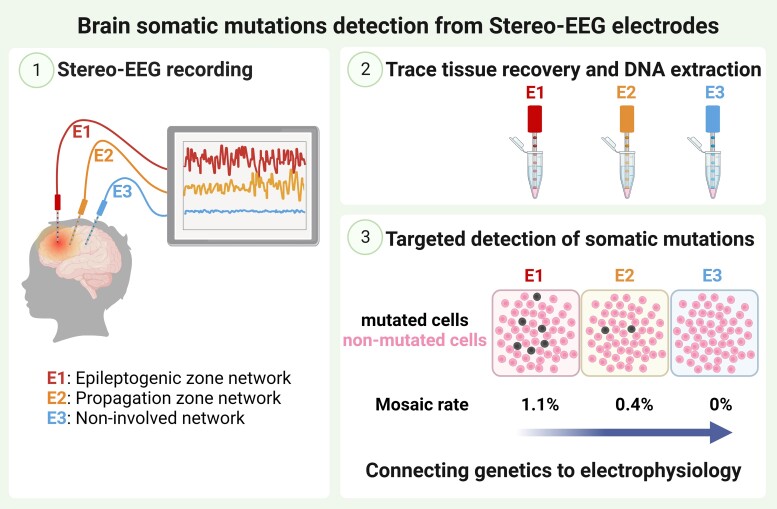
Abstract
Brain-restricted somatic variants in genes of the mechanistic target of rapamycin signalling pathway cause focal epilepsies associated with focal cortical dysplasia type II. We hypothesized that somatic variants could be identified from trace tissue adherent to explanted stereoelectroencephalography electrodes used in the presurgical epilepsy workup to localize the epileptogenic zone.
We investigated three paediatric patients with drug-resistant focal epilepsy subjected to neurosurgery. In the resected brain tissue, we identified low-level mosaic somatic mutations in AKT3 and DEPDC5 genes. We collected stereoelectroencephalography depth electrodes in the context of a second presurgical evaluation and identified 4/33 mutation-positive electrodes that were either located in the epileptogenic zone or at the border of the dysplasia.
We provide the proof-of-concept that somatic mutations with low levels of mosaicism can be detected from individual stereoelectroencephalography electrodes and support a link between the mutation load and the epileptic activity. Our findings emphasize future opportunities for integrating genetic testing from stereoelectroencephalography electrodes into the presurgical evaluation of refractory epilepsy patients with focal cortical dysplasia type II to improve the patients’ diagnostic journey and guide towards precision medicine.
Keywords: somatic mutations, focal cortical dysplasia, mTOR pathway, epilepsy surgery, stereo-EEG
Checri et al. report the detection of somatic mutations at low allele frequency from brain-derived trace-tissue DNA recovered from stereo-EEG electrodes used in the presurgical evaluation of patients with epileptogenic focal cortical dysplasia. This proof-of-concept study illustrates a minimally invasive approach for detecting mosaic variants in refractory epilepsies.
Graphical Abstract
Graphical Abstract.
Introduction
Focal cortical dysplasia type II (FCDII) is a type of cortical malformation manifesting with intractable epilepsy in childhood. Surgical resection of the epileptogenic zone (defined as the cortical area generating seizures) is often required to control FCDII-associated seizures.1
The International League Against Epilepsy (ILAE) has provided a classification system to define FCD lesions according to neuropathological findings in the resected brain specimens.2,3 FCDII are characterized by focal disruption of cortical cytoarchitecture and the presence of dysmorphic neurons (in FCDIIa and FCDIIb) and balloon cells (only in FCDIIb).
Postzygotic variants (or somatic variants) that arise during cortical development have emerged as important causes of FCDII.4 Brain somatic variants at variable levels of mosaicism have been identified in several genes (AKT3, DEPDC5, MTOR, PIK3CA, RHEB and TSC1/2) within the mechanistic target of rapamycin (mTOR) signalling pathway in individuals with FCDII.5–12 Yet, the genetic diagnosis depends on access to surgical brain tissue as mosaic mutations are present in only a small fraction of brain cells (mosaic level usually less than 5% in FCDII).13 Genetic investigation of surgical brain specimens has gained increasing interest in refractory focal epilepsy and has recently been integrated into the updated ILAE classification of FCD.3,4
Presurgical assessment of patients with drug-resistant focal epilepsy includes medical and neuropsychological evaluation, FDG-PET, high-resolution 3 T MRI and long-term scalp-video-EEG. In some cases, intracranial recording using stereo-EEG (SEEG) is required to accurately identify the epileptogenic zone. SEEG is an invasive technique based on the stereotactic implantation of multiple depth electrodes in the brain to record intracerebral EEG. SEEG allows anatomo-electrical correlations and tailored surgeries and may offer a therapeutic option by thermocoagulation.14,15 In the context of epilepsy surgery, the epileptogenic zone corresponds to the brain regions generating seizures on SEEG recordings. Recently, the regions involved in seizure production and propagation have been defined into different brain networks: epileptogenic zone network (EZN), propagation zone network (PZN) and non-involved network (NIN).14
Here we investigated three FCDIIa patients in which we identified somatic mutations in AKT3 and DEPDC5 in the DNA extracted from the resected tissue. Our findings indicate that it is possible to detect low-level somatic mutations by analysing brain-derived DNA extracted from residual tissue attached to SEEG electrodes for the presurgical evaluation of focal epilepsies associated with FCDII. We further report an association between the mutation load and the electrophysiological findings, with a higher level of mosaicism in the epileptogenic zone network.
Materials and methods
Patient cohort
We investigated three paediatric patients (SE1, SE2 and SE3) with refractory focal epilepsy subjected to neurosurgery at Rothschild Foundation Hospital (Paris, France), with a poor surgery outcome (Engel score Class II or III). The three patients had a neuropathological diagnosis of FCDIIa according to the ILAE classification for cortical malformations.2,3 A search for brain somatic variants was performed by deep-targeted panel sequencing of paired blood/brain resected tissue from the first surgery, as previously described.6 None of the patients were previously reported. The patients underwent SEEG in the context of presurgical evaluation for a complementary resection. An informed consent was obtained from all patients. The study protocol received approval from the ethical committee of CPP Ile de France II (N° ID-RCB/EUDRACT-2015-A00671-48).
Histological analyses
Standard hematoxylin–eosin staining was performed on 4 μm formalin-fixed paraffin-embedded (FFPE) sections and 20 μm cryosections from frozen postoperative tissues. Slides were scanned in brightfield with a NanoZoomer scanner (Hamamatsu) at a 40X objective, and images were captured with the NDP.view2 software. Fluorescent immunostaining was performed according to standard procedures on 20 μm cryosections with primary antibody against SMI311 (1:500, BioLegend #837801), followed by incubation with conjugated secondary antibody (anti-mouse Alexa-555, 1:1000) and DAPI counterstain. Slides were scanned with an Axioscan (Zeiss) with a 40X objective. Whole slide scans (CZI format) were loaded on QuPath (v.0.3.2),16 and a pixel-trained thresholding (resolution = 0.66 µm/px, classifier = artificial neural network) was applied to measure SMI311 pixel intensity across the entire tissue section (three sections per cortex block).
Stereoelectroencephalography sampling and analysis
Intracranial SEEG electrodes were implanted under general anaesthesia with the assistance of the robotic stereotactic assistance (ROSA) system. Intracerebral EEG was recorded using 11–14 depth electrodes (with 8–18 contacts for each electrode, 0.8 mm diameter), allowing simultaneous recordings from multiple sites. The EZN was defined by abnormal electrical discharges and dramatic changes in brain rhythms, most commonly low voltage fast discharge (LFD). The LFD may be preceded by EEG changes in the form of preictal epileptic spikes, train of spikes, or slow-wave complexes. The propagation zone network was defined by a sequential progression of abnormal neuronal activity, including the appearance of spikes or sharp waves in new brain regions, as well as changes in the frequency or amplitude of existing spikes or sharp waves. The NIN refers to the cortical network not implicated in seizures and without interictal abnormalities. SEEG-guided radiofrequency thermocoagulation (RFTC) of the epileptogenic zone network contacts was performed in patients SE1 and SE2.
Genomic DNA extraction
Genomic DNA from trace tissue adherent to SEEG electrodes was extracted as follows: each electrode was immediately immersed in 5 ml of ice-cold PBS for cell resuspension for 24 h at 4°C. The resuspended cells/trace tissue samples were centrifuged at 3000 rpm for 5 min at 4°C, and the supernatant was removed. The samples were incubated overnight at 56°C in lysis buffer (50 mM Tris pH 7.4, 0.5% Tween-20, 200 ng/µl proteinase K) and quantified using the Qubit dsDNA high sensitivity assay kit (Thermo Fisher Scientific). Bulk DNA from the resected brain tissue was extracted as previously reported.6
Droplet digital PCR
The commercial droplet digital PCR (ddPCR) mutation detection assay FAM + HEX was used to detect the AKT3 p.E17K variant in the DNA samples from patients SE1 and SE2 with the QX200 ddPCR system (Bio-Rad Laboratories). The reactions were prepared using the ddPCR supermix for probes with 5 U of HindIII according to the manufacturer’s protocol. Data were analysed with the QuantaSoft Analysis Pro software (version 1.0.596). Wells with less than 10 000 accepted droplets were discarded. For each sample, 5 ng of genomic DNA was tested and divided into two wells (i.e. 2.5 ng of DNA per reaction) that were subsequently merged for analysis using the ‘merge wells’ function of the QuantaSoft Analysis Pro software. All experiments were performed in duplicates for statistical analyses, and variant allele frequencies (VAFs) were calculated as a mean of the fractional abundance obtained in the two duplicates. The DNA samples extracted from the bulk brain resected tissues for both SE1 and SE2 were used as mutation-positive controls. We used 20 DNA samples extracted from the blood of healthy individuals as mutation-negative controls.
Deep-targeted amplicon sequencing
For the detection of the DEPDC5 p.R843* variant in patient SE3, we used a deep-targeted (4000 × mean coverage) amplicon sequencing approach since no commercial ddPCR probes were available. Genomic DNA extracted from the bulk brain resected tissues of SE3 was used as a mutation-positive control. To account for possible sequencing artifacts, we used 10 DNA samples extracted from the blood of healthy individuals as mutation-negative controls. We obtained PCR amplicons (243 bp) from 10 ng of genomic DNA from mutation-positive and mutation-negative controls and 6/11 electrodes, or between 1.2 and 6.7 ng of genomic DNA for the remaining 5/11 electrodes. Libraries were run on a MiSeq sequencer (2 × 250 bp) at the iGenSeq sequencing facility at ICM. Variant calling was performed by the Data Analysis Core facility at ICM following standard procedures: reads were aligned to the hg38 reference human genome, and single nucleotide variants (SNVs) were called using the GATK pipeline and SAMtools mpileup.
Statistical analysis
For the ddPCR sample analysis, we defined mutation-positive or mutation-negative samples based on an experimental limit of the blank (LOB) and limit of detection (LOD) values as previously described for ΛFP >0.05 [ΛFP corresponding to the average false-positive count, i.e. mutation-positive droplets (FAM+) in the mutation-negative control samples].17 Briefly, we first calculated the LOB as follows: . Then, the LOD was obtained from the LOB as follows: . The non-integers were rounded up to the nearest integer. The electrodes presenting a FAM + droplets count above the experimental LOD were considered mutation-positive. A two-tailed t-test (equal variance) was used to assess if the VAF was statistically different between the SEEG-derived DNAs and the mutation-negative controls in both ddPCR and targeted deep amplicon experiments.
Results
Three paediatric patients (SE1, SE2 and SE3) that underwent neurosurgery to treat drug-resistant focal seizures were included in this study. All three cases received a diagnosis of FCDIIa (with dysmorphic neurons) according to the ILAE criteria based on neuropathological findings.2,3 The first resection was considered incomplete based on the persistence of similar seizure types, abnormal postsurgical EEG and MRI. Because of persisting seizures, a new presurgical evaluation using depth SEEG recording was conducted. We hypothesized that traces of brain tissue adherent to SEEG electrodes could be used to detect somatic brain mutations.
Patient SE1
Patient SE1 is a 3-year-old girl with drug-resistant infantile spasms and focal seizures starting at 15 days of life (Table 1). Surgical resection of the epileptogenic zone network in the precentral left frontal lobe was performed at 9 months, but seizures relapsed immediately after surgery (Engel III surgical outcome: worthwhile improvement, but persistence of clusters of seizures). A new presurgical evaluation that included SEEG analysis was conducted at 2.6 years of age. Eight of the 13 SEEG electrodes implanted were thermocoagulated (Table 1). Because of the persistence of daily seizures (although at decreased frequency after thermocoagulation), the patient underwent a functional hemispherotomy (consisting of the disconnection of the two hemispheres, with left frontal lobe cortectomy). The patient is now Engel 1 (7 months follow-up).
Table 1.
Clinical, genetic, neuropathological and electrophysiological features
| Patient | SE1 | SE2 | SE3 |
|---|---|---|---|
| Neuropathology | FCDIIa | FCDIIa | FCDIIa |
| Genetic findings | AKT3 p.E17K somatic | AKT3 p.E17K somatic | DEPDC5 2-hit |
| Sex | Female | Male | Female |
| Age at seizure onset | 15d | 4y | 6m |
| Age at latest follow-up | 3y | 12y | 7y |
| Number of surgeries | 2 | 3 | 1 |
| Seizure types | Infantile spasms, focal seizures | Focal seizures, loss of consciousness | Focal seizures, auras |
| Seizure frequency | Daily | Daily | Daily |
| Antiseizure medications | TPM, VGB, OXC | CBZ, CLB, VPA | LTG, CLB, VPA, LEV |
| 3 T MRI | Diffuse frontal lobe FCD | Postcentral and superior parietal FCD | Superior frontal lobe and precentral gyrus blurring |
| Scalp EEG (ictal) | L frontal rapid discharge | R fronto-central rapid discharge | R fronto-central rapid discharge |
| Stereo-EEG (ictal) | Rhythmic activation/rapid discharge in supramarginal and postcentral gyri; rapid discharge in superior frontal gyrus and motor operculum | Rapid discharges in pre- and postcentral and premotor cortex | Discharges in precentral gyrus |
| EZN | Pre- and postcentral gyri, premotor frontal cortex, insula, and motor operculum | Mesial central-SMA and postcentral gyrus | Precentral gyrus |
| Electrodes in MRI lesion | AS, FA, FS, IA, MS, OM, PA, PI, PS | FA, FS, MS, PA | LP, FA, FS |
| Electrodes in EZN | FA, PI, PS, IA OM | FS, MS, PA | LP, FS |
| Electrodes in PZN | AS, FS, IA, OM, OP, MS, PA, PP, | PA, MS, FS, OM, PA | FA, OM, PS, LP, FS |
| Electrodes in NIN | TA, TS, TP | AS, BL, BM, CA, FI, LS, OM, OP, PI, PP, | BL, BM, FB, FI, FM |
| Thermocoagulated contacts | AS3-10, FA7-16, IA2-18, MS1-7, OM1-4, PA4-6, PI1-11, PS1-7 | FS1-5, MS1-5, PA1-5, PA15-18 | None |
| Follow-up post-RFTC | NA | Electroclinical improvement for 2m | NA |
| SEEG-guided surgery | Yes | No | No |
d, days; m, months, y, years; FU, follow-up; EZN, epileptogenic zone network; PZN, propagation zone network; NIN, non-involved network; RFTC, radiofrequency thermocoagulation; SMA, supplementary motor area; TPM, topiramate, VGB, vigabatrin, OXC, oxcarbazepine, CBZ, carbamazepine; VPA, sodium valproate; CLB, clobazam; LTG, lamotrigine; LEV, levetiracetam. Following a medication change to felbamate post-SEEG, seizure frequency was reduced in patient SE3 (1-year follow-up period).
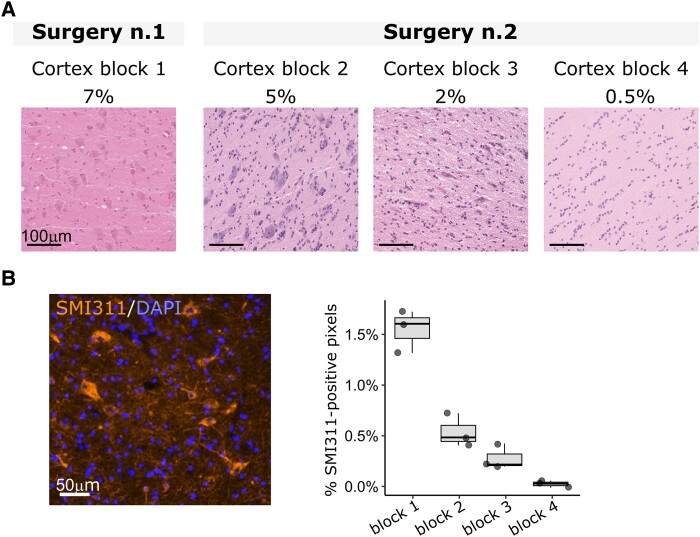
We performed targeted panel sequencing on matched blood/brain resected tissues from the first resection and identified a brain-restricted (absent from the blood-derived DNA) recurrent variant in AKT3 (NM_005465.7, p.E17K), with a variant allele frequency (VAF) of 7% in the fronto-lateral region (cortex block 1) (Fig. 1A). After the second surgery, we assessed the mutation distribution in three additional frozen tissue specimens by ddPCR, an ultra-sensitive and allele-specific approach for variant detection. We identified a mutation gradient from 5% in the premotor cortex (cortex block 2) to 2% in the periventricular region (cortex block 3) and 0.5% in the temporal cortex adjacent to the lesion (cortex block 4) (Fig. 1A). We performed SMI311 immunostaining, a canonical marker of dysmorphic neurons, and observed the highest signal intensity in the cortical area with the highest mosaic level (Fig. 1B).
Figure 1.
Neuropathology and genetic findings in patient SE1. (A) Hematoxylin–eosin staining on 20 µm frozen brain sections showing the variable density of dysmorphic neurons across the resected tissues with variable mutation mosaic rate from two surgeries in patient SE1. Cortex block 1: frontal-lateral cortex; Cortex block 2: premotor cortex; Cortex block 3: periventricular region; Cortex block 4: temporal cortex. (B) Left: representative image of SMI311-positive dysmorphic neurons (orange) in 20 µm frozen sections (DAPI-positive nuclei are indicated in blue); Right: boxplot indicating the percentage of SMI311-positive pixels in the sequenced tissue blocks (three whole sections were analysed per block). An increased percentage of SMI311-positive pixels characterized the cortex block 1 with the highest variant allele frequency.
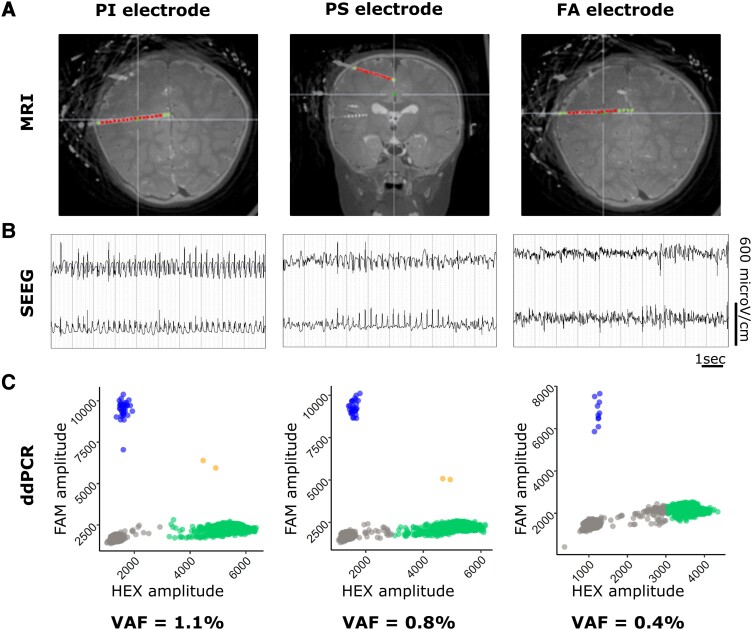
We extracted the DNA from each of the 13 SEEG explanted electrodes and obtained DNA concentrations ranging from 0.2 to 2.9 ng/µl and DNA quantities ranging from 40 to 500 ng (including the eight thermocoagulated samples), indicating that the thermocoagulation procedure does not affect DNA recovery. We assessed the distribution of the AKT3 p.E17K hotspot mutation in 9/13 electrodes by ddPCR [four electrodes were excluded due to low DNA amount (<20 ng)]. We defined an experimental LOD of n = 8 mutation-positive (FAM+) droplets based on mutation-negative controls (see methods). In mutation-negative controls, the average technical false-positive VAF was 0.02% (range 0–0.06%). We identified 3/9 mutation-positive electrodes (P < 1.12 × 10−16), with VAFs ranging from 0.4% to 1.1% (electrodes PI, PS and FA) (Table 2, Fig. 2). All three electrodes were in contact with the epileptogenic zone network within the premotor cortex, and located in the MRI-defined lesion. The mutation-negative electrodes were either implanted in the propagation zone network or the NIN.
Table 2.
Genetic findings from SEEG electrodes
| Electrode ID | Genomic DNA amount (ng) | Electrode location | Thermocoagulated contacts | VAF |
|---|---|---|---|---|
| SE1 (AKT3, p.E17K) | ||||
| PI | 66 | EZN, MRI lesion | Yes | 1.1% |
| PS | 208 | EZN, MRI lesion | Yes | 0.8% |
| FA | 91 | EZN, MRI lesion | Yes | 0.4% |
| AS | 188 | PZN, MRI lesion | Yes | ns |
| IA | 127 | PZN, MRI lesion | Yes | ns |
| OM | 501 | PZN, MRI lesion | Yes | ns |
| MS | 104 | PZN, MRI lesion | Yes | ns |
| PA | 85 | PZN, MRI lesion | Yes | ns |
| TS | 240 | NIN | No | ns |
| SE3 (DEPDC5, p.R843*) | ||||
| LP | 7.4 | EZN, MRI lesion | No | ns |
| FS | 113 | EZN, MRI lesion | No | ns |
| CA | 105 | PZN | No | 0.4% |
| OM | 87 | PZN | No | ns |
| FA | 245 | PZN, MRI lesion | No | ns |
| PS | 17 | PZN | No | ns |
| FI | 7.2 | NIN | No | ns |
| FB | 157 | NIN | No | ns |
| BL | 62 | NIN | No | ns |
| BM | 5.4 | NIN | No | ns |
| FM | 33 | NIN | No | ns |
Electrode CA from patient SE3 was placed at the border of the MRI lesion. VAF, variant allele frequency; ddPCR, droplet digital PCR; ns, not significant; EZN, epileptogenic zone network; PZN, propagation zone network; NIN, non-involved network.
Figure 2.
Identification of mutation-positive SEEG electrodes in patient SE1. (A) Location of mutation-positive electrodes on MRI images from patient SE1. Green plots are outside the MRI lesion; red plots are inside the lesion. (B) Representative SEEG traces of contacts within the epileptogenic zone network from mutation-positive electrodes. (C) Representative ddPCR 2D fluorescence amplitude plots showing mutant droplets of mutation-positive electrodes in patient SE1. Wild-type droplets (HEX) are indicated in green, mutant droplets (FAM) are indicated in blue, double-positive HEX/FAM droplets are indicated in orange and double-negative HEX/FAM droplets are indicated in grey. Plots were generated in R using the ggplot2 package. Fractional abundance of mutation-positive droplets (corresponding to the VAF) in mutation-negative control DNAs ranged from 0% to 0.06% (mean 0.02%, false-positive rate). FA, ascendant frontal; FAM, 6-carboxyfluorescein; HEX, 5’-hexachlorofluorescein; PI, inferior parietal; PS, superior parietal; SEEG, stereo-EEG; VAF, variant allele frequency.
Patient SE2
Patient SE2 is a 12-year-old boy with drug-resistant focal seizures since the age of 4 years. Surgical resection in the right parietal lobe was first performed at the age of 8 years, but seizures relapsed immediately after surgery (Engel III). A complementary resection was performed at 10 years without seizure control, and the patient underwent a third presurgical evaluation, including SEEG exploration. The patient had seizure improvement after thermocoagulation for 2 months (Engel 2) and is now a candidate for laser interstitial thermal therapy. We recovered brain DNA from the 14 explanted depth electrodes (among which three were thermocoagulated); we obtained DNA concentrations ranging from 0.2 to 1.2 ng/µl and DNA quantities from 1 to 44 ng.
By targeted gene panel sequencing, we identified the somatic AKT3 (p.E17K) hotspot mutation at an ultra-low VAF of 0.25% in a small tissue specimen with few hypertrophic neurons (from the second surgery). We then searched for the mutation in the DNA extracted from 12/14 SEEG electrodes (including the three thermocoagulated, two electrodes with low DNA concentrations were excluded) but failed to detect it probably because of the ultra-low level of mosaicism (below the ddPCR sensitivity limit).
Patient SE3
Patient SE3 is a 7-year-old girl with drug-resistant focal seizures from 6 months of age. Surgical resection in the right precentral region was performed at 6 years of age, with an Engel 2 surgical outcome (persistence of rare seizures immediately after surgery). The patient was therefore subjected to a second presurgical evaluation using SEEG recording. No thermocoagulation was performed, and following a medication change to felbamate, seizure frequency was reduced (1-year follow-up). We collected 11 individual SEEG electrodes for DNA extraction (none were thermocoagulated): we obtained between 3 and 228 ng of DNA, with a concentration ranging from 0.1 to 3.9 ng/µl per sample.
By targeted gene panel sequencing on bulk DNA from the resected brain tissue, we identified two-hit loss-of-function mutations in DEPDC5 (NM_001242896.3): a germline heterozygous start-loss mutation c.2_6delAGATG (p.? ) and a somatic loss-of-function p.R843* variant. The germline variant was classified as ‘likely pathogenic’ based on the American College of Medical Genetics (ACMG) guidelines,18 and the somatic variant was classified as ‘pathogenic’ based on the recently published guidelines for pathogenicity classification of somatic alterations in tumors.19 The VAF of the somatic p.R843* ranged from 7% in the precentral cortex to 5% in the cingulate gyrus.
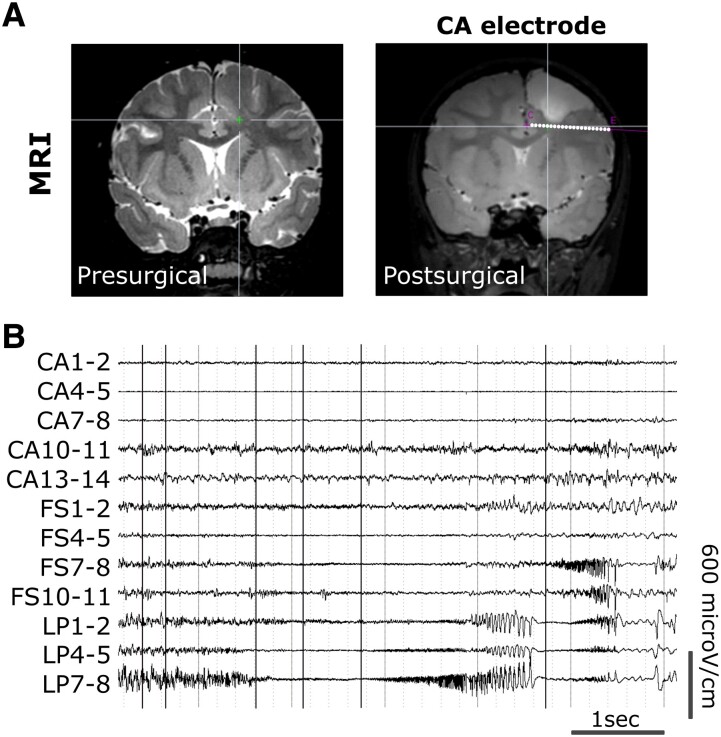
We subsequently assessed the presence of the somatic DEPDC5 p.R843* variant in each electrode by targeted amplicon sequencing (without genomic pre-amplification), and tested mutation-negative controls to account for possible sequencing artifacts. In mutation-negative controls, we calculated a technical false-positive VAF of 0.19% (range 0.11–0.28%). Only one electrode (CA) was considered mutation-positive, with a VAF of 0.39% (significantly above the false-positive rate, P = 0.002). The electrode CA was in a cortical area involved in the propagation zone network and adjacent to the border of the previously resected dysplasia (Table 2, Fig. 3). However, we did not confidently identify the DEPDC5 somatic mutation on the two electrodes that were in the epileptogenic zone network (the VAF was below the confidence threshold).
Figure 3.
Mutation-positive electrode (CA) in patient SE3. (A) Pre- and postsurgical 3T MRI of patient SE3, showing the trajectory of the mutation-positive electrode (CA) at the bottom of the dysplastic region not resected during the first surgery (white dots). (B) Representative SEEG traces of the contacts within the epileptogenic zone network from LP and FS mutation-negative electrodes and the CA mutation-positive electrode in the propagation zone network. CA, anterior cingular; FS, superior frontal; LP, posterior limit.
Discussion
Brain mosaicism is increasingly being recognized as a significant cause of drug-resistant focal epilepsy associated with FCDII.4 Because genetic diagnosis is hindered by direct access to brain tissue (either from surgical resection or autopsy), novel strategies have emerged to identify brain mosaic variants in biofluids. We and others provided evidence that brain variants can be detected from the circulating cell-free DNA (cfDNA) from the cerebrospinal fluid (CSF) of patients with refractory focal epilepsy.20,21 However, this cutting-edge approach is technically challenging due to the minimal amount of cfDNA present in the CSF. In 2019, Montier et al. conducted a pioneering study that identified a somatic mutation with a high VAF (16.7%) in the MEN1 gene from the DNA extracted from trace tissues adhering to SEEG electrodes in a patient with focal epilepsy and bilateral periventricular nodular heterotopia.22 A second case report study identified a KCNT1 mutation at a mosaic level of ∼25% from one patient with non-lesional focal epilepsy.23 Both studies were conducted on whole genome-amplified DNA and pooled SEEG electrodes.
In this study, we aimed to detect mTOR pathway somatic mutations in the context of the presurgical workup for FCDII and assess the level of mosaicism on each individual SEEG electrode. To validate this approach, we focused on three FCDIIa patients for which somatic mutations in AKT3 and DEPDC5 had been previously identified from the surgically resected tissues. Using targeted sequencing approaches, we detected low-level mosaic variants in brain DNA isolated from trace tissue adherent to individual SEEG electrodes from two FCDIIa patients. All mutation-positive electrodes in patient SE1 were in contact with the epileptogenic zone network and within the MRI-defined lesion, suggesting a correlation between genetics, electrophysiology, and brain pathology. This assumption is also corroborated by previous studies reporting the existence of a mutation gradient in the resected tissues, with higher mosaicism levels in the most epileptogenic area,24–26 and that dysmorphic neurons in FCDII tissues carry the disease-causing somatic variants.6,26,27 Moreover, Rampp et al. recently provided evidence for the contribution of dysmorphic neurons to interictal spikes, fast gamma activity and ripples, thus linking neuropathological and electrophysiological abnormalities in FCDII.28
This study has a limitation in its interpretation because the multiple SEEG contacts of a single electrode pass through several brain zones not always involved in the epileptogenic network. Therefore, the DNA extracted from an individual electrode could represent a combination of these different regions. As a result, the sensitivity and specificity of the study may be reduced, which could lead to false-negative results (e.g. in patient SE3 with two mutation-negative electrodes in contact with the epileptogenic zone network). Despite this ‘dilution effect’, we could detect the pathogenic mutation from the DNA extracted from the electrodes with contacts in the epileptogenic zone network.
FCD is among the most frequent malformations encountered in the paediatric epilepsy surgery population.1 While we here applied a targeted genetic approach to detect known variants, our work shows that genetic testing from SEEG electrode-recovered tissue can be applied to small brain lesions such as FCDII, which are caused by somatic mutations at low mosaic levels (<5%). Moreover, our findings suggest that such genetic studies could be integrated as part of the presurgical workup in FCDII to guide the localization of the resection area by targeted panel sequencing, as recently described.9 Since >50% of FCDII are caused by variants in mutational hotspots in MTOR, PIK3CA and AKT3 genes, implementing presurgical genetic testing using multiplex ddPCR could also be a valuable approach, as previously showed in the resected tissues.29,30 On average, we extracted 60 ng of DNA from the electrodes of the epileptogenic zone network, which would be theoretically sufficient to perform capture gene panel sequencing as performed in routine settings.6
Currently, the resection of the epileptogenic zone is the unique therapeutic option for patients with FCD and drug-resistant epilepsy. The opportunity for a genetic diagnosis in cases that are not eligible for surgery should allow better clinical management with the use of precision-medicine therapeutic approaches, for example, targeting the mTOR signalling pathway.31 Integration of genetic, neuroimaging, electrophysiological or neuropathological data from patients with FCDII may also provide insights for the selection of surgical candidates and the prognostic value of the postsurgical outcome.32–34
This study paves the way towards integrating genetic diagnosis into the multidisciplinary epilepsy presurgical assessment by detecting mosaic mutations. Moreover, an SEEG-based approach has the potential to be applied to other conditions (with confirmed or suspected mosaic mutations) that are treated using deep brain stimulation through intracranial recording electrodes, most notably psychiatric (treatment-resistant depression and obsessive-compulsive disorder, Tourette’s syndrome) or neurologic diseases (Parkinson’s disease, genetic dystonia and tremor).35
Acknowledgements
We thank the patients and their families for their participation, the neuropaediatricians who referred the patients, the ERN EpiCARE consortium, the ICM core facilities (iGenSeq, Data Analysis Core and the DNA and cell bank), Eric Noé for the technical assistance, Valerie Taly for the advice on ddPCR experiments, Angel Escudero for contributing to the preparation of the graphical abstract (created with BioRender.com) and Fabrice Bartolomei for the helpful comments.
Abbreviations
- ACMG =
American College of Medical Genetics
- ASM =
antiseizure medication
- 3T =
3 Tesla
- CBZ =
carbamazepine
- cfDNA =
cell-free DNA
- CLB =
clobazam
- CZI =
Carl Zeiss image
- ddPCR =
droplet digital PCR
- EZN =
epileptogenic zone network
- FCDII =
focal cortical dysplasia type II
- FDG-PET =
fluorodeoxyglucose-positron emission tomography
- FFPE =
formalin-fixed paraffin-embedded
- FU =
follow-up
- ILAE =
International League Against Epilepsy
- LEV =
levetiracetam
- LFD =
low voltage fast discharge
- LOB =
limit of the blank
- LOD =
limit of detection
- LTG =
lamotrigine
- mTOR =
mechanistic target of rapamycin
- NIN =
non-involved network
- OXC =
oxcarbazepine
- PZN =
propagation zone network
- ROSA =
robotic stereotactic assistance
- RFTC =
radiofrequency thermocoagulation
- SEEG =
stereo-EEG
- SMA =
supplementary motor area
- SNV =
single nucleotide variant
- TPM =
topiramate
- VAF =
variant allele frequency
- VGB =
vigabatrin
- VPA =
sodium valproate.
Contributor Information
Rayann Checri, Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
Mathilde Chipaux, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France.
Sarah Ferrand-Sorbets, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France.
Emmanuel Raffo, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France; Unité de recherche 3450 DevAH, Développement, Adaptation et Handicap, Campus Brabois-Santé, Université de Lorraine, 54505, Vandoeuvre-lès-Nancy, France.
Christine Bulteau, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France; Université de Paris Cité, MC2Lab, Institut de Psychologie, F-92100 Boulogne-Billancourt, France.
Sarah Dominique Rosenberg, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France.
Marion Doladilhe, Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
Georg Dorfmüller, Department of Pediatric Neurosurgery, Rothschild Foundation Hospital EpiCARE, 75019, Paris, France.
Homa Adle-Biassette, Université de Paris Cité, service d’Anatomie Pathologique, APHP, Hôpital Lariboisière, DMU DREAM, UMR 1141, INSERM, 75010, Paris, France.
Sara Baldassari, Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
Stéphanie Baulac, Sorbonne Université, Institut du Cerveau—Paris Brain Institute—ICM, Inserm, CNRS, Hôpital de la Pitié Salpêtrière, 75013, Paris, France.
Funding
This work was funded by the Agence Nationale de la Recherche—Programme d'Investissements d'Avenir (ANR-18-RHUS-0005 to SBau), the Association Robert Debré pour la Recherche Médicale (to Sbau) and Biobank BB-0033-00064 (to H.A-.B.).
Competing interests
The authors report no competing interests.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Blumcke I, Spreafico R, Haaker G, et al. . Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med. 2017;377(17):1648–1656. [DOI] [PubMed] [Google Scholar]
- 2. Blumcke I, Thom M, Aronica E, et al. . The clinico-pathological spectrum of focal cortical dysplasias: A consensus classification proposed by an ad hoc task force of the ILAE diagnostic methods commission. Epilepsia. 2011;52(1):158–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Najm I, Lal D, Alonso Vanegas M, et al. . The ILAE consensus classification of focal cortical dysplasia: An update proposed by an ad hoc task force of the ILAE diagnostic methods commission. Epilepsia. 2022;63(8):1899–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blumcke I, Budday S, Poduri A, Lal D, Kobow K, Baulac S. Neocortical development and epilepsy: Insights from focal cortical dysplasia and brain tumours. Lancet Neurol. 2021;20(11):943–955. [DOI] [PubMed] [Google Scholar]
- 5. D’Gama AM, Woodworth MB, Hossain AA, et al. . Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21(13):3754–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baldassari S, Ribierre T, Marsan E, et al. . Dissecting the genetic basis of focal cortical dysplasia: A large cohort study. Acta Neuropathol. 2019;138(6):885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sim NS, Ko A, Kim WK, et al. . Precise detection of low-level somatic mutation in resected epilepsy brain tissue. Acta Neuropathol. 2019;138(6):901–912. [DOI] [PubMed] [Google Scholar]
- 8. Lai D, Gade M, Yang E, et al. . Somatic variants in diverse genes leads to a spectrum of focal cortical malformations. Brain. 2022;145(8):2704–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pirozzi F, Berkseth M, Shear R, et al. . Profiling PI3K-AKT-MTOR variants in focal brain malformations reveals new insights for diagnostic care. Brain. 2022;145(3):925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. López-Rivera JA, Leu C, Macnee M, et al. . The genomic landscape across 474 surgically accessible epileptogenic human brain lesions. Brain. 2023;146(4):1342–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chung C, Yang X, Bae T, et al. . Comprehensive multi-omic profiling of somatic mutations in malformations of cortical development. Nat Genet. 2023;55(2):209–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujita A, Kato M, Sugano H, et al. . An integrated genetic analysis of epileptogenic brain malformed lesions. Acta Neuropathol Commun. 2023;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baldassari S, Baulac S. Genetics of FCD: An emerging scenario. In: Chassoux F, Palmini A, eds. Focal cortical dysplasias: New advances for curing epilepsy. France and England: John Libbey Eurotext Ltd-Arnette-Doin-Pradel; 2022:41-49. [Google Scholar]
- 14. Bartolomei F, Lagarde S, Wendling F, et al. . Defining epileptogenic networks: Contribution of SEEG and signal analysis. Epilepsia. 2017;58(7):1131–1147. [DOI] [PubMed] [Google Scholar]
- 15. Isnard J, Taussig D, Bartolomei F, et al. . French guidelines on stereoelectroencephalography (SEEG). Neurophysiol Clin. 2018;48(1):5–13. [DOI] [PubMed] [Google Scholar]
- 16. Bankhead P, Loughrey MB, Fernández JA, et al. . Qupath: Open source software for digital pathology image analysis. Sci Rep. 2017;7(1):16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Milbury CA, Zhong Q, Lin J, et al. . Determining lower limits of detection of digital PCR assays for cancer-related gene mutations. Biomol Detect Quantif. 2014;1(1):8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richards S, Aziz N, Bale S, et al. . Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koeppel F, Muller E, Harlé A, et al. . Standardisation of pathogenicity classification for somatic alterations in solid tumours and haematologic malignancies. Eur J Cancer. 2021;159:1–15. [DOI] [PubMed] [Google Scholar]
- 20. Kim S, Baldassari S, Sim NS, et al. . Detection of brain somatic mutations in cerebrospinal fluid from refractory epilepsy patients. Ann Neurol. 2021;89(6):1248–1252. [DOI] [PubMed] [Google Scholar]
- 21. Ye Z, Chatterton Z, Pflueger J, et al. . Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain. Brain Commun. 2021;3(1):fcaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Montier L, Haneef Z, Gavvala J, et al. . A somatic mutation in MEN1 gene detected in periventricular nodular heterotopia tissue obtained from depth electrodes. Epilepsia. 2019;60(10):e104–e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye Z, Bennett MF, Neal A, et al. . Somatic mosaic pathogenic variant gradient detected in trace brain tissue from stereo-EEG depth electrodes. Neurology. 2022;99(23):1036–1041. [DOI] [PubMed] [Google Scholar]
- 24. Mirzaa GM, Campbell CD, Solovieff N, et al. . Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol. 2016;73(7):836–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ribierre T, Deleuze C, Bacq A, et al. . Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128(6):2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee WS, Stephenson SEM, Howell KB, et al. . Second-hit DEPDC5 mutation is limited to dysmorphic neurons in cortical dysplasia type IIA. Ann Clin Transl Neurol. 2019;6(7):1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee WS, Baldassari S, Chipaux M, et al. . Gradient of brain mosaic RHEB variants causes a continuum of cortical dysplasia. Ann Clin Transl Neurol. 2021;8(2):485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rampp S, Rössler K, Hamer H, et al. . Dysmorphic neurons as cellular source for phase-amplitude coupling in focal cortical dysplasia type II. Clin Neurophysiol. 2021;132(3):782–792. [DOI] [PubMed] [Google Scholar]
- 29. Chen WL, Pao E, Owens J, et al. . The utility of cerebrospinal fluid-derived cell-free DNA in molecular diagnostics for the PIK3CA-related megalencephaly-capillary malformation (MCAP) syndrome: a case report. Cold Spring Harb Mol Case Stud. 2022;8(3):a006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee WS, Leventer RJ, Lockhart PJ. Droplet digital PCR as a first-tier molecular diagnostic tool for focal cortical dysplasia type II. Brain. 2022;145(12):e119–e121. [DOI] [PubMed] [Google Scholar]
- 31. D’Gama AM, Poduri A. Precision therapy for epilepsy related to brain malformations. Neurotherapeutics. 2021;18(3):1548–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Benova B, Sanders MWCB, Uhrova-Meszarosova A, et al. . GATOR1-related focal cortical dysplasia in epilepsy surgery patients and their families: A possible gradient in severity? Eur J Paediatr Neurol. 2021;30:88–96. [DOI] [PubMed] [Google Scholar]
- 33. Specchio N, Pepi C, De Palma L, Trivisano M, Vigevano F, Curatolo P. Neuroimaging and genetic characteristics of malformation of cortical development due to mTOR pathway dysregulation: Clues for the epileptogenic lesions and indications for epilepsy surgery. Expert Rev Neurother. 2021;21(11):1333–1345. [DOI] [PubMed] [Google Scholar]
- 34. Jehi L, Braun K. Does etiology really matter for epilepsy surgery outcome? Brain Pathol. 2021;31(4):e12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kangas SM, Teppo J, Lahtinen MJ, et al. . Analysis of human brain tissue derived from DBS surgery. Transl Neurodegener. 2022;11(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.