Abstract
The lack of specificity of cancer treatment causes damage to normal cells as well, which limits the therapeutic range. To circumvent this problem one would need to use an absolute difference between normal cells and cancer cells as therapeutic target. Such a difference exists in the genome of all individuals suffering from a tumor that is characterized by loss of genetic material [loss of heterozygosity (LOH)]. Due to LOH, the tumor is hemizygous for a number of genes, whereas the normal cells of the individual are heterozygous for these genes. Theoretically, polymorphic sites in these genes can be utilized to selectively target the cancer cells with an antisense oligonucleotide, provided that it can discriminate the alleles and inhibit gene expression. Furthermore, the targeted gene should be essential for cell survival, and 50% gene expression sufficient for the cell to survive. This will allow selective killing of cancer cells without concomitant toxicity to normal cells. As an initial step in the experimental test of this putative selective cancer cell therapy, we have developed a set of antisense phosphorothioate oligonucleotides which can discriminate the two alleles of a polymorphic site in the gene encoding the large subunit of RNA polymerase II. Our data show that the exact position of the antisense oligonucleotide on the mRNA is of essential importance for the oligonucleotide to be an effective inhibitor of gene expression. Shifting the oligonucleotide position only a few bases along the mRNA sequence will completely abolish the inhibitory activity of the antisense oligonucleotide. Reducing the length of the oligonucleotides to 16 bases increases the allele specificity. This study shows that it is possible to design oligonucleotides that selectively target the matched allele, whereas the expression level of the mismatched allele, that differs by one nucleotide, is only slightly affected.
INTRODUCTION
The central problem in cancer therapy is to kill cancer cells without damaging normal cells. In order to solve this problem one needs to identify a consistent and absolute difference between cancer cells and normal cells, and be able to utilize this difference to selectively target and kill cancer cells. Such a difference can be found in the variation that exists in the human genome, combined with the fact that regions of loss of heterozygosity (LOH) are created in cancer cells during tumorigenesis.
The loss of large chromosomal regions, or even whole chromosomes, is an early event in the clonal evolution of cancers. LOH can involve >20% of the total genome in certain cancers (1). Therefore, the genetic difference between normal cells and cancer cells extends beyond the loss of a tumor suppressor gene. Many genes will be reduced to hemizygosity in cancer cells due to LOH, and some of these genes may be essential for cell survival.
We have identified a large number of single nucleotide polymorphisms (SNPs) in genes that are essential for cell survival and are localized in chromosomal regions often involved in LOH in various cancer types (A.L.M.A.ten Asbroek, D.Housman, V.Stanton Jr and F.Baas, manuscript in preparation). Many of these SNPs have a high degree of heterozygous expression in a panel of normal individuals. Theoretically these polymorphisms can be selectively targeted by drugs such as antisense oligonucleotides (ODNs) that will discriminate between the two alleles. Allele-specific inhibition (ASI) of the allele of an essential gene that is left in the cancer cells, as a result of the LOH, will lead to cell death. The heterozygous normal cells, however, will lose only 50% of the expression of the gene by this ODN and, provided that 50% gene activity is enough for cell survival, will not be damaged by this antisense ODN. This hypothetical therapy, also known as Variagenic Targeting, has been described in more detail by Basilion et al. (2).
One of the genes in which we have found many SNPs with a high frequency of heterozygous expression in normal individuals, is the large subunit of RNA polymerase II (POLR2A). RNA polymerase II is a multisubunit enzyme that transcribes protein-coding genes in eukaryotes (3,4). POLR2A encodes a 220 kDa protein which is the central heart of the transcription machinery and is localized on chromosome 17p13.1, in close proximity to p53 (5). LOH of tumor suppressor gene p53 occurs very frequently in many tumor types (6). Consequently it is expected that only one allele of POLR2A is present in many tumors. We have analyzed a set of glioma samples, a tumor type in which LOH of 17p is a frequent event. We could indeed show LOH of POLR2A in the samples that were also hemizygous for p53 (unpublished results).
The POLR2A gene product is essential for cell survival as has been demonstrated by the use of the mushroom toxin α-amanitin. This inhibitor of RNA polymerase II binds to the 220 kDa protein subunit and inhibits the chain elongation reaction of the enzyme. This leads to cell death already at a low dose of α-amanitin (7,8). Therefore, POLR2A is a good candidate for the approach of tumor-specific cell kill by ASI therapy.
A number of reports have shown that antisense ODNs can be successfully used to knock out and inhibit gene expression (9,10 and references therein). The mechanism by which this occurs is an RNase H-mediated degradation of the mRNA moiety in the DNA–RNA duplex that is formed between the ODN and the mRNA. To prevent degradation of the oligonucleotides by endo- or exonucleolytic activities in the cell, the antisense ODNs usually contain modified DNA analogues, such as phosphorothioate (PS) instead of the regular phosphodiester (PO) DNA structure (11,12). PS-ODNs can display some cellular toxicity, so caution has to be taken with respect to non-specific side-effects and other effects that cannot be accounted for by specific action of the antisense ODN and RNase H (13).
This report shows the development of a set of antisense phosphorothioate oligonucleotides that can discriminate the two alleles of a polymorphic site in POLR2A. The oligonucleotides selectively target the matched allele, whereas the expression of the mismatched allele, that differs in one nucleotide, is only slightly affected.
MATERIALS AND METHODS
Cell lines
The pancreatic carcinoma cell line MiaPacaII was maintained by serial passage in Dulbecco’s modified Eagle’s medium (DMEM) and the prostate cancer cell line 15PC3 in RPMI 1640 medium. Cells were grown at 37°C and 5% CO2. Media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
Transfections
One day prior to transfection cells were plated at a density of 105 cells per well in a 6-well culture plate. Cells were gently washed once in serum-free medium, and 1 ml serum-free medium containing transfection reagent DAC-30 (Eurogentec) was added per well; then the PS-ODN was added to the well. DAC-30 was used at a concentration of 1 µl DAC-30 per ml serum-free medium per 100 nM PS-ODN used. After 5 h incubation of the cells at 37°C and 5% CO2, the ODN-containing medium was aspirated off and replaced with complete medium for overnight growth. Scaling-up or -down of the transfection protocol to a different plating size was done according to the surface area of the plates.
For fluorescence microscopy of transfections with FITC-labeled ODNs the cells were plated on glass coverslips in a 6-well culture plate. At time of analysis, the cells were fixed on the glass in PBS containing 4% paraformaldehyde and sealed on microscopic glass in Vectashield Mounting Medium (Vector Laboratories Inc.). Fluorescence microscopy was done with a Vanox microscope and appropriate filters.
All oligonucleotides (PS and PO) were purchased from Isogen (The Netherlands).
RNA analysis
Cells were harvested in Trizol (Gibco BRL) after removing the growth medium 20 h post-transfection. The RNA isolation was according to the manufacturer’s procedure. RNA was separated on glyoxal gels containing 1% agarose, following standard protocols. RNA was subsequently transferred to Hybond-N+ membrane (Amersham) in 20× SSC. Following transfer the RNA was UV-crosslinked and the membrane was baked for 2 h at 80°C. Hybridizations and post-hybridization washes were according to Church and Gilbert (14).
Probes
The following cDNA fragments of POLR2A were generated by RT–PCR and cloned into the pGEM-T Easy vector (Promega). 5′POL: forward primer F7, AGGGGGAACAGTCAGTACCC (position 1222–1241); reverse primer R8, CAGGAGGTTC-ATCACTTCACC (position 1692–1672). 3′POL, forward primer F17: ACCTGCCACCCAGATGAC (position 3291–3308); reverse primer R20, GTACAGCTCCCGCTCCAG (position 4149–4132). All positions are relative to the start codon (GenBank accession no. X63564).
For the 28S ribosomal RNA, a cDNA fragment was generated using: forward primer, TGGTTCCCTCCGAAGTTTC (position 1635–1653 of GenBank accession no. M11167); reverse primer, CGGATTCCGACTTCCATG (position 1973–1956).
EcoRI fragments containing the inserts from the pGEM-T Easy clones were purified from gel and used as probes. Radioactive signal on the blots was visualized with a Phosphorimager (Molecular Dynamics) and signal intensities were quantified.
RESULTS
FITC-labeled PS-ODNs were used to optimize the transfection procedure of cultured cells. The use of liposomal transfection reagent DAC-30 and serum-free medium allowed reproducible transfection of >95% of the cells in a plate, irrespective of the cell line and the PS-ODN used, without major cellular toxicity problems.
MiaPacaII cells were transfected with PS-ODNs targeted to one of the highly frequent SNPs in POLR2A, a C/T polymorphism located at position 2673 relative to the ATG in the POLR2A mRNA sequence. MiaPacaII cells are homozygous for a T at the target position. Figure 1 shows the sequence around the polymorphic site and the antisense ODNs tested. Since we want to target a particular site in the mRNA, we are restricted to a limited number of available positions for the antisense ODN. We assayed the POLR2A mRNA level using northern blots, since these ODNs are presumed to result in mRNA degradation via RNase H activity (Fig. 2). No difference in mRNA level was detected when the cells were transfected with a 20mer containing the polymorphic nucleotide at position 11 in the PS-ODN (Cas20 or Tas20) in comparison to the missense (Cms20 or Tms20) or mock-transfected controls (Fig. 2, lanes 1–6). When the position of the 20mer antisense ODN was shifted 5 nucleotides towards the 3′ end (R5as20) or the 5′ end (L5as20) in the POLR2A mRNA sequence, a decreased mRNA level was detected (Fig. 2, lanes 7–12). The R5 ODNs display only ∼50% of the potency of the L5 ODNs. The corresponding sense control ODNs (R5s20 and L5s20, respectively) have no effect on the POLR2A mRNA level.
Figure 1.
The sequence surrounding the polymorphic C/T site at position 2673 relative to the startcodon in the mRNA sequence (GenBank accession no. X63564) and antisense PS-ODN sequences used to target this polymorphism. Each PS-ODN exists in two versions complementary to the two alleles; X in the ODN name is a C or T, corresponding to X in the ODN sequence containing a G or A, respectively.
Figure 2.
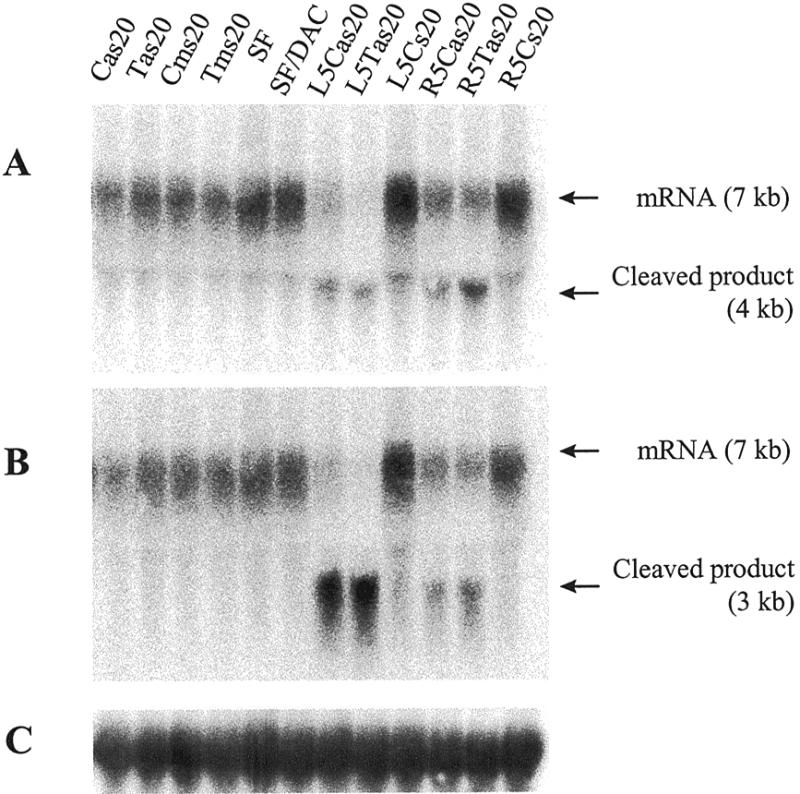
Northern blot analysis of MiaPacaII cells transfected with 800 nM PS-ODN. Hybridizations were with (A) a 3′POL probe, which is located downstream of the ODN target site; (B) a 5′POL probe located upstream of the ODN target site; (C) a 28S RNA control probe for RNA loading. Indicated are the POLR2A mRNA, as well as the RNA products that result from cleavage of the mRNA by RNase H at the site of the ODN. The PS-ODNs used in the transfections (all at 800 nM) are indicated on top of the lanes. Cms20 and Tms20 are the missense controls (antisense sequence in reverse orientation) of Cas20 and Tas20, respectively; L5s20 and R5s20 are the sense controls (sequence as in the mRNA) of L5as20 and R5as20, respectively; SF, serum-free medium only; SF/DAC, serum-free medium containing DAC-30.
The ODNs L5as20 and R5as20, showed an efficient cleavage of the POLR2A target mRNA. Besides the 7 kb mRNA, the 3′POL probe (Fig. 2A), located downstream of the ODN position in the mRNA, detects a 4 kb RNA (masked partly by the 28S RNA on this blot), whereas the 5′POL probe (Fig. 2B), located upstream of the ODN position in the gene, detects a 3 kb cleaved RNA product. The size of these products is compatible with the expected size of the cleavage products. Some degradation of the cleaved RNA products does occur, as is demonstrated by the smear of the cleaved product detected with the 5′POL probe.
The reduction of the POLR2A mRNA level is dependent on the concentration of the antisense ODN L5Tas20 (Fig. 3). The mRNA level of the antisense ODN treated cells compared to the sense ODN control situation (corrected for RNA loading with the 28S RNA control hybridization) is 77 ± 3% (100 nM), 48 ± 1% (200 nM), 24 ± 3% (400 nM) and <10% (800 nM). The cleaved 5′ RNA product is 18 ± 2%, 37 ± 2%, 70 ± 3% and 98 ± 5%, respectively. Although one has to bear in mind that this may be an underestimation of the true reduction, since the cleaved products apparently are subjected to some degradation, the relative differences are valid.
Figure 3.
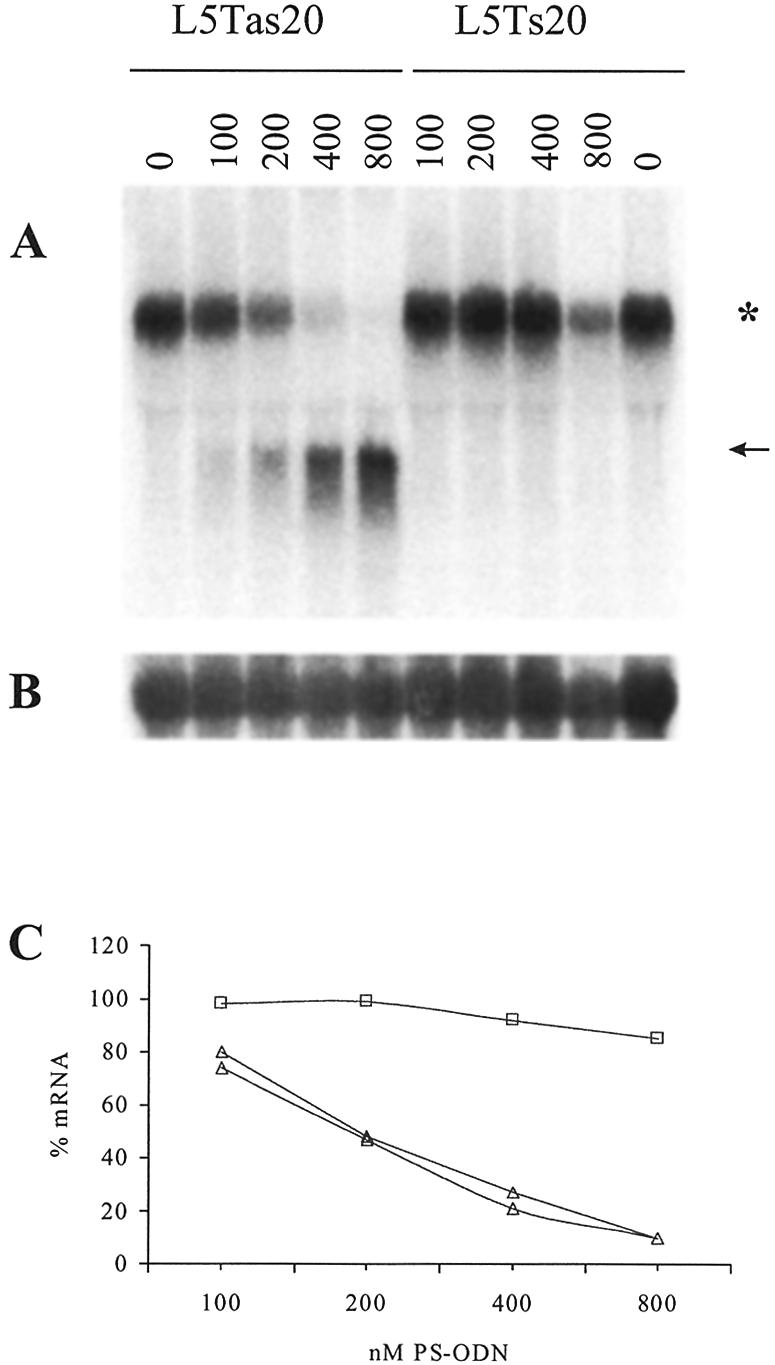
Northern blot analysis of a dose–response series of L5Tas20 on MiaPacaII cells. Cells were transfected with a concentration series of antisense ODN L5Tas20 and the sense control ODN L5Ts20. RNA was extracted 20 h post-transfection. Concentration of ODNs (in nM) are given on top of the lanes. (A) 5′POL probe, the asterisk indicates the mRNA, the arrow indicates the cleaved 5′ product RNA; (B) 28S RNA control hybridization; (C) graphical presentation of the POLR2A mRNA levels, quantified and corrected for RNA loading using the 28S RNA signal, as percentage of the POLR2A mRNA level in the mock-treated cells: squares, sense ODN values; triangles, antisense ODN values of two independent experiments.
For ASI therapy to be successful it is necessary to discriminate the two alleles of a polymorphic site. However, both L5Cas20 and L5Tas20 worked efficiently on the homozygous (T/T) MiaPacaII cells. Therefore, we designed new ODNs to increase the specificity for the individual alleles by reducing the length of the ODNs. Two series of PS-ODNs, named L5as20→16 and L4as18→15, were generated in a C- and T-version (see Fig. 1 for sequences and positions of the ODNs with respect to the polymorphic target mRNA).
These L4 and L5 PS-ODN series were assayed in MiaPacaII cells at a concentration of 800 nM (Fig. 4). In both ODN series the size of the PS-ODN influences the specificity for a particular allele. The shorter T-version ODNs are more efficient than the corresponding C-versions on the target (T/T) mRNA. Judged from the intensity of the cleaved product bands, the L4 antisense ODNs seem more potent inhibitors than their L5 size counterparts. However, they display less allele specificity, as was revealed when the ratio of the resultant mRNA levels with the Tas ODN (matched) and that with the corresponding Cas ODN (mismatched) were calculated. Similar results were obtained with an in vitro RNase H assay with run-off RNA template containing a C or T at the target position and PO-ODNs of the L4 or L5 series of the C or T type (data not shown). The highest allelic specificity (lowest mRNA ratio of matched/mismatch ODN) was obtained in both assays with the ODN couple L5as16.
Figure 4.
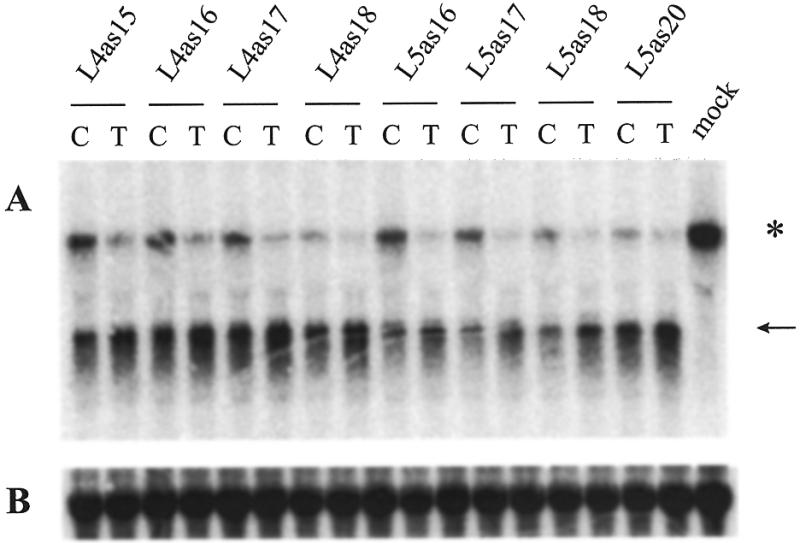
Northern blot analysis of MiaPacaII cells transfected with 800 nM of the L4 and L5 PS-ODN series. On top of the lanes is given if the C- or T-version of the indicated ODN is used. Hybridizations are with the 5′POL probe (A) and the 28S RNA control probe (B). The asterisk indicates the POLR2A mRNA, the arrow indicates the cleaved 5′ product RNA.
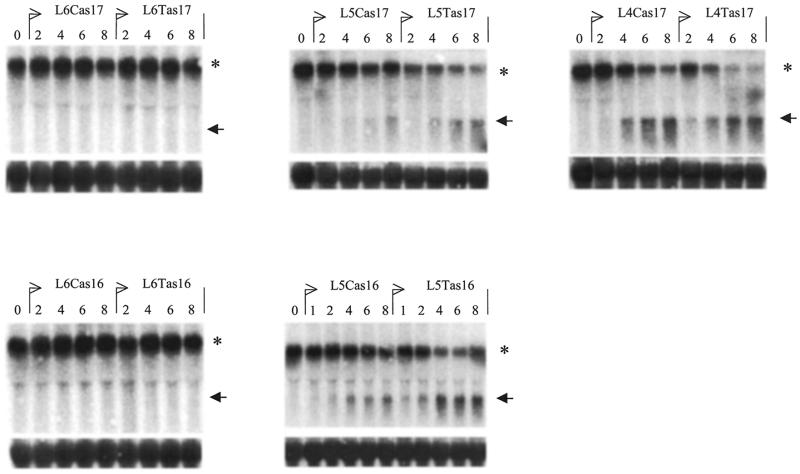
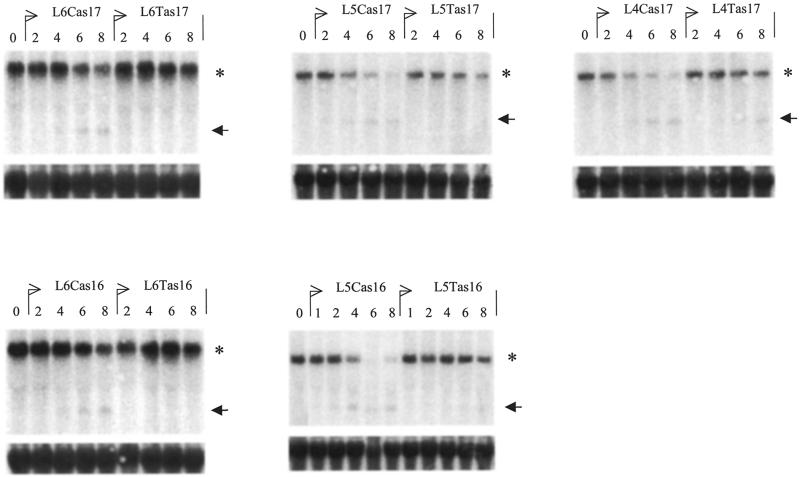
We examined the two allelic versions of L5as16, as well as those of L5as17, L4as17, L6as17 and L6as16 (Fig. 1) in two cell lines with different genotypes: the pancreatic carcinoma cell line MiaPacaII which is T/T at position 2673, and the prostate cancer cell line 15PC3 which is C/C at this position. With FITC-labeled ODNs we could demonstrate that both cell types could be transfected equally well, in terms of percentage of positive cells as well as intensity per cell and staining pattern within the cell.
Each ODN couple was tested in both cell lines with a concentration series. Figure 5 shows the northern analysis of the MiaPacaII cells, and Figure 6 shows that of the 15PC3 cells. Comparison of the IC50 of these allelic PS-ODN couples (Table 1) indicates that L5as17 is the most specific ODN for the T-genotype target mRNA, whereas the 16mer ODN couple L5as16 is a better discriminator of the C-genotype. The 15PC3 cells, containing the C-target sequence, yield in all cases a more efficient targeting with the matched (C-version) than the mismatched (T-version) PS-ODN. The L6as16 and L6as17 PS-ODNs are the least efficient inhibitors of the POLR2A mRNA; they are, in fact, not at all able to target the T-target sequence in the MiaPacaII cells. All PS-ODNs tested provide less specificity for the T-genotype than for the C-genotype target sequence. Analysis of the data indicates that this cannot solely be attributed to the lower melting temperature (Tm) of the T-version ODN compared to the corresponding C-version. Rather, it suggests that the secondary structure is less advantageous for ODN-binding when this SNP target contains a T.
Figure 5.
Northern blot analysis of a concentration series of various ODNs on MiaPacaII cells. The top panels show hybridizations with the 5′POL probe (the asterisk indicates the POLR2A mRNA, the arrow indicates the cleaved 5′ product RNA); the bottom panels show the corresponding 28S RNA control hybridizations. The values on top of the lanes show the concentration of the indicated PS-ODN used: 0, mock; 1, 100 nM; 2, 200 nM; 4, 400 nM; 6, 600 nM; 8, 800 nM.
Figure 6.
Northern blot analysis of a concentration series of various ODNs on 15PC3 cells. The top panels show hybridizations with the 5′POL probe (the asterisk indicates the POLR2A mRNA, the arrow the cleaved 5′ product RNA); the bottom panels show the corresponding 28S RNA control hybridizations. The values on top of the lanes show the concentration of the indicated PS-ODN used: 0, mock; 1, 100 nM; 2, 200 nM; 4, 400 nM; 6, 600 nM; 8, 800 nM.
Table 1. IC50 (nM) of the ODN series shown in Figures 5 and 6.
| ODN couple | MiaPacaII (T/T) | 15PC3 (C/C) | |
|---|---|---|---|
| L6as17 | :C | >800 | >800 |
| :T | >800 | >800 | |
| L6as16 | :C | >800 | >800 |
| :T | >800 | >800 | |
| L5as17 | :C | >800 | 600 |
| :T | <200 | >800 | |
| L5as16 | :C | >800 | 300 |
| :T | 300 | >800 | |
| L4as17 | :C | 600 | 600 |
| :T | 400 | >800 |
To test whether the reduction of POLR2A mRNA levels would be cytotoxic, MiaPacaII cells and 15PC3 cells were treated with the most potent inhibitor L5as20 and the allele-specific ODN couple L5as16. Despite the gross reduction of the mRNA level observed 20 h post-transfection, no effect on cell growth was observed after prolonged incubation. We followed the mRNA reduction caused by L5Tas20 in time, following a single transfection (unpublished results). Twenty-four hours post-transfection the level was <10%, at 48 h it was ∼50% and at 72 h it was back to normal. Despite the fact that we could microscopically detect the ODN inside the cells 3 days post-transfection, the effect on the mRNA reduction is apparently only temporary. Maybe due to the dilution of the ODN over the cells upon cell division, as well as loss that will undoubtedly occur in time, the concentration in the cells needed to effectively lower the mRNA level over a longer period in order to affect the RNA polymerase II protein level sufficiently, is not reached in time. A second transfection at 48 h after the first transfection prolonged the antisense inhibitory effect on the POLR2A mRNA, but apparently not long enough to result in a protein reduction to a level that would allow cytostatic or cytotoxic effects.
DISCUSSION
We have analyzed a number of different antisense PS-ODNs targeted to a frequently occurring single nucleotide polymorphism in the mRNA of the large subunit of RNA polymerase II. The POLR2A mRNA levels can be efficiently reduced with some of the ODNs we have tested. The decrease in POLR2A mRNA is most likely due to RNase H-mediated cleavage of the mRNA, since additional RNA fragments (presumed cleaved RNA products) of the expected size, can be detected with probes localized 5′ or 3′ in the mRNA sequence with respect to the ODN target sequence position. The sequence specificity and the dose-dependent effect observed with several of the antisense ODNs support a true antisense ODN effect.
The different amounts of cleaved RNA product detected in the MiaPacaII cells and the 15PC3 cells is striking. The reduction of the mRNA level in 15PC3 cells is more pronounced than in MiaPacaII cells in several cases; however, the cleaved RNA products are present at a much higher level in MiaPacaII cells than in 15PC3 cells. In MiaPacaII cells, the concentration series show that the increase in the amount of cleaved product is related to the decrease observed in the mRNA level, suggesting stability of the cleaved product. The signal intensities of the mRNA and the cleaved product more or less add up to a constant value in the dose-escalation studies. In the 15PC3 cells, however, the mRNA level decreases with increased ODN concentration, with only marginal increase of the cleaved product RNA. This suggests that the stability of the cleavage products might be cell-type dependent. We are currently investigating this.
The large effect of shifting the ODN along the mRNA suggests that the sequence surrounding the polymorphic site forms a secondary or tertiary structure that restricts the binding possibilities of an antisense ODN. The most potent inhibitor (L5as20) reduces the mRNA level to <10%. This inhibitory activity is completely abolished when we shift the 20mer ODN 5 nucleotides along the mRNA sequence to the 3′ end (C/Tas20, containing the polymorphic base in the middle of the ODN). Shifting the position another 5 nucleotides to the 3′ end (R5as20) results in ∼50% inhibition of expression. This is not due to different ODN concentration or localization in the cell, since this effect was observed over a range of ODN concentrations (unpublished results), and since we could not detect any difference in the fluorescence staining pattern or intensity when we used the ODNs in an FITC-labeled form in transfections.
The 20mer ODN couple L5C/Tas20 does not discriminate the two alleles of the POLR2A polymorphism, but rather is an efficient inhibitor of both alleles. We therefore generated a series of shortened ODNs. Northern analyses of transfected cells with these PS-ODNs show that the specificity for either allele is increased by reducing the length of the ODNs. The 16mer couple L5as16 gives the highest allele specificity for a C in the target sequence, while the T-genotype is best targeted by L5as17. Our data suggest that this is not only due to the higher Tm of the C-versions ODN, but also that a T in this SNP target sequence provides a secondary structure to the mRNA and/or the ODN that is less advantageous for antisense ODN binding. The analysis of the complementary panel MiaPacaII/15PC3 shows that allele specificity of an ODN is really due to an antisense effect (Table 1).
Despite the potential for POLR2A as ASI target, in particular the requisite for the cell’s survival (as demonstrated with α-amanitin, a drug that acts directly at the protein level), the localization in a chromosomal region that is frequently affected by LOH (17p13.1) and the presence of highly frequent SNPs, it may be difficult to realize this potential with antisense PS-ODNs in vitro. The gross reduction of the POLR2A mRNA level that was observed with the successful ODNs did not result in inhibition of cell growth. Apparently the intracellular pool of RNA polymerase II protein is sufficient to overcome the temporarily reduced mRNA level. It may still be feasible to utilize POLR2A as ASI-target in vivo, however, since in an animal model the delivery of the ODN can be done in a continuous fashion using ODN infusion. Furthermore, advanced chemistry of antisense ODNs might allow a more precise targeting of the two alleles of the gene with a prolonged effect on the target mRNA level. In particular the use of 2′-methoxy-ethoxy residues flanking the PS-ODN SNP target sequence is expected to provide more stability, and thus more specificity, to the RNA–DNA hybrid (15,16). Too little is known about the exact requirements and mode of action of RNase H-mediated inhibition of mRNAs, in vitro as well as in vivo, to make general statements about the effect of antisense ODN on reducing mRNA levels in cultured cells in relation to inhibition of cell growth in vivo. Individual parameters playing a role in these processes have to be further analyzed in order to obtain predictive values.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our colleagues for support and Drs P. Borst, W. Scheper and L. Valentijn for critical reading of the manuscript. K.F. is funded by the Stichting Kindergeneeskundig Kankeronderzoek.
REFERENCES
- 1.Lengauer C., Kinzler,K.W. and Vogelstein,B. (1998) Nature, 396, 643–649. [DOI] [PubMed] [Google Scholar]
- 2.Basilion J.P., Schievella,A.R., Burns,E., Rioux,P., Olson,J.C., Monia,B.P., Lemonidis,K.M., Stanton,V.P.Jr and Housman,D.E. (1999) Mol. Pharmacol., 56, 359–369. [DOI] [PubMed] [Google Scholar]
- 3.Cho K.W.Y., Khalili,K., Zandomeni,R. and Weinmann,R. (1985) J. Biol. Chem., 260, 15204–15210. [PubMed] [Google Scholar]
- 4.Buratowski S. (1994) Cell, 77, 1–3. [DOI] [PubMed] [Google Scholar]
- 5.Cannizzaro L.A., Emanuel,B.S., Cho,K.W. and Weinmann,R. (1986) Am. J. Hum. Genet., 38, 812–818. [PMC free article] [PubMed] [Google Scholar]
- 6.Kirsch D.G. and Kastan,M.B. (1998) J. Clin. Oncol., 16, 3158–3168. [DOI] [PubMed] [Google Scholar]
- 7.Lindell T.J., Weinberg,F., Morris,P.W., Roeder,R.G. and Rutter,W.J. (1970) Science, 170, 447–449. [DOI] [PubMed] [Google Scholar]
- 8.Cochet-Meilhac M. and Cambon,P. (1974) Biochim. Biophys. Acta, 353, 160–184. [DOI] [PubMed] [Google Scholar]
- 9.Flanagan W.M. (1998) Cancer Metastasis Rev., 17, 169–176. [DOI] [PubMed] [Google Scholar]
- 10.Agrawal S. and Zhao,Q. (1998) Curr. Opin. Chem. Biol., 2, 519–528. [DOI] [PubMed] [Google Scholar]
- 11.Stein C.A., Subasinghe,C., Shinozuka,K. and Cohen,J.S. (1988) Nucleic Acids Res., 16, 3209–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma S. and Eckstein,F. (1998) Annu. Rev. Biochem., 67, 99–134. [DOI] [PubMed] [Google Scholar]
- 13.Stein C.A. (1999) Nature Biotech., 17, 209. [DOI] [PubMed] [Google Scholar]
- 14.Church G.M. and Gilbert,W. (1984) Proc. Natl Acad. Sci. USA, 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 16.McKay R.A., Miraglia,L.J., Cummins,L.L., Owens,S.R., Sasmor,H. and Dean,N.M. (1999) J. Biol. Chem., 274, 1715–1722. [DOI] [PubMed] [Google Scholar]