Abstract
Rose (Rosa hybrida) is one of most famous ornamental plants in the world, and its commodity value largely depends on its flower color. However, the regulatory mechanism underlying rose flower color is still unclear. In this study, we found that a key R2R3-MYB transcription factor, RcMYB1, plays a central role in rose anthocyanin biosynthesis. Overexpression of RcMYB1 significantly promoted anthocyanin accumulation in both white rose petals and tobacco leaves. In 35S:RcMYB1 transgenic lines, a significant accumulation of anthocyanins occurred in leaves and petioles. We further identified two MBW complexes (RcMYB1-RcBHLH42-RcTTG1; RcMYB1-RcEGL1-RcTTG1) associated with anthocyanin accumulation. Yeast one-hybrid and luciferase assays showed that RcMYB1 could active its own gene promoter and those of other EBGs (early anthocyanin biosynthesis genes) and LBGs (late anthocyanin biosynthesis genes). In addition, both of the MBW complexes enhanced the transcriptional activity of RcMYB1 and LBGs. Interestingly, our results also indicate that RcMYB1 is involved in the metabolic regulation of carotenoids and volatile aroma. In summary, we found that RcMYB1 widely participates in the transcriptional regulation of ABGs (anthocyanin biosynthesis genes), indicative of its central role in the regulation of anthocyanin accumulation in rose. Our results provide a theoretical basis for the further improvement of the flower color trait in rose by breeding or genetic modification.
Introduction
Anthocyanins are secondary metabolites of flavonoid biosynthetic pathways and are present in most flowering plants [1]. Anthocyanins not only give flowers and other organs rich colors, but also prevent damage from drought, ultraviolet radiation, low temperature stress, diseases, pests, and other adverse factors [2–5].
Anthocyanins range in color from orange-red (pelargonidin, cyanidin) to purple and pink-magenta (peonidin, malvidin, delphinidin, and petunidin) [6]. These pigments are produced by a branch of the phenylpropanoid metabolic pathway that is a flavonoid metabolic pathway. The genes for original phenylpropanoid biosynthesis include phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL). The anthocyanin early biosynthesis genes (EBGs) include chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), and flavonoid 3′-hydroxylase (F3′H), and the anthocyanin late biosynthesis genes (LBGs) include dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), and flavonoid 3-O-glucosyltransferase (UFGT). The enzymes encoded by these genes work sequentially to generate specific anthocyanins [7].
The MYB transcription factors (TFs) are the most important transcription level regulatory genes of anthocyanins in the plant phenylpropane metabolism pathway. MYB-TFs can be divided into four groups according to the number of (1R, 2R, 3R, 4R) repeats and repeat sequence variation: 1R-MYB, R2R3-MYB, 3R-MYB, and 4R-MYB. In anthocyanin biosynthesis, some MYB-TFs function as activators (R2R3-MYB) and some function as repressors (R2R3-MYB and R3-MYB) [8]. For example, in sweet cherry (Prunus avium), PavMYB10.1 and PavMYB75 activate the cascade of anthocyanin downstream regulators and structural genes by increasing the expression level of anthocyanin biosynthesis genes (ABGs) [9]. A similar pattern of regulation has been reported for MYBs in peach (Prunus persica) (PpMYB7/10.1/10.4/9) [10], blueberry (Vaccinium myrtillus) (VmMYBA1 and VmMYBA2) [11], apple (Malus domestica) (MdMYB1/10/11 and MdMYBA) [12, 13], Arabidopsis (Arabidopsis thaliana) (AtMYB75/90) [14], eggplant (Solanum melongena) (SmMYB113) [15] and celery (Apium graveolens) (AgMYB1) [16]. The MYB-TFs that function as repressors of anthocyanin synthesis include Fragaria × ananassa FaMYB1 [17], Arabidopsis AtMYB4 [18], Petunia hybrida PhMYBx [19], peach PpMYB140 [19], PpMYB17–20 [21], Populus tremula PtrMYB182 [22], and apple MdMYB16 [23].
The MYB-TFs usually form MYB-bHLH-WD40 (MBW) complexes with bHLH (basic helix–loop–helix) and WDR (WD40-repeat) proteins, and these complexes can promote or inhibit anthocyanin biosynthesis [18]. The R3 repeats [with a conserved motif (D/E) LX2 (R-K) X3LX6LX3R] in MYB-TFs interact with the N-terminal MYB interacting region (MIR) of bHLHs, and together with WDR proteins form the MBW complex [18, 24]. In anthocyanin synthesis, there are usually multiple sets of MBW complexes that synergize or antagonize anthocyanin synthesis. In Arabidopsis, the four MYB-TFs PAP1–4 can form MBW complexes with bHLHs (TT8/GL3/EGL3) and WDR (TTG1) to further enhance anthocyanin biosynthesis synergistically [25]. In orchid (Phalaenopsis equestris), PeMYB2/11/12 independently activate the expression of PeDFR, and the presence of PebHLH1 further activates the expression of PeDFR and enhances red pigmentation [26]. When MdMYB10 was co-transformed with bHLHs in apple, the activity of MdDFR promoter was significantly enhanced. [13]. In woodland strawberry (Fragaria vesca), co-expression of FvMYB10 and FvbHLH33 can more strongly activate the promoters of FvDFR and FvUFGT to promote anthocyanin biosynthesis [27]. The small R3-MYB, which has a single R3 domain and lacks a putative activation domain, can inhibit MBW activity, and in general MYB inhibitors can competitively bind bHLH to reduce the activity of the MBW complex and decrease anthocyanin synthesis [28]. For example, IbMYB44 in Ipomoea batatas inhibits its own transcriptional activity by competitively binding bHLH2 in the MBW (MYB340-bHLH2-NaC56) complex, thereby inhibiting anthocyanin biosynthesis [29]. The MYB inhibitor MYBL2 can competitively bind to bHLHs (MYC1/EGL3/GL3/TT8), and these interactions can disrupt the MBW complex or the binding of the complex to the promoters of ABGs, resulting in decreased anthocyanin accumulation [30].
Only a few studies have reported that MYB-TFs may be involved in the anthocyanin biosynthesis of rose. RrMYB113 and RhMYB10 were supposed to play a role in anthocyanin synthesis [31, 32]. RrMYB5 and RrMYB10 are induced by wounding, and their expression leads to increased accumulation of anthocyanins and proanthocyanidins [33]. Therefore, the MYB-TFs and the MBW complex regulating flower color in roses have not been studied in depth.
Previously, we performed a detailed analysis of R. hybrid ‘Rhapsody in Blue’ petal RNA-seq data (accession number: PRJNA885821) and identified genes involved in rose petal color regulation. We noticed that the FPKM value of RcMYB1 increased during flower coloration, suggesting thatzit may promote anthocyanin accumulation in rose flowers. Here, we analysed the regulation of RcMYB1 and two MBW complexes on rose pigment formation to explore the molecular regulation mechanism of rose color formation. These results provide new information for the regulation of rose color and will be useful for breeding roses and possibly other flowering plants with a wider range of flower colors.
Results
RcMYB1 encodes an SG6 R2R3 MYB TF
A phylogenetic tree of RcMYB1 and other members of the MYB transcription factor family in Arabidopsis and some Rosaceae species was constructed. In the tree, RcMYB1 was in subgroup 6 (SG6), whose members are known to promote anthocyanin accumulation in plants (Fig. S1, see online supplementary material). In addition, RcMYB1 showed high homology with RrMYB113 of rugosa rose (Rosa rugosa), RiMYB10 of red raspberry (Rubus idaeus), FaMYB10 of strawberry (F. × ananassa), and FvMYB10 of woodland strawberry (F. vesca) (Fig. S1, see online supplementary material). Multiple sequence alignment showed that RcMYB1 contained the typical bHLH interaction motif, the ANDV motif, and the conserved R2R3 MYB DNA-binding domain [R/K]Px[P/A/R]xx[F/Y] motif (Fig. S2, see online supplementary material). The results indicate that RcMYB1 may be an activator of anthocyanin biosynthesis in rose.
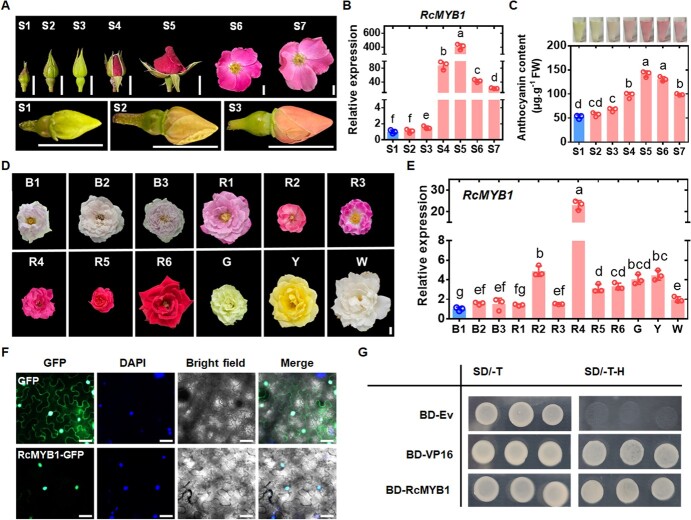
To study the transcript profile of RcMYB1 during petal coloration of rose, we selected the petals of seven developmental stages of ‘Old Blush’ flowers (Figs S1–S7, see online supplementary material) (Fig. 1A) and determined the RcMYB1 transcript levels and anthocyanin contents. The transcript levels of RcMYB1 gradually increased as anthocyanins accumulated, with the highest transcript level at S5, and then gradually decreased as the anthocyanins degraded (Fig. 1B). Thus, the transcriptional profile of RcMYB1 was consistent with the trends in anthocyanin accumulation during Figs S1–S7, see online supplementary material (Fig. 1C).
Figure 1.
Identification of the transcription factor RcMYB1. A Seven stages of of Rosa chinensis ‘Old Blush’ development (S1, S2, S3, S4, S5, S6, and S7). Images were digitally extracted for comparison. The scale bar is 1 cm. B Relative transcript levels of RcMYB1 in rose flowers at seven stages of development. C Anthocyanin content in petals of rose flowers at seven stages of development. D Differently colored rose flowers. B stands for blue-purple rose varieties: B1 is Rosa hybrida ‘Lavender Bouquet’; B2 is R. hybrida ‘Libellula’; B3 is R. hybrida ‘Kong Meng’. R stands for red rose varieties: R1 is R. hybrida ‘Muriel Robin’; R2 is R. hybrida ‘Angela’; R3 is R. hybrida ‘Yan Li’; R4 is R. hybrida ‘Ren Yue’; R5 is R. hybrida ‘Cherry Bonica’; R6 is R. hybrida ‘Black Magic’. G stands for green rose, and G is R. hybrida ‘Duo Lei’. Y stands for yellow rose, Y is R. hybrida ‘Yellow Leisure Liness’. W stands for white rose, W is R. hybrida ‘Gabriel’. Images were digitally extracted for comparison. The scale bar is 1 cm. E Relative transcript levels of RcMYB1 in differently colored rose flowers. F Subcellular localization of RcMYB1-GFP transiently expressed in Nicotiana benthamiana leaf cells. Note that GFP fluorescence overlaps with that of DAPI (nuclear marker). The scale bar is 50 μm. G Transcriptional activation associated with RcMYB1 in yeast cells. SD/−T, SD-Leu-Trp medium; SD-T-H, SD-Trp-His medium. Values are means ± SDs (n = 3). Lowercase letters (a–f) in B, (a–d) in C, and (a–g) in F indicate significantly different values (Student’s t test, P < 0.05).
Next, we determined the transcript levels of RcMYB1 in rose flowers with different colors, including blue-purple, red, green, yellow, and white (Fig. 1D). Interestingly, although RcMYB1 showed relatively high transcript levels in red roses, they were equally high in green, yellow, and white roses, but low in several blue-purple roses (Fig. 1E). The yellow flower is conferred by carotenoids, and the white rose studied here had a strong fragrance. Thus, we speculate that RcMYB1 may also be involved in the biosynthesis of volatile aromatic compounds and carotenoids.
To determine where RcMYB1 functions in cells and whether it has transcriptional activation activity, we performed a subcellular localization analysis and a trans-acting activity assay using a yeast system. In subcellular localization analysis, the RcMYB1-GFP signal was localized in nucleus cells of Nicotiana benthamiana leaves, consistent with its role as transcription factor (Fig. 1F). In the yeast system, RcMYB1-BD yeast cells and positive control VP16-BD also grew well on selective medium (SD/−Trp/-His), while negative control (BD) could not grow on selective medium (Fig. 1G). These results provided further evidence that RcMYB1 functions as an activating transcription factor in the nuclei.
RcMYB1 promotes anthocyanin accumulation in rose petals and tobacco (Nicotiana tabacum) leaves
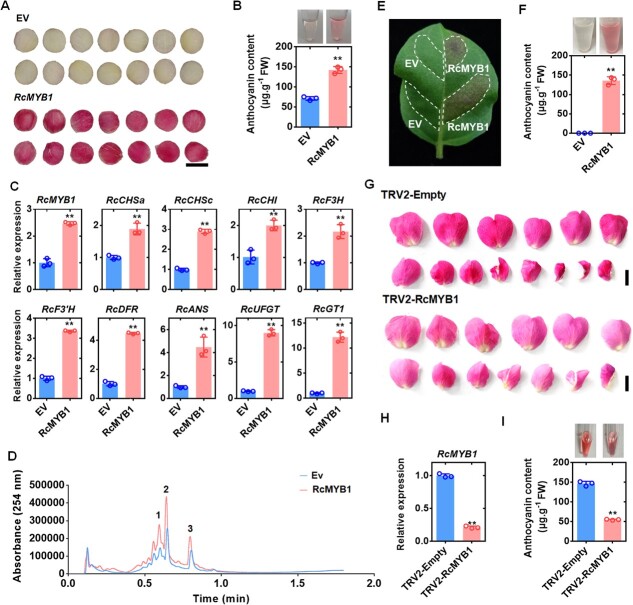
To clarify the function of RcMYB1 in anthocyanin accumulation, we performed transient overexpression experiments in Rosa hybrida ‘Gabriel’ petals and tobacco (N. tabacum) leaves. At 5 d after transformation, the white petals in the control (harboring the empty vector, EV) showed a slight red color, while those transiently overexpressing RcMYB1 had changed to deep red (Fig. 2A) and their anthocyanin content was significantly increased (Fig. 2B). The groups overexpressing RcMYB1 also showed significantly increased transcript levels of ABGs (RcCHSa, RcCHSc, RcCHI, RcF3H, RcF3′H, RcDFR, RcANS, RcUFGT, and RcGT1) (Fig. 2C). To determine the major anthocyanin components in red petals upon RcMYB1 overexpression, we examined the anthocyanin composition by liquid chromatography–mass spectrometry (LC–MS). The red color was mainly due to the increased contents of cyanidin and cyanidin derivatives (Fig. 2D). In tobacco leaves, the leaf areas injected with 35S:RcMYB1 showed a red color and accumulated anthocyanins (Fig. 2E and F). Further, we used virus-induced gene silencing (VIGS) to silence RcMYB1 in ‘Old Blush’. Our results showed that petal color was significantly lightened and anthocyanin content was significantly reduced after the silencing of RcMYB1 (Fig. 2G, H, I).
Figure 2.
Transient overexpression of RcMYB1 in rose petals and tobacco (Nicotiana tabacum) leaves. A Rose petals transformed with empty vector (EV) or vector containing RcMYB1. Images were digitally extracted for comparison. The scale bar is 1 cm. B Anthocyanin contents in rose petals expressing EV or RcMYB1. C Relative transcript levels of RcMYB1 and anthocyanin-related genes in rose petals expressing EV or RcMYB1. D LCMS analysis of the anthocyanin composition after overexpression of RcMYB1: 1 and 3 indicate cyanidin derivatives, 2 indicate cyanidin. E Tobacco leaf transformed with EV or RcMYB1. White dotted lines show infiltrated zones. F Anthocyanin content in parts of tobacco leaf transformed with EV or RcMYB1. G The phenotype of rose petals after silencing RcMYB1. The scale bar is 2 cm. H The relative expression of RcMYB1 in rose petals after silencing RcMYB1. I Anthocyanin contents in rose petals after silencing RcMYB1. Values are means ± SDs (n = 3). Asterisks in B, C, F, H, and I indicate significantly different values (Student’s t test, **P < 0.01).
Significant accumulation of anthocyanins in RcMYB1 transgenic lines in rose
To further clarify the function of RcMYB1 in rose anthocyanin biosynthesis, we transferred the full-length coding sequence of RcMYB1 into the pCAMBIA2300 vector to obtain 35S:RcMYB1 transgenic rose lines (OE-1 and OE-2) (Fig. 3A). The rose transgenic lines were confirmed by amplifying partial sequences of the pCAMBIA2300 vector with RcMYB1 by polymerase chain reaction (PCR) from genomic DNA, as well as by western blotting with an anti-GFP antibody (Fig. 3B and C). In the RcMYB1-OE lines, the leaves, petioles and stems significantly accumulated anthocyanins (Fig. 3D, E, F; Fig. S3, see online supplementary material). Further RT-qPCR analysis revealed up-regulation of RcMYB1 and ABGs (Fig. 3G), consistent with the results of the transient expression experiments. These results suggested that RcMYB1 has extensive involvement in the regulation of anthocyanin biosynthesis to mediate anthocyanin accumulation.
Figure 3.
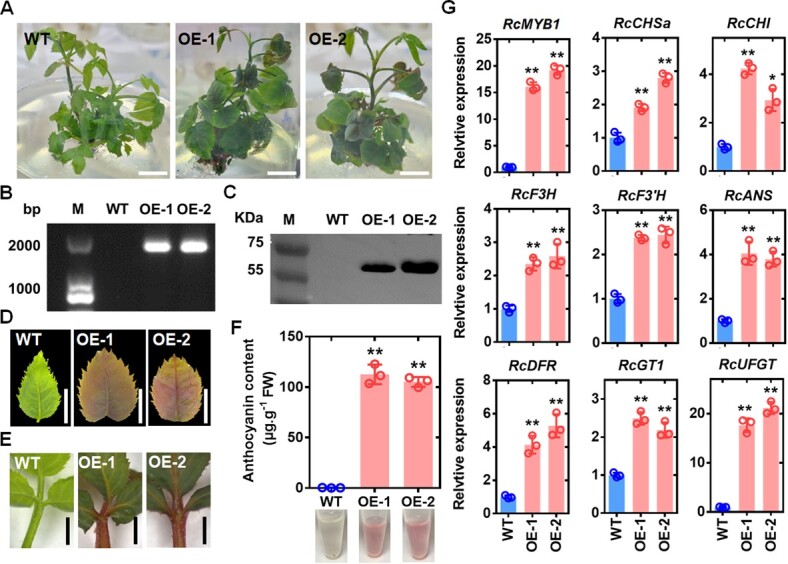
Effects of overexpression of RcMYB1 in transgenic rose plants. A Phenotypes of wild type (WT) and two RcMYB1-overexpressing lines (OE-1 and OE-2). The scale bar is 1 cm. B Confirmation of OE lines by PCR amplification. C Confirmation of OE lines by western blot analysis with GFP antibody. D WT and OE lines leaf phenotypes. The scale bar is 1 cm. E Phenotypes of petioles of WT and OE lines. The scale bar is 1 cm. F Anthocyanin content in WT and OE lines. G Relative transcript levels of RcMYB1 and anthocyanin-related genes in WT and OE lines. Values are means ± SDs (n = 3). Asterisks in F and G indicate significantly different values (Student’s t test, *P < 0.05 and **P < 0.01).
RcMYB1 physically interacts with RcTTG1 and RcbHLH42 or RcEGL1 to form MBW complexes
The MBW complex plays a core role in regulating anthocyanin biosynthesis. Therefore, we further identified the possible MBW complexes related to anthocyanin biosynthesis in rose. The phylogenetic tree was constructed using rose bHLH family proteins and reported bHLH proteins (AtMYC1, AtEGL3, AtGL3, AtTT8) related to anthocyanin biosynthesis in Arabidopsis. The tree showed that RcbHLH42 and RcEGL1 were closely related to these four bHLH proteins in Arabidopsis (Fig. S4, see online supplementary material). Another phylogenetic tree was constructed using RcbHLH42, RcEGL1 and other reported bHLH proteins related to anthocyanin biosynthesis in other species. That tree showed that RcbHLH42 and RcEGL1 were closely related to bHLH3 and bHLH33 in strawberry, respectively (Fig. S5A, see online supplementary material). Multiple sequence alignment analysis revealed conserved bHLH motifs in RcbHLH42 and RcEGL1 (Fig. S6, see online supplementary material). According to the amino acid sequence of AtTTG1 in Arabidopsis, we selected the WDR gene, RcTTG1, with the highest homology from rose. A phylogenetic tree was constructed with the TTG1 proteins related to anthocyanin biosynthesis reported for different plant species and showed that RcTTG1 was closely related to FaTTG1 in strawberry (Fig. S5B, see online supplementary material). Multiple sequence alignment analysis showed that RcTTG1 contained a conserved tandem repeat WD motif (Fig. S7, see online supplementary material).
To further determine whether RcbHLH42, RcEGL1, and RcTTG1 are related to anthocyanin biosynthesis, we examined their transcript levels in flowers at different stages of development. The transcript levels of RcbHLH42 and RcTTG1 increased during the early stage of anthocyanin accumulation, while RcEGL1 showed a decreasing trend (Fig. S5C, see online supplementary material). Nevertheless, we found that the transcript levels of these three genes were significantly elevated in the transgenic lines (Fig. S5D).
To determine where RcbHLH42, RcEGL1, and RcTTG1 function in the cells, we performed subcellular localization. The results showed that the GFP signal of RcEGL1-GFP fusion protein was localized in the nucleus, while RcbHLH42-GFP and RcTTG1-GFP fusion proteins were localized in the nucleus and cytoplasm of N. benthamiana epidermal cells (Fig. S5E, see online supplementary material).
To determine whether RcMYB1 could form an MBW complex with RcTTG1, RcbHLH42, or RcEGL1, we performed yeast two-hybrid assays (Y2H) assays. Directed Y2H assays validated that RcMYB1 interacted with the two bHLH proteins (RcbHLH42 and RcEGL1) and the WDR protein RcTTG1 (Fig. 4A), and RcTTG1 also interacted with the two bHLH proteins (RcbHLH42 and RcEGL1).
Figure 4.
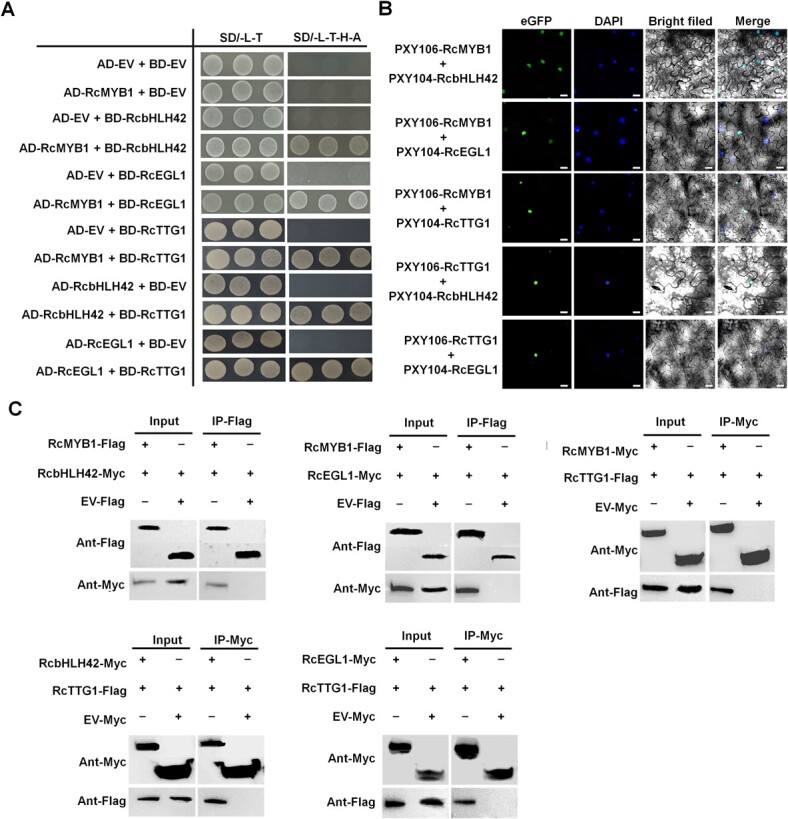
Analyses of potential of RcMYB1 to form MBW complexes with RcTTG1, RcbHLH42, or RcEGL1. A Yeast two-hybrid analyses demonstrating interaction between RcMYB1 and other components of MBW complexes. BD, DNA-binding domain; AD, activation domain; SD/−L-T, SD-Leu-Trp medium; SD-L-T-H-A medium SD-Leu-Trp-His-Ade medium. B Bimolecular fluorescence complementation analyses of demonstrating interaction between RcMYB1 and other components of MBW complexes in N. benthamiana leaves. Nuclei were counterstained with DAPI. Scale bar is 25 μm. C Coimmunoprecipitation assays demonstrating interactions among RcMYB1, two bHLHs (RcbHLH42 and RcEGL1), and RcTTG1.
To verify whether RcMYB1, the two bHLHs (RcbHLH42 or RcEGL1) and RcTTG1 HAT1 interact with each other in vivo, we conducted bimolecular fluorescence complementation (BiFC) assays. When RcMYB1-nYFP was coinfiltrated with RcbHLH42-cYFP or RcbEGL1-cYFP or RcTTG1-cYFP, and when RcTTG1-nYFP was coinfiltrated with RcbHLH42-cYFP or RcbEGL1-cYFP into N. benthamiana leaves, the strong YFP fluorescence was located in the nucleus (Fig. 4B). The interactions between RcMYB1 and RcbHLH42 or RcEGL1 or RcTTG1, and between RcTTG1 and RcbHLH42 or RcEGL1 were also confirmed by Co-IP assays (Fig. 4C). These results indicated that RcMYB1, two bHLHs (RcbHLH42 and RcEGL1) and RcTTG1 can interact with each other to form MBW complexes.
Two MBW complexes positively regulate anthocyanin accumulation in a functionally redundant manner
To determine whether RcbHLHs (RcbHLH42, RcEGL1), RcTTG1, and the two MBW complexes were involved in anthocyanin accumulation, we performed transient expression and transgenic assays in tobacco. Transient expression of RcbHLHs (RcbHLH42, RcEGL1) and RcTTG1 alone did not promote anthocyanin accumulation in N. tabacum leaves (Fig. S8A and B, see online supplementary material), while tobacco leaves co-infected with genes encoding components of two MBW complexes, RcMYB1, RcbHLH42 and RcTTG1 (MBT) and RcMYB1, RcEGL1 and RcTTG1 (MET) accumulated more anthocyanins compared with those transiently expressing RcMYB1 alone. There was no significant difference in anthocyanin accumulation between two lines containing different MBW complexes (Fig. S8A and B, see online supplementary material).
We further verified this result in the white petals of rose flowers. Transient overexpression of RcbHLH42 and RcEGL1 alone did not promote anthocyanin accumulation (Fig. 5A and B), Those expressing each of the MBW complexes accumulated more anthocyanins than did the line expressing RcMYB1 alone, but there was no significant difference in anthocyanin content between the two MBW complex lines (Fig. 5A and B). In contrast to the results of transient overexpression in tobacco leaves, white petals transiently expressing RcTTG1 alone showed a significant increase in anthocyanin accumulation (Fig. 5A and B). Further analyses revealed that transient overexpression of RcTTG1, but not RcbHLHs (RcbHLH42 or RcEGL1), increased the transcript levels of ABGs (Fig. 5C). The transcript levels of ABGs were higher in the groups expressing the two MBW complexes than in those expressing RcMYB1 and RcTTG1 alone (Fig. 5C). We detected higher transcript levels of both EBGs and LBGs in the groups expressing the two MBW complexes than in the group expressing RcMYB1 alone. Previous studies have shown that MBW complexes generally regulate LBGs. Thus, we hypothesized that the MBW complexes could enhance the expression of RcMYB1 to regulate the expression of EBGs. In addition, there were no significant differences in phenotype and regulation of ABGs between the two lines expressing MBW complexes, indicating that the two complexes are functionally redundant.
Figure 5.
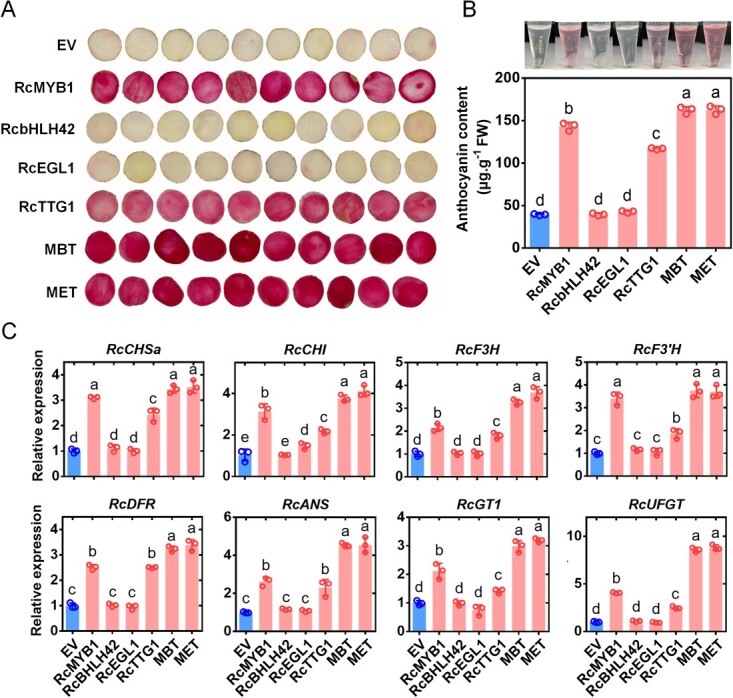
Transient overexpression of MBW complexes alone or in combination in rose petals. A Phenotypes of petals expressing different constructs. EV, empty vector. Other expressed rose genes encoding MYB1, bHLH42, EGL1, TTG1, and two MBW complexes (MBT and MET). B For (A) anthocyanin content determination. C Relative expression of anthocyanin-related genes after vacuuming for 5 d. Values are means ± SDs (n = 3). Lowercase letters, (a–d) in B and (a–e) in C, indicate significantly different values (Student’s t test, P < 0.05).
The phenotypes of transgenic tobacco lines also confirmed our results. The 35S:RcbHLH42 and 35S:RcEGL1 transgenic lines had the same flower color as the wild-type (WT), with no significant accumulation of anthocyanins (Fig. S8C and D, see online supplementary material). The 35S:RcMYB1 transgenic line had deeper flower color and significant accumulation of anthocyanins (Fig. S8C and D, see online supplementary material). Unlike the transient expression results, the 35S:RcTTG1 transgenic line had deeper flower color compared with that of WT (Fig. S8C and D, see online supplementary material). There was no significant difference in anthocyanin accumulation between the two transgenic lines expressing the MBW complexes, but both of them had deeper flower color and accumulated more anthocyanins compared with the 35S:RcMYB1 and 35S:RcTTG1 transgenic lines (Fig. S8C and D, see online supplementary material).
RcMYB1 alone or in MBW complexes activates its own promoter and those of other anthocyanin biosynthesis genes
To further investigate the role of RcMYB1 in regulating ABGs, we performed Y1H and dual-luciferase reporter (LUC) assays. When pLacZi-proRcMYB1, proRcCHSa, proRcCHSc, proRcCHI, proRcF3H, proRcF3′H, proRcDFR, proRcANS, proRcUFGT or proRcGT1 fusion constructs were co-expressed with pB42AD-RcMYB1 in yeast cells, the yeast cells turned blue when screened on the selective medium (SD/−Trp/-Ura) supplemented with X-gal. The negative control (BD) failed to turn blue on the selective medium (Fig. 6A). These results showed that RcMYB1 is able to bind not only to its own promoter but also to those of a wide range of ABGs (EBGs and LBGs).
Figure 6.
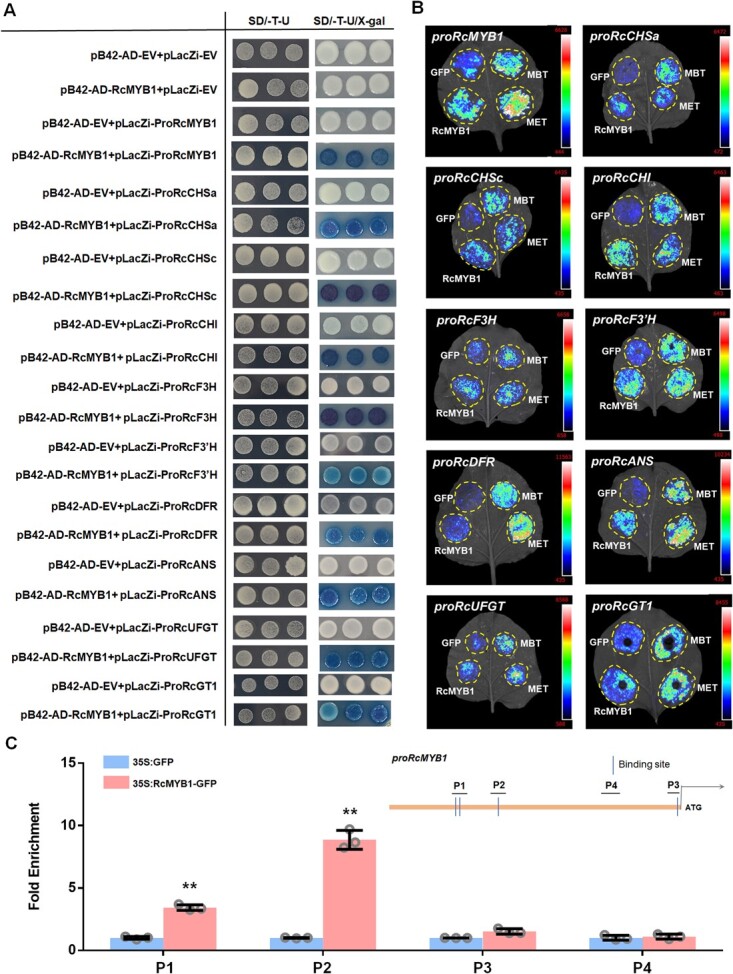
Analyses of interactions between MYB1 and promoters of various genes involved in anthocyanin biosynthesis. A Y1H assays showing the binding of RcMYB1 proteins to the promoters of RcMYB1, RcCHSa, RcCHSc, RcCHI, RcF3H, RcF3′H, RcDFR, RcANS, RcUFGT, RcGT1. Yeast cells were cultured on selective medium (SD-Trp-Ura/80 mg/L X-gal). The blue precipitate represents cumulative beta-galactosidase activity resulting from LacZ reporter binding activation. Three representative colonies are shown for each binding. B LUC assays verified the transcriptional activation ability of RcMYB1 and two rose MBW complexes (MBT and MET) towards the RcMYB1 and ABGs. CCD imaging system was used to capture luminous images. Color scale indicates LUC signal intensity (red, strong; blue, weak). C ChIP-qPCR assay for the direct binding of RcMYB1 to RcMYB1 promoter. Values are means ± SDs (n = 3). Asterisks indicate significantly different values (Student’s t test, **P < 0.01).
Next, we performed LUC experiments to determine whether RcMYB1 and the two MBW complexes activate the promoters of RcMYB1 and ABGs. The results showed that RcMYB1 can activate its own promoter, and the transcriptional activity was further enhanced by the two MBW complexes (Fig. 6B). ChIP-qPCR further confirmed the interaction between RcMYB1 and its own promoter in vivo (Fig. 6C). In addition, RcMYB1 also significantly activated the promoters of all of EBGs and LBGs. The two MBW complexes further enhanced the promoter activity of LBGs, but not those of EBGs (Fig. 6B). These results suggested that RcMYB1 can activate its own promoter and those of a wide range of ABGs, while the two MBW complexes enhance the promoter activity of RcMYB1 and LBGs.
RcMYB1 participate in carotinoids and volatile aroma metabolism but not in PA metabolism
Some studies have indicated that MYB-TFs can regulate the accumulation of anthocyanins and proanthocyanidins (PA), and also play an important regulatory role in a variety of secondary metabolic pathways. We found that RcMYB1 has a high expression level in yellow flowers and fragrant white flowers (Fig. 1D and E). Furthermore, we found potential MYB binding sites in these metabolic pathways genes promoters. Therefore, we studied whether RcMYB1 was involved in these metabolic pathways. Our results showed that the expression of Leucidin reductase (LAR) or Anthin reductase (ANR), key enzymes involved in PA synthesis [30], were not increased after overexpression of RcMYB1 and two MBW complexes (Fig. S9A, see online supplementary material). Y1H and Luc assays also indicated that RcMYB1 is not involved in transcriptional regulation of RcLAR and RcANR (Fig. S10A and B, see online supplementary material).
Lycopene beta cyclase (LCYB) and lycopene εcyclase (LCYE) are key branching points in the carotenoid synthesis pathway [34]. Interestingly, we found that the expression level of RcLYCB, RcLYCE-1, and RcLYCE-2 were significantly increased after overexpression of RcMYB1, and further enhanced by two MBW complexes (Fig. S9B, see online supplementary material). In order to further study the relationship between RcMYB1 and carotenoid synthesis, transient overexpression was performed in R. hybrida ‘Lady of Shalott’, which had light orange petals. The results showed that the petals showed a deeper orange-red color after overexpression of RcMYB1 (Fig. 7A), and the contents of carotenoids and anthocyanins in petals were significantly increased (Fig. 7B). Further, UPLC was used to determine the changes of α-carotene and β-carotene contents after overexpression RcMYB1. The results showed that both α-carotene and β-carotene contents were significantly increased after RcMYB1 overexpression in R. hybrida ‘Lady of Shalott’ (Fig. 7C). To further verify whether RcMYB1 directly regulated the transcription of RcLYCB, RcLYCE-1 and RcLYCE-2, Y1H, Luc, and ChIP-qPCR assays were conducted. Our results showed that RcMYB1 could directly bind and activate the promoter of RcLYCB, RcLYCE-1, and RcLYCE-2 (Fig. 7D and E). ChIP-qPCR also showed that RcMYB1 could significantly enrich the binding sites on promoters of RcLYCB, RcLYCE-1, and RcLYCE-2 (Fig. 7F, G, H).
Figure 7.
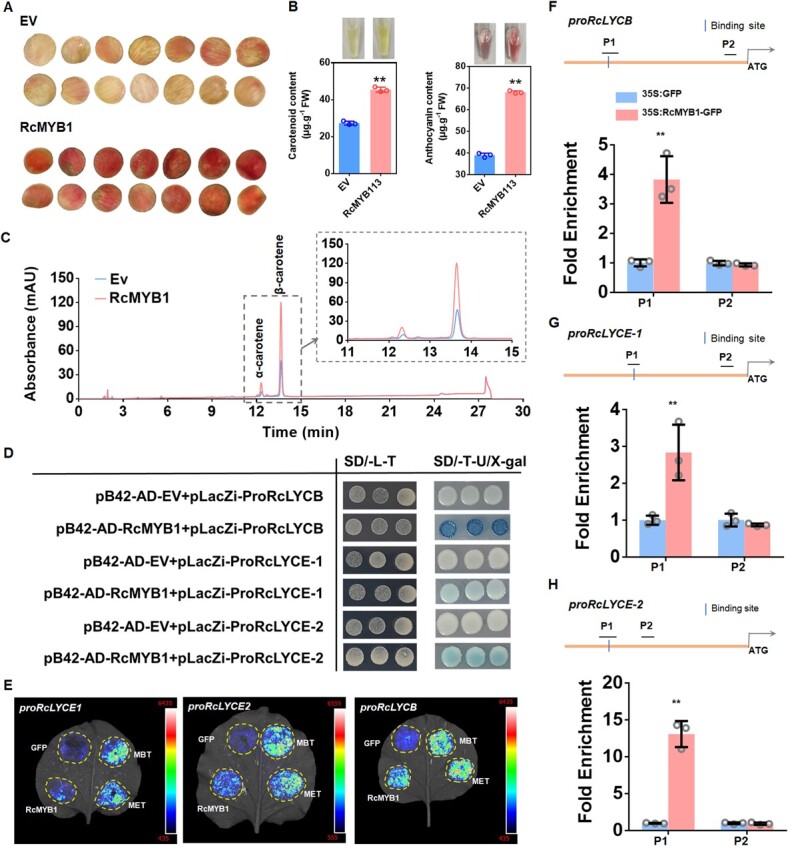
RcMYB1 regulated the biosynthesis of α-carotene and β-carotene. A Phenotypes of Rosa hybrida ‘Lady of Shalott’ petals after over-expressing RcMYB1. B Carotenoids and anthocyanins content determination. C The determination of α-carotene and β-carotene contents after overexpression RcMYB1 by UPLC. D Y1H assays showing the binding of RcMYB1 proteins to the promoters of RcLYCB, RcLYCE-1 and RcLYCE-2. Yeast cells were grown on selective medium (SD-Trp-Ura/80 mg/L X-gal). The blue precipitate represents cumulative beta-galactosidase activity resulting from LacZ reporter binding activation. Three representative colonies are shown for each binding. E LUC assays verified the transcriptional activation ability of RcMYB1 and two rose MBW complexes (MBT and MET) towards the RcLYCB, RcLYCE-1, and RcLYCE-2. CCD imaging system was used to capture luminous images. Color scale indicates LUC signal intensity (red, strong; blue, weak). F ChIP-qPCR assay for the direct binding of RcMYB1 to RcLYCB promoter. G ChIP-qPCR assay for the direct binding of RcMYB1 to RcLYCE-1 promoter. H ChIP-qPCR assay for the direct binding of RcMYB1 to RcLYCE-2 promoter. Values in B, F, G, H are means ± SDs (n = 3). Asterisks in B, F, G, H indicate significantly different values (Student’s t test, **P < 0.01).
In modern roses, RhNUDX1 is involved in the formation of geraniol, the main rose fragrance compound [35]. RcEGS1 is responsible for eugenol biosynthesis in rose [36]. The expression of RcEGS1 and RcNUDX1 was also increased after overexpression of RcMYB1 and two MBW complexes (Fig. S9C, see online supplementary material). In order to further investigate whether RcMYB1 is involved in the metabolism of aromatic compounds, GC-TOFMS analysis was performed on the transient overexpression RcMYB1 in R. hybrida ‘Gabriel’. After overexpression of RcMYB1, the relative contents of the main aromatic substances phenylpropanoids and monoterpenoids were significantly increased (Fig. 8A and B). Geraniol belongs to monoterpenoids, which is synthesized by RcNUDX1. In our results, we also found a significant increase in the relative content of geraniol (Fig. 8C). However, eugenol was not found in our determination results. Eugenol is in phenylpropanoids, which commonly present in rose stamens but not in rose petals [36]. Further, Y1H, Luc, and ChIP-qPCR assays were conducted to determine whether RcMYB1 directly regulated RcNUDX1 and RcEGS1. Our results showed that RcMYB1 could directly bind and activate the promoter activity of RcNUDX1 and RcEGS1 (Fig. 8D, E, F, G).
Figure 8.
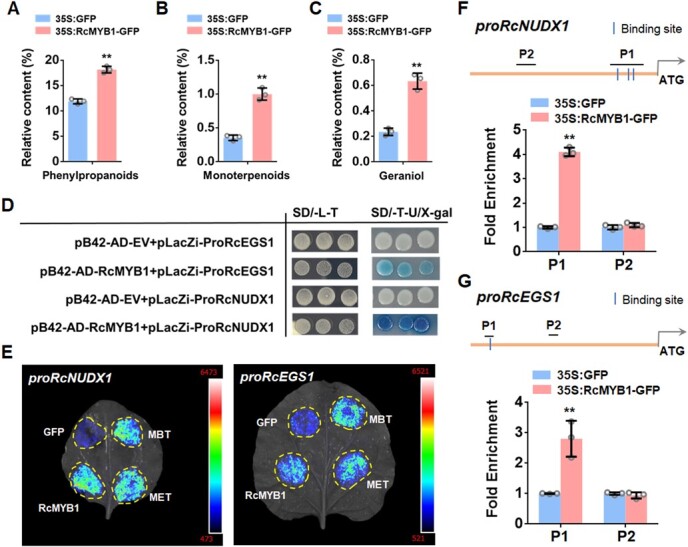
RcMYB1 regulated the geraniol and eugenol biosynthesis in rose. A GC-TOFMS analysis of the relative contents of phenylpropanoids after overexpression of RcMYB1. B GC-TOFMS analysis of the relative contents of monoterpenoids after overexpression of RcMYB1. C GC-TOFMS analysis of the relative contents of geraniol after overexpression of RcMYB1. D Y1H assays showing the binding of RcMYB1 proteins to the promoters of RcNUDX1 and RcEGS1. Yeast cells were grown on selective medium (SD-Trp-Ura/80 mg/L X-gal). The blue precipitate represents cumulative beta-galactosidase activity resulting from LacZ reporter binding activation. Three representative colonies are shown for each binding. E LUC assays verified the transcriptional activation ability of RcMYB1 and two rose MBW complexes (MBT and MET) towards the RcNUDX1 and RcEGS1. CCD imaging system was used to capture luminous images. Color scale indicates LUC signal intensity (red, strong; blue, weak). F ChIP-qPCR assay for the direct binding of RcMYB1 to RcNUDX1 promoter. G ChIP-qPCR assay for the direct binding of RcMYB1 to RcEGS1 promoter. Values in A, B, C, F, and G are means ± SDs (n = 3). Asterisks in A, B, C, F, and G indicate significantly different values (Student’s t test, **P < 0.01).
Discussion
RcMYB1 plays an important role in regulating anthocyanin accumulation in rose
R2R3-MYBs are the most important in anthocyanin synthesis, and many R2R3-MYBs that positively regulate anthocyanin biosynthesis have been identified in various plant species. In Arabidopsis, PAP1 makes seedlings produce anthocyanins, but MYB113/114 and PAP2 are partially redundant in anthocyanin biosynthesis [25]. Anthocyanin biosynthesis is activated by SlAN2-like tomatoes (Solanum lycopersicum) [37]. In addition, PhAN2 in petunia [38]; MdMYB1/10/110a, and in apple [12, 13, 39, 40]; PpMYB10.1/10.2/ /108 in peach [41, 42]; SmMYB1 in eggplant [43]; AaMYB2 in Anthurium andraeanum [44]; VvMYBA1 in common grape (Vitis vinifera) [45]; FhPAP1 in Freesia hybrida [46]; and PcMYB10 in Chinese white pear (Pyrus bretschneideri) [47] have also shown a positive role in regulating anthocyanin biosynthesis. In our study, we identified the RcMYB1 (R2R3 MYB) as a key activator that promotes anthocyanin biosynthesis in rose. Overexpression of RcMYB1 strongly promoted the accumulation of anthocyanins in white petals, leaves, and petioles (Figs 2 and 3).
MYB-TFs can directly bind to the promoters of ABGs to activate their transcription. PpMYB108 regulates peach anthocyanin biosynthesis by binding to PpDFR promoter [41]. AaMYB2 promotes anthocyanin accumulation in A. andraeanum by regulating the expression of AaCHS, AaF3H, and AaANS [44]. In golden kiwifruit (Actinidia chinensis), AcMYBF110 binds to the AcCHS, AcF3′H, AcANS, AcUFGT3a/T6b, but not to the promoters of AcCHI, AcF3H, or AcDFR [48]. However, in our study, we found that RcMYB1 broadly binds and activates the promoters of EBGs (RcCHSa, RcCHSc, RcCHI, RcF3H, and RcF3’H) and LBGs (RcDFR, RcANS, RcUFGT, and RcGT1) in the anthocyanin pathway (Fig. 6).
In our study, we found that RcMYB1 is involved in an autoregulatory feedback loop to regulate the accumulation of anthocyanins (Fig. 6). A similar autoregulatory feedback loop has also been reported in apple MdMYB10 and monkeyflower (Mimulus lewisii) PELAN (Petal Lobe Anthocyanin) [13, 49]. Some inhibitory R2R3-MYBs have also been shown to be under autoregulatory control. Arabidopsis MYB4 can inhibit its own transcription by binding to its own promoter as part of a negative autoregulatory loop [50]. Similar to rice (Oryza sativa) OsMYB4 [51] and petunia MYB27 [19].
RcbHLH42, RcEGL1, and RcTTG1 are involved in the regulation of anthocyanin accumulation by interacting with RcMYB1
In this study, we constructed a phylogenetic tree containing all rose bHLH family proteins as well as documented bHLH proteins (MYC1/EGL3/GL3/TT8) related to anthocyanin biosynthesis in Arabidopsis. We found that only RcbHLH42 and RcEGL1 were closely related to these four bHLH proteins in Arabidopsis (Fig. S4, see online supplementary material). bHLHs typically play a role in the formation of MBW complexes and enhance MYB activity but do not promote anthocyanin biosynthesis alone [52]. Consistent with the finding that FhGL3L and FhTT8L of F. hybrida cannot regulate anthocyanin biosynthesis independently of endogenous MYB proteins [53], we did not observe anthocyanin accumulation after overexpressing RcbHLH42 and RcEGL1 alone in tobacco leaves and rose petals (Fig. 5A; Fig. S8A, see online supplementary material). However, in Arabidopsis, the mutation of TT8 reduced anthocyanin and made seed coat colorless [54]. As RcMYB1 can independently activate EBGs and LBGs and can strongly promote the accumulation of anthocyanin in tobacco leaves and white rose petals, we think that the accumulation process of anthocyanin in rose petals mediated by RcMYB1 may not depend on RcbHLH42 and RcEGL1. In addition, after forming the MBW complex with RcMYB1 and RcTTG1, the transcriptional activity of RcMYB1 was enhanced by two MBW complexes (MBT, MET), so RcbHLH42 and RcEGL1 may play a role as enhancers—mediated by RcMYB1—in anthocyanin accumulation in rose petals.
A few WDR are involved in anthocyanin synthesis; on example is the WD40 protein TTG1, which has been isolated from a number of plant species [55]. In Arabidopsis, lack of seed coat pigment and decrease of anthocyanin accumulation in ttg1 mutant [56]. In rice, the anthocyanin content in the osttg1 mutant was significantly reduced in various organs [57]. Overexpression of CsWD40 in tobacco resulted in significant increase of anthocyanin content in petals [4]. In our study, anthocyanins accumulated significantly after RcTTG1 was overexpressed in white rose petals (Fig. 5A), which showed that RcTTG1 played an important role in the accumulation of anthocyanins in rose.
In the MBW complex, bHLH proteins usually interact with both MYB and WDR as linkers, while WDR may not interact with MYB. In apple, MdTTG1 interacts with bHLH but not with MYB proteins [58]. In Paeonia qiui, PqMYB113 interacts with PqbHLH1 but not with PqWD40 [52]. In tomato, SlAN11 (WDR) interacts with bHLH but not with the MYB protein in the MBW complex [59]. However, in our study, we found that RcTTG1 can interact not only with RcbHLHs but also with the RcMYB1 protein (Fig. 5). The WD40 protein interaction with MYB protein has also been reported in recent studies. For example, in woodland strawberry, FcTTG1 can interact with FcMYB114, FcMYB123, and FcbHLH42 [60]; in eggplant SmMYB86 interacts with SmTTG1 [61]; in Medicago truncatula MtWD40–1 interacts with MtPAR or MtLAP1 [62]; and in kiwifruit AcWDR1 interacts with AcMYBF110 [48].
The MBW complex is the key factor in the regulation of LBGs [28]. In Arabidopsis, EBGs are activated by coactivator-independent and functionally redundant R2R3-MYBs (MYB11/12/111), whereas activation of LBGs requires a MBW complex [25]. Similar to our results, we found that two MBW complexes significantly enhanced the activity of LBGs but had only a slight effect on EBGs activity (Fig. 6B). However, some studies have revealed that all ABGs are regulated by MBW complexes, such as in maize (Zea mays) [63], F. hybrida [46], rice (O. sativa) [64], and sweet potato [65]. These results suggest that MBW complexes may have different regulatory mechanisms for anthocyanin accumulation.
RcMYB1-RcbHLH42-RcTTG1 (MBT) and RcMYB1-RcEGL1-RcTTG1 (MET) are functionally redundant in anthocyanin accumulation in rose
The MBW complex has been shown to have functional redundancy for anthocyanidin biosynthesis in many plants [55]. Examples include AtTT8, AtGL3, and AtEGL3 in Arabidopsis [25], MdbHLH3/33 in apple [13], LcbHLH1/3 in Litchi chinensis [66], and FhGL3L and FhTT8L in F. hybrida [53]. In this study, we identified two MBW complexes (MBT, MET) that can promote anthocyanin accumulation. Based on ABGs expression level and anthocyanin content (Fig. 5A), we found that these two complexes may have functional redundancy in the regulation of anthocyanidin biosynthesis.
In addition, we found that the transcript levels of RcbHLH42 and RcEGL1 were not associated with anthocyanin accumulation during rose flower development, consistent with previous observations for LcbHLH1/2/3 in L. chinensis [66]; for MdbHLH33/3 in apple [13]; for VvMYC1 in grape [45]; and for FhGL3L in F. hybrida [53]. RcEGL1 was only highly expressed at the S1 stage and gradually decreased with the development of flower buds, while RcbHLH42 was highly expressed at the S3 and S4 stages (Fig. S5C, see online supplementary material), so we speculated that the MET complex might play a role in anthocyanin synthesis at an earlier stage than the MBT complex. In addition, we also found that the expression of RcEGL1 increased after MBT complex overexpression, and the expression of RcbHLH42 also increased after MET overexpression (Fig. S11, see online supplementary material). Therefore, we speculated that there might be cooperation in the regulatory mechanism in these two MBW complexes.
RcMYB1 is involved in other secondary metabolic pathways
So far, the roles of R2R3-MYBs in plants are mainly (∼70%) in regulating plant metabolic processes, including the benzenoid, phenylpropanoid, terpenoid, and glucosinolate (GSL) pathways [67]. In this study, we found that RcMYB1 is involved in multiple secondary metabolic pathways. Previous studies have found that MYB regulates anthocyanin and proanthocyanidin biosynthesis. Such as MtMYB5/14 in M. truncatula [68], MdMYB9/11 in apple [69], RrMYB5/10 in R.rugosa [33]. However, in this study, we found that RcMYB1 did not regulate the transcription of RcLAR and RcANR (Figs S8A and S9, see online supplementary material).
Interestingly, we found that RcMYB1 regulates the biosynthesis of carotenoids and aromatic volatiles. Carotenoids are closely related to the coloration of yellow, orange red or red flowers [70]. Lycopene beta cyclase (LCYB) and lycopene εcyclase (LCYE) are key branching points in the carotenoid synthesis pathway [34]. In this study, we transient overexpressed RcMYB1 in R. hybrida ‘Lady of Shalott’, and found that petals changed from light orange to orange-red (Fig. 7A), further we confirmed that RcMYB1 can directly regulate the transcription of RcLYCB, RcLYCE-1, and RcLYCE-2, thereby mediating the accumulation of α-carotene and β-carotene (Fig. 7). Many studies have reported that R2R3 MYB TFs can directly regulate carotenoid biosynthesis. For instance, in M. truncatula, WHITE PETAL1 (WP1) directly regulates the expression of MtLYCe and MtLYCb [71]. AdMYB7 can activate the promoter of AdLCYB in A. chinensis [34]. Similarly, in Camellia sinensis [72], in Rhyncholaeliocattleya [73], and in M. lewisii [74] have also been reported.
In addition, many R2R3 MYB-TFs have also been found to contribute to volatile aroma metabolism and control the emission of volatile substances [75]. A regulatory network of three R2R3 MYB-TFs, ODORANT1 (ODO1), EMISSION OF BENZENOIDS I (EOBI), and EOBII, can directly activate the transcription of many genes in the shikimic acid pathway and its downstream floral synthesis pathway [76, 77]. Expression of AtPAP1 not only promotes the synthesis of phenylpropanoid and terpenoid odour compounds but also promotes the accumulation of surfactants in R. hybrida, tobacco, and petunia [78]. Aroma components such as aldehydes, phenylpropanoids, and terpenes increased significantly in SlMYB75-OE tomato fruits [79]. In A. thaliana, MYB3 binds to the promoter of the gene cinnamate 4-hydroxylase (C4H) to negatively regulate gene expression and inhibit phenylpropanoid synthesis [80]. PhMYB4 has also been reported to inhibit the expression of PhC4H in P. hybrida [81]. In rose, a 1R-MYB TF, RhMYB1 may be a putative gene involved in the biosynthesis of rose scent [82]. Here, we provide evidence that a R2R3 MYB TF, RcMYB1, is involved in the biosynthesis of eugenol and geraniol in rose (Fig. 8). Phenylpropanoids and monoterpenoids are the main floral compounds of rose. After overexpression of RcMYB1, we found that the content of phenylpropanoids and monoterpenoids showed significant increases, and the content of geraniol, catalyzed by RcNUDX1, also significantly increased. Our determination results did not detect eugenol, catalyzed by RcEGS1, possibly because eugenol is normally present in the stamens of rose [36]. Interestingly, the contents of many phenylpropanoids and monoterpenoids were significantly increased after overexpression of RcMYB1, suggesting that RcMYB1 may also be widely regulated in the metabolism of aromatic compounds.
Conclusion
In conclusion, we revealed that R2R3-MYB RcMYB1 plays a central role in anthocyanin biosynthesis and also contributes to the metabolism of carotenoids and aromatic volatiles. Two bHLH proteins (RcbHLH42 and RcEGL1) can bind RcMYB1 and RcTTG1 to form the MBW complex, mainly regulating LBGs to regulate anthocyanin biosynthesis.
Materials and methods
Plant materials
Rosa chinensis and R. hybrida roses cultivars were planted at the Germplasm Resource Center of Shanghai Normal University, Shanghai, China. Petals were sampled at seven developmental stages (S1–S7) from R. chinensis ‘Old Blush’. Petals were also sampled on the day of flowering from the following: three blue-purple rose cultivars (R. hybrida ‘Lavender Bouquet’, R. hybrida ‘Libellula’, and R. hybrida ‘Kong Meng’); six red rose cultivars (R. hybrida ‘Muriel Robin’, R. hybrida ‘Angela’, R. hybrida ‘Yan Li’, R. hybrida ‘Ren Yue’, R. hybrida ‘Cherry Bonica’, and R. hybrida ‘Black Magic’); one green rose cultivar (R. hybrida ‘Duo Lei’); one yellow rose cultivar (R. hybrida ‘Yellow Leisure Liness’); and one white rose cultivar (R. hybrida ‘Gabriel’). The petals were divided into three replicates, frozen in liquid nitrogen and stored at −80°C.
Cloning of the RcMYB1, RcbHLHs (RcbHLH42 and RcEGL1), and RcTTG1 genes and the promoters of RcMYB1 and anthocyanin biosynthesis genes
The full-length sequences of RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 were cloned from the cDNA of R. chinensis ‘Old Blush’. The promoter sequences of RcMYB1 (2230 bp), RcCHSa (987 bp), RcCHSc (578 bp), RcCHI (826 bp), RcF3H (615 bp), RcF3′H (557 bp), RcDFR (522 bp), RcANS (2128 bp), RcGT1 (2053 bp), and RcUFGT (1986 bp) were also cloned from the DNA of R. chinensis ‘Old Blush’. The primers used are shown in Table S1 (see online supplementary material).
Sequence alignment and phylogenetic analysis
The full-length amino acid sequences of RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 in rose were aligned using CLUSTAL OMEGA. The amino acid sequences of MYB, bHLHs, and WD40 previously identified were from NCBI GenBank. RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 were functionally classified by phylogenetic tree analysis. MEGA11 (https://megasoftware.net/) and iTOL (https://itol.embl.de/) were used to build a maximum likelihood tree to analyse the phylogenetic relationship between protein.
RT–qPCR
Total RNA was extracted using the SteadyPure Plant RNA Extraction Kit (Accurate Biology, Hunan, ChangSha, China). qPCR was performed using Hifair® III 1st Strand cDNA Synthesis SuperMix (Yeasen, Shanghai, China) PerfectStart® Green qPCR SuperMix (Transgen Biotech, Beijing, China) was used for RT-qPCR. The 2−ΔΔCt method was used, with RcActin as the internal parameter. The primers of RT-qPCR are shown in Table S2 (see online supplementary material).
Subcellular localization
The ORFs of RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 (without stop codons) were cloned into pCAMBIA 2300 and transformed into Agrobacterium tumefaciens. The solution (10 mM MgCl2, 10 mM methylester sulfonate, and 200 μM acetosyringone, pH 5.7) was used to prepare an OD600 = 1.0 infiltration buffer to infiltrate 6-week-old N. benthamiana leaves. After 3 d, with 150 mu g/mL DAPI (4',6-diamidino-2-phenylindole) staining. Fluorescence images of GFP and DAPI were observed under 488 nm and 340 nm excitation light (Olympus FV3000 confocal scanning microscope, Olympus, Tokyo, Japan), respectively.
Transient overexpression in R. hybrida ‘Gabriel’, R. hybrida ‘Lady of Shalott’, and N. tabacum
For the transient assay in R. hybrida ‘Gabriel’ and R. hybrida ‘Lady of Shalott’ petal discs, A. tumefaciens-mediated vacuum infiltration was used. OD600 = 1.0, as described above. The petals were immersed in the infection solution, then treated in a vacuum of −80 kPa for 5 minutes, twice, placed in a filter paper petri dish containing 1% sucrose at 18°C for 6 d. Each treatment contained three experimental replicates, and approximately 100 petal discs were used from five flowers.
For the transient assay in N. tabacum leaves, the above different combinations of strains infected leaves, 7 days sampling.
Transformation of R. hybrida ‘Novalis’ and N. tabacum
Transformation of R. hybrida ‘Novalis’ was performed via A. tumefaciens-mediated transformation using leaf-derived embryogenic callus [83]. Briefly, sterile young leaves were used to induce callus on the callus induction medium (CIM) and further induce somatic embryos on the embryos induction medium (EIM). A. tumefaciens cells containing pCAMBIA 2300-RcMYB1 were collected and suspended at OD600 = 0.5. Then they were washed three times in an infiltrating solution at −80 kPa for 15 min and cocultured on a co-culture medium (CM) at 22°C dark for 4 days. Somatic embryos were cultured on screening medium supplemented with kanamycin and timentin, and positive transgenic plants were identified by genomic PCR, RT-q PCR, and Western blotting.
A. tumefaciens cells containing RcMYB1, RcbHLH42, RcEGL1, or RcTTG1 were transformed into N. tabacum leaves using the leaf disc method [84]. Transgenic plants were selected for each construct in a medium containing kanamycin and timentin. Then transfer to the greenhouse until flowering. Positive transgenic (genomic PCR and RT-q PCR) T1 progeny tobacco plants were identified.
YIH
The CDS of RcMYB1 was inserted into the pB42AD vector, and the promoter and anthocyanin synthesis genes of RcMYB1 were inserted into the pLacZi vector. Strain Y1HGold (Clontech, Mountain View, CA, USA) was used. PEG/LiAc method was used for conversion. The transformed yeast cells were inoculated into (SD/−Trp/-Ura) medium with or without X-Gal. Use the primers in Table S3 (see online supplementary material).
Y2H
Insert CDS of RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 into the pGADT7 or pGBKT7 carrier. The plasmid was transformed into Y2HGold (Clontech, Mountain View, CA, USA) cells and then grown on SD/−Leu/−Trp medium. The potential physical interactions between proteins were further tested on SD/−Trp/−Leu/-His/−Ade medium. The primers used are shown in Table S3 (see online supplementary material).
BiFCassay
The coding sequences of RcMYB1 and RcTTG1 were constructed into PXY106, and the coding sequences of RcbHLH42, RcEGL1, and RcTTG1 were constructed into PXY104. The primers are in Table S3 (see online supplementary material). As mentioned above, infect the N. benthamiana leaves. After 3 d infection, the cells were stained with 150 μg/mL DAPI. Yellow fluorescent protein (YFP) fluorescence and DAPI fluorescence (Olympus FV3000 confocal scanning microscope) were observed at excitation wavelengths of 505 nm and 340 nm, respectively.
Co-IP assay
RcMYB1 and RcTTG1 were cloned into PEG104 with Flag tag by Gateway™ LR Clonase™ II Enzyme mix. RcMYB1, RcbHLH42, and RcEGL1 were cloned into PEG104 with Myc tag. Primers are in Table S3 (see online supplementary material). In Co-IP, A. tumefaciens cells containing different structures were collected, as described above, OD600 = 1.0, and then infiltrated N. benthamiana leaves. After 3 days, N. benthamiana leaves were ground at a low temperature to extract protein, then incubated at 4°C for 4 h in the presence of anti-Flag or anti-Myc antibody conjugated beads. They were rinsed four times in the wash buffer. The protein was isolated in 8% SDS-PAGE gel and analysed by Western blot.
Anthocyanin extraction and quantification
The anthocyanin measurements followed the method described by Khazaei et al [85]: 2 mL extract [1% (v/v) HCl/methanol] plus a sample (FW, 0.2 g) at 4°C for 24 h under light protection, at 4°C, 12000 rpm for 1 min. Absorbance was determined by spectrophotometer (Techcomp UV-2600, Beijing, China) under A530 and A657. The calculation formula of anthocyanin relative content is as follows: ((A530 – A657) × dilution factor/mg FW tissue) × 1000.
For the determination of anthocyanins, liquid chromatography–mass spectrometry (LCMS) assays were performed on a Shimadzu LCMS-2020 (Shimadzu, Kyoto, Japan). Chromatographic separation was performed on an Agilent Zorbax SB-C18 column (4.6*250 mm, 3 μm). Eluent A (0.1% formic acid solution): Eluent B (0.1% formic acid acetonitrile solution) = 95:5. The flow rate was 1.2 mL/min, the column temperature was 30°C for 3 min, and the detection wavelength was 254 nm. Electrospray ionization (ESI) voltage was set at 0.9 KV, capillary temperature at 350°C, jacket gas pressure at 45 psi, and auxiliary gas at 10 psi. According to the retention time, the anthocyanin composition was determined, and then the anthocyanin equivalent was quantified.
Carotenoid extraction and quantification
A 0.5 g petal sample was crushed and 2 mL anhydrous ethanol (containing 0.1%BHT) was added, then a water bath at 80°C for 5 min, adding 100 μL KOH solution (80%w/v). In a water bath at 80°C for 15 min, 1 mL purified water and 1 mL n-hexane were added, after vortex mixing, centrifuge at 3000 rpm for 5 min, transfer the supernatant, add 1 mL n-hexane into the residue again for extraction, combine the two supernatants after centrifugation, and then steam dry at 30°C under mild nitrogen. Redissolve with 0.2 mL methanol solution for detecting. The sample extracts were analysed at 450 nm using an UPLC system with DAD detector (UPLC, U3000; Thermo, Karlsruhe, Germany). The analytical conditions were as follows, UPLC: column, YMC Carotenoid S-3 μm (150*4.6 mm), column temperature, 40°C; flow rate, 1.0 mL/min; injection volume, 2 μL; solvent system, MeOH: (MeOH: MTBE: H2O = 20: 75: 5); gradient program, 100:0 V/V at 0 min, 39:61 V/V at 15 min, 0:100 V/V at 25 min, 100:0 V/V at 25.1 min, 100:0 V/V at 30 min.
Identification of the volatile compounds
Volatiles were collected from petal discs by solid-phase micro-extraction (SPME) methods. Incubate 10 min at 50°C, extract 30 min, and deadsorb 5 min in GC injection mouth. After extraction, the compounds were analyzed with GC-TOFMS (Pegasus BT, Leco, St. Joseph, MI, USA). Conditions of GC-TOFMS: the temperature of DB-WAX (30 m × 250 μm × 0.25 μm) is 250°C, the interface temperature is 290°C and the ion source temperature is 230°C. The initial temperature was 40°C for 3 min, then increased to 70°C at 3°C/min, increased to 180°C at 5°C/min, and finally increased to 240°C at 10°C/min for 7 min.
Dual-luciferase reporter assay
The LUC analysis was performed with minor modifications according to the method described by He et al. [87]. CDS of RcMYB1, RcbHLH42, RcEGL1, and RcTTG1 were inserted into pCAMBIA 2300 vector to form effectors. The promoters of RcMYB1 and ABGs are inserted into pGreen II-0800 to form the reporter vector. The primers used are in Table S3 (see online supplementary material). The plasmid was introduced into A. tumefaciens strain (containing the pSoup helper plasmid). Different A. tumefaciens cells composed of effector and reporter are infected into the N.benthamiana leaves. Three day, 20 mg/mL D-lucifin (Goldbio, St Louis, MO, USA) was sprayed on the leaves and images were collected using a cooled low-light CCD imager (Tanon-4200, Shanghai, China).
ChIP-qPCR
ChIP-qPCR assays were performed as described by Bowler et al. [86]. The 5 g leaf tissues from RcMYB1-GFP-OE1 and GFP-OE plants were treated with 1% (v/v) formaldehyde to cross-link the protein-DNA complexes. The protein-DNA complexes were immunoprecipitated with an anti-GFP antibody (Cat. no. ab290; Abcam, Cambridge, UK). The immunoprecipitated DNA was purified and analysed using RT-qPCR. Use primers in Table S3 (see online supplementary material).
Accession numbers
Gene accession numbers used in this study: RcMYB1 (LOC112193894), RcbHLH42 (LOC112170399), RcEGL1 (LOC112182591), RcTTG1 (LOC112184646), RcCHSa (LOC112175474), RcCHSc (LOC112200102), RcCHI (LOC112182551), RcF3H (LOC112186620), RcF3′H (LOC112178545), RcDFR (LOC112173668), RcANS (LOC112179310), RcGT1 (LOC112184328), RcUFGT (LOC112198073), RcLAR (LOC112199978), RcANR (LOC112202296), RcLYCB (LOC112164457), RcLYCE-1 (LOC112188431), RcLYCE-2 (LOC112189564), RcEGS1 (LOC112201825) and RcNUDX1 (LOC112189710).
Acknowledgments
We acknowledge Wenqiu Wang (College of Agriculture and Biotechnology, Zhejiang University, China) for the technical assistance. We thank Jennifer Smith, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn/) for editing the English text of a draft of this manuscript.
This work was supported by Shanghai Special Project of Capacity Construction for Local Colleges and Universities, No.20070502500; Shanghai Science and Technology Agriculture Program, No.2022-02-08-00-12-F01146; Science and Technology Commission of Shanghai Municipality, No.18DZ2260500; and Shanghai Plant Germplasm Resources Engineering Research Center, 17DZ2252700.
Author contributions
F.M. designed the research and is the author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors. G.H., R.Z., S.J., and H.W. conducted the experiments. G.H. and R.Z. analysed the data and wrote the manuscript. All authors read and approved the manuscript.
Data availability
All relevant data in this study are provided in the article and its supplementary file.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Guoren He, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
Ren Zhang, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
Shenghang Jiang, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
Huanhuan Wang, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
Feng Ming, Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China.
References
- 1. Rudall PJ. Colourful cones: how did flower colour first evolve? J Exp Bot. 2020;71:759–67. [DOI] [PubMed] [Google Scholar]
- 2. Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao DQ, Tao J. Recent advances on the development and regulation of flower color in ornamental plants. Front Plant Sci. 2015;6:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu YJ, Hou H, Jiang XLet al. A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB−bHLH−WD40 ternary complexes. Int J Mol Sci. 2018;19:1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Y, Xue XX, Zhao CLet al. Cloning and functional characterization of chalcone isomerase genes involved in anthocyanin biosynthesis in Clivia miniata. Ornam Plant Res. 2021;1:2. [Google Scholar]
- 6. Patra S, Makhal PN, Jaryal Set al. Anthocyanins: plant-based flavonoid pigments with diverse biological activities. Intl J Plant Based Pharma. 2022;2:118–27. [Google Scholar]
- 7. Saigo T, Wang T, Watanabe Met al. Diversity of anthocyanin and proanthocyanin biosynthesis in land plants. Curr Opin Plant Biol. 2020;55:93–9. [DOI] [PubMed] [Google Scholar]
- 8. Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol. 1999;41:577–85. [DOI] [PubMed] [Google Scholar]
- 9. Qi XL, Liu CL, Song LLet al. A sweet cherry glutathione S-transferase gene, PavGST1, plays a central role in fruit skin coloration. Cell. 2022;11:1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou H, Lin-Wang K, Wang Fet al. Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 2019;221:1919–34. [DOI] [PubMed] [Google Scholar]
- 11. Karppinen K, Lafferty DJ, Albert NWet al. MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 2021;232:1350–67. [DOI] [PubMed] [Google Scholar]
- 12. Ban Y, Honda C, Hatsuyama Yet al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–70. [DOI] [PubMed] [Google Scholar]
- 13. Espley RV, Hellens RP, Putterill Jet al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mondal SK, Roy S. Genome-wide sequential, evolutionary, organizational and expression analyses of phenylpropanoid biosynthesis associated MYB domain transcription factors in Arabidopsis. J Biomol Struct Dyn. 2018;36:1577–601. [DOI] [PubMed] [Google Scholar]
- 15. Yang GB, Li LJ, Wei Met al. Smmyb113 is a key transcription factor responsible for compositional variation of anthocyanin and color diversity among eggplant peels. Front Plant Sci. 2022;13:843996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng K, Xing GM, Liu JXet al. AgMYB1, an R2R3-MYB factor, plays a role in anthocyanin production and enhancement of antioxidant capacity in celery. Veg Res. 2021;1:2. [Google Scholar]
- 17. Aharoni A, DeVos CH, Wein Met al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–32. [DOI] [PubMed] [Google Scholar]
- 18. Wang XC, Wu J, Guan MLet al. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020;101:637–52. [DOI] [PubMed] [Google Scholar]
- 19. Albert NW, Lewis DH, Zhang Het al. Members of an R2R3-MYB transcription factor family in petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–84. [DOI] [PubMed] [Google Scholar]
- 20. Ni J, Premathilake AT, Gao Yet al. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2021;105:167–81. [DOI] [PubMed] [Google Scholar]
- 21. Zhou H, Lin-Wang K, Wang HLet al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–21. [DOI] [PubMed] [Google Scholar]
- 22. Yoshida K, Ma D, Constabel CP. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 2015;167:693–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu HF, Wang N, Liu JXet al. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol Biol. 2017;94:149–65. [DOI] [PubMed] [Google Scholar]
- 24. Rodrigues JA, Espley RV, Allan AC. Genomic analysis uncovers functional variation in the C-terminus of anthocyanin-activating MYB transcription factors. Hortic Res. 2021;8:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonzalez A, Zhao MZ, Leavitt JMet al. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27. [DOI] [PubMed] [Google Scholar]
- 26. Hsu CC, Chen YY, Tsai WCet al. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 2015;168:175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin-Wang K, McGhie TK, Wang Met al. Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci. 2014;5:651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan HL, Pei XN, Zhang Het al. MYB-mediated regulation of anthocyanin biosynthesis. Int J Mol Sci. 2021;22:3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wei ZZ, Hu KD, Zhao DLet al. MYB44 competitively inhibits the formation of the MYB340-bHLH2-NAC56 complex to regulate anthocyanin biosynthesis in purple-fleshed sweet potato. BMC Plant Biol. 2020;20:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–85. [DOI] [PubMed] [Google Scholar]
- 31. Zou K, Wang Y, Zhao MYet al. Cloning and expression analysis of RrMYB113 gene related to anthocyanin biosynthesis in Rosa rugose. Am J Plant Sci. 2018;09:701–10. [Google Scholar]
- 32. Lin-Wang K, Bolitho K, Grafton Ket al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen YX, Sun TT, Pan Qet al. RrMYB5-and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol J. 2019;17:2078–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ampomah-Dwamena C, Thrimawithana AH, Dejnoprat Set al. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019;221:309–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conart C, Saclier N, Foucher Fet al. Duplication and specialization of NUDX1 in Rosaceae led to geraniol production in rose petals. Mol Biol Evol. 2022;39:msac002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yan HJ, Baudino S, Caissard JCet al. Functional characterization of the eugenol synthase gene (RcEGS1) in rose. Plant Physiol Biochem. 2018;129:21–6. [DOI] [PubMed] [Google Scholar]
- 37. Sun CL, Deng L, Du MMet al. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol Plant. 2020;13:42–58. [DOI] [PubMed] [Google Scholar]
- 38. Quattrocchio F, Wing J, Woude Ket al. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takos AM, Jaffé FW, Jacob SRet al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chagné D, Lin-Wang K, Espley RVet al. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013;161:225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khan IA, Cao K, Guo Jet al. Identification of key gene networks controlling anthocyanin biosynthesis in peach flower. Plant Sci. 2022;316:111151. [DOI] [PubMed] [Google Scholar]
- 42. Zhou H, Peng Q, Zhao JBet al. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front Plant Sci. 2016;7:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Docimo T, Francese G, Ruggiero Aet al. Phenylpropanoids accumulation in eggplant fruit: characterization of biosynthetic genes and regulation by a MYB transcription factor. Front Plant Sci. 2016;6:1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li CH, Qiu J, Yang GSet al. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.). Plant Cell Rep. 2016;35:2151–65. [DOI] [PubMed] [Google Scholar]
- 45. Azuma A, Kobayashi S, Goto-Yamamoto Net al. Color recovery in berries of grape (Vitis vinifera L.) ‘Benitaka’, a bud sport of ‘Italia’, is caused by a novel allele at the VvmybA1 locus. Plant Sci. 2009;176:470–8. [DOI] [PubMed] [Google Scholar]
- 46. Li YQ, Shan XT, Tong LNet al. The conserved and particular roles of the R2R3-MYB regulator FhPAP1 from Freesia hybrida in flower anthocyanin biosynthesis. Plant Cell Physiol. 2020;61:1365–80. [DOI] [PubMed] [Google Scholar]
- 47. Wang ZG, Meng D, Wang Aet al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red Bartlett pear. Plant Physiol. 2013;162:885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu YF, Ma KX, Qi YWet al. Transcriptional regulation of anthocyanin synthesis by MYB-bHLH-WDR complexes in kiwifruit (Actinidia chinensis). J Agric Food Chem. 2021;69:3677–91. [DOI] [PubMed] [Google Scholar]
- 49. Espley RV, Brendolise C, Chagne Det al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao JF, Zhang WH, Zhao Yet al. SAD2, an importin -like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell. 2007;19:3805–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baldoni E, Genga A, Medici Aet al. The OsMyb4 gene family: stress response and transcriptional auto-regulation mechanisms. Biol Plant. 2013;57:691–700. [Google Scholar]
- 52. Liu XK, Duan JJ, Huo Det al. The Paeonia qiui R2R3-MYB transcription factor PqMYB113 positively regulates anthocyanin accumulation in Arabidopsis thaliana and tobacco. Front Plant Sci. 2022;12:810990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li YQ, Shan XT, Gao RFet al. Two IIIf clade-bHLHs from Freesia hybrida play divergent roles in flavonoid biosynthesis and Trichome formation when ectopically expressed in Arabidopsis. Sci Rep. 2016;6:30514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nesi N, Debeaujon I, Jond Cet al. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell. 2000;12:1863–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang BP, Schrader A. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plan Theory. 2017;6:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Walker AR, Davison PA, Bolognesi-Winfield ACet al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang XH, Wang JR, Xia XZet al. OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 2021;107:198–214. [DOI] [PubMed] [Google Scholar]
- 58. An XH, Tian Y, Chen KQet al. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol. 2012;169:710–7. [DOI] [PubMed] [Google Scholar]
- 59. Gao YF, Liu JK, Chen YFet al. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic Res. 2018;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fan ZY, Zhai YL, Wang Yet al. Genome-wide analysis of anthocyanin biosynthesis regulatory WD40 gene FcTTG1 and related family in Ficus carica L. Front Plant Sci. 2022;13:948084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li LZ, He YJ, Ge HYet al. Functional characterization of SmMYB86, a negative regulator of anthocyanin biosynthesis in eggplant (Solanum melongena L.). Plant Sci. 2021;302:110696. [DOI] [PubMed] [Google Scholar]
- 62. Li PH, Chen BB, Zhang GYet al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016;210:905–21. [DOI] [PubMed] [Google Scholar]
- 63. Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–29. [DOI] [PubMed] [Google Scholar]
- 64. Zheng J, Wu H, Zhu HBet al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019;223:705–21. [DOI] [PubMed] [Google Scholar]
- 65. Zhu ZX, Wang HL, Wang YTet al. Characterization of the cis elements in the proximal promoter regions of the anthocyanin pathway genes reveals a common regulatory logic that governs pathway regulation. J Exp Bot. 2015;66:3775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lai B, Du LN, Liu Ret al. Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front Plant Sci. 2016;7:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu Y, Wen J, Xia YPet al. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Hortic Res. 2022;9:uhac058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu CG, Jun JH, Dixon RA. MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 2014;165:1424–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. An XH, Tian Y, Chen KQet al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–62. [DOI] [PubMed] [Google Scholar]
- 70. Iorizzo M, Ellison S, Senalik Det al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet. 2016;48:657–66. [DOI] [PubMed] [Google Scholar]
- 71. Meng YY, Wang ZY, Wang YQet al. The MYB activator WHITE PETAL1 associates with MtTT8 and MtWD40-1 to regulate carotenoid-derived flower pigmentation in Medicago truncatula. Plant Cell. 2019;31:2751–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li PH, Xia EH, Fu JMet al. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 2022;110:1144–65. [DOI] [PubMed] [Google Scholar]
- 73. Li BJ, Zheng BQ, Wang JYet al. New insight into the molecular mechanism of colour differentiation among floral segments in orchids. Commun Biol. 2020;3:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sagawa JM, Stanley LE, LaFountain AMet al. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016;209:1049–57. [DOI] [PubMed] [Google Scholar]
- 75. Muhlemann JK, Maeda H, Chang CYet al. Developmental changes in the metabolic network of snapdragon flowers. PLoS One. 2012;7:e40381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Verdonk JC, Haring MA, van-Tunen AJet al. ODORANT1 regulates fragrance biosynthesis in petunia flowers. Plant Cell. 2005;17:1612–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Spitzer-Rimon B, Farhi M, Albo Bet al. The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell. 2012;24:5089–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xie DY, Sharma SB, Wright Eet al. Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J. 2006;45:895–907. [DOI] [PubMed] [Google Scholar]
- 79. Jian W, Cao HH, Yuan Set al. SlMYB75, an MYB-type transcription factor, promotes anthocyanin accumulation and enhances volatile aroma production in tomato fruits. Hortic Res. 2019;6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou ML, Zhang KX, Sun ZMet al. LNK1 and LNK2 Corepressors interact with the MYB3 transcription factor in Phenylpropanoid biosynthesis. Plant Physiol. 2017;174:1348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Colquhoun TA, Kim JY, Wedde AEet al. PhMYB4 fine-tunes the floral volatile signature of petunia × hybrida through PhC4H. J Exp Bot. 2011;62:1133–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yan HJ, Zhang H, Wang QGet al. Isolation and identification of a putative scent-related gene RhMYB1 from rose. Mol Biol Rep. 2011;38:4475–82. [DOI] [PubMed] [Google Scholar]
- 83. Liu GQ, Yuan Y, Jiang Het al. Agrobacterium tumefaciens-mediated transformation of modern rose (Rosa hybrida) using leaf-derived embryogenic callus. Hortic Plant J. 2021;7:359–66. [Google Scholar]
- 84. Horsch RB, Fry JE, Hoffmann NLet al. A simple and general method for transferring genes into plants. Science. 1985;227:1229–31. [DOI] [PubMed] [Google Scholar]
- 85. Khazaei KM, Jafari SM, Ghorbani Met al. Optimization of anthocyanin extraction from saffron petals with response surface methodology. Food Anal Methods. 2016;9:1993–2001. [Google Scholar]
- 86. Bowler C, Benvenuto G, Laflamme Pet al. Chromatin techniques for plant cells. Plant J. 2004;39:776–89. [DOI] [PubMed] [Google Scholar]
- 87. He GR, Cao YW, Wang Jet al. WUSCHEL-related homeobox genes cooperate with cytokinin to promote bulbil formation in Lilium lancifolium. Plant Physiol. 2022;190:387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data in this study are provided in the article and its supplementary file.