Abstract
Matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) has been applied to increase the informational output from DNA sequence analysis. It has been used to analyze DNA by hybridization with microarrays of gel-immobilized oligonucleotides extended with stacked 5mers. In model experiments, a 28 nt long DNA fragment was hybridized with 10 immobilized, overlapping 8mers. Then, in a second round of hybridization DNA–8mer duplexes were hybridized with a mixture of 10 5mers. The stability of the 5mer complex with DNA was increased to raise the melting temperature of the duplex by 10–15°C as a result of stacking interaction with 8mers. Contiguous 13 bp duplexes containing an internal break were formed. MALDI MS identified one or, in some cases, two 5mers contiguously stacked to each DNA–8mer duplex formed on the microchip. Incorporating a mass label into 5mers optimized MALDI MS monitoring. This procedure enabled us to reconstitute the sequence of a model DNA fragment and identify polymorphic nucleotides. The application of MALDI MS identification of contiguously stacked 5mers to increase the length of DNA for sequence analysis is discussed.
INTRODUCTION
The main objective of the Human Genome Program, determining the sequence of the 3 billion nt of the human genome, will be completed in a few years. The Sanger method has been a key tool in this effort. The next important step in the program will be to compare the structures of the genomes from a large number of individuals and from different organisms. A few million nucleotides may differ from each other in human genome polymorphic sites. Comparative analyses of the polymorphic sites should provide a wealth of information that could help biomedical researchers understand the functions of genes and identify the genes responsible for numerous physiological traits.
Using the Sanger sequencing method to screen polymorphic nucleotides in huge populations of people and compare the genomes of related organisms would be too expensive, slow and cumbersome. Therefore, a great deal of effort is being directed towards the development of alternative approaches (1).
Monitoring the hybridization of DNA with a high density array of immobilized oligonucleotides on oligonucleotide microchips (2) using matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) (3) is considered a highly promising approach for DNA sequencing and sequence analysis. The direct hybridization of fluorescently labeled DNA with a set of complementary oligonucleotides on a custom-made microchip (4–6) is an efficient procedure for identifying nucleotide polymorphisms in DNA. This microchip hybridization can be combined with enzymatic single base extension of the immobilized oligonucleotides (7). However, such an analysis of DNA polymorphism requires multiple redundancies in the complementary microchip oligonucleotides for each nucleotide change to be identified. It also demands the manufacture of a custom microchip for each gene. The hybridization of DNA with a generic set containing all possible 4n oligonucleotides of length n and immobilized as an array has been proposed as a general approach to sequencing (8–10). However, there are still essential hurdles to sequencing by hybridization with oligonucleotides on microchips. Discrimination between perfect and mismatched duplexes is not completely reliable and there are ambiguities in sequencing due to the presence of repeats and to significant variations in the stability of different duplexes, in particular, G-C- and A-T-rich duplexes.
Extending the length of the immobilized oligonucleotides on the generic microchip by 1 nt increases the size of the analyzed DNA linearly by about two times, but at the same time the size of the microchip increases exponentially by four times. Available technologies allow the manufacture of high density generic oligonucleotide arrays or microchips containing from 4096 6mers (11) to 262 144 9mers (12). Generic 6mer and 9mer microchips have been used for polymorphism analysis of DNA strands that were ~100 and 1000 nt in length, respectively. To obviate the low stability of 8 and 9 bp long duplexes formed on glass-immobilized oligonucleotides, the oligonucleotides were extended from the 3′-end with a 20 base long anchor and from the 5′-end with two inosine nucleotides, and the interrogated DNA was ligated to the anchor complement (12). The stability of the duplexes formed by 6mers on gel-based generic microchips has been enhanced in two ways: (i) by using high concentrations of immobilized oligonucleotides within the three-dimensional polyacrylamide gel pads; and (ii) by extending the immobilized 6mers on both ends with either an equimolar mixture of four bases or a universal base able to pair with any of the four bases (11,13).
However, to analyze larger segments of DNA of thousands of nucleotides in length, researchers would need to use extremely complex generic microchips containing, for example, 413 13mers or 418 18mers. Contiguous stacking hybridization (CSH) (9,14,15) was proposed to circumvent the resultant increase in microchip complexity that would be disproportional with respect to sequencing efficiency. In this approach, the microchip-immobilized 8mers or longer oligonucleotides were hybridized with DNA in the presence of a mixture of 5mers. The 5mers themselves do not form stable duplexes with DNA. However, their base pairing with DNA is stabilized when one or two of them are stacked to the hybridized microchip 8mers; in this way the 8 bp long duplexes are extended with the hybridized stacked 5mers to form 13 and 18 bp long duplexes. This stabilization results from the stacking interactions of the terminal bases in the 5mer and the immobilized 8mer. Because of their short length, contiguously stacked 5mers are much more sensitive to the presence of mismatches in their duplexes than longer probes. This sensitivity enhances the accuracy of the hybridization tests. The contiguously stacked 5mers were earlier identified by rather tedious procedures in which they were labeled with different fluorescent dyes or by using only one 5mer in each hybridization assay. Several rounds of successive CSH followed by ligation of the 5mers with the immobilized oligonucleotides were applied in a model experiment to sequence DNA repeats (16).
MALDI MS appears to be a fast and precise method for multiplex analysis of mixtures containing from up to 500 rather short DNA molecules of different molecular masses (3). Its efficiency has been demonstrated for reading Sanger sequencing ladders (17) and for identifying polymorphisms by extending primers with a single base (18) or by directly comparing the masses of PCR-amplified fragments (19).
This article describes the use of MALDI MS to identify which 5mers in a mixture participate in CSH. Microchip-immobilized 8mers were hybridized with DNA in the presence of a mixture of 5mers. In a model experiment, a 22 nt sequence of 28 nt long DNA was reconstructed and polymorphic nucleotides were analyzed by identifying each 5mer contiguously stacked to each microchip 8mer as a result of hybridizing with the DNA. The incorporation of a mass label into the 5mers increased the MALDI resolution. The results were corroborated by fluorescence monitoring of the CSH with individual fluorescently labeled 5mers. The application of MALDI MS analysis of contiguously stacked short 5mers significantly increased the efficiency with which mismatched duplexes were discriminated and the accuracy of custom-made microchips in hybridization tests. This approach may be used to extend the efficiency of a generic microchip containing 4n nmers to the level of microchips containing 4(n + 5) n+5mers and 4(n + 10) n+10mers upon CSH of a nmer with one or two 5mers, respectively, for analyzing the sequence and polymorphism of much longer DNA.
MATERIALS AND METHODS
Solvents and reagents were obtained from commercial suppliers and were used without further purification.
Oligonucleotides
The 5mers, 8mers and 28mers were synthesized on an ABI-394 DNA-RNA synthesizer (Applied Biosystems, Foster City, CA). A 3′-amino group was introduced into 8mers and 28mers using 3′-amino-modifier C7 CPG (Glen Research, Sterling, VA) during synthesis. A 5′-amino group was introduced into some 5mers using 5′-amino-modifier C6 (Glen Research). Labeling of the 3′-amino group of 28mers with fluorescein isothiocyanate (FITC) and the 5′-amino group of 5mers with Texas red was performed by following the standard protocol (20). Unlabeled and fluorescently labeled oligonucleotides were purified by either reverse phase liquid chromatography (HPLC) on a C-18 Nucleosil (Sigma, St Louis, MO) column or by electrophoresis in denaturing polyacrylamide gels.
Microchip manufacture
Microchips containing immobilized 8mers within polyacrylamide gel pads were manufactured as described (14). The microchips for mass spectrometry monitoring were in a linear arrangement of ten 1 × 1 × 0.02 mm gel pads spaced 1.5 mm apart on a 500 × 10 × 1 mm electrically conductive silicon chip. These dimensions and geometry were determined by the parameters of the KRATOS mass spectrometer. The standard microchips for fluorescence monitoring were in a rectangular arrangement of 0.1 × 0.1 × 0.02 mm gel pads spaced 0.2 mm apart and bonded to a glass slide.
Microchip hybridization and its fluorescence monitoring
The hybridization of 100 µl of a 10 mM fluorescein-labeled 28mer DNA solution in hybridization buffer (1 M NaCl, 1 mM EDTA, 1% Tween 20, 5 mM Na phosphate, pH 7.0) (14) with microchip-immobilized 8mers was carried out in a hybridization chamber at 15°C for 2 h. The chamber was washed with hybridization buffer and the fluorescence was monitored with filters for fluorescein in order to measure DNA hybridization. The chamber was then filled with 100 µl of a 1 mM solution of one of 10 Texas red-labeled 5mers in the hybridization buffer, and microchip hybridization was carried out at 5°C for 1 h. After washing the microchip with buffer at 5°C, the fluorescence of Texas red was monitored with a proper filter to measure 5mer hybridization and to discriminate between it and DNA hybridization. The hybridized 5mer was washed off with buffer at 60°C for 2 min and CSH was repeated successively with the other nine Texas red-labeled 5mers.
Fluorescence measurements of microchip hybridization were carried out in real time on an automatic set-up. The set-up consisted of a two-wavelength fluorescence microscope, CCD camera (Princeton Instruments, Trenton, NJ), Peltier thermotable, temperature controller, and a computer equipped with a data acquisition board and data processing software. The microscope had FITC and Texas red switchable bandpass and bandstop filters (21).
Mass spectral monitoring of microchip hybridization
Mass spectra analysis was carried out on MALDI-TOF mass spectrometer KOMPACT MALDI 4 (Kratos Analytical, Chestnut Ridge, NY). The hybridization of 100 µl of a 10 µM solution of unlabeled DNA was carried out with the microchip in 1 M triethylammonium acetate (TEAA; Fluka) buffer at pH 7.0 and 10°C for 2 h. The microchip was then hybridized with the mixture of the 5mers, 1 µM each in 100 µl of hybridization buffer at 0°C for 1 h. After hybridization, the buffer and remaining unhybridized oligonucleotides were removed and 1 µl of matrix solution was placed on each gel pad. The matrix solution (22) consisted of 0.5% ammonium citrate (Fluka) in a saturated water solution of 2-amino-5-nitropyridine (Sigma). The microchip was incubated over a water layer at 60°C in a Petri dish for 30 min to avoid evaporation of the matrix solution. The 5mers were dissociated from the duplexes under these conditions and distributed between the gel pads and the drop of matrix solution above the pad. The microchip was dried at 60°C and placed into the mass spectrometer. Alternatively, the drop of solution above the pad was transferred with a pipette to a standard KRATOS sample slide to be placed in the mass spectrometer. Mass spectra were obtained in the linear mode, with pulsed extraction and negative ion registration.
RESULTS
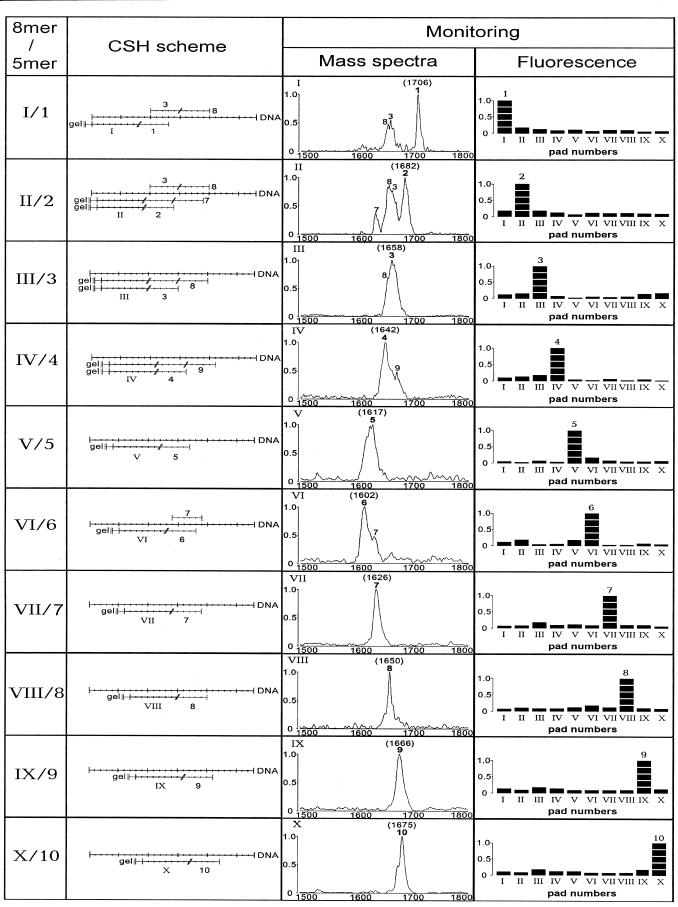
Figure 1 shows the sequences for a strand of a synthetic 28 nt long DNA fragment, complementary and overlapping 5mers and microchip 8mers. Ten 8mers (I–X) were immobilized through their 3′-terminal amino groups within 10 polyacrylamide gel pads of a microchip that was 1 × 1 × 0.02 mm for mass spectrometry experiments and 0.1 × 0.1 × 0.02 mm for fluorescence analysis. Ten 5mers (1–10) contained 5′-amino-modifier C6 at the 5′-end. Each of the 5mers can be contiguously stacked to a corresponding microchip 8mer forming a duplex with the DNA fragment. In fluorescence monitoring experiments, Texas red was attached to the amino group of the 5mers and fluorescein was attached to the 28mer. The sequence of DNA was selected so that the stacked 5mers could be easily discriminated between according to their different masses without any mass tags and other additional procedures required for some native DNA fragments.
Figure 1.
The sequence of the DNA fragment, microchip gel-immobilized 8mers and contiguously stacked 5mers. A 28 nt long DNA fragment is shown in the upper line (capital letters). Ten gel-immobilized oligonucleotides (I–X, upper case letters) and 10 contiguously stacked 5mers (1–10, lower case letters) are shown under the corresponding DNA complementary sequences. Microchip-immobilized 8mers and contiguously stacked 5mers in solution are separated by a backslash (/). R = -(CH2)6-NH2 (5′-amino-modifier C6) or R = -(CH2)6-NH-TR were used in the 5mers for mass spectral and fluorescence monitoring, respectively. R1 = H or R = 3′-amino-modifier C7 CPG with FITC were used for mass spectral and fluorescence monitoring, respectively.
MALDI MS monitoring
The extraction of 5mers from the gel pads to make them available for mass spectrometric analysis was carried out either by drying the excess matrix solution over the gel pads or by transferring the matrix solution with a pipette to a standard KRATOS sample slide. The second method produced peaks that were about two to three times higher, but was more tedious to carry out. Figure 2 shows the results of an analysis of the hybridization of DNA with 10 immobilized 8mers within 10 gel pads of a microchip in the presence of a mixture of 10 5mers. In all cases, a specific 5mer that contiguously stacked to the corresponding 8mer in its duplex with the DNA was identified. In some cases, two 5mer peaks could be seen in the mass spectra: a stronger one for a 5mer stacked to an 8mer and a weaker one for the second 5mer stacked to the first one. MALDI MS does not provide for quantitative measurements of 5mers. However, it can be used for comparative assessments of a predominant 5mer in a mixture.
Figure 2.
CSH monitored by MALDI MS and fluorescence microscopy. Ten 8mers (I–X) were immobilized within gel pads. In a MALDI-TOF experiment, an oligonucleotide chip was hybridized with DNA in the presence of a mixture of 10 5mers (1–10). Mass spectra of the contiguously stacked 5mers were registered for each 8mer in a corresponding gel pad. The intensities of the signals varied up to 10 times in different experiments due to uncontrolled variations in crystallization and elution of the 5mers from the gels. The molecular mass of a 5mer is shown near its peak. In 10 fluorescence experiments, the microchip was hybridized successively with a mixture of DNA and one of 10 fluorescently labeled 5mers each time. Fluorescence of contiguously stacked 5mers was measured in each experiment in parallel for all immobilized 8mers in the corresponding gel pads of the microchip with a fluorescence microscope.
Two contiguously stacked 5mers were observed in cases where there was enough space on the DNA for an 8mer and two 5mers: II + 2 + 7; III + 3 + 8; IV + 4 + 9. No simultaneous stacking of two 5mers, 1 + 6 and 5 + 10, with the respective 8mers was observed, probably because the stacking interaction between T and C (their 5′- and 3′-terminal bases) is weak (see Fig. 1). Two 5mers, 3 and 8, were observed in pads I and II, most probably because they stack to each other, independently of the immobilized 8mers. It appears that the highly G-C-rich 5mer 3 (80% G-C) is able to form a duplex with DNA in the absence of a stacked 8mer when it is stabilized by the second stacked 5mer, 8.
It appears that MALDI MS can be efficiently used to detect specific contiguously stacked 5mers within the duplexes formed by DNA and microchip-immobilized 8mers. Hybridization of the 28 nt long DNA with the microchip 8mers was not directly detected by MALDI MS, probably because of the lower sensitivity of the analysis for such large molecules under the conditions used and less efficient extraction of the 28mer from the gel. However, DNA hybridization with immobilized oligonucleotides is deduced from the presence of contiguously stacked 5mers in the mass spectra.
This MALDI-TOF analysis of CSH of the DNA with 10 microchip-immobilized 8mers in the presence of the mixture of 10 5mers provides enough information to reconstitute the sequence of a 22 nt long DNA region.
Fluorescence analysis
In 10 fluorescence monitoring experiments, only one fluorescently labeled 5mer was hybridized each time on the microchip with DNA–8mer duplexes, as was described earlier (15). In all cases, the label was found only in the gel pad that contained a 5mer contiguously stacked to a proper 8mer (Fig. 2). These data support the high specificity of the CSH experiments monitored with MALDI-TOF and carried out with the 5mer mixture rather than with a single 5mer. The presence of the fluorescence label in the 5mers did not affect the pattern of hybridization.
Polymorphism analysis with mass-labeled 5mers
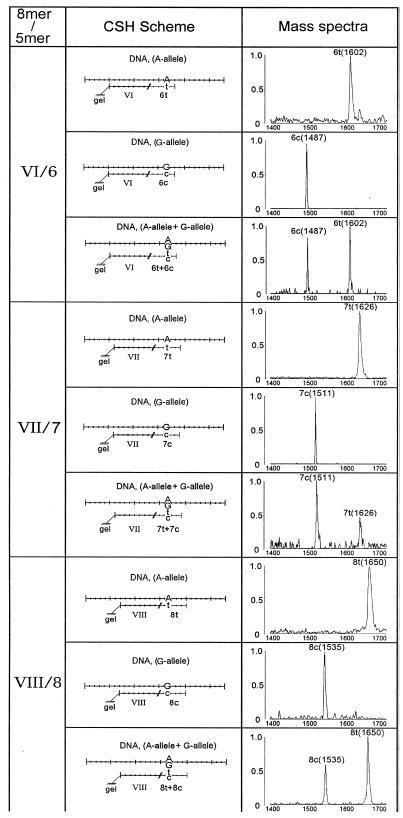
Two polymorphic strands of DNA having either nucleotide A, ‘A-DNA’ (Fig. 1), or G, ‘G-DNA’ (Fig. 3), at the 17th position were hybridized separately or in mixture with a microchip containing three immobilized 8mers (VI, VII and VIII; see Fig. 1) complementary to both DNAs. A mixture of 13 5mers in two sets was used in CSH. The first set contained 10 5mers, 1t–10t (1–10 in Fig. 1), complementary to ‘A-DNA’. The second set contained three additional 5mers, 6c–8c, that differed by substituting C for T, thereby making them complementary to ‘G-DNA’. The first set of 5mers, 1t–10t, also contained a mass label 5′-amino-modifier C6, while the second polymorphic set, 6c–8c, did not. Figure 3 shows the MALDI MS analysis of the CSH of A- and G-DNAs separately (in imitation of homozygous DNA) and together (heterozygous DNA) with the microchip 8mers VI–VIII in the presence of two sets of 5mers. The labeling of only one set of 5mers, 1t–10t, increased their mass difference from the second set, 6c–8c, by 100 Da. This enhanced the reliability of discrimination of the allelic 5mers by MALDI MS and the identification of two homozygotes and their heterozygous DNA allele.
Figure 3.
Polymorphism detection in DNA with MALDI MS CSH. Microchip-immobilized 8mers (VI–VIII) were hybridized with each of two DNA samples (A allele and G allele) containing an A→G base change, separately or in a mixture, and then with a mixture of 5mers containing T and C base substitutions (1t–10t) and (6c–8c). CSH of 5mers was monitored by mass spectrometry with the gel pads containing 8mers VI–VIII.
The 5mers 6t–8t as well as 6c–8c upon CSH with G- or A-DNA formed duplexes containing G-T and A-C mispairs in the central second, third and fourth positions from their 5′-ends. The G-T mismatch is the least destabilizing one and the most difficult to discriminate from perfect base pairs in the duplexes (3). However, no G-T or A-C mismatched 5mer duplexes were revealed in contiguous stacking experiments. This finding confirms that there is an essentially higher efficiency in discrimination of mismatched duplexes for contiguously stacked 5mers than for longer directly hybridized oligonucleotides (15).
DISCUSSION
MALDI MS is a highly effective method for identifying 5mers in a mixture used in DNA sequence analysis by CSH.
In sequencing by hybridization, shorter oligonucleotide probes result in more efficient discrimination of perfect and mismatched duplexes and more reliable sequencing. However, at the same time, with shorter DNA, the probability of internal repetitions of the length of the probe is increased, thus decreasing the length of DNA that can be sequenced. This incompatibility between the accuracy and the length of analyzed DNA is partly overcome by the use of CSH. In this method, extended oligonucleotide probes are assembled from two pieces: longer microchip-immobilized oligonucleotides and shorter 5mers present in solution and stacked to the immobilized ones after the latter have formed duplexes with the interrogated DNA.
The stacking interactions of adjacent bases in DNA are the main forces in maintaining duplexes even when one DNA strand is broken. As a result of such interactions, the rather weak base pairings of 5mers with DNA are significantly stabilized, their melting temperature increasing by ~10–15°C on stacking with adjacent duplexes (15). One or, in some cases, two 5mers were contiguously stacked with gel-immobilized 8mers to form, upon hybridization with single-strand DNA, 13 or 18 bp long duplexes with one or two internal breaks, respectively (Fig. 2). Such contiguously stacked and assembled probes can increase the efficiency of the sequencing by hybridization of the microchip nmers to the level of n+5mers and n+10mers. A higher fidelity of hybridization with short 5mers is achieved in this method and it offers the possibility of analyzing longer DNA.
The use of a gel support for oligonucleotide immobilization provides some essential advantages for CSH (14). More than 100 times higher the number of molecules of immobilized probes on a microchip surface unit can be achieved with a three-dimensional gel structure then with two-dimensional glass surface. This increases the apparent stability of the duplexes formed on gel-immobilized oligonucleotides and facilitates the use of short, contiguously stacked 5mers (9,13). The immobilized oligonucleotides are well spaced and thus avoid interfering with each other and hybridized DNA. The gel-immobilized oligonucleotides occur in a more homogeneous water environment than that associated with heterophase glass-immobilized oligonucleotides. Therefore, the sharpness of the melting curves and, as a result, the discrimination of matched and mismatched duplexes is higher for gel-based microchips.
Identification of stacked 5mers with a MALDI-TOF mass spectrometer is an effective approach for multiplex analysis of polymorphic nucleotides in DNA by CSH. Several samples of analyzed DNA can be hybridized to many microchip-immobilized oligonucleotides in the presence of 5mer mixtures. Molecular mass differences between any of the four nucleotides dAp (313.2 Da), dCp (289.2 Da), dGp (329.2 Da) and dTp (304.2 Da) constitute at least 9 Da, which is enough to allow MALDI MS to resolve all four 5mers complementary to a polymorphic site and differing by a base change in one position. Introducing mass labels, as shown in Figure 3, can enhance the difference. The discrimination between matched and mismatched 5mers is rather high: no mismatched signals were observed even when a poorly identified G-T mispair was located in the second, third or fourth position of the hybridized 5mers (Fig. 3). It has been shown that the first 3′-terminal stacked nucleotide is the most sensitive to mispairing, whereas the last one is the least sensitive (15). A variable nucleotide in DNA can be hybridized with all five overlapped complementary and stacked 5mers, increasing the reliability of the polymorphism analysis.
CSH of DNA on a generic microchip with one or two 5mer molecules in the presence of all 1024 5mers could be an attractive method for significantly increasing the power of sequencing by hybridization. The sequencing efficiency of a microchip containing all 4n nmers could approach that of generic microchips containing 4(n + 5) n+5mers or 4(n + 10) n+10mers. This increased efficiency could allow a many-fold increase in the length of DNA to be analyzed for polymorphism, proofreading and sequence. The simultaneous use of four fluorescence labels, substitution of bases in different positions by a universal base and five rounds of hybridization have been suggested for identifying any 5mers in a complete set of 5mers (15). MALDI MS seems to be much more efficient than the fluorescence measurement procedures for monitoring stacked 5mers. The masses of 5mers vary from 1.385 to 1.585 kDa, which corresponds to the optimal mass range for MALDI analysis. However, application of the method for de novo sequencing with generic microchips would require the resolution of all 1024 5mers, including many that have very similar masses or the same composition and mass. The incorporation of mass labels (Fig. 3) could help in discriminating all 5mers.
Some 5mers, in particular G-C-rich ones, were shown to form rather stable duplexes when stacked with each other as well as with immobilized oligonucleotides. This 5mer self-stacking interfered with the interpretation of the results. It has been shown that a fluorescence dye bound to the terminal position of 5mers that is involved in the stacking interaction behaves like a terminator and prevents them from stacking (15). These and other terminators could be applied to prevent stacking of 5mers with each other when needed.
Another drawback of hybridization methods is that they result in significant differences in the stability of duplexes, depending on duplex length, composition and sequence. Oligonucleotide extension with one or two universal bases (13), as well as the incorporation of modified nucleotides (23), could equalize the stability of the 5mer duplexes or even stabilize them (24).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr S. Surzhikov for synthesizing and purifying oligonucleotides, Drs V. Shick and A. Zasedatelev for some useful suggestions, V. V. Chupeeva for manufacturing mass spectral microchips, E. Novikova for providing editorial assistance and Dr J. Zlatanova for critically reviewing the manuscript. This work was supported by the Defense Advanced Research Project Agency under Interagency Agreement no. AO-E428 and by the Russian Human Genome Program.
REFERENCES
- 1.Landegren U., Nilsson,M. and Kwok,P.Y. (1998) Genome Res., 8, 769–776. [DOI] [PubMed] [Google Scholar]
- 2.Editorial (1999) Nature Genet., 21. [Google Scholar]
- 3.Graber J.H., Smith,C.L. and Cantor,C.R. (1999) Genet. Anal., 14, 215–219. [DOI] [PubMed] [Google Scholar]
- 4.Lipshutz R.J., Moriss,D., Chee,M., Hubbell,E., Kozal,M.J., Shah,N., Shen,N., Yang,R. and Fodor,S.P. (1995) Biotechniques, 19, 442–447. [PubMed] [Google Scholar]
- 5.Chee M., Yang,R., Hubbell,E., Berno,A., Huang,X.C., Stern,D., Winkler,J., Lockhart,D.J., Morris,M.S. and Fodor,S.P. (1996) Science, 274, 610–614. [DOI] [PubMed] [Google Scholar]
- 6.Drobyshev A., Mologina,N., Shick,V., Pobedimskaya,D., Yershov,G. and Mirzabekov,A. (1997) Gene, 188, 45–52. [DOI] [PubMed] [Google Scholar]
- 7.Dubiley S., Kirillov,E. and Mirzabekov,A. (1999) Nucleic Acids Res., 27, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lysov Y., Florentiev,V., Khorlin,A., Khrapko,K., Shick,V. and Mirzabekov,A. (1988) Proc. Acad. Sci. USSR, 303, 1508–1511. [Google Scholar]
- 9.Khrapko K., Lysov,Y., Khorlin,A., Shick,V., Florentiev,V. and Mirzabekov,A. (1989) FEBS Lett., 256, 118–122. [DOI] [PubMed] [Google Scholar]
- 10.Southern E., Markos,U. and Elder,R. (1992) Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 11.Proudnikov D., Kirillov,E., Chumakov,K., Donlon,J., Rezapkin,G. and Mirzabekov,A. (2000) Biologicals, submitted for publication. [DOI] [PubMed] [Google Scholar]
- 12.Gunderson K.L., Huang,X.C., Morris,M.S., Lipshutz,R.J., Lockhart,D.J. and Chee,M.S. (1998) Genome Res., 8, 1142–1153. [DOI] [PubMed] [Google Scholar]
- 13.Fotin A., Drobyshev,A., Proudnikov,D., Perov,A. and Mirzabekov,A. (1998) Nucleic Acids Res., 26, 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yershov G., Barsky,V., Belgovskiy,A., Kirillov,E., Kreindlin,E., Ivanov,I., Parinov,S., Guschin,D., Drobyshev,A., Dubiley,S. and Mirzabekov,A. (1996) Proc. Natl Acad. Sci. USA, 93, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parinov S., Barsky,V., Yershov,G., Kirrillov,E., Timofeyev,E., Belgovsky,A. and Mirzabekov,A. (1996) Nucleic Acids Res., 24, 2998–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubiley S., Kirillov,E., Lysov,Y. and Mirzabekov,A. (1997) Nucleic Acids Res. 25, 2259–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu D., Tang,K., Braun,A., Reuter,D., Darnhofer-Demar,B., Little,D.P., O’Donnel,M.J., Cantor,C.R. and Koster,H. (1998) Nature Biotechnol., 16, 381–384. [DOI] [PubMed] [Google Scholar]
- 18.Fei Z., Ono,T. and Smith,L.M. (1998) Nucleic Acids Res., 26, 2827–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haff L.A. and Smirnov,I.P. (1997) Nucleic Acids Res., 25, 3749–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugland R.P. (1996) Handbook of Fluorescent Probes and Research Chemicals, 6th Edn. Molecular Probes Inc., Eugene, OR.
- 21.Barsky Y., Grammatin,A., Ivanov,A., Kreindlin,E., Kotova,E., Barskii,V. and Mirzabekov,A. (1998) J. Opt. Technol., 65, 938–941. [Google Scholar]
- 22.Fitzgerald M.C., Parr,G.R. and Smith,L.M. (1993) Anal. Chem., 65, 3204–3211. [DOI] [PubMed] [Google Scholar]
- 23.Hacia J.G., Woski,S.A., Fidanza,J., Edgemon,K., Hunt,N., McGall,G., Fodor,S.P. and Collins,F.S. (1998) Nucleic Acids Res., 26, 4975–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiler J., Gausepohl,H., Hauser,N., Jensen,O.N. and Hoheisel,J.D. (1997) Nucleic Acids Res., 25, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]