Abstract
48With the growing number of biomaterials and printing technologies, bioprinting has brought about tremendous potential to fabricate biomimetic architectures or living tissue constructs. To make bioprinting and bioprinted constructs more powerful, machine learning (ML) is introduced to optimize the relevant processes, applied materials, and mechanical/biological performances. The objectives of this work were to collate, analyze, categorize, and summarize published articles and papers pertaining to ML applications in bioprinting and their impact on bioprinted constructs, as well as the directions of potential development. From the available references, both traditional ML and deep learning (DL) have been applied to optimize the printing process, structural parameters, material properties, and biological/mechanical performance of bioprinted constructs. The former uses features extracted from image or numerical data as inputs in prediction model building, and the latter uses the image directly for segmentation or classification model building. All of these studies present advanced bioprinting with a stable and reliable printing process, desirable fiber/droplet diameter, and precise layer stacking, and also enhance the bioprinted constructs with better design and cell performance. The current challenges and outlooks in developing process-material-performance models are highlighted, which may pave the way for revolutionizing bioprinting technologies and bioprinted construct design.
Keywords: Bioprinting, Machine learning, Deep learning, Biomaterials, Bioprinted constructs
1. Introduction
Three-dimensional (3D) bioprinting can precisely manipulate biomaterials or bioinks and fabricate constructs with well-defined microstructures in a controllable and reproducible manner. Such constructs can provide 3D environments for in vitro studies in cell biology, tissue engineering, and drug screening[1-3]. A growing number of biomaterials and printing technologies are available to fabricate such constructs[3,4]. This creates a tremendous 49workload when researchers are trying to optimize printing materials and process parameters and evaluate their impacts on bioprinted constructs. For example, biomaterial/bioink should be formulated with the desired performance, the printing process should be quantified with consistent printing results, and the performance of bioprinted constructs should be purposely linked to the material, structure, and process. It is extremely difficult to conduct these studies by merely using mathematical models or experimental equations. To cope with such complicated scenarios, both traditional machine learning (ML) and deep learning (DL) methods have been adopted, which could potentially provide cost-effective solutions.
In the following section, we overview the working principles of the most popular bioprinting technologies, such as droplet-based bioprinting (DBB), extrusion-based printing (EBB) and electrohydrodynamic printing (EHD), and their printed constructs. In addition, general strategies for applying traditional ML and DL methods to make bioprinting more powerful for fabricating custom-made structures are discussed.
1.1. Bioprinting technologies for construct fabrication
Printing technologies such as DBB, EBB, stereolithography, EHD bioprinting, and laser-assisted bioprinting can be used to fabricate constructs with micro/nanoscale features for 3D cell culture systems to establish in vitro models[1-5]. These technologies have been fully investigated and many commercial machines have been launched[6,7].
As shown in Figure 1a, DBB dispenses droplets from a nozzle using thermal, pneumatic, or sonic actuation. This technology can precisely control the volume and position of biomaterial/bioinks, growth factors, and drugs to produce microstructures for tissue engineering, regenerative medicine, high-throughput screening, and cancer research[1]. This technology is only applicable to a narrow range of printable materials with good biocompatibility and easy crosslinking mechanisms. Bioprinted constructs may have weak mechanical and structural integrity. Moreover, the dispensing process induces cell damage at substantial levels, which places additional limitations when fabricating cell-laden structures.
Figure 1.
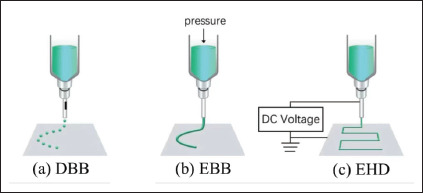
Bioprinting technologies. Abbreviations: DBB, droplet-based bioprinting; EBB, extrusion-based printing; EHD, electrohydrodynamic printing.
As shown in Figure 1b, EBB uses a pressure-controlled reservoir and nozzle to spatially pattern hydrogel constructs layer by layer with varied pore sizes and compositional gradients[2,3]. The combination of relevant printing parameters including needle diameter, extrusion rate, printer head speed, and temperature of the nozzle and material-related factors, such as viscoelastic properties and curing mechanism, play critical roles in determining the shape fidelity and biocompatibility of constructs[8,9]. As low-viscosity materials are used for extrusion, EBB cannot be used to fabricate high-resolution bioprinted constructs.
During EHD bioprinting (Figure 1c), a high voltage is applied between the nozzle and the collecting substrate to electrically eject biomaterial/bioink flows. EHD bioprinting uses viscous synthetic polymer solutions or melts to produce well-oriented structures with precisely stacked micro/nanoscale fibers[4]. The polymer inks adopted include poly-ε-caprolactone (PCL)[6,10,11], polylactic acid (PLA)[12], and polyethylene oxide (PEO)[13]. EHD bioprinting can produce fibers ranging from hundreds of nanometers to a few micrometers, which can regulate cellular behaviors[3,14,15]. This process is controlled by the properties of the biomaterial ink (viscosity, surface tension, and electrical conductivity), the environmental factors (temperature and humidity), and the process parameters (nozzle-to-substrate distance, solution feeding rate, and nozzle dimensions). EHD bioprinting can be disturbed by environmental factors or inhomogeneous material properties and then becomes unstable during the stacking of printed fiber structures[15]. In addition to printing fiber structures, EHD bioprinting can also be used to pattern 2D structures. For example, a drop-on-demand EHD inkjet can use low-viscosity solutions to print droplets with organized patterns and form micro/nanoscale dot arrays.
50Hydrogel scaffolds fabricated using EBB and fibrous scaffolds fabricated using EHD can mimic extracellular matrix (ECM) components from the native environment and influence cell behaviors and outcomes[11,14,16]. Numerous experiments have been conducted to investigate material properties, process parameters, and their effects on scaffold building. However, researchers in cell biology or drug screening may not be available or capable of experimenting with these factors. This has impeded the widespread adoption of these technologies across multiple disciplines.
In addition, the concept of customized constructs to specifically tailor cell responses in 3D cultures has drawn ongoing research interest. Such constructs require the precise control of biomaterial compositions, structural designs, and printing technologies, which cannot be realized using experimental or mathematical models. ML has proven its capability to model complex processes with multiperformance characteristics. It is therefore introduced to systematically model materials and parameters, as well as to quantitatively link process-material-performance in bioprinted constructs[17].
1.2. Common ML methods used in bioprinting
ML has experienced rapid progress over the past two decades and has demonstrated outstanding capability in pattern identification and parameter optimization for metal machining and printing[17,18]. As an emerging technology, it has the potential to streamline the current bioprinting workflow through process-material-performance modeling.
As shown in Figure 2, the current ML applications in bioprinting include material property optimization for reliable printability and shape fidelity, process optimization with the desired fiber or droplet diameter, in situ process monitoring for stable fabrication and process adjustment, and bioprinted construct optimization for better cell-microenvironment interactions. Both traditional ML and DL methods are applied to develop prediction, segmentation, and detection models[19].
Figure 2.
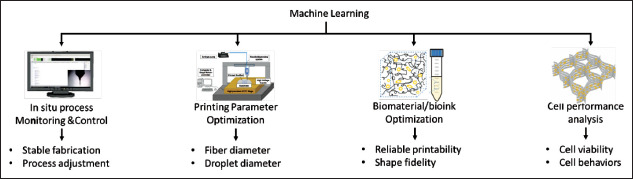
ML applications in bioprinting.
The traditional ML methods shown in Figure 3a use extracted and selected numerical features as inputs for prediction tasks. Using the collected dataset or images, 51many features can be extracted based on the expert users’ prior knowledge and practical experience. For numerical data, statistical analysis, threshold methods, and frequency analysis are typically used for feature extraction. By contrast, shape, edge, color, and texture detection are widely used for image feature extraction. To achieve better predictions, the informative relevance of these features should be carefully chosen for each task[20]. Traditional ML methods, such as Support Vector Machines (SVM) and K-Nearest Neighbors, are typically used to build classification or regression models for such prediction tasks. For example, classification models can classify Taylor cone shapes in EHD bioprinting, identify the reliability of printability, and judge the shape fidelity. Regression models can be used to optimize the printing parameters and material properties for the desired fiber diameter. One of the most commonly used ML methods is SVM, and a few powerful SVM toolboxes have been launched for prediction models built in MATLAB or other platforms with user-friendly interfaces and simple instructions[21].
Figure 3.
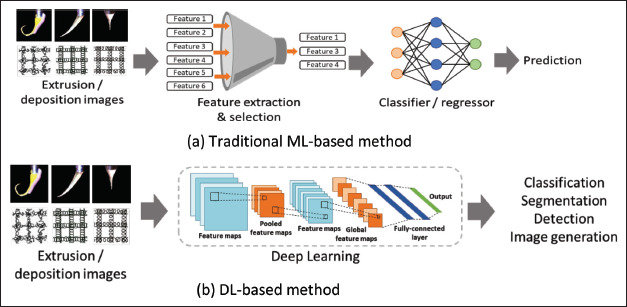
ML methods.
As illustrated in Figure 3b, DL methods can automatically discover underlying patterns and identify the most descriptive and salient features in image-recognition tasks. Feature extraction and selection steps are omitted in this type of learning method. DL methods have achieved significant success in many image application scenarios, and convolutional neural networks (CNN) are the most popular DL methods[15]. Unlike traditional ML methods, DL methods can handle more complex tasks, such as classification, segmentation, and object detection, which require relatively large-scale datasets.
Traditional ML and DL methods used in bioprinting applications can also be categorized into supervised and unsupervised learning[21]. Supervised ML methods can establish mathematical models between inputs and outputs using labeled datasets, such as multilayer perceptron (MLP)[24], SVM[22,25,26], CNN[15,22], and backpropagation neural network (BPNN)[27]. Because of the complicated nature of images, some objects are difficult to label, such as nuclei, morphological phenotypes, and cell shapes during proliferation and migration. Unsupervised ML methods have been proposed to explore unlabeled objects as patterns or clusters and to identify hidden patterns or similarities through self-taught rules[28,29]. It was believed that ML applications in process optimization, dimensional accuracy analysis, manufacturing defect detection, and material property prediction may accelerate the perspectives of bioprinting development[22].
In short, traditional ML methods are fully transparent, and researchers can transfer their insights and knowledge to domain-specific tasks. DL methods are superior in terms of flexibility in discovering hidden patterns and relationships in complex images. For both methods, the parameters could be adjusted to improve the classification and regression models. Labeled/unlabeled datasets, dataset size, and task complexity are key factors in ML method selection.
Researchers worldwide have explored ML applications in bioprinting from fundamental perspectives to performance modeling. The potential adoptions of DL in design and fabrication of patient-specific 3D tissue-engineered constructs were reviewed, such as image-processing and segmentation, optimization and in situ correction of printing parameters and refinement of the tissue maturation process[5]. The authors also reported some relevant practical applications and summarized that the availability of huge training datasets and well-defined evaluation metrics are the key factors to accelerate the corresponding research areas.
Shin et al.[23] discussed the supervised ML, unsupervised ML, semi-supervised ML and reinforcement ML methods, and their applications in preprinting, printing, and postprinting. They concluded that ML can optimize printing parameters and bioinks, save printing time, and detect the anomalies. The identified bottlenecks are the limited amount of data and the transferability of current models, since these models heavily depend on mathematical features of the training data and may suffer from inconsistency when dealing with data from other sources.
The aim of this work is to collect and summarize the publications and present a state-of-the-art review, and highlight the emerging scientific potential of ML when applied to bioprinting. First, ML applications in process monitoring, printing parameters, and biomaterial/bioink design are discussed. Second, ML applications in cell performance analysis are investigated. After that, a literature-based analysis of challenges and outlook is given.
2. ML applications in process, parameter, and material optimization
The published papers on ML applications in bioprinting are summarized in Table 1, along with the application areas, specific tasks, and proposed ML algorithms. Current applications are divided into four categories: image analysis-based in situ process monitoring, printing parameter optimization, biomaterial/bioink optimization, and cell performance analysis. Various properties are considered for parameter optimization such as fiber resolution, integrity of the printed constructs, cell viability after extrusion and the integration of them. Even though diverse tasks are listed, the focus is on building process–material–performance models. It can be seen that both traditional ML and DL methods can be applied, and the ML method selection often considers the task complexity 52and available dataset. Moreover, multiple methods have been compared to identify a competitive model with better performance[24,25]. One task can also be organized into either a classification or regression model, and then solved accordingly based on data processing strategies[26,30-34].
Table 1. Summary of ML algorithms in bioprinting.
| Application area | Tasks | ML methods | Ref. |
|---|---|---|---|
| Image analysisbased in situ monitoring | Identify cone mode in scaffold fabrication process in EHD jetting | CNN | [14] |
| Identify deposit fibers’ continuity, uniformity, and regularity in EBB | Four-layer CNN, ResNeXt-50 network, linear SVM classifier | [25] | |
| Extract the flow pattern and droplet evolution in DBB | Deep recurrent neural network (DRNN) | [29] | |
| Printing parameter optimization | Predict the electrospun diameter of PCL/Gt nanofibers | Multiple regression, multilayer perceptron ANN | [24] |
| Identify suitable printing conditions for PPF scaffold in EBB | Random forest classifiers (RFc), random forest regression (RFr) | [26] | |
| Optimize ink composition and printing parameters in EBB | SVM classifier | [27] | |
| Predict the droplet diameter in EHD inkjet printing | Statistical regression analysis, GA-NN, BPNN | [28] | |
| Optimize printing parameters for GelMA and HAMA bioinks | Bayesian optimization (BO) | [30] | |
| Optimize the droplet size and printing frequency in EHD inkjet | Desirability function analysis | [31] | |
| Biomaterial/bioink optimization | Achieve high shape fidelity in EBB | Inductive logic programming, multiple regression | [32] |
| Achieve ideal linewidth and shape fidelity in EBB | Hierarchical machine learning | [33] | |
| Predict filament diameter in EBB | RFr, linear regression, intrastudy linear regression | [34] | |
| Cell performance analysis | Predict cell viability | SVM regression, linear regression, RFr, SVM classifier, RFc, logistic regression classifier | [34] |
| Detect the impact of scaffold morphology on cell shape phenotypes | SVM classifier | [35] | |
| Analyze cell-scaffold interaction | AD-GAN | [36] | |
| Predict cell-material interactions in fibrous scaffold | RFr model | [37] | |
| Associate cell morphologies with diverse microenvironment | SVM classifier | [38] |
These studies aimed to enhance the reliability and stability of the bioprinting process as well as the mechanical and biological performance of bioprinted constructs. In the following section, ML methods are discussed with regard to application areas.
2.1. Image-based in situ process monitoring
To maintain the stability and reliability of bioprinting in large-scale and long-term fabrication, the development of in situ process monitoring is necessary. As such, the quality of bioprinted constructs relies on intelligent printing process control rather than operator experience.
Similar to the manufacturing process, bioprinting process monitoring often collects real-time extrusion and deposition images, as shown in Figure 4. Subsequently, image preprocessing methods are utilized to extract key features from the captured images, which are the inputs of the corresponding ML models. These features reflect the difference between the standard cone and identified cone in EHD and deliver critical information to the ML model for identification purposes. The extrusion parameters were adjusted based on model outputs. Similarly, the deposited images can be compared with predefined patterns using the extracted features to depict the fiber quality and pattern. The identification model built using such features can be directly linked to the adjustment strategy for the deposition parameters. Generally, extrusion and deposition images are used to monitor EHD[15] and EBB[25], respectively.
Figure 4.
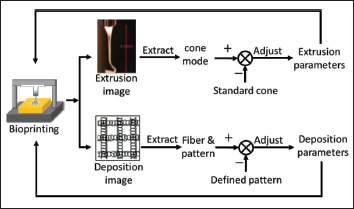
Image-based in situ process monitoring.
Both traditional ML and DL methods can be used to analyze the collected images. The former extracts image 53features as inputs for model building and the latter uses images directly to discover underlying patterns for the same task.
One of the most dominant DL-based methods is CNN. This method has been used to identify either the standard cone mode for stable fabrication in EHD bioprinting or deformed cones for the adaptive tuning of printing parameters[15]. To compare the performances of the CNN algorithms, two CNN algorithms (self-designed four-layer CNN network and pretrained ResNeXt-52 network) were constructed to evaluate the fiber quality, printout pattern, and location information of the deposition layers in the EBB[25]. For benchmarking purposes, a linear SVM model was built using the histogram of oriented gradients from the deposition images. The performances of the three methods were compared, and the pretrained ResNeXt-52 network achieved the best detection accuracy on fiber continuity, regularity, and surface uniformity in the overall anomaly cases. The identification of printing defects, printout patterns, and location information can facilitate the implementation of dynamic parameter tuning.
Traditional ML classification and regression methods have also been applied to evaluate the spacing and pore size when building EBB scaffolds[26]. With layer-by-layer imaging, the random forest classifiers (RFCs) can categorize the deviation of fiber spacing and diameter as “low” or “high” in deposition monitoring. For the same task, the regression models used the quantitative deviation of the fiber spacing and diameter as inputs. Both proved their capability in identifying suitable scaffold-printing conditions. However, these models have not been integrated with EBB for adaptive-parameter control.
For in situ process monitoring, it is relatively easy to label the captured images quantitatively or qualitatively, such as fiber quality, diameter, and interfiber spacing. However, it is difficult to label flow patterns and droplet evolution. Unsupervised ML methods such as DRNN have been introduced to predict the spatial and temporal information of the flow pattern and droplet evolution in droplet-based inkjet printing[29]. As expected, the prediction task in the droplet forming, motion, and jetting behaviors is computationally expensive as a self-learning process using unlabeled data.
2.2. Printing parameter optimization
The droplet size or fiber diameter reflects the bioprinting resolution and governs the mechanical properties of the bioprinted constructs. High-resolution droplets or fibers can be obtained by optimizing the printing parameters. Owing to the intricate biomaterial/bioink properties, researchers cannot mathematically model the relationship between process parameters and printing resolution in an effective manner. To solve this problem, ML has been applied as an alternative choice for model building.
Before using ML to handle this task, a dataset is collected, as shown in Figure 5, where the printing parameters were taken as inputs and the printing performance indices were chosen as outputs. Using this dataset, ML can model the printing process and optimize the relevant printing parameters to achieve a desirable fiber or droplet size.
Figure 5.
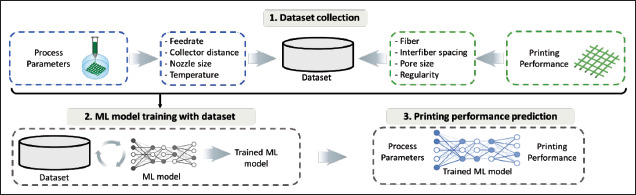
ML applications in printing parameter optimization.
Traditional ML methods, such as SVM, linear regression and random forest, have been used to develop prediction models for the fiber diameter[26,33,34]. The polymer weight fraction, solvent concentration, feed rate, applied voltage, and collector distance were the inputs of the prediction models[34]. The SVM method has also been used to study other printing parameters, such as nozzle temperature and diameter, ink composition, and path height when extruding Pluronic F128 in EBB[27]. The ink composition, nozzle temperature, and printing path height were identified as key parameters to determine the shape fidelity of the deposited filaments and the corresponding structural printability. In fact, the SVM method can not only reveal the complex relationship between inputs and outputs but also optimize the relevant parameters for high-quality prints. Only 12 experimental samples were collected to build this SVM model, and the effectiveness 54of the developed model for a wide range of material properties was questionable. Hence, fine-tuning of the optimized printing parameters may be utilized to further improve the bioprinting performance.
In addition to printing resolution, bioprinting often requires good regularity in EBB layer stacking. Two of these can be incorporated into one scoring metric for printing quality evaluation. To optimize this metric, Bayesian optimization (BO) has been used to explore the printing parameter space for various bioinks[30]. The inputs of this BO model consisted of gelatin methacryloyl (GelMA) inks with three concentrations, three inks with mixed concentrations of GelMA and hyaluronic acid methacrylate (HAMA), and printing parameters (bioink reservoir temperature, extrusion pressure, print-head speed, and platform temperature). Compared to trial-and-error experiments, this ML application can systematically accelerate the retrieval speed of printing parameters and bioinks for high-quality printing.
Traditional ML methods have also been applied to optimize process parameters for EHD inkjet printing and electrospinning. For example, statistical regression analysis, neural networks (NNs) trained with genetic algorithms (GA-NN), and backpropagation NNs have been compared when predicting droplet diameter in EHD inkjet printing[28]. The standoff height, applied voltage, and ink flow rate were the inputs to the prediction models. The GA-NN model outperformed the other two models in most cases when predicting droplet diameter. Similarly, multiple regression (MLR), multilayer perceptron artificial neural network (ANN), and SVM models have been used to predict the electrospun diameter of PCL/Gt nanofibers[24]. With the key input parameters identified by saliency analysis, the ANN model demonstrated the best performance compared with other models[24,39].
Theoretically, the printing process parameters usually have conflicting multiperformance characteristics. For example, a higher applied voltage may increase the biomaterial/bioink printability but lower the printing resolution. A statistical-based method named “desirability function analysis” has been reported to simplify the process parameter optimization in multicriteria objectives[31]. Furthermore, this statistical method that relies on composite desirability may not be able to handle the delicate relationship between the process parameters in EHD inkjet printing.
As discussed above, traditional ML algorithms have been used to rank input feature relevance, establish printing performance models, and optimize the process parameters. This may simplify the process-material-performance model building with limited in-depth knowledge, experiments, and empirical experience. It also provides a simulation tool to model the impact of the printing parameters on the filament/droplet diameter, fiber quality, shape fidelity, and layer stacking. Although the current models are developed under a controllable range of material properties, they may suffer from poor generalization and robustness under varied material properties or modified experimental protocols.
2.3. ML in biomaterial/bioink optimization
An increase in cell culture applications in tissue engineering and drug screening has resulted in a growing demand for biomaterial/bioinks[14,16]. Higher concentrations of bioink/biomaterial may yield good shape fidelity, but poor printability during extrusion.
Increasing the viscosity of biomaterial/bioink can improve shape fidelity, but may compromise printability. Because traditional governing equations cannot effectively and efficiently handle such bioink/biomaterial optimization tasks, manual calibration with numerous experiments was conducted.
To support a new material design workflow, several ML methods have been implemented to optimize the composition and viscosity of biomaterial/bioink to digitally manipulate printability and shape fidelity[40-42]. For example, inductive logic programming has been applied to investigate the rheological properties of hydrogel inks and their corresponding printing qualities[32]. Rheological properties, such as elastic modulus and yield stress, were classified into three classes, and extrusion capability and shape fidelity were classified into two classes. The analysis results from the ML models indicate that printable ink should have a high elastic modulus for high shape fidelity and low yield stress for extrusion. Based on this, a multiple regression model was proposed to quantify the relationship between ink formulations and printability. Another regression model, hierarchical machine learning (HML), was reported to determine the material properties of sodium alginate prints with high/low fidelity[33]. The middle layer in the HML was constructed based on acquired knowledge of the flow-gelation process of alginate. The dimensional similarity between the deposited structure and original design was used to evaluate the HML model. This method can effectively guide high-fidelity EBB scaffold fabrication by optimizing biomaterial formulation and printing parameters. This may be a promising way to scale up the biomaterial/bioink shape fidelity study through the combination of a small number of iterative tests, practical experience, and theoretical knowledge.
In addition, hydrogel inks may require crosslinking to maintain the shape and retain the printed structures 55after deposition. ML is expected to optimize process-related parameters, such as the type of crosslinking and the density of the formed crosslinks, and to explore the trade-off between the stiffness of the deposited structures and the printability of hydrogel inks. However, to date, no such studies have been reported.
It is often assumed that ML methods can accelerate biomaterial/bioink studies by optimizing formulations, viscosity, and the consequent rheological properties to control the biological and mechanical performance. The integration of multiple properties can also be considered as the evaluation metrics for the optimization tasks. To develop a stable Col-I bioink which is a mixture of 15 compositions[43], the bioink concentration should be optimized in terms of both mechanical properties and transparency. Such research would significantly advance the manipulation of cell proliferation, migration, and differentiation in bioprinted constructs. Moreover, ML models are expected to link biomaterial/bioink with complex biological functions, such as biodegradability, and even their effectiveness in in vivo experiments. These studies will create new avenues for bioprinted construct applications in tissue engineering and drug screening. Leveraging ML modeling capabilities would also drive informative biomaterial/bioink designs with more flexibility.
3. ML applications in cell performance studies
ML applications in cell performance include cell viability prediction and cell-microenvironment interaction analysis.
3.1. Cell viability in printing
EBB can incorporate cells for cell-laden construct building, and the extrusion force may damage the incorporated cells when they pass through an extrusion nozzle. Since the printability and cell viability are two critical issues in EBB, the selection of appropriate process parameters and bioink properties would help fragile and sensitive cells survive during and after extrusion, and benefit cell growth in culture. ML has been applied to search for these parameters with the aim of minimizing the negative influence of this extrusion force.
The workflow of ML applications in cell viability analysis is shown in Figure 6, where the dataset is prepared with the inputs from the extrusion parameters and bioink properties, and the output, that is, the corresponding cell viability. Various cell viability prediction models can be constructed using this workflow. The prediction methods for cell viability studies are determined by the available data. When the actual value of cell viability is known, three traditional ML regression models were built, including SVM, linear regression, and random forest, when printing alginate/gelatin-based hydrogels loaded with cells[34]. For the same task, classification methods can be applied when using a threshold to judge the cell viability. Three binary classifiers including random forest, logistic regression classification, and SVM, were constructed to classify cell viability using a threshold of 80%. All of them are also capable of ranking the impact of bioink properties and process parameters and establishing their individual cell viability prediction models.
Figure 6.
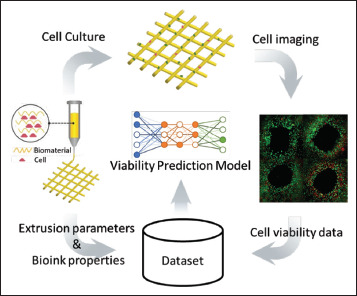
ML application in cell viability analysis.
In addition to the extrusion parameters, the nozzle geometry was further investigated in terms of its influence on shear stress and resultant cell viability. The Gaussian process was utilized to identify the key geometric parameters among the radius of the middle and exit of the nozzle and the nozzle length[44]. As a result, the influence of nozzle geometry on cell viability was quantitatively assessed using a relatively small number of preliminary experiments when extruding the hydrogel bioink.
With the aid of traditional ML methods, cell viability can be optimized by selecting appropriate biomaterial/bioink and printing parameters. Researchers may doubt the overall performance of a single method or model when handling different material properties and printing conditions. Thus, an ensemble learning algorithm was proposed by combining neural networks, ridge regression, K-nearest neighbors, and random forest (RF) to predict cell viability in constructs printed by stereolithography[45]. The experimental dataset consisted of the UV intensity, UV exposure time, gelatin methacrylate concentration, layer thickness, and associated cell viability. By exploiting 56the advantages of multiple ML methods, this learning algorithm used the weighted average of multiple ML models to improve its robustness. This model demonstrated a high prediction accuracy under various printing conditions. Although the necessity of this cell viability study is high, the available datasets for this research topic are limited[46]. This is in part caused by the tedious and expensive cell viability data collection process.
DL methods, such as CNN, can advance cell viability studies by segmenting cells directly in fluorescence images, which can rapidly screen the images with less manpower and effort. A dataset comprising images of 4974 single spheroids with corresponding labels was used to build the prediction model. The developed CNN model successfully categorized cell viability into three classes with a balanced accuracy of 78.7%. CNN has also demonstrated good generalization performance when predicting the cell viability of bioprinted renal spheroids under varied inhibitory concentrations as well as experimental settings[47]. This may be a new pathway to prompt efficient cell viability studies by segmenting cell/nuclei in fluorescence images and then counting live/dead cells using the developed DL models. Nevertheless, cell assays using multidimensional fluorescence images are required for more comprehensive and accurate analysis.
3.2. Cell-microenvironment interaction
Cell-microenvironment interactions are crucial for immune response and tissue regeneration. It is well known that the cell response varies with both materials and their forms. Even for the same material, the cell response may vary significantly when interacting with nanoparticles, scaffolds, coatings, or films. Some cell types may experience the benefits/risks associated with particular material compositions or forms. To investigate the needs and preferences of cell growth in bioprinted constructs, their behavior should be digitalized for in situ analysis.
There has been a growing interest in applying ML to identify cell types, phenotypes, and shapes. ML has outperformed experts in segmenting and classifying cell/nuclei in biological images across various tasks[48,49]. Motivated by this progress, researchers have initiated studies applying ML for cell-microenvironment interaction analysis, that is, cell-scaffold interaction, cell-cell interaction, and cell-material interaction[35,36].
Traditional ML methods can model the relationship between cell proliferation and the physicochemical properties of electrospun scaffolds with regard to the fiber diameter, pore diameter, water contact angle, and Young’s modulus[50]. Six regression algorithms, namely, linear regression, SVM regression, RFr, lasso regression, decision tree regression, and k-nearest neighbor (KNN) regression, were developed and compared. The RFr model yielded the highest accuracy when predicting the proliferation rate of L929 fibroblast cells after a 7-day culture, and this prediction model also coincided with the data collected from in vivo studies. This demonstrates the potential of ML for discovering the impact of fiber diameter on angiogenesis. However, the generalization performance of the developed model has not been verified for more cell types and scaffold structures.
Traditional ML algorithms, including KNN, logistic regression, RFc, SVM, and ANN, have also been utilized to evaluate delta permittivity in a scaffold-based cell culture environment. Four cell types were cultured on the PCL scaffolds, and the corresponding delta permittivity was measured over time using dielectric impedance spectroscopy to build the dataset. Input features were extracted from the relative permittivities at different frequencies, time points, and cell types[50]. Among the developed ML models, KNN yielded the best accuracy in classifying cell types and culture durations.
Biological images, such as confocal laser scanning microscopy (CLSM), are widely used to visualize cell behavior and reveal cell-microenvironment interactions[38]. However, the cell shape and morphological features of these images were difficult to extract using statistical methods. ML algorithms have been proposed to quantitatively translate and deliver this information to researchers. SVM can quantify the diverse cell morphologies of hBMSCs populations using CLSM images and associate them with specific microenvironments, such as PCL fibrous substrates and PCL spin-coated films[51]. SVM can also detect the impact of bioprinted construct morphology on cell-shape phenotypes[35]. Using the extracted metrics from cellular and subcellular morphometry, the developed SVM model successfully classified the substrates into either woven PCL mesh with precision-stacked microscale fibers or nonwoven mesh with randomly oriented fibers. As pioneering research has linked cellular and subcellular morphometry with substrate topology, the quality of the training dataset is critical for reliable identification. However, the size and diversity of the datasets used in the aforementioned studies were not mentioned. This could raise concerns regarding the robustness and convergence of the proposed SVM models when dealing with images collected from different substrate morphologies.
Owing to the complicated nature of biological images, it is difficult to prepare datasets with labeled nuclei, morphological phenotypes, or labeled cell shapes for proliferation and migration. Therefore, unsupervised methods have been applied to model the objects of interest. 57For example, the nuclei segmentation method AD-GAN was proposed to segment cell/nuclei in an unlabeled CLSM image dataset[52], which consisted of 40 images from cultured A549, 3T3, and HeLa cells. This DL method can distinguish nuclei with preserved shape and location information so as to rapidly screen cell-microenvironment interactions[36]. In addition, self-label clustering has been used to cluster and identify distinct morphological phenotypes of a single cell type using low-resolution brightfield images[53]. Compared with supervised ML methods using datasets with concrete labels, unsupervised DL methods can utilize prior knowledge from human beings at abstract levels and explore raw data with unknown structures or ideas.
In general, the ML workflow in image-based cell–microenvironment interaction analysis is summarized in Figure 7, which consists of biological image collections and subsequent cell/nuclei segmentation, cell phenotype identification, and cell type classification. This analysis can visually depict the in situ biological performance of bioprinted constructs and intuitively illustrate the influence of physicochemical properties on cell behavior. In addition, the quantitative indication of the applied material, morphology, and structural design on biological performance provides crucial insights into the environmental impact on cell behavior.
Figure 7.
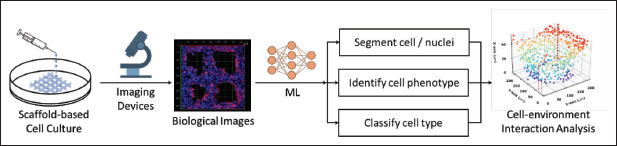
Cell-microenvironment interaction analysis in bioprinted constructs.
As previously discussed, both traditional ML and DL methods can identify cell shapes and phenotypes on bioprinted constructs with varied nanotopography and diverse structures. Segmented cells may serve as candidate templates to offer more effective in situ cell morphology analysis, efficient cell counting, and growth pattern discovery. This investigation may potentially link cell shape and functionalities in the next step. Meanwhile, there is still much space to discover methodologies for nuclei identification under high cell densities, diverse cell morphologies, or multiple cell types, which can be observed in dynamic cell phenotype transformations such as differentiation, migration, and proliferation. In fact, DL methods are more competent in biological image analysis than traditional ML methods, considering their ability to identify underlying patterns or salient features. This may lead to the discovery of additional unknown cell responses or functions in the future, which have not been observed and analyzed in current laboratory practices.
4. Challenges and outlook
ML methods can be used to develop classification, regression, and segmentation models for bioprinting processes and bioprinted constructs. It provides a systematic solution to diagnose uncontrollable factors, maintains the reliability of the bioprinting process, and optimizes biomaterial/bioink and process parameters. Studies have offered the potential to customize bioprinted constructs and manipulate their cell culture responses, as expected. However, several challenges and concerns must be addressed prior to further exploration.
4.1. Dataset quality and size for ML model building
The performance of ML models is determined by the quality of accessible datasets. Comprehensive and consistent datasets may benefit the development of ML models such that they can be scaled up for wider material properties or scaled out for diverse bioprinting technologies. However, the quality of the current datasets is far below this requirement. First, the datasets collected from diverse bioprinters and operational protocols include a large amount of noise and bias. Even if the datasets are collected under the same experimental setup, material composition, and process parameters, they can be easily interrupted by uncontrollable factors in micro/nanoscale bioprinting. Using such datasets directly may reduce the effectiveness and reliability of the developed ML models. However, no study has explored dataset cleaning and data normalization in bioprinting.
In addition to dataset quality, the size of the datasets is another issue. Owing to the expensive and tedious data collection process, only a small dataset is currently available for ML applications in bioprinting, particularly in material optimization and cell performance analysis. For example, Tian et al.[34] built a dataset consisting of 617 instances regarding cell viability and 339 instances regarding filament diameter using 75 published research 58papers by searching the Web of Science database. A nuclei segmentation dataset, Scaffold-A549, was built by collecting human lung cancer cell images for cell-scaffold interaction studies. This dataset consisted of 20 high-resolution unlabeled CLSM images and one fully labeled image with approximately 800 segmented nuclei. Although data augmentation can artificially enlarge a dataset by adding noise and interpolating or extrapolating between samples in a feature space, its application in bioprinting has not yet been reported.
To advance bioprinting using ML, it is essential to make additional experimental data publicly available or accessible upon request. Using similar open-source software for bioprinters and establishing a worldwide data-sharing network could be promising[46]. However, researchers may not wish to publish the collected data because of expenses related to the materials, manpower, and facilities. This has impeded the advancement of ML applications for bioprinting.
4.2. Information integration and coordination from multiple sources
Apart from dataset quality, information integration from multiple formats and sources is an ongoing research issue. In current studies, the collected numerical data or images in bioprinting are used individually for specific prediction tasks. For example, features extracted from the process parameters or material properties can be conveniently used to develop prediction models[20,39,54]. However, it is difficult to deliver a prediction task when both image and numerical data are input. They are equally important and should be synchronously used to develop process-material-performance models or refine bioprinted construct designs for better cell performance. Further strategic studies should be conducted to integrate the diverse data formats when developing ML prediction models. In addition, the emerging technologies, such as big data and digital twin, may also contribute to the information integration and coordination. One way is to build ML training databases using big data curation, and the other is to build digital twins of human tissue/organs with cellular resolution and properties[17]. As such, a standard bioprinting simulation practice is expected to balance virtual and physical experiments and maximize bioprinting resource utilization.
Information integration and coordination is of increasing interest in multi-material printing. To achieve a cost-effective and time-efficient printing, ML is expected to integrate and coordinate diverse information in this process so as to enhance printing resolution, printing path planning, G-code error detection, and structural stability of bioprinted construct. ML can also facilitate the material selection and the subsequent crosslinking at different degrees to build biological constructs with desired shape fidelity.
4.3. Unlabeled data in cell performance analysis
The application of supervised ML methods relies on a carefully and precisely labeled dataset. However, it is difficult to label nuclei or cells on bioprinted constructs using biological images. First, several overlapping cells or thousands of nuclei with irregular or deformed shapes are observed. Varied cell shapes, phenotypes, and types cause this labeling task to worsen. Moreover, a single-cell type may exhibit a wide range of morphologies in terms of adhesion, proliferation, and migration. Thus, the interest in applying unsupervised ML methods to model such complex unlabeled datasets has continued to grow. Such studies aim to automatically identify cell types or behaviors, assist in the discovery of biological phenomena at the single cell level, and upgrade cell laboratory workflow.
4.4. Digitalized in situ performance analysis
The bioprinted constructs are biologically evaluated based on the overall performance of comparative samples. This gap has motivated researchers to explore in situ evaluation methods. As the biological images of these constructs are collected, DL can digitalize them by segmenting cell/nuclei, classifying cell types, and identifying phenotypes. This is valuable when analyzing bioprinted constructs with multilayer heterogeneous or gradient structures. Currently, overall performance evaluation cannot provide in situ information about the biological response of a structure. The digitalized cell distribution can also present an intuitive biological response to the structural design, layer-specific fiber diameter and pore size, and biomaterial composition. Consequently, the cell number and spatial behavior of bioprinted constructs can be subjectively and quantitatively evaluated. This would impel researchers to combine diverse materials and structures and support functional scaffold design with better biological performance and revolutionize construct-based organ or disease model studies. More studies using ML to predict tissue formation and create organoid and tumoroid models are expected, although these topics seem significantly challenging.
5. Conclusion
Bioprinting has demonstrated its ability to produce constructs for cell culture in tissue engineering and drug screening. These applications are still primitive because of the limited fundamental research on process-material-performance.
In this review, the current status, challenges, and outlook of ML applications in bioprinting are discussed. 59Both the traditional ML and DL methods can empower bioprinting by manipulating and optimizing micro/nanostructures, materials, and printing parameters. This capability, when applied to bioprinted constructs, can generate more advanced concepts, enhance their biological and mechanical performance, and prompt effective cell-microenvironment interactions. These customized bioprinted constructs with controlled material compositions as well as specific micro/nanostructures would establish a solid foundation for developing organoid and tumoroid models from a technical perspective. It is no doubt that ML methods would expand their applications to facilitate diverse printing scenarios and application topics in the near future.
Acknowledgments
None.
Funding
This work was financially supported by Xi’an Jiaotong-Liverpool University’s Key Program Special Fund under Grant KSF-E-37.
Conflict of interest
The authors declare no conflicts of interest.
Author contributions
Conceptualization: Kaizhu Huang, Dejian Huang
Investigation: Jia An, Linzhi Jing
Supervision: Kaizhu Huang, Dejian Huang
Writing – original draft: Jie Sun, Kai Yao
Writing – editing & review: Jia An, Linzhi Jing
All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data
Not applicable.
References
- 1.Gudapati H, Dey M, Ozbolat I. 2016. A comprehensive review on droplet-based bioprinting: Past, present and future Biomaterials 102:20 42 https://doi.org/10.1016/j.biomaterials.2016.06.012 [DOI] [PubMed] [Google Scholar]
- 2.Placone JK, Engler AJ. Recent advances in extrusion-based 3D printing for biomedical applications. Adv Healthc Mater. 2018;7(8):1701161. doi: 10.1002/adhm.201701161. https://doi.org/10.1002/adhm.201701161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaioannou TG, Manolesou D, Dimakakos E, et al. 3D bioprinting methods and techniques: Applications on artificial blood vessel fabrication. Acta Cardiol Sinica. 2019;35(3):284. doi: 10.6515/ACS.201905_35(3).20181115A. https://doi.org/10.6515/ACS.201905_35(3).20181115A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Zhang B, Li Z, et al. High-resolution electrohydrodynamic bioprinting: A new biofabrication strategy for biomimetic micro/nanoscale architectures and living tissue constructs. Biofabrication. 2020;12(4):042002. doi: 10.1088/1758-5090/aba1fa. https://doi.org/10.1088/1758-5090/aba1fa. [DOI] [PubMed] [Google Scholar]
- 5.Ng WL, Chan A, Ong YS, et al. Deep learning for fabrication and maturation of 3D bioprinted tissues and organs. Virtual Phys Prototyp. 2020;15(3):340–358. https://doi.org/10.1080/17452759.2020.1771741. [Google Scholar]
- 6.RegenhuThe R-GEN 100 bioprinter [EB/OL]. 2022. https://www.regenhu.com/3dbioprinting-solutions/r-gen-100-3dbioprinter (Accessed November 8, 2022)
- 7.3dsman EnvisionTEC: 3D-Bioplotter [EB/OL] 2022 https://3dsman.com/envisiontec-3d-bioplotter (Accessed November 8, 2022) [Google Scholar]
- 8.Ozbolat IT, Hospodiuk M. Current advances and future perspectives in extrusion-based bioprinting. Biomaterials. 2016;76:321–343. doi: 10.1016/j.biomaterials.2015.10.076. https://doi.org/10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 9.Ning L, Chen X. A brief review of extrusion-based tissue scaffold bio-printing. Biotechnol J. 2017;12(8):1600671. doi: 10.1002/biot.201600671. https://doi.org/10.1002/biot.201600671. [DOI] [PubMed] [Google Scholar]
- 10.Brown TD, Dalton PD, Hutmacher DW. Direct writing by way of melt electrospinning. Adv Mater. 2011;23(47):5651–5657. doi: 10.1002/adma.201103482. https://doi.org/10.1002/adma.201103482. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Fuh J, Wong Y, et al. Fabrication of 3D scaffolds via E-jet printing for tendon tissue repair. International Manufacturing Science and Engineering Conference. 2015 in. 56833, V002T03A005. [Google Scholar]
- 12.Zhang B, Seong B, Nguyen V, et al. 3D printing of high-resolution PLA-based structures by hybrid electrohydrodynamic and fused deposition modeling techniques. J Micromech Microeng. 2016;26(2):025015. https://doi.org/10.1088/0960-1317/26/2/025015. [Google Scholar]
- 13.He J, Xu F, Cao Y, et al. Towards microscale electrohydrodynamic three-dimensional printing. J Phys D Appl Phys. 2016;49(5):055504. https://doi.org/10.1088/0022-3727/49/5/055504. [Google Scholar]
- 14.60Jing L, Sun J, Liu H, et al. Using plant proteins to develop composite scaffolds for cell culture applications. Int J Bioprint. 2021;7(1):66–77. doi: 10.18063/ijb.v7i1.298. https://doi.org/10.18063/ijb.v7i1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, Jing L, Fan X, et al. Electrohydrodynamic printing process monitoring by microscopic image identification. Int J Bioprint. 2019;5(1):1–9. doi: 10.18063/ijb.v5i1.164. https://doi.org/10.18063/ijb.v5i1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun J, Jing L, Liu H, et al. Generating nanotopography on PCL microfiber surface for better cell-scaffold interactions. Proc Manuf. 2020;48:619–624. https://doi.org/10.1016/j.promfg.2020.05.090. [Google Scholar]
- 17.An J, Chua CK, Mironov V. Application of machine learning in 3D bioprinting: Focus on development of big data and digital twin. Int J Bioprint. 2021;7(1):1–6. doi: 10.18063/ijb.v7i1.342. https://doi.org/10.18063/ijb.v7i1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Hong G, Rahman M, et al. Improved performance evaluation of tool condition identification by manufacturing loss consideration. Int J Prod Res. 2005;43(6):1185–1204. https://doi.org/10.1080/00207540412331299701. [Google Scholar]
- 19.Sun J, Yao K, Huang K, et al. Machine learning applications in scaffold based bioprinting. Mater Today Proc. 2022;70(2022):17–23. [Google Scholar]
- 20.Jie S, Hong GS, Rahman M, et al. Feature extraction and selection in tool condition monitoring system. Australian Joint Conference on Artificial Intelligence. 2002:487–497. in. [Google Scholar]
- 21.MathWorks Support vector machine classification [EB/OL] 2022 https://www.mathworks.com/help/stats/support-vector-machine-classification.html (Accessed November 8, 2022) [Google Scholar]
- 22.Yu C, Jiang J. A perspective on using machine learning in 3D bioprinting. Int J Bioprint. 2020;6(1):4–11. doi: 10.18063/ijb.v6i1.253. https://doi.org/10.18063/ijb.v6i1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin J, Lee Y, Li Z, et al. Optimized 3D bioprinting technology based on machine learning: A review of recent trends and advances. Micromachines. 2022;13(3):363. doi: 10.3390/mi13030363. https://doi.org/10.3390/mi13030363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalantary S, Jahani A, Jahani R. MLR and Ann approaches for prediction of synthetic/natural nanofibers diameter in the environmental and medical applications. Sci Rep. 2020;10(1):1–10. doi: 10.1038/s41598-020-65121-x. https://doi.org/10.1038/s41598-020-65121-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin Z, Zhang Z, Shao X, et al. Monitoring anomalies in 3D bioprinting with deep neural networks. ACS Biomater Sci Eng. 2021 doi: 10.1021/acsbiomaterials.0c01761. https://doi.org/10.1021/acsbiomaterials.0c01761. [DOI] [PubMed] [Google Scholar]
- 26.Conev A, Litsa EE, Perez MR, et al. Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Tissue Eng Part A. 2020;26(23-24):1359–1368. doi: 10.1089/ten.tea.2020.0191. https://doi.org/10.1089/ten.tea.2020.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Z, Angeline V, Sun W. Evaluation of printing parameters on 3D extrusion printing of pluronic hydrogels and machine learning guided parameter recommendation. Int J Bioprint. 2021;7(4):179–189. doi: 10.18063/ijb.v7i4.434. https://doi.org/10.18063/ijb.v7i4.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ball AK, Das R, Roy SS, et al. Modeling of EHD inkjet printing performance using soft computing-based approaches. Soft Comput. 2020;24(1):571–589. https://doi.org/10.1007/s00500-019-04202-0. [Google Scholar]
- 29.Huang J, Segura LJ, Wang T, et al. Unsupervised learning for the droplet evolution prediction and process dynamics understanding in inkjet printing. Addit Manuf. 2020;35:101197. https://doi.org/10.1016/j.addma.2020.101197. [Google Scholar]
- 30.Ruberu K, Senadeera M, Rana S, et al. Coupling machine learning with 3D bioprinting to fast track optimisation of extrusion printing. Appl Mater Today. 2021;22:100914. https://doi.org/10.1016/j.apmt.2020.100914. [Google Scholar]
- 31.Das R, Ball AK, Roy SS. Optimization of E-jet based micro-manufacturing process using desirability function analysis. Industry Interactive Innovations in Science, Engineering and Technology Springer. 2018:477–484. in. [Google Scholar]
- 32.Lee J, Oh SJ, An SH, et al. Machine learning-based design strategy for 3D printable bioink: Elastic modulus and yield stress determine printability. Biofabrication. 2020;12(3):035018. doi: 10.1088/1758-5090/ab8707. https://doi.org/10.1088/1758-5090/ab8707. [DOI] [PubMed] [Google Scholar]
- 33.Bone JM, Childs CM, Menon A, et al. Hierarchical machine learning for high-fidelity 3D printed biopolymers. ACS Biomater Sci Eng. 2020;6(12):7021–7031. doi: 10.1021/acsbiomaterials.0c00755. https://doi.org/10.1021/acsbiomaterials.0c00755. [DOI] [PubMed] [Google Scholar]
- 34.Tian S, Stevens R, McInnes BT, et al. Machine assisted experimentation of extrusion-based bioprinting systems. Micromachines. 2021;12(7):780. doi: 10.3390/mi12070780. https://doi.org/10.3390/mi12070780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tourlomousis F, Jia C, Karydis T, et al. Machine learning metrology of cell confinement in melt electrowritten three-dimensional biomaterial substrates. Microsyst Nanoeng. 2019;5(1):1–19. doi: 10.1038/s41378-019-0055-4. https://doi.org/10.1088/1758-5090/ab8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao K, Huang K, Sun J, et al. Scaffold-A549: A benchmark 3D fluorescence image dataset for unsupervised nuclei segmentation. Cognit Comput. 2021;13(6):1603–1608. https://doi.org/10.1007/s12559-021-09944-4. [Google Scholar]
- 37.61Sujeeun LY, Goonoo N, Ramphul H, et al. Correlating in vitro performance with physico-chemical characteristics of nanofibrous scaffolds for skin tissue engineering using supervised machine learning algorithms. R Soc Open Sci. 2020;7(12):201293. doi: 10.1098/rsos.201293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paddock SW. Confocal laser scanning microscopy. Biotechniques. 1999;27(5):992–1004. doi: 10.2144/99275ov01. https://doi.org/10.2144/99275ov01. [DOI] [PubMed] [Google Scholar]
- 39.Kalantary S, Jahani A, Pourbabaki R, et al. Application of ANN modeling techniques in the prediction of the diameter of PCL/gelatin nanofibers in environmental and medical studies. RSC Adv. 2019;9(43):24858–24874. doi: 10.1039/c9ra04927d. https://doi.org/10.1039/C9RA04927D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kievit FM, Florczyk SJ, Leung MC, et al. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials. 2010;31(22):5903–5910. doi: 10.1016/j.biomaterials.2010.03.062. https://doi.org/10.1016/j.biomaterials.2010.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee F, Kurisawa M. Formation and stability of interpenetrating polymer network hydrogels consisting of fibrin and hyaluronic acid for tissue engineering. Acta Biomater. 2013;9(2):5143–5152. doi: 10.1016/j.actbio.2012.08.036. https://doi.org/10.1016/j.actbio.2012.08.036. [DOI] [PubMed] [Google Scholar]
- 42.Fitzgerald KA, Guo J, Tierney EG, et al. 2015. The use of collagen-based scaffolds to simulate prostate cancer bone metastases with potential for evaluating delivery of nanoparticulate gene therapeutics Biomaterials 66:53 66 https://doi.org/10.1016/j.biomaterials.2015.07.019 [DOI] [PubMed] [Google Scholar]
- 43.Song Y, Hua S, Sayyar S, et al. Corneal bioprinting using a high concentration pure collagen I transparent bioink. Bioprinting. 2022;28:e00235. https://doi.org/10.1016/j.bprint.2022.e00235. [Google Scholar]
- 44.ReinaRomo E, Mandal S, Amorim P, et al. Towards the experimentally-informed in silico nozzle design optimization for extrusion-based bioprinting of shear-thinning hydrogels. Front Bioeng Biotechnol. 2021;9:701778. doi: 10.3389/fbioe.2021.701778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Liu Q, Casillas J, et al. Prediction of cell viability in dynamic optical projection stereolithography-based bioprinting using machine learning. J Intel Manuf. 2020;33:1–11. [Google Scholar]
- 46.Malekpour A, Chen X. Printability and cell viability in extrusion-based bioprinting from experimental, computational, and machine learning views. J Funct Biomater. 2022;13(2):40. doi: 10.3390/jfb13020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tröndle K, Miotto G, Rizzo L, et al. Deep learning-assisted nephrotoxicity testing with bioprinted renal spheroids. Int J Bioprint. 2022;8(2):164–173. doi: 10.18063/ijb.v8i2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.AlKofahi Y, Zaltsman A, Graves R, et al. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinf. 2018;19(1):1–11. doi: 10.1186/s12859-018-2375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McQuin C, Goodman A, Chernyshev V, et al. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16(7):e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shohan S, Harm J, Hasan M, et al. Non-destructive quality monitoring of 3D printed tissue scaffolds via dielectric impedance spectroscopy and supervised machine learning. Proc Manuf. 2021;53:636–643. [Google Scholar]
- 51.Chen D, Sarkar S, Candia J, et al. Machine learning based methodology to identify cell shape phenotypes associated with microenvironmental cues. Biomaterials. 2016;104:104–118. doi: 10.1016/j.biomaterials.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao K, Sun J, Huang K, et al. Analyzing cell-scaffold interaction through unsupervised 3D nuclei segmentation. Int J Bioprint. 2021;8(1):167–181. doi: 10.18063/ijb.v8i1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao K, Rochman ND, Sun SX. Cell type classification and unsupervised morphological phenotyping from low-resolution images using deep learning. Sci Rep. 2019;9(1):1–13. doi: 10.1038/s41598-019-50010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun J, Hong GS, Rahman M, et al. The application of nonstandard support vector machine in tool condition monitoring system. Proceedings. DELTA 2004. Second IEEE International Workshop on Electronic Design, Test and Applications. 2004:295–300. in. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


