Summary
The authors of a recent study identified noncanonical peptides (NCP) presented by cancer cells’ HLA and observed lack of reactivity to these antigens by endogenous tumor-reactive T cells. In vitro sensitization generated NCP-reactive T cells that recognized epitopes shared by a majority of cancers tested, providing opportunities for novel therapies to shared antigens.
In this issue of Clinical Cancer Research, Lozano-Rabella and colleagues (1) identified noncanonical peptides (NCP) presented by the HLA class I molecules of human tumor cell lines and demonstrated that these peptides were immunogenic (1). While other publications support these findings of HLA presentation of noncanonical, aberrantly translated peptides derived from upstream open reading frame (uORF), 5′UTRs, ncRNAs, endogenous retroelements (ERE), pseudogenes and out-of-frame transcripts, from tumor cells, there has been little evidence that these rapidly degraded NCPs, or Dark Matter, were targets of the endogenous anticancer immune response (2–5).
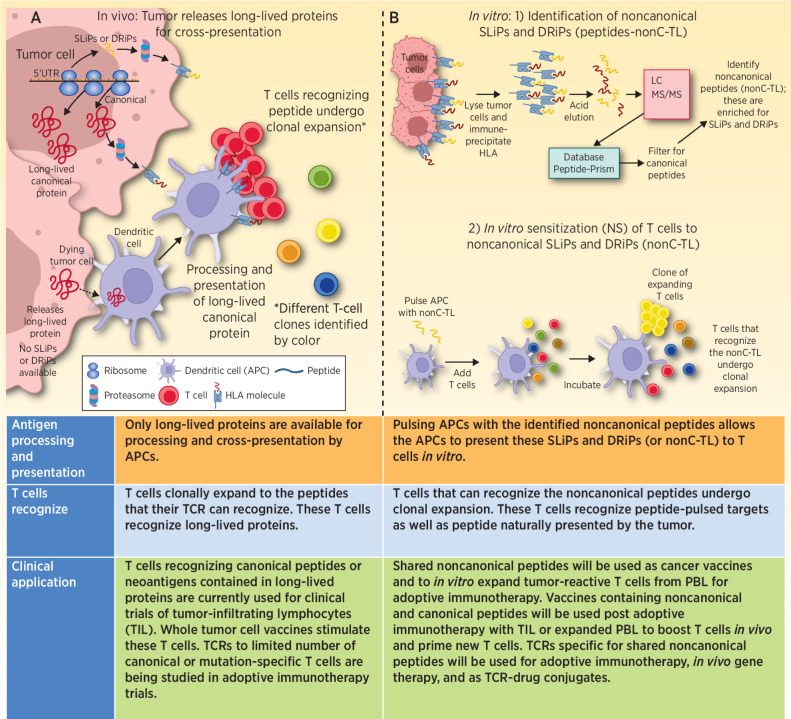
The authors’ explore this question by determining whether tumor-reactive T cells from the peripheral blood or tumor-infiltrating lymphocytes (TIL) recognized this HLA-bound Dark Matter, which they characterized as noncanonical tumor ligands (nonC-TL). While there was no evidence of an endogenous immune response to the NCPs, in some instances peripheral blood lymphocytes (PBL) in vitro sensitized (IVS) to the nonC-TL recognized nonC-TL pulsed onto antigen-presenting cells (APC) and secreted IFNγ or upregulated 4–1BB (Fig. 1). These sensitized T cells also recognized the NCP naturally presented by the patient's tumor cell line as well as other cancer cell lines that shared the same HLA restriction element but represented different tumor types. Upon transfection with the appropriate restricting HLA molecule that presented the specific nonC-TL, two of three nonC-TL–reactive T cell cultures recognized a majority of the 20+ tumor cell lines tested. This included endometrial, ovarian, gastrointestinal, and melanoma cancer cell lines and provided evidence for this HLA-bound Dark Matter to be shared by a spectrum of different cancers.
Figure 1.
A, Progressively growing tumors are transcribing and translating canonical proteins. At the same time, NCPs derived from uORFs, 5′UTRs, ncRNAs, EREs, pseudogenes, and out-of-frame transcripts may also be generated, some of which can contribute to malignant properties of the cancer. The majority of these NCPs are SLiPs and DRiPs that will be rapidly degraded, and presented by HLA, but because of their short-lived nature are unable to be picked up, processed, and presented by APCs. Thus, the patient is unable to generate an immune response to these short-lived noncanonical proteins. When tumor cells die, the proteins that are available to be picked up and processed are predominantly long-lived proteins. Peptides derived from these long-lived proteins are the dominant antigens that the patient makes a T-cell response against. B, 1) In vitro isolated tumor cells are lysed, antibody to HLA class I is incubated with the lysate, and HLA molecules are immunoprecipitated. The peptides are next eluted from the HLA molecule and run through LC/MS-MS to identify peptide sequences. These data are then queried using a database (Peptide-PRISM) that identifies the peptides that were bound to HLA. These data are then filtered for canonical proteins. The result is a list of NCPs presented by HLA. 2) These NCPs (nonC-TL) were used in IVS assays to prime and expand T cells able to recognize the NCPs. In some cases, the expanded T cells recognized the NCP pulsed onto APC. These also recognized naturally presented peptide on tumor cells and secreted IFNγ or upregulated 4–1BB expression. TCRs isolated from these expanded T cells also recognize the NCP.
The inability of short-lived noncanonical proteins to be picked-up, processed and cross-presented by professional APCs has been described previously (4). Most of the noncanonical proteins/peptides are short-lived proteins (SLiP) or defective ribosomal products (DRiP), not subject to thymic selection, typically degraded within 30 minutes, but efficiently captured by MHC class I and thereby widely expressed on the surface of tumor cells (Fig. 1; refs. 4, 6). While the stability provided by MHC binding allows the peptide to be recognized by LC/MS-MS and a spectra of amino acids generated, decoding the LC/MS-MS spectra requires either a computational approach, which the authors used, or a database of expected sequences determined by whole transcriptome or ribo-seq (7–9). Both analytical methods are in a state of evolution and it is expected that additional nonC-TL will be identified as these processes mature.
The authors’ data help explain why we are only now discovering the immunogenic nature of these nonC-TL. For instance, the detection of the first MHC-restricted tumor antigen in mice was made using cytotoxic T lymphocytes from spleen cells of animals vaccinated with P815 tumor cells (10). Similarly, tumor-reactive PBL or TIL were used to facilitate the cloning of the early human melanoma antigens (11, 12). On the basis of Lozano-Rabella's work, neither of these sources of tumor-reactive T cells would be expected to harbor T cells capable of recognizing nonC-TL derived from SLiPs or DRiPs. In retrospect, these findings may also explain the work of Prehn and Main, who more than 60 years ago demonstrated that vaccination with chemically induced tumors was exquisitely specific, providing protection against a live tumor challenge with the same tumor used as the vaccine, but not syngeneic sarcomas induced by the same carcinogen (13). Today we postulate that only the neoantigens contained in the long-lived proteins are retained in the tumor cells used as a vaccine, the nonC-TL, anticipated to be present, because of their short-lived nature, are not cross-presented and thus fail to induce a therapeutic anticancer immune response. However, Twitty and colleagues employing the same chemically induced sarcoma model developed a vaccine enriched for SLiPs and DRiPs and found it could provide significant cross-protection in 8 of 9 combinations tested (14). Their vaccine strategy blocks the proteasome and thereby shunts the SLiPs and DRiPs to the autophagosomes, which are harvested and used as a vaccine. Thus, we postulate that Twitty's vaccine actually enriched for nonC-TL, allowed for cross-presentation and ultimate induction of an immune response to these noncanonical antigens. Currently we are characterizing the NCPs contained in the human version of this vaccine, which has moved into a combination immunotherapy trial for patients with advanced cancer (NCT04470024; refs. 15–17)
Are these nonC-TL relevant cancer antigens? It is becoming increasingly clear that many of the uORFs, 5′UTRs, and ncRNAs unexpectedly code for peptides associated with malignant processes, providing a basis for their presence in cancer and strengthening the rationale for developing therapies that target these peptides (18). Some of these nonC-TL might be compared with truncal cancer neoantigens that are driver mutations for cancer development. In this case, an increasing number of NCPs are being found to facilitate malignant processes. Targeting cancer cells that express these NCPs, with expression restricted to or substantially increased in cancer, has a solid footing for translation into humans. These findings have relevance for both vaccine and adoptive cell immunotherapy approaches.
In addition to the short-lived nonC-TL described above, NCPs can also be derived from alternative splicing junctions of transposable elements and exons in proteins (19). In some cases these are selectively expressed in cancer and, unlike nonC-TL derived from uORFs, 5′UTRs, and ncRNAs, are apparently long-lived proteins that can be cross-presented and prime an endogenous immune response to this class of NCPs. It will be interesting to see which class of nonC-TL is most effective and whether T cells targeting these long-lived peptides will represent a more exhausted pool of T cells as compared with the short-lived nonC-TL.
As tumor-specific nonC-TL are identified, vaccine strategies should be developed and trials initiated. In acute myeloid leukemia (AML), Ehx and colleagues have already identified 58 AML-specific nonC-TL; approximately one-half represent intron retention (20).They predict that a vaccine with these 58 antigens would provide at least one tumor-specific antigen to 99% of patients with AML. One should expect to see additional reports of nonC-TL that are shared by cancers of the same, and possibly different histologies in the near future.
TCR adoptive immunotherapy or the generation of nonC-TL-reactive T cells by IVS and expansion ex vivo, represent two additional strategies that can take advantage of these findings. The preliminary data shown for HLA-A 11.01 suggests that multiple tumor types could be targeted with a single TCR. While it will take some time, it is not unreasonable to expect that clinical adoptive immunotherapy with peripheral blood T cells transduced with three or more TCRs specific for nonC-TL will be attempted. Similar to advances in CAR T cells, these strategies might involve short-term in vitro transduction and reinfusion in 24 to 48 hours. Alternatively, in vivo transduction strategies with these TCRs could also change the landscape of cancer treatment while reducing costs.
It is clear that we have entered a watershed period for cancer immunotherapy. The discovery of NCPs appears certain to grow as improved libraries of the transcribable genome become available. This also represents a watershed period for pharmacology approaches to target noncanonical uORFs, 5′UTRs, and ncRNAs that are involved in driving the malignant properties of cancer cells. This realm need not be restricted to immunotherapists; there is space for multiple approaches to address the apparent broad universe of targets presented by Cancer's Dark Matter.
Acknowledgments
B.A. Fox received support from Steve and Cindy Harder, Nancy Lematta, Robert and Elsie Franz, Lynn and Jack Loacker, The Chiles Foundation, The Providence Medical Foundation, and Shimadzu Scientific.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authors' Disclosures
B.A. Fox reports grants and nonfinancial support from Shimadzu Scientific, Incyte, and Bristol Myers Squibb; personal fees, nonfinancial support, and other support from UbiVac; nonfinancial support from Akoya, Lunaphore, and NanoString; personal fees from Calidi, Hookipa, Neogenomics, Pfizer, PrimeVax, and Turnstone; and grants from Macrogenics and Merck outside the submitted work; in addition, B.A. Fox has a patent for autophagosome vaccine issued and licensed to UbiVac and a patent for ubiquitinylated proteins issued and licensed to UbiVac. D.B. Page reports personal fees from Genentech, Novartis, Oncocyte, Lilly, Sanofi, Biothernostics, AstraZeneca/D-S, Gilead, NGM Bio, and Sanford Burnham Prebys and grants from Bristol Myers Squibb, Merck, Brooklyn Immunotherapeutics, and WindMIL outside the submitted work. B.D. Curti reports personal fees from Merck and Sanofi and grants and nonfinancial support from Clinigen outside the submitted work. R.E. Sanborn reports personal fees from AstraZeneca, Targeted Oncology, Janssen, EMD Serono, Illumina, GSK, GameOn!, Sanofi Aventis, Daiichi Sankyo, OncLive, Regeneron Pharmaceuticals, G1 Therapeutics, Blueprint Medicines, and Mirati Therapeutics outside the submitted work. R.S. Leidner reports grants from Incyte, grants and personal fees from BMS, and nonfinancial support from Clinigen and Celldex outside the submitted work. No disclosures were reported by the other authors.
References
- 1. Lozano-Rabella M, Garcia-Garijo A, Palomero J, Yuste-Estevanez A, Erhard F, Martin-Liberal J, et al. Exploring the immunogenicity of noncanonical HLA-I tumor ligands identified through proteogenomics. Clin Cancer Res 2023;29:2250–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chong C, Müller M, Pak H, Harnett D, Huber F, Grun D, et al. Integrated proteogenomic deep sequencing and analytics accurately identify noncanonical peptides in tumor immunopeptidomes. Nat Commun 2020;11:1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Apavaloaei A, Hardy M-P, Thibault P, Perreault C. The origin and immune recognition of tumor-specific antigens. Cancers 2020;12:2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MV Ruiz Cuevas, Hardy M-P, Hollý J, É Bonneil, Durette C, Courcelles M, et al. Most non-canonical proteins uniquely populate the proteome or immunopeptidome. Cell Rep 2021;34:108815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ouspenskaia T, Law T, Clauser KR, Klaeger S, Sarkizova S, Aguet F, et al. Unannotated proteins expand the MHC-I-restricted immunopeptidome in cancer. Nat Biotechnol 2022;40:209–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dersh D, Hollý J, Yewdell JW. A few good peptides: MHC class I–based cancer immunosurveillance and immunoevasion. Nat Rev Immunol 2021;21:116–28. [DOI] [PubMed] [Google Scholar]
- 7. Erhard F, Dölken L, Schilling B, Schlosser A. Identification of the cryptic HLA-I immunopeptidome. Cancer Immunol Res 2020;8:1018–26. [DOI] [PubMed] [Google Scholar]
- 8. Yewdell JW. MHC class I immunopeptidome: past, present, and future. Mol Cell Proteomics. 2022;21:100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo X, Huang Y, Li H, Luo Y, Zuo Z, Ren J, et al. SPENCER: a comprehensive database for small peptides encoded by noncoding RNAs in cancer patients. Nucleic Acids Res 2022;50:D1373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Plaen E, Lurquin C, Van Pel A, Mariamé B, Szikora JP, Wölfel T, et al. Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum- antigen P91A and identification of the tum- mutation. Proc Natl Acad Sci USA 1988;85:2274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991;254:1643–7. [DOI] [PubMed] [Google Scholar]
- 12. Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, et al. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA 1994;91:3515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst 1957;18:769–78. [PubMed] [Google Scholar]
- 14. Twitty CG, Jensen SM, Hu H-M, Fox BA. Tumor-Derived autophagosome vaccine: induction of cross-protective immune responses against short-lived proteins through a p62-dependent mechanism. Clin Cancer Res 2011;17:6467–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Page DB, Hulett TW, Hilton TL, Hu H-M, Urba WJ, Fox BA. Glimpse into the future: harnessing autophagy to promote antitumor immunity with the DRibbles vaccine. J Immunother Cancer 2016;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patel JM, Cui Z, Wen Z-F, Dinh CT, Hu H-M. Peritumoral administration of DRibbles-pulsed antigen-presenting cells enhances the antitumor efficacy of anti-GITR and anti-PD-1 antibodies via an antigen presenting independent mechanism. J Immunother Cancer 2019;7:311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwamoto N, Shimada T, Moudgil T, Meng R, Christie T, Dowell A, et al. 113 Uncovering the dark immunopeptidome of head and neck squamous cell carcinoma (HNSCC): relevance for universal cancer vaccines, immunological monitoring and TIL therapy. J Immunother Cancer [Internet]. BMJ Specialist Journals; 2022;10. Available from: https://jitc.bmj.com/content/10/Suppl_2/A125 [Google Scholar]
- 18. Setrerrahmane S, Li M, Zoghbi A, Lv X, Zhang S, Zhao W, et al. Cancer-related micropeptides encoded by ncRNAs: promising drug targets and prognostic biomarkers. Cancer Lett 2022;547:215723. [DOI] [PubMed] [Google Scholar]
- 19. Merlotti A, Sadacca B, Arribas YA, Ngoma M, Burbage M, Goudot C, et al. Noncanonical splicing junctions between exons and transposable elements represent a source of immunogenic recurrent neo-antigens in patients with lung cancer. Sci Immunol 2023;8:eabm6359. [DOI] [PubMed] [Google Scholar]
- 20. Ehx G, Larouche J-D, Durette C, Laverdure J-P, Hesnard L, Vincent K, et al. Atypical acute myeloid leukemia-specific transcripts generate shared and immunogenic MHC class I–associated epitopes. Immunity 2021;54:737–52. [DOI] [PubMed] [Google Scholar]