operon) is spliced at intergenic regions and the ndhA intron is removed without a preferential order. Sites I, II and IV are probably edited before any splicing has taken place. Site III is only edited after splicing of the ndhA intron frees the site III C base of pairing and/or brings it close to cis-acting elements upstream of the 3′-end of the ndhA first exon.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr R. M. Maier for helpful discussions and suggestions during the preparation of this manuscript and C. Díaz-Sala, M. D. Abarca and P. Hauke for kind discussions and suggestions. This work has been supported by the Spanish DGICYT (grant PB96-0675).
DDBJ/EMBL/GenBank accession no. AJ011848
REFERENCES
- 1.Maier R.M., Neckermann,K., Igloi,G.L. and Kössel,H. (1995) J. Mol. Biol., 251, 614–628. [DOI] [PubMed] [Google Scholar]
- 2.Matsubayashi T., Wakasugi,T., Shinozaki,K., Yamaguchi-Shinozaki,K., Zaita,N., Hidaka,T., Meng,B.Y., Ohto,C., Tanaka,M., Kato,A., Maruyama,T. and Sugiura,M. (1987) Mol. Gen. Genet., 218, 385–393. [DOI] [PubMed] [Google Scholar]
- 3.Schantz R. and Bogorad,L. (1988) Plant Mol. Biol., 11, 239–247. [DOI] [PubMed] [Google Scholar]
- 4.Woodbury N.W., Roberts,L.L., Palmer,J.D. and Thompson,W.F. (1988) Curr. Genet., 14, 75–89. [Google Scholar]
- 5.Kanno A. and Hirai,A. (1993) Curr. Genet., 23, 166–174. [DOI] [PubMed] [Google Scholar]
- 6.Meurer J., Berger,A. and Westhoff,P. (1996) Plant Cell, 8, 1193–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinkins R.D., Bandaranayake,H., Baeza,L., Griffiths,A.J.F. and Green,B.R. (1997) Plant Physiol., 113, 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez C., Freyer,R., Guera,A., Maier,R.M., Martin,M. and Sabater,B. (1997) Plant Physiol., 115, 313.9380777 [Google Scholar]
- 9.Jenkins B.D., Kulhanek,D. and Barkan,A. (1997) Plant Cell, 9, 283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier R.M., Zeltz,P., Kossel,H., Bonnard,G., Gualberto,J.M. and Grienenber,J.M. (1996) Plant Mol. Biol., 32, 343–365. [DOI] [PubMed] [Google Scholar]
- 11.Neckermann K., Zelt,P., Igloi,G.L., Kössel,H. and Maier,R.M. (1994) Gene, 146, 177–182. [DOI] [PubMed] [Google Scholar]
- 12.Del Campo E.M., Albertazzi,F., Freyer,R., Maier,R.M., Sabater,B. and Martín,M. (1998) Plant Physiol., 117, 718. [Google Scholar]
- 13.Maier R.M., Hoch,B., Zeltz,P. and Kössel,H. (1992) Plant Cell, 4, 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freyer R., Lopez,C., Maier,R.M., Martin,M., Sabater,B. and Kössel,H. (1995) Plant Mol. Biol., 29, 679–684. [DOI] [PubMed] [Google Scholar]
- 15.Hirose T., Fan,H., Suzuki,J.Y., Wakasugi,T., Tsudzuki,T., Kössel,H. and Sugiura,H. (1996) Plant Mol. Biol., 30, 667–672. [DOI] [PubMed] [Google Scholar]
- 16.Freyer R., Hoch,B., Neckermann,K., Maier,R.M. and Kössel,H. (1993) Plant J., 4, 621–629. [DOI] [PubMed] [Google Scholar]
- 17.Ruf S., Zeltz,P. and Kössel,H. (1994) Proc. Natl Acad. Sci. USA, 91, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (1989) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY, Vol. I, pp. 2.3.1–2.3.3.
- 19.Martínez P., Lopez,C., Roldan,M., Sabater,B. and Martin,M. (1997) Plant Sci., 123, 113–122. [Google Scholar]
- 20.Jones J.D.G., Dunsmuir,D. and Bedbrook,J. (1985) EMBO J., 4, 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eker J.R. and Davis,R.W. (1987) Proc. Natl Acad. Sci. USA, 84, 5202–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Habor, NY.
- 23.Zuker M. and Stiegler,P. (1981) Nucleic Acids Res., 9, 133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohyama K., Fukuzawa,H., Kohchi,T., Shirai,H., Sano,T., Sano,S., Umesono,K., Shiki,Y., Takeuchi,M., Chang,Z., Aota,N.S., Inokuchi,H. and Ozeki,H. (1986) Nature, 322, 572–574. [Google Scholar]
- 25.Shinozaki K., Ohme,M., Tanaka,M., Wakasugi,T., Hayashida,N., Matsubayashi,T., Zaita,N., Chunwongse,J., Obokata,J., Yamaguchi-Shinozaki,K., Ohto,C., Torazawa,K., Meng,B.Y., Sugita,M., Deno,H., Kamogashira,T., Yamada,K., Kusuda,J., Takaiwa,F., Kato,A., Tohdoh,N., Shimada,H. and Sugiura,M. (1986) EMBO J., 5, 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugiura M. (1992) Plant Mol. Biol., 19, 149–168. [DOI] [PubMed] [Google Scholar]
- 27.Hiratsuka J., Shimada,H., Whitier,R., Ishibashi,T., Sakamoto,M., Mori,M., Kondo,C., Honji,Y., Sun,C.R., Meng,B.Y., Li,Y.Q., Kanno,A., Hirai,A., Shinozaki,K. and Sugiura,M. (1989) Mol. Gen. Genet., 217, 185–194. [DOI] [PubMed] [Google Scholar]
- 28.Del Campo E.M., Sabater,B. and Martín,M. (1997) Plant Physiol., 114, 748. [Google Scholar]
- 29.Martín M., Casano,L.M. and Sabater,B. (1996) Plant Cell Physiol., 37, 293–298. [DOI] [PubMed] [Google Scholar]
- 30.Michel F. and Ferat,J.L. (1995) Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- 31.Kim J.-K. and Hollingsworth,M.J. (1993) Curr. Genet., 23, 175–180. [DOI] [PubMed] [Google Scholar]
- 32.Saldanha R., Mohr,G., Belfort,M. and Lambowitz,A.M. (1993) FASEB J., 7, 15–24. [DOI] [PubMed] [Google Scholar]
- 33.Vogel J., Hess,W.R. and Borner,T. (1997) Nucleic Acids Res., 25, 2030–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bock R., Hermann,M. and Kössel,H. (1996) EMBO J., 15, 5052–5059. [PMC free article] [PubMed] [Google Scholar]
- 35.Reed M.M. and Hanson,M.R. (1997) Mol. Cell. Biol., 17, 6948–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]