Abstract
The use of doubled haploids is one of the most efficient breeding methods in modern agriculture. Irradiation of pollen grains has been shown to induce haploids in cucurbit crops, possibly because it causes preferential fertilization of the central cell over the egg cell. Disruption of the DMP gene is known to induce single fertilization of the central cell, which can lead to the formation of haploids. In the present study, a detailed method of creating a watermelon haploid inducer line via ClDMP3 mutation is described. The cldmp3 mutant induced haploids in multiple watermelon genotypes at rates of up to 1.12%. These haploids were confirmed via fluorescent markers, flow cytometry, molecular markers, and immuno-staining. The haploid inducer created by this method has the potential to greatly advance watermelon breeding in the future.
Introduction
Generating haploid plants to obtain pure doubled haploid (DH) lines is one of the most efficient breeding strategies in modern agriculture. The simultaneous genetic fixation of every locus within a single generation enables breeders to avoid the time-consuming conventional requirement for extensive selfing or backcrossing before homozygous breeding-ready lines can be obtained. Currently, haploids can be obtained through either in vitro or in vivo methods. In vitro methods are mainly based on culturing haploid gametophytic tissues or organs, including anthers, microspores or ovaries, to obtain haploid plants. However, in vitro haploid methods are often affected by genotype, developmental stage, culture condition, physiological status and stress pretreatment of the plant material. In vivo induction generates haploids through interspecific or intraspecific crosses but results in a low frequency of haploid induction. Parthenogenesis, on the other hand, has naturally occurred in over 400 plant species and is characterized by the development of egg cells into haploid embryos without fertilization [1]. Additionally, haploids can sometimes be induced after pollination with pollen treated with radiation, chemicals or high temperatures [2].
Haploid inducer lines are increasingly used to simplify haploid induction process and new techniques are constantly being developed to generate such lines as the understanding of reproductive biology increases. Generally, haploid inducer lines induce parental genome elimination during the first division of zygotic embryos or parthenogenesis of egg cells during fertilization [2]. For example, the maize inbred line STOCK6 has been reported to induce haploids at rates of 2.3–3.2% [3]. Map-based cloning revealed that a 4-bp insertion in a phospholipase gene, MATRILINEAL/PHOSPHOLIPASE A1/NOT LIKE DAD (MTL/PLA1/NLD), was responsible for the haploid induction ability of STOCK6 [4–6]. MTL protein was shown to localize to the sperm-cell membrane, indicating that it is likely involved in the interaction of sperm cell and egg cell during fertilization. Mutation of Zea mays PHOSPHOLIPASE D3 (ZmPLD3), which encodes a member of the phospholipase D subfamily, was also shown to trigger maternal haploids during fertilization in maize. Furthermore, the zmpld3 mutant was found to synergistically increase the efficiency of haploid induction mediated by the zmpla1 mutant [7]. Additionally, manipulation of the centromeric histone protein CenH3 can cause haploid induction through parental genome elimination, possibly because modified CenH3 protein results in slower chromosome movement during the first division of zygotic embryos, leading to the elimination of one parental genome [8]. Point mutations in the histone folding domain or centromere-targeting domain of CenH3 protein were also shown to induce haploids in Arabidopsis [9–11]. The CenH3-mediated haploid induction system has been successfully applied to many different monocot crops, including maize, wheat and others [12–14]. BABY BOOM (BBM), an AP2 transcription factor family protein, can initiate maternal parthenogenic embryogenesis without fertilization and leads to haploid formation in rice, maize, millet and Arabidopsis [15–18]. By combining a Meiosis instead of Mitosis (MiMe) system with BBM, researchers have shown that hybrid vigor can be efficiently fixed across generations through synthetic apomixis in rice [19, 20]. However, ectopic expression of the BBM gene in egg cells is a prerequisite, so BBM-mediated parthenogenic haploids are transgenic and have limited application in plant breeding.
Watermelon (Citrullus lanatus) is an important fruit crop worldwide that is consumed for its nutritional value and flavor [21]. Long-term artificial selection of watermelon had led to genetic narrowing, and a haploid induction system is urgently needed to advance traditional breeding by enabling the creation of valuable pure DH lines [22, 23]. Irradiated pollen grains have been demonstrated to induce haploid plants in cucurbit crops with relatively low efficiencies [24–26]. This may be because irradiation causes sperm cells to lose the ability to fertilize the egg cell, while maintaining the ability to fuse with the central cell to give rise to the endosperm. In rare cases, unfertilized egg cells could then further develop into parthenogenic haploid embryos. Therefore, we hypothesized that the preferential fertilization of the central cell over the egg cell could possibly induce haploid formation in watermelon.
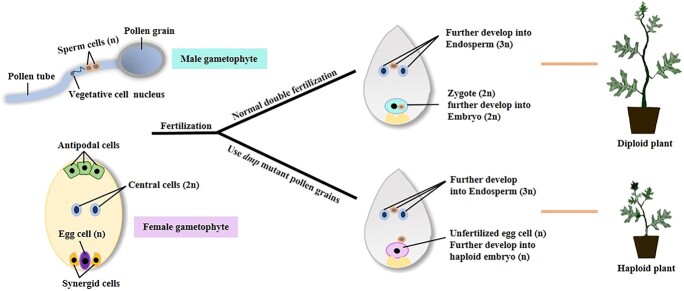
To date, several genes have been reported to be involved in sperm-egg fusion, and their mutations can result in single fertilization of the central cell. The LlDMP9 gene, which encodes a DOMAIN OF UNKNOWN FUNCTION 679 (DUF679) protein localized at the sperm membrane in lily (Lilium longiflorum), is involved in the fusion of gametes. Earlier work found that knocking down the Arabidopsis DMP9 gene resulted in 17.2 ± 1.8% single central cell fertilization events [27]. DMP8 is another DUF679 gene that is specifically expressed in pollen, and the Arabidopsis dmp8/9 double mutant led to single fertilization of the central cell at a rate of 19.04 ± 5.68% [28, 29]. Consistent with our hypothesis that single fertilization of the central cell induces haploids in plant species, the dmp8/9 double mutant was found to induce haploids in Arabidopsis [30]. The maize zmdmp mutant was initially identified as an enhancer of the haploid inducer zmpla1, which typically induces haploids at low efficiency [31]. The DMP-based haploid induction system has been successfully applied to several plant species, including tomato, tobacco, potato, Brassica napus, and Medicago truncatula, which prompted us to explore the possibility of inducing haploids in watermelon with this same approach [32–36] (Fig. 1).
Figure 1.
Schematic illustration of the hypothesis of DMP mediated haploid induction
In the present study, we describe a method for creating a haploid inducer line in watermelon by employing mutations in the ClDMP3 gene. Mutation of ClDMP3 caused single fertilization of the central cell over the egg cell, and the cldmp3 mutant could induce haploids in several different watermelon genotypes, with rates ranging from 0.51% to 1.12%. The generated haploids were confirmed via fluorescent markers, flow cytometry, molecular markers and immuno-staining. These findings have the potential to significantly accelerate future watermelon breeding efforts.
Results
Identification of the ClDMP3 gene in watermelon
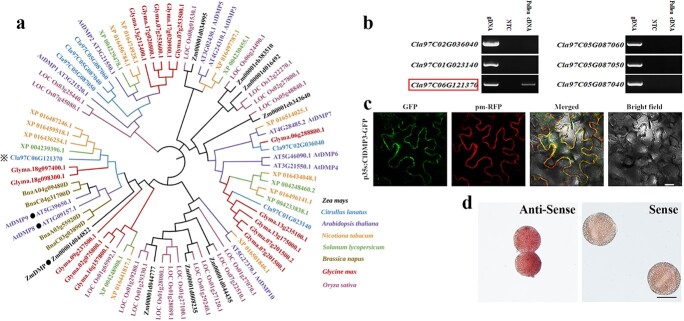
Six putative DUF679 proteins were identified in the watermelon genome, among which the protein encoded by Cla97C06G121370 exhibited the highest homology to Arabidopsis AtDMP8/9 and maize ZmDMP proteins (Fig. 2a, Table S2). Subsequent expression analysis revealed that ClDMP3 was the only DUF679 gene that was specifically expressed in pollen (Fig. 2b, Fig. S1), which was consistent with our pollen RNA-seq data (unpublished data). Subcellular localization analysis indicated that ClDMP3 was localized to the cell membrane (Fig. 2c), which is consistent with a possible role in sperm-egg cell membrane fusion. A whole-mount RNA in situ experiment further confirmed the presence of ClDMP3 transcripts in mature pollen (Fig. 2d). Therefore, ClDMP3 was chosen for mutagenesis mediated by CRISPR/Cas9 gene editing technology.
Figure 2.
Identification of the ClDMP3 gene in watermelon. a. Phylogenetic analysis of DMP homologs in eight green plant species. ClDMP3 proteins are highlighted with asterisks, and the DMPs in Arabidopsis and Zea mays are noted with dots. b. Expression of watermelon DUF679 in mature pollen. Genomic DNA (gDNA) and non-template control (NTC) were used as controls for RT-PCR. The ClDMP3 gene is noted in the red box. c. Subcellular localization of ClDMP3-GFP proteins in tobacco leaf. pm-RFP is a plasma membrane marker. d. Whole-mount RNA in situ hybridization analysis of ClDMP3 in mature watermelon pollen. Scale bars: 20 μm (c), 50 μm (d).
Generation of cldmp3 mutants
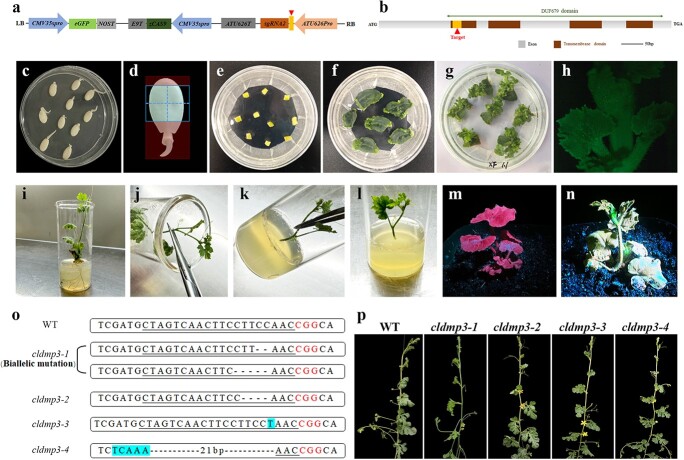
A single target was designed against the ClDMP coding region and cloned into the CRISPR/Cas9 gene editing vector pBSE402, which contained a 35S-driven Cas9, an AtU6–26 driven SgRNA cassette and a GFP fluorescent marker driven by the 35S promoter (Fig. 3a and b). Stable watermelon transformation was carried out using Agrobacterium tumefaciens strain EHA105, and watermelon cotyledon sections were utilized as explants. Additionally, 6-benzyladenine (6-BA) and Timentin were supplied in the medium during the shoot elongation process, and Timentin alone was added to the medium to inhibit the growth of bacteria during the rooting process. Healthy transgenic plants were transferred to soil for further analysis (Fig. 3c-n).
Figure 3.
Generation of the cldmp3 mutants through CRISPR/Cas9 gene editing technology. a. The CRISPR/Cas9 editing vector for ClDMP3 with a single guide RNA. The red triangle indicates the ClDMP3 target. b. Schematic diagram of the ClDMP3 gene. c-n. Watermelon transformation protocol: (c) Germination of watermelon seeds; (d) Schematic diagram of the cotyledon explant cutting method. The red area represents the part not suitable for severing as explants, which should be removed and discarded. The blue area represents the cutting area; (e) Explants cultivated on the recovery medium; (f) Explants cultivated on the shoot generation medium; (g) New shoots formed after one month of cultivation on the shoot generation medium; (h) Positive transgenic shoots with GFP marker expression; (i) Positive transgenic seedling on the rooting medium; (j-l) Asexual reproduction of positive shoots; (m) Control watermelon seedling without GFP fluorescence; (n) Transgenic positive seedling with green fluorescence. o. Target sequence and the actual editing results in different cldmp mutant plants. The PAM sequence is shown in red font and the 20 bp target sequence is underlined. Insertions are highlighted in blue. Deletions are shown with black dashed lines. p. Comparison of WT and cldmp3 mutant plants.
After obtaining the potential transgenic plants, primers were designed to amplify the region spanning the target site (Table S3), and Sanger sequencing was used to further confirm the editing events. Subsequently, Hi-TOM deep sequencing was utilized to reveal the editing types and ratios [37]. Out of 11 positive transgenic plants, 4 plants were identified with valid edits that were either biallelic or homozygous mutations. cldmp3–1 contained −2/−5 biallelic mutations in the target, while cldmp3–2 was homozygous for a 4-bp deletion, and cldmp3–3 was homozygous for a 1-bp insertion at the target site. Additionally, cldmp3–4 contained a homozygous 21-bp deletion and a 5-bp insertion inside the target region (Fig. 3o). All mutations caused pre-termination of the ClDMP3 protein. Phenotypic analysis did not reveal any obvious abnormalities in the mutant plants during vegetative growth (Fig. 3p).
Phenotypic analysis of the cldmp3 mutants
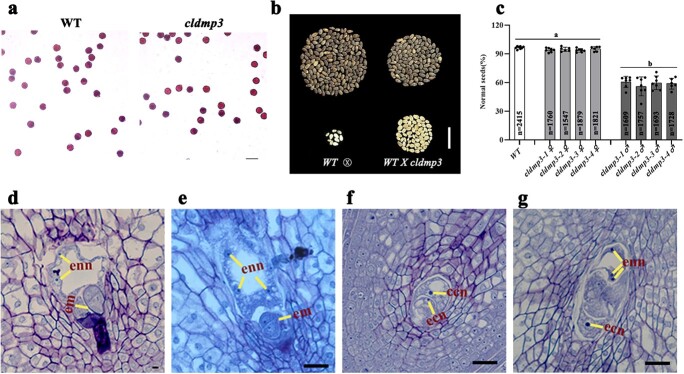
cldmp3 pollen grains were fully viable when stained by Alexander solution, which ruled out the possibility of defective male gametophyte development (Fig. 4a). To further identify potential defects in male and female gametes of the mutants, reciprocal crosses were carried out between cldmp3 and wild type (WT) plants. When the cldmp3 mutants were used as male parents, there was a significant reduction in the number of normal seeds produced compared to selfed WT plants (Fig. 4b and c). However, the seed number was not affected when the mutants were pollinated with WT pollen (Fig. 4c), indicating that the mutation in ClDMP gene caused partial male sterility. The above results implied that the reduced seed-set while crossing to wild type could be caused by defective double fertilization. Since all mutants showed similar defects, the homozygous cldmp3–2 mutant was chosen for further analysis.
Figure 4.
Phenotypic analysis of the cldmp3 mutants. a. Pollen viability comparison of wild-type and cldmp3 mutant stained by Alexander staining. b. Watermelon seeds from selfing WT and a WT x cldmp3 cross. Undeveloped seeds (white) are shown at the bottom of the image. c. Percentage of normal seeds when cldmp3 mutants were used as female (♀) and male (♂) parents, respectively. Data are the means ± SD. P < 0.0001 (two-sided Student’s t-test). n represents the total number of seeds. d-g. Semi-thin paraffin sections of WT (d) and the cross progeny (WT x cldmp3–2) ovules (e-g) three days after pollination. Ecn, egg cell nuclei; ccn, central cell nuclei; em, embryo; enn, endosperm nuclei. Scale bars: 100 μm (a), 3 cm (b), 20 μm (d-g).
To better understand the developmental process of double fertilization occurred in the mutants, semi-thin paraffin sectioning was carried out on WT ovules pollinated by the cldmp3–2 mutant pollen grains. Three days after pollination (DAP), some ovules developed normal embryos and endosperm, which were indistinguishable from the wild type (Fig. 4d and e). However, some embryo sacs contained undeveloped egg cells and central cells, representing a complete failure of fertilization of both gametes (Fig. 4f). Notably, some embryo sacs contained only endosperm without detectable developing embryos at 3DAP (Fig. 4g), indicating single fertilization of the central cell by the sperm cell. Consistent with previous studies, our results showed that absence of the ClDMP3 protein resulted in single fertilization events of central cells in watermelon. It is therefore possible that a small portion of unfertilized egg cells could sense the abnormal sperm-egg cell interaction, and initiate parthenogenic embryogenesis to develop into maternal haploid embryos.
Haploid screening process in watermelon
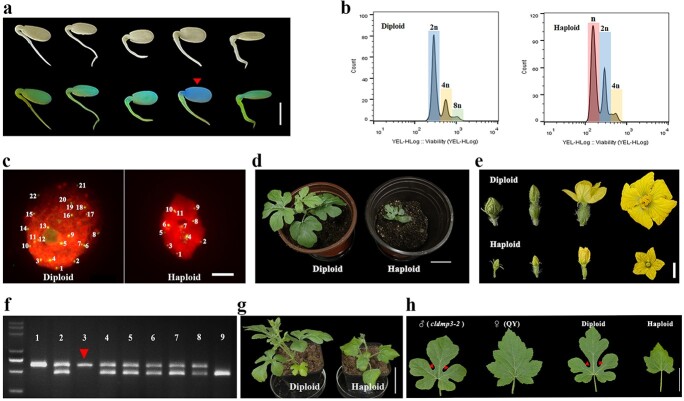
To confirm the haploid induction ability of cldmp mutants, the cldmp3–2 mutant was used as a pollen inducer that was crossed with the hybrid “linglongwang” (LLW). Since the GFP expression cassette was integrated into the cldmp3 mutant genome along with the T-DNA of CRISPR/Cas9 vector, we utilized it as a marker for hybrid seeds. Seeds with no fluorescence were therefore considered to be potential haploids, due to the fact that haploids induced by the cldmp3 mutant should be maternal and not inherit GFP from the male side (Fig. 5a). Flow cytometry was then utilized to check the ploidy level of the plants. Compared to the 2C, 4C and 8C peaks in the WT plants, haploid plants contained 1C, 2C and 4C peaks (Fig. 5b), which was indicative of a reduction of DNA ploidy. Immunofluorescence staining experiments were carried out with a centromere-specific CenH3 protein antibody to further confirm the chromosome number in haploids. This analysis detected 22 signals in the diploid WT watermelon plants (2n = 22) but only 11 fluorescence signals in the haploid plants (n = 11) (Fig. 5c), which further confirmed that chromosome number of the haploid was half that of the WT. Phenotypic analysis indicated that the haploids grew more slowly than the diploids, and had smaller leaves and floral organs (Fig. 5d and e). Additionally, haploid plants were sterile due to the uneven segregation of the chromosomes during meiosis. A total of four haploid plants were identified from 356 F1 hybrid seeds, representing a 1.12% haploid induction rate (HIR).
Figure 5.
Identification of haploids induced by the cldmp3 inducer line. a. Germinated seeds under bright field and green fluorescence. The red arrow indicates the potential haploid seed with no fluorescence. b. Flow cytometry verification of diploid and haploid plants. c. Immuno-staining of diploid and haploid nuclei using a watermelon centromere-specific CenH3 antibody. DNA was counterstained with DAPI (in red). Centromere signals (in green) indicate the chromosome number (diploid 2n = 22; haploid n = 11). d. Diploid and haploid plants in the F1 progeny of the “LLW” x cldmp3–2 crosses. e. Flower buds of diploid and haploid plants at different stages. f. Genotyping of diploid and haploid plants using the CAPS maker on Chromosome 3. Lane 1 is the female parent and lanes 2–8 are the progeny plants of “97 103” x cldmp3–2. Lane 9 is the male parent. The red triangle indicates the haploid. g. Diploid and haploid plants identified from F1 progeny of “QY” x cldmp3–2 crosses. h. Leaf shape comparison of diploid and haploid plants identified from the F1 progeny of cldmp3–2 x “QY” crosses. The red triangles indicate the lobed edge of the leaves. Scale bars: 1 cm (a and e), 5 μm (c), 5 cm (d and g), 3 cm (h).
To further test the haploid induction ability of cldmp3 in different genotypes, cldmp3–2 was used as the male parent in a cross with “97 103” and non-lobed leaf watermelon material “QY” [38]. Cleaved amplified polymorphic sequence (CAPS) markers were developed on all 11 chromosomes of “97 103” (Table S1). During haploid screening of the F1 generation, the hybrids displayed two bands, whereas the haploids contained only one maternal band (Fig. 5f). Additionally, analysis of all 11 chromosomal markers confirmed the haploids. The non-lobed leaf trait is known to be controlled by a single recessive gene, and cldmp3–2 was created in the inbred “TC”, which has lobed leaves. Therefore, in the F1 progeny of “QY” x cldmp3–2 crosses, the leaves of maternal haploids were round and non-lobed, which were visually distinguishable from the hybrids with lobed leaf edges at the seedling stage (Fig. 5g and h). The above visual recessive marker greatly simplified the haploid screening process. Analysis of this phenotype revealed that cldmp3–2 induced haploids at rates of 0.60% and 0.51% in “97 103” and “QY”, respectively, demonstrating a broad range of DMP-mediated haploid induction across different watermelon genotypes (Table 1).
Table 1.
Haploid induction rate (HIR) of the cldmp3–2 inducer line
| Female parent | Total seeds | Haploids | Haploid Induction Rate (HIR) |
|---|---|---|---|
| LLW | 356 | 4 | 1.12% |
| 97 103 | 498 | 3 | 0.60% |
| QY | 592 | 3 | 0.51% |
Discussion
Haploid induction systems have the potential to greatly accelerate plant breeding processes through the generation of homozygous DH lines in one generation. Since irradiated pollen grains can induce parthenogenic haploids in cucurbits crops at low frequencies, we hypothesized that the irradiated pollen grains cause preferential fertilization of the central cell over the egg cell, which leads to haploid formation. In this study, we have shown that mutation of the ClDMP3 gene, which encodes a DUF679 protein that is localized to the sperm membrane, can induce watermelon haploids at rates of 0.51 to 1.12%. Recently, Xu’s group also developed a haploid induction system in watermelon by utilizing cldmp mutants, with a similar haploid induction rate (1.08%) [39]. Both Xu’s and our results clearly show that the DMP-mediated haploid induction technique is feasible in watermelon, with significant implications for the development of new breeding strategies in watermelon that could lead to the production of valuable watermelon DH lines.
Further optimization of the watermelon transformation system
In order to successfully generate haploid inducer lines in watermelon at large scales, the transformation system, which facilitates the generation of mutant plants, needs further optimizations. Since the establishment of transformation systems in cucurbit crops, significant effort has been put into optimizing these processes [40–42]. Plant growth regulator genes, including BBM, GRF, WUS and WOX5, have been proven to boost transformation efficiencies in many plant speces [43–47], and the chimeric GRF4-GIF1 gene has been shown to increase transformation efficiency up to nine fold in a genotype-independent manner in watermelon [48]. Although constitutive overexpression of GRF4-GIF1 by the 35S promoter increases regeneration rates, it also results in malformed tissue during transformation (data not shown). It is possible that switching to a less active promoter could fix this issue, but additional research is needed to identify candidates. Callus or tissue-culture specific promoters are other possible strategies to avoid pleiotropic effects in later developmental stages caused by the overexpression of the growth regulator genes [49]. However, no callus-specific promoters have been reported in dicot plants. In addition to low efficiency, watermelon transformation also has a long cycle time, requiring an average of four to five months to obtain transgenic plants. Furthermore, rooting is one of the limiting steps of this process, taking up to 1.5 months. The development of more efficient rooting protocols or grafting positive transgenic shoots onto rooted stock plants have the potential to dramatically accelerate this process.
Precision genome editing tools are needed in watermelon
Since the establishment of the first CRISPR/Cas9 system in watermelon, it has been widely applied in breeding and gene function characterization [40, 50, 51]. More recently, an herbicide-resistant watermelon variety was created by employing the CRISPR/Cas9 system to edit the ALS gene [41]. With the rapid development of genomics and pan-genomics, more essential SNPs for agronomic traits have been identified and more efficient base editors are therefore needed to genetically engineer crops. For haploid induction, amino acid substitutions in the CenH3 protein have proven to be an effective strategy to induce haploids in model plants [9–11]. Efficient base editors and the PRIME editing system have the potential to significantly accelerate the development of inducer lines in watermelon [52].
The haploid screening process needs to be simplified in watermelon
Currently, the haploid screening process is dependent on fluorescent protein markers to identify haploids during the seed or seedling stage. Identifying the haploid at the seed or seedling stage could greatly simplify the process and result in time and labor savings. However, the 35S-driven GFP construct cannot stably propagate across generations in watermelon (data not shown), likely due to silencing of the GFP-cassette. A new marker system is therefore needed if extensive haploid screening is to be carried out. Fluorescent protein markers driven by seed-specific promoters represent one possible solution [53]. Another strategy is to use recessive phenotypic or molecular markers to simplify the process, such as the non-lobed leaf marker and CAPS markers used in this work. However, the ultimate goal of this process is to induce haploids in the hybrid backgrounds, which makes the above strategies less useful. Chromosome number confirmation is also necessary for haploid identification because aneuploids can be generated during some induction processes, such as the CenH3-based approach. Currently, there are two major methods to determine chromosome number, including chromosome counting by cytological staining [39] and immuno-staining. Immuno-staining is a two-day process and does not require pretreatment of the samples, which makes it possible to simultaneously handle a large number of samples.
In this work, we present a detailed method of establishing the haploid induction system in watermelon. Additional work is still required to increase the efficiency of this system and make it more applicable to commercial DH lines used in watermelon breeding.
Materials and methods
Plant material
The wild watermelon variety “TC” used in this experiment was collected in Tongchuan (Shaanxi province). “97 103” was provided by Dr. Yong Xu’s lab (Beijing Academy of Agricultural and Forestry Sciences) and inbred for at least four generations. “LLW” is a hybrid variety and “QY” is a self-crossing offspring identified from the selfing progeny of the commercial watermelon hybrid “Lingxiu”. All the above materials were planted in experimental fields of Northwest A&F University, Yangling.
Phylogenetic analysis
The DMP protein sequences from Nicotiana tabacum, Solanum lycopersicum (https://www.ncbi.nlm.nih.gov/), C. lanatus (http://cucurbitgenomics.org/), Arabidopsis thaliana (https://www.arabidopsis.org/), B. napus (http://www.brassicadb.cn/), Glycine max (https://www.soybase.org/), Z. mays (https://www.maizegdb.org/) and Oryza sativa (https://rapdb.dna.affrc.go.jp/) were used in the construction of phylogenetic trees. Protein IDs are listed in Table S2. Multiple sequence alignment of the DMP proteins was conducted using ClustalW with default parameters. The misaligned proteins portions before and after the central protein sequence were removed manually. A neighbor-joining tree was constructed with 1000 bootstrap replicates using MEGA 11 software [54].
Subcellular localization of ClDMP3
The cDNA sequence of ClDMP3 was amplified using gene-specific primers (Table S3) and ligated into the pGREEN vector to fuse with a green fluorescent protein (GFP) gene. The fusion protein ClDMP3-GFP was driven by a 35S promoter. AtCBL-RFP was also driven by a 35S promoter and was used as the membrane maker [55]. The two constructs were transiently co-transformed into tobacco (Nicotiana benthamiana) leaves through infiltration with Agrobacterium strain GV3101 (containing helper plasmid pSoup). After 48 hours, a confocal laser scanning microscope (Leica TCS-SP8 SR, Germany) was used to observe fluorescence.
Whole-mount RNA in situ hybridization of pollen grains
Whole-mount RNA in situ hybridization was performed as described previously, with minor modifications [56]:
The full-length cDNA of ClDMP3 was used to prepare the RNA probes.
A DIG RNA Labeling Kit (Roche, Rotkreuz, Switzerland) was used to label the sense and antisense probes of ClDMP3, according to the manufacturer’s instructions.
Fixation: Pollen grains were collected or germinated in an Eppendorf tube containing 4% paraformaldehyde in PBS (pH 7.4) at room temperature for 2–3 hours.
Samples were dehydrated with rinses of 3:7, 1:1 and 7:3 ME: PP (ME = 90% methanol-10% 0.5 M EGTA; PP = 4% formaldehyde in PBS containing 130 mM NaCl-10 mM NaH2PO4, pH 7.4) and 2 x 100% ethanol. Each rinse was carried out for 30 minutes, followed by 30 minutes of 100% xylene rinsing.
Rehydration was carried out with an 85%, 70%, 50%, 30% and 10% water series, 10 minutes each.
Proteinase treatment: The sample was incubated in Proteinase K buffer: 1 μg/mL proteinase K, 100 mM Tris–HCl (pH 7.5), 50 mM EDTA at 37°C for 35 minutes.
The sample was washed with PBS + 2 mg/mL Glycine for 2 x 5 minutes.
Refixation: The sample was incubated in 4% formaldehyde in PBS for 20 minutes and washed for 2 x 5 minutes in PBS.
Prehybridization: Samples were incubated in prehybridization buffer: 6 x SSC, 0.1% SDS, 50% formamide, 100 μg/mL tRNA at 42°C for 3 hours.
Hybridization: DIG-labeled RNA probes were added to a final concentration of 500 ng/mL and incubated at 42°C overnight.
Samples were washed in 2 x SSC, 0.1% SDS at 42°C for 10 minutes.
RNase treatment: Samples were incubated in 150 μL 10 mM Tris–HCl (pH 8.0), 0.5 M NaCl, 1 mM EDTA and 40 μg/mL RNase A at 37°C for 30 minutes.
Samples were washed with 1 x SSC, 0.1% SDS and 0.5 x SSC-0.1% SDS for 3 x 10 minutes at 42°C.
Blocking of the samples was carried out with 1% blocking reagent and 0.3% Triton X-100 for 1 hour at room temperature.
The sample was incubated in PBS-3% BSA with anti-DIG-AP conjugate for 1–2 hours and washed 3 x 20 minutes in PBS.
For detection, the sample was incubated with BCIP/NBT solution until a signal appeared.
Vector construction
The pBSE402 gene editing vector was used to create the cldmp3 mutant. A single guide RNA was designed by CRISPR-P v2.0 (http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR) and cloned into the pBSE402 vector [48]. The colony-PCR-positive clones were further confirmed by Sanger sequencing. The plasmid was isolated and transformed into the Agrobacterium strain EHA105 for watermelon transformation.
Watermelon transformation
Healthy watermelon seeds were soaked in distilled water at 50–55°C for 30 minutes, then deshelled. The deshelled seeds were placed on germination medium (BM, breeding medium; agar 4.43 g/L) and incubated in a petri dish at 28°C in the dark for 1–2 days.
After the seeds were evenly germinated, two cotyledons were cut into eight pieces to serve as explants. Simultaneously, a single EHA105 agrobacteria colony was grown to OD600 = 0.8–1.0. Excised cotyledons were rinsed in diluted bacterial cultures for 15 minutes (OD600 = 0.08–0.1), then transferred onto a filter paper to drain out the extra liquid. The explants were then transferred and cultivated in medium (MS519 4.43 g/L, sucrose 30 g/L, agar 3 g/L, 6-BA 1.5 mg/L) in the dark at 28°C for three days before being moved to normal growth conditions with 18 hours of daylight and 6 hours of darkness.
Explants were transferred to shoot generation medium (MS519 4.43 g/L, sucrose 30 g/L, agar 3 g/L, 6-BA 1.5 mg/L, 200 mg/L Timentin) and placed at 28°C for 10–12 days. Fluorescence was visualized with a stereoscopic fluorescence microscope (MZ10F, Leica, Germany), and positive shoots that contained GFP fluorescence were labeled. When the positive shoots had grown to 1–2 cm, they were transferred to new shoot generation medium.
Healthy positive shoots were transferred onto rooting medium (MS519 4.43 g/L, sucrose 30 g/L, agar 3 g/L, 200 mg/L Timentin). When 4–5 fresh leaves had formed on the shoots, the transgenic plants were transferred to the soil to continue growth.
DNA was extracted from positive shoots. After amplifying the target region with gene-specific primers, Sanger sequencing and Hi-TOM deep sequencing were used to confirm the editing types and ratios.
Alexander staining
Fresh flowers were collected to spread pollen evenly onto glass slides with approximately 20 μL Alexander’s staining solution, and incubated for 2–3 hours after the application of cover slips [57]. An Axiocam light microscope (Zeiss) was used to observe the pollen viability.
Semi-thin paraffin sectioning
Semi-thin paraffin sectioning was carried out using 7100 and 3040 embedding kits purchased from the Kulzer Technovit company, with the following steps:
Ovules were collected three days after pollination and fixed in FAA solution for infiltration at 4°C for 24 hours.
Samples were dehydrated by washing them with a 20%, 40%, 60%, 80% and 100% ethanol series for 30 minutes per wash.
100% ethanol: 7100 base liquid (1:1) mixed solution was used to pre-infiltrate the samples for 2–3 days.
One gram of hardener I was diluted into 100 mL of 7100 base solution, and the infiltrate solution was added to the sample after pouring out the pre-infiltrated solution from the previous step.
A total of 1 mL of hardener II was diluted into 15 mL of infiltrate solution, and the solution was added to each sample after the tissues were placed into the mold and covered with parafilm to exclude air for overnight incubation.
The parafilm was removed when the samples had settled. A glue solution (1:2 volume of 3040 liquid: 3040 powder) was used to attach the embedded tissues and the sectioning blocks.
The ovules were sectioned to a thickness of 3 μm using a glass knife. After stretching the samples out on water, they were placed on a 60°C hotplate for two hours. Then, 1% TBO was added to stain the samples for 1–2 minutes.
Flow cytometric analysis
Fresh true leaves were collected from watermelon seedlings, then a razor blade was used to chop the leaves (1 cm x 1 cm) in nuclei isolation buffer. The suspension solution was filtered through a 35 μm cell filter, and the flow-through nuclei were collected. The nuclei solution was treated with 50 μg/mL RNaseA, then stained by adding 50 μg/mL PI (Propidium Iodide) for 30 minutes [58]. Flow cytometry analysis was performed using a Muse Cell Analyzer (Luminex), according to the manufacturer’s direction.
Marker development
The whole genome sequence of “TC” was combined with data from the Cucurbit Genomics Database (CuGenDB, http://cucurbitgenomics.org/) to identify SNP (single nucleotide polymorphism) loci on each chromosome. CAPS markers were then developed for reliable SNP loci confirmed by PCR and sequencing. Primers were designed using Geneious software (http://www.geneious.com). The primers of the markers are shown in Table S1.
Immuno-staining
CenH3 protein antibody was raised against the N terminus protein sequence, which was manufactured by Genscript (Hangzhou).
A total of 10 mg of leaf tissue was fixed in 10 mL ice-cold 4% paraformaldehyde in Tris-buffer for 1 x 5 minutes under vacuum. Fixation was carried out for 20 minutes in ice-cold fixative.
Two 10-minute rinses with ice-cold Tris buffer were carried out.
The tissue was sliced in 1000 μL ice-cold LB01 buffer in a pre-cooled Petri-dish with a razor blade.
A 15 μL suspension was added to microscope slides with a pipet tip in circular movements. Slides were then left to dry at room temperature overnight.
The slide glass was washed twice with 1 x PBS for five minutes at room temperature.
60 μL of 1st antibody solution (2% BSA in 1 x PBS with 0.1% Tritonex-100:1st antibody = 200:1) was dropped onto the slide, which was then covered with parafilm. The slide was kept at high humidity overnight at 4°C.
The slide glass was washed two times with 1 x PBS for five minutes at room temperature.
60 μL of 2nd antibody solution (2% BSA in 1 x PBS with 0.1% Tritonex-100:2st antibody = 200:1) was dropped onto the slide, then left at high humidity at 37°C for one hour.
The slide glass was then washed two times with 1 x PBS for five minutes at room temperature.
The sample was dehydrated with two-minute sequential washes of 70%, 90% and 100% ethanol at room temperature. The slide was then left to dry at room temperature.
Samples were stained with DAPI.
Supplementary Material
Contributor Information
Xiner Chen, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China.
Yuxiu Li, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China.
Gongli Ai, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China.
Jinfan Chen, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China.
Dalong Guo, College of Horticulture and Plant Protection Henan University of Science and Technology, 471000, Luoyang, China .
Zhonghou Zhu, Luoyang Nongfa Agricultural Technology Co., LTD, 471100, Luoyang, China .
Xuejie Zhu, Luoyang Nongfa Agricultural Technology Co., LTD, 471100, Luoyang, China .
Shujuan Tian, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China; Shenzhen Research Institute, Northwest A&F University, Shenzhen, 518000, Guangdong, China.
Jiafa Wang, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China; Shenzhen Research Institute, Northwest A&F University, Shenzhen, 518000, Guangdong, China.
Man Liu, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China; Shenzhen Research Institute, Northwest A&F University, Shenzhen, 518000, Guangdong, China.
Li Yuan, State Key Laboratory of Crop Stress Biology for Arid Areas, College of Horticulture, Northwest A&F University, Yangling, 712100, Shanxi, China; Shenzhen Research Institute, Northwest A&F University, Shenzhen, 518000, Guangdong, China.
Acknowledgements
This work was supported by the National Youth Talent Program (A279021801), Earmarked Fund for China Agriculture Research System (CARS-25), Key-Area R&D Program of Guangdong Province (2022B0202060001), Key R&D Program of Shaanxi province (2023-YBNY-008), the Natural Science Foundation of Shaanxi Province (2022JM-112) and the Science and Technology Innovation Team of Shaanxi (2021TD-32). We would like to thank A&L Scientific Editing (www.alpublish.com) for its linguistic assistance during the preparation of this manuscript.
Author’s contributions
L.Y. conceived the study and designed the experimental scheme. X.E.C., Y.X.L., G.L.A. and J.F.C. executed the experiments. S.J.T., J.F.W., M.L., D.L.G., Z.H.Z. and X.J.Z. provided technical suggestions. L.Y. and X.E.C. wrote the manuscript.
Data availability statement
All data generated in this study are presented in the paper. Additional data related to this paper may be requested from the authors.
Conflict of interests
The authors declare no competing interests.
Supplementary Data
Supplementary data is available at Horticulture Research online.
References
- 1. Leon-Martinez G, Vielle-Calzada JP. Apomixis in flowering plants: developmental and evolutionary considerations. Curr Top Dev Biol. 2019;131:565–604. [DOI] [PubMed] [Google Scholar]
- 2. Kalinowska K, Chamas S, Unkel Ket al. State-of-the-art and novel developments of in vivo haploid technologies. TAG Theoretical and applied genetics Theoretische und angewandte Genetik. 2019;132:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Coe EH. A line of maize with high haploid frequency. Am Nat. 1959;93:381–2. [Google Scholar]
- 4. Kelliher T, Starr D, Richbourg Let al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature. 2017;542:105–9. [DOI] [PubMed] [Google Scholar]
- 5. Liu C, Li X, Meng Det al. A 4-bp insertion at ZmPLA1 encoding a putative phospholipase a generates haploid induction in maize. Mol Plant. 2017;10:520–2. [DOI] [PubMed] [Google Scholar]
- 6. Gilles LM, Khaled A, Laffaire JBet al. Loss of pollen-specific phospholipase NOT LIKE DAD triggers gynogenesis in maize. EMBO J. 2017;36:707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Lin Z, Yue Yet al. Loss-of-function alleles of ZmPLD3 cause haploid induction in maize. Nature plants. 2021;7:1579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–8. [DOI] [PubMed] [Google Scholar]
- 9. Kuppu S, Ron M, Marimuthu MPAet al. A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol J. 2020;18:2068–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Karimi-Ashtiyani R, Ishii T, Niessen Met al. Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc Natl Acad Sci U S A. 2015;112:11211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuppu S, Tan EH, Nguyen Het al. Point mutations in Centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet. 2015;11:e1005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lv J, Yu K, Wei Jet al. Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat Biotechnol. 2020;38:1397–401. [DOI] [PubMed] [Google Scholar]
- 13. Kelliher T, Starr D, Su Xet al. One-step genome editing of elite crop germplasm during haploid induction. Nat Biotechnol. 2019;37:287–92. [DOI] [PubMed] [Google Scholar]
- 14. Wang N, Gent JI, Dawe RK. Haploid induction by a maize cenh3 null mutant. Sci Adv. 2021;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Z, Conner J, Guo Yet al. Haploidy in tobacco induced by PsASGR-BBML transgenes via parthenogenesis. Genes (Basel). 2020;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Khanday I, Skinner D, Yang Bet al. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature. 2019;565:91–5. [DOI] [PubMed] [Google Scholar]
- 17. Conner JA, Podio M, Ozias-Akins P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 2017;30:41–52. [DOI] [PubMed] [Google Scholar]
- 18. Conner JA, Mookkan M, Huo Het al. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc Natl Acad Sci U S A. 2015;112:11205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wei X, Liu C, Chen Xet al. Synthetic apomixis with normal hybrid rice seed production. Mol Plant. 2023;16:489–92. [DOI] [PubMed] [Google Scholar]
- 20. Vernet A, Meynard D, Lian Qet al. High-frequency synthetic apomixis in hybrid rice. Nat Commun. 2022;13:7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tian S, Ge J, Ai Get al. A 2.09 Mb fragment translocation on chromosome 6 causes abnormalities during meiosis and leads to less seed watermelon. Hortic Res. 2021;8:256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo S, Zhang J, Sun Het al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45:51–8. [DOI] [PubMed] [Google Scholar]
- 23. Guo S, Zhao S, Sun Het al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat Genet. 2019;51:1616–23. [DOI] [PubMed] [Google Scholar]
- 24. Dong YQ, Zhao WX, Li XHet al. Androgenesis, gynogenesis, and parthenogenesis haploids in cucurbit species. Plant Cell Rep. 2016;35:1991–2019. [DOI] [PubMed] [Google Scholar]
- 25. Gonzalo MJ, Claveria E, Monforte AJet al. Parthenogenic haploids in melon: generation and molecular characterization of a doubled haploid line population. J Am Soc Hortic Sci. 2011;136:145–54. [Google Scholar]
- 26. Sari N, Abak K, Pitrat Met al. Induction of parthenogenetic haploid embryos after pollination by irradiated pollen in watermelon. HortScience. 1994;29:1189–90. [Google Scholar]
- 27. Takahashi T, Mori T, Ueda Ket al. The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development. 2018;145. [DOI] [PubMed] [Google Scholar]
- 28. Wang W, Xiong H, Zhao Pet al. DMP8 and 9 regulate HAP2/GCS1 trafficking for the timely acquisition of sperm fusion competence. Proc Natl Acad Sci U S A. 2022;119:e2207608119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cyprys P, Lindemeier M, Sprunck S. Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nature plants. 2019;5:253–7. [DOI] [PubMed] [Google Scholar]
- 30. Zhong Y, Chen B, Li Met al. A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nature plants. 2020;6:466–72. [DOI] [PubMed] [Google Scholar]
- 31. Zhong Y, Liu C, Qi Xet al. Mutation of ZmDMP enhances haploid induction in maize. Nature plants. 2019;5:575–80. [DOI] [PubMed] [Google Scholar]
- 32. Zhang J, Yin J, Luo Jet al. Construction of homozygous diploid potato through maternal haploid induction. aBIOTECH. 2022;3:163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong Y, Chen B, Wang Det al. In vivo maternal haploid induction in tomato. Plant Biotechnol J. 2022;20:250–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li Y, Li D, Xiao Qet al. An in planta haploid induction system in Brassica napus. J Integr Plant Biol. 2022;64:1140–4. [DOI] [PubMed] [Google Scholar]
- 35. Wang N, Xia X, Jiang Tet al. In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol J. 2021;20:22–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhong Y, Wang Y, Chen Bet al. Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J Integr Plant Biol. 2022;64:1281–94. [DOI] [PubMed] [Google Scholar]
- 37. Liu Q, Wang C, Jiao Xet al. Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci China Life Sci. 2019;62:1–7. [DOI] [PubMed] [Google Scholar]
- 38. Wei C, Chen X, Wang Zet al. Genetic mapping of the LOBED LEAF 1 (ClLL1) gene to a 127.6-kb region in watermelon (Citrullus lanatus L.). PLoS One. 2017;12:e0180741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tian S, Zhang J, Zhao Het al. Production of double haploid watermelon via maternal haploid induction. Plant Biotechnol J. 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian S, Jiang L, Gao Qet al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017;36:399–406. [DOI] [PubMed] [Google Scholar]
- 41. Tian S, Jiang L, Cui Xet al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018;37:1353–6. [DOI] [PubMed] [Google Scholar]
- 42. Liu B, Santo Domingo M, Mayobre Cet al. Knock-out of CmNAC-NOR affects melon climacteric fruit ripening. Front Plant Sci. 2022;13:878037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones T, Lowe K, Hoerster Get al. Maize transformation using the Morphogenic genes Baby Boom and Wuschel2. Methods Mol Biol. 2019;1864:81–93. [DOI] [PubMed] [Google Scholar]
- 44. Lowe K, La Rota M, Hoerster Get al. Rapid genotype "independent" Zea mays L. (maize) transformation via direct somatic embryogenesis. In vitro cellular & developmental biology. Plant : journal of the Tissue Culture Association. 2018;54:240–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Debernardi JM, Tricoli DM, Ercoli MFet al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol. 2020;38:1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Che P, Wu E, Simon MKet al. Wuschel2 enables highly efficient CRISPR/Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun Biol. 2022;5:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang K, Shi L, Liang Xet al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nature plants. 2022;8:110–7. [DOI] [PubMed] [Google Scholar]
- 48. Feng Q, Xiao L, He Yet al. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J Integr Plant Biol. 2021;63:2038–42. [DOI] [PubMed] [Google Scholar]
- 49. Zhou J, Li D, Zheng Cet al. Targeted transgene expression in Rice using a callus strong promoter for selectable marker gene control. Front Plant Sci. 2020;11:602680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin A, Troadec C, Boualem Aet al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–8. [DOI] [PubMed] [Google Scholar]
- 51. Ren Y, Guo S, Zhang Jet al. A tonoplast sugar transporter underlies a sugar accumulation QTL in watermelon. Plant Physiol. 2018;176:836–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anzalone AV, Randolph PB, Davis JRet al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chenarani N, Emamjomeh A, Rahnama Het al. Characterization of sucrose binding protein as a seed-specific promoter in transgenic tobacco Nicotiana tabacum L. PLoS One. 2022;17:e0268036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tamura K, Stecher G, Peterson Det al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Batistic O, Waadt R, Steinhorst Let al. CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 2010;61:211–22. [DOI] [PubMed] [Google Scholar]
- 56. Li XR, Li HJ, Yuan Let al. Arabidopsis DAYU/ABERRANT PEROXISOME MORPHOLOGY9 is a key regulator of peroxisome biogenesis and plays critical roles during pollen maturation and germination in planta. Plant Cell. 2014;26:619–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alexander MP. Differential staining of aborted and nonaborted pollen. Stain Technol. 1969;44:117–22. [DOI] [PubMed] [Google Scholar]
- 58. Galbraith DW, Harkins KR, Maddox JMet al. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are presented in the paper. Additional data related to this paper may be requested from the authors.